Bacteria use weapons to deliver effectors into target cells. One of these weapons, the type VI secretion system (T6SS), assembles a contractile tail acting as a spring to propel a toxin-loaded needle. Its expression and activation therefore need to be tightly regulated. Here, we identified an internal promoter within the sci1 T6SS gene cluster in enteroaggregative E. coli. We show that this internal promoter is controlled by Fur and Dam-dependent methylation. We further demonstrate that Fur and Dam compete at the −10 transcriptional element to finely tune the expression of T6SS genes. We propose that this elegant regulatory mechanism allows the optimum production of the T6SS in conditions where enteroaggregative E. coli encounters competing species.
KEYWORDS: type VI secretion, epigenetism, methylation, microbial communities, regulation, repression
ABSTRACT
The type VI secretion system (T6SS) is a weapon for delivering effectors into target cells that is widespread in Gram-negative bacteria. The T6SS is a highly versatile machine, as it can target both eukaryotic and prokaryotic cells, and it has been proposed that T6SSs are adapted to the specific needs of each bacterium. The expression of T6SS gene clusters and the activation of the secretion apparatus are therefore tightly controlled. In enteroaggregative Escherichia coli (EAEC), the sci1 T6SS gene cluster is subject to a complex regulation involving both the ferric uptake regulator (Fur) and DNA adenine methylase (Dam)-dependent DNA methylation. In this study, an additional, internal, promoter was identified within the sci1 gene cluster using +1 transcriptional mapping. Further analyses demonstrated that this internal promoter is controlled by a mechanism strictly identical to that of the main promoter. The Fur binding box overlaps the −10 transcriptional element and a Dam methylation site, GATC-32. Hence, the expression of the distal sci1 genes is repressed and the GATC-32 site is protected from methylation in iron-rich conditions. The Fur-dependent protection of GATC-32 was confirmed by an in vitro methylation assay. In addition, the methylation of GATC-32 negatively impacted Fur binding. The expression of the sci1 internal promoter is therefore controlled by iron availability through Fur regulation, whereas Dam-dependent methylation maintains a stable ON expression in iron-limited conditions.
IMPORTANCE Bacteria use weapons to deliver effectors into target cells. One of these weapons, the type VI secretion system (T6SS), assembles a contractile tail acting as a spring to propel a toxin-loaded needle. Its expression and activation therefore need to be tightly regulated. Here, we identified an internal promoter within the sci1 T6SS gene cluster in enteroaggregative E. coli. We show that this internal promoter is controlled by Fur and Dam-dependent methylation. We further demonstrate that Fur and Dam compete at the −10 transcriptional element to finely tune the expression of T6SS genes. We propose that this elegant regulatory mechanism allows the optimum production of the T6SS in conditions where enteroaggregative E. coli encounters competing species.
INTRODUCTION
The fate of microbial communities is governed by communication, cooperation, and competition mechanisms between microorganisms (1–9). Bacteria have therefore developed an arsenal of signaling, sensing, and antagonistic activities. To eliminate competitors, bacteria evolved distinct mechanisms for release of antibiotics or bacteriocins in the extracellular medium, as well as delivery of toxins directly into the target cell (10–12). One of the delivery apparatuses, the type VI secretion system (T6SS), transports effectors into competing bacteria using a mechanism similar to that used by contractile injection systems such as bacteriophages and R-pyocins (13–19). This secretion apparatus comprises an ∼800-nm long cytoplasmic needle-like structure composed of an inner tube tipped by a spike complex that is used to penetrate the membrane of the target cell (12, 14, 19). The inner tube is wrapped by an outer sheath that is assembled under an extended metastable conformation (20, 21). The tail tube/sheath complex is built on a baseplate that is anchored to the cell envelope by a membrane complex (22–29). Tail tube/sheath assembly, which can be visualized in vivo by fluorescence microscopy, is completed in a few tens of seconds (30–32). Contraction of the sheath powers the propulsion of the inner tube to deliver effectors into the target cell (15, 17, 31, 33–35). Effectors are usually charged within the inner tube lumen or loaded onto the spike complex via direct interactions with the VgrG/PAAR spike or via adaptor proteins (36–45).
The T6SS is a very efficient mechanism and hence is an important player in the regulation of microbiota (7, 46). Bacteria equipped with this apparatus colonize an environmental niche more efficiently and hence have better access to resources (47–51). Most of the T6SS gene clusters are not constitutively expressed and T6SS-dependent antagonistic activities are usually deployed once cells experience stress or nutrient starvation conditions (52–57). T6SS gene clusters are therefore subjected to a tight regulation that involves sensing of the environmental conditions (52, 53, 55). Most known regulatory mechanisms are hijacked by T6SSs for their own regulation, including transcriptional activators and repressors, alternate sigma factors, histone-like proteins, two-component transduction cascades, or quorum-sensing systems (52, 53). In addition, a number of T6SSs are posttranslationally activated by a threonine phosphorylation pathway in response to cell damage or envelope stress (58).
Enteroaggregative Escherichia coli (EAEC) is equipped with two functional T6SSs, named Sci1 (T6SS-1 subfamily) and Sci2 (T6SS-3 subfamily) (59, 60). These two T6SSs confer antagonistic activities but are not expressed under the same conditions, suggesting that T6SS-mediated antibacterial activities are required in two conditions that EAEC may encounter during its life cycle (31, 44). The sci2 gene cluster is expressed during infection conditions and is activated in laboratory conditions when cells are grown in a synthetic medium mimicking the macrophage environment (59). This sci2 gene cluster is under the control of the AraC-like AggR transcriptional regulator (59), which also modulates the expression of most biofilm determinants (59, 61), suggesting that the Sci2 T6SS is required for eliminating competing bacteria during aggregation, a phenomena that occurs during host colonization. In contrast, the sci1 gene cluster is expressed in minimal synthetic media and has been shown to be under the dual control of the ferric uptake repressor (Fur) and Dam-dependent methylation (62).
To better understand the organization of the sci1 gene cluster, we defined its operon structure. Reverse transcriptase polymerase chain reaction (RT-PCR) experiments showed that all genes are contiguous, suggesting that all the genes are present on a single mRNA or on several overlapping mRNAs. Using +1 transcriptional mapping, we confirmed the existence of a promoter region upstream of the first gene of the cluster and revealed an additional promoter located upstream to the EC042_4532 gene, within the EC042_4531 coding sequence. We further identified a Fur-binding sequence overlapping with the −10 transcriptional box and demonstrated that Fur binds this sequence with high affinity, thereby preventing RNA polymerase from gaining access to the promoter. Sequence analyses showed that this Fur box overlaps with a GATC Dam methylation site, GATC-32. In vivo, we showed that Fur prevents methylation of the GATC-32 site when cells were grown in iron-replete conditions. In vitro competition experiments confirmed that Fur prevents GATC-32 methylation. In addition, we observed that Dam-dependent methylation of GATC-32 decreases the affinity of Fur for its Fur box. Taken together, our results demonstrate that a second functional, internal promoter controls the expression of T6SS sci1 genes and that this promoter is under a regulatory mechanism similar to the main promoter.
RESULTS AND DISCUSSION
Operon structure of the sci1 T6SS gene cluster.
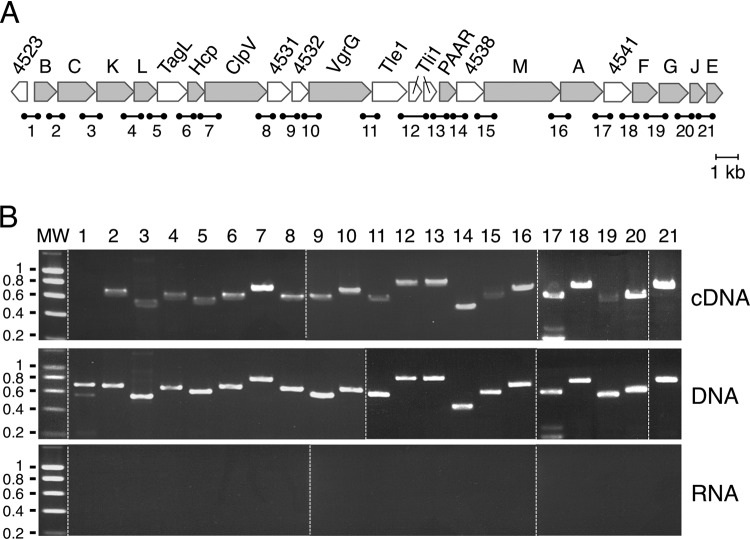
We previously reported that the promoter located upstream of the tssB gene, i.e., the first gene of the EAEC sci1 T6SS gene cluster, contains operator sequences for the ferric uptake regulator (Fur), as well as an overrepresentation of GATC motifs that are targets of the DNA adenine methylase (Dam). Using in vivo and in vitro Fur binding and methylation assays, we delineated the contribution of these two regulators on the expression of the tssB gene (62). However, whether additional or internal promoters exist and whether the entire gene cluster is subjected to this regulatory control remained undetermined. The EAEC sci1 gene cluster is an ∼26-kb DNA fragment on the pheU pathogenicity island (Fig. 1A) (59). Prediction of the open reading frames (ORFs) within this fragment shows that it likely encodes 21 gene products, including the 14 T6SS core components, a toxin-immunity pair, and accessory genes or of unknown function (genes tssB to tssE [Fig. 1A]). With the exception of a large intergenic sequence (162 bp between the hcp and the clpV genes), most of the start and stop codons of contiguous genes overlap or are separated by few (<8) nucleotides (see Fig. S1 in the supplemental materials). This genomic organization suggests that translational coupling must occur and that the expression of these genes must be coordinated. To test whether the sci1 gene cluster is organized as a single genetic unit or constituted of several operons, we performed reverse transcriptase polymerase chain reactions (RT-PCR) using oligonucleotides designed for the amplification of each gene junction (numbered 1 to 21 [Fig. 1A]). RT-PCR experiments were performed on purified total RNAs extracted from cells grown in Sci1-inducing medium (SIM) (Fig. 1B, top panel). As controls, RT-PCRs were performed on purified genome DNA (Fig. 1B, middle panel), as well as on the total RNA preparation in the absence of reverse transcriptase, to test for DNA contamination (Fig. 1B, bottom panel). As shown in Fig. 1B, RT-PCR products with the expected sizes were obtained for each gene junction of the sci1 gene cluster from DNA or cDNA but not from RNA (Fig. 1B, lanes 2 to 21), suggesting that the 21 genes are cotranscribed. As expected, the Ec042_4523 ORF, upstream of the first gene of the sci1 cluster and in the reverse orientation compared to the tss genes, is not cotranscribed with tssB (Fig. 1B, lane 1). These results suggest that all the sci1 genes are present on a unique polycistronic mRNA or that overlapping mRNAs are expressed from internal promoters.
FIG 1.
Operon structure of the EAEC sci1 T6SS gene cluster. (A) Schematic organization of the EAEC sci1 T6SS gene cluster (EC042_4524 to EC042_4545). Genes encoding T6SS core components are indicated in gray. Accessory genes or genes of unknown function are represented in white. The fragments corresponding to gene junctions and amplified in the RT-PCR experiments are indicated below (1, 692 bp; 2, 672 bp; 3, 550 bp; 4, 618 bp; 5, 586 bp; 6, 643 bp; 7, 748 bp; 8, 629 bp; 9, 575 bp; 10, 654 bp; 11, 581 bp; 12, 768 bp; 13, 762 bp; 14, 459 bp; 15, 600 bp; 16, 673 bp; 17, 576 bp; 18, 720 bp; 19, 552 bp; 20, 591 bp; 21, 678 bp). (B) Operon structure of the EAEC sci1 T6SS gene cluster. Agarose gel analyses of the indicated gene junctions (numbered 1 to 21 in panel A) amplified by PCR from cDNA, genomic DNA (middle panel; positive control), and total RNA (negative control). The presence of PCR fragments in the cDNA gels demonstrates cotranscription of the genes located 5′ and 3′ of the amplified region. Molecular weight markers (MW, in kilobases) are indicated on the left. Dashed lines separate different gels combined into a single image.
An additional promoter is located upstream of EC042_4532.
To identify a potential internal promoter(s), we used an in silico approach. Analysis of the T6SS sci1 gene cluster using the BProm algorithm (Softberry; http://linux1.softberry.com/berry.phtml) suggested the existence of an additional promoter with a σ70 −10 element upstream of the EC042_4532 gene. To test whether an internal promoter was present upstream of Ec042_4532, we used a 5' rapid amplification of cDNA ends (5′ RACE) assay. mRNAs were extracted from EAEC cells grown in Sci1-inducing medium (SIM) and subjected to primer extension. The putative tssB promoter was also included in this assay. The results showed that transcription of the tssB mRNA starts at the A base located 73 bases upstream of the ATG start codon of tssB (colored red in Fig. 2A). The tssB transcription starts are therefore compatible with the putative −10 and −35 transcription boxes identified through in silico analyses in our previous study (62) (Fig. 2A). A transcriptional start was also detected upstream of the EC042_4532 gene, suggesting the existence of an active internal promoter. The position of the identified transcriptional start (base G, located 117 bases upstream of the ATG of EC042_4532 [colored red in Fig. 2B]) is compatible with the location of the −10 element predicted by the BProm algorithm (Fig. 2B).
FIG 2.
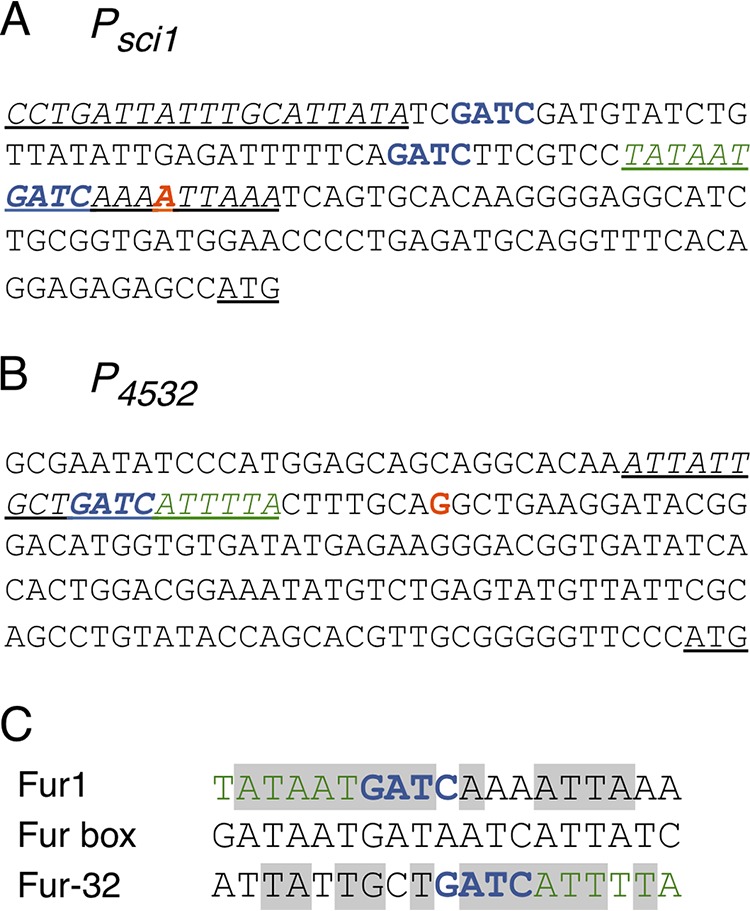
Regulatory elements of the sci1 and 4532 promoters. (A and B) Nucleotide sequences of the sci1 (A) and EC042_4532 (B) promoters highlighting overlaps between the transcriptional elements, Fur binding boxes, and Dam methylation motifs. The +1 transcriptional site identified by 5′RACE is indicated in red. GATC Dam methylation sites are indicated in blue. The −10 elements are indicated in green. The underlined sequences indicate Fur binding boxes (italics) and translational start codons. (C) Sequence alignment of the fur1 (sci1 promoter) and fur-32 (EC042_4532 promoter) boxes with the E. coli Fur box consensus sequence. Identical bases are framed in gray. The −10 elements (green) and GATC motifs (blue) are indicated.
In silico sequence analyses of the EC042_4532 promoter region identify Fur and Dam sites overlapping with the −10 element.
Interestingly, the BProm computer program also identified a putative Fur-binding box in the EC042_4532 promoter region (hereafter called Fur-32). This putative operator sequence overlaps with the −10 of transcription (Fig. 2B and C). This situation is reminiscent of the main promoter, which is repressed by the Fur protein in an iron-dependent manner (62). One of the Fur boxes contained in the tssB promoter contains a Dam-dependent methylation site (Fig. 2A), and we previously reported that Fur and Dam compete at this specific site to fine-tune the expression of the sci1 gene cluster (62). Strikingly, a GATC motif is also found within the putative Fur-32 box of the EC042_4532 promoter (Fig. 2C) (hereafter called GATC-32). Taken together, the in silico sequence analyses raised the question of whether the internal promoter was under a similar regulatory mechanism as the tssB main promoter.
The P4532-lacZ translational fusion is responsive to iron limitation and Fur.
To test whether the expression of the internal promoter was regulated by Fur, we engineered a low-copy-number plasmid-borne translational fusion of a 570-bp fragment comprising the EC042_4532 promoter (from −450 to +120 relative to the transcriptional +1, called here P4532) to lacZ. The β-galactosidase activity of this P4532-lacZ translational fusion was monitored in the EAEC lacZ strain or its fur isogenic mutant in the presence or absence of the iron chelator 2,2’-dipyridyl (dip). Figure 3 shows that the expression of the P4532 translational fusion increased ∼6-fold in the wild-type (WT) strain upon treatment with the iron chelator. Compared to the WT strain in the absence of iron chelator, the activity of the translational fusion increased ∼13-fold in the fur isogenic background. Treatment of the fur mutant strain with 2,2’-dipyridyl had no additional effect on the activity of the P4532-lacZ translational reporter fusion (data not shown). From these activities, we concluded that the expression from the P4532 promoter is repressed by the Fur transcriptional regulator in an iron-dependent manner.
FIG 3.
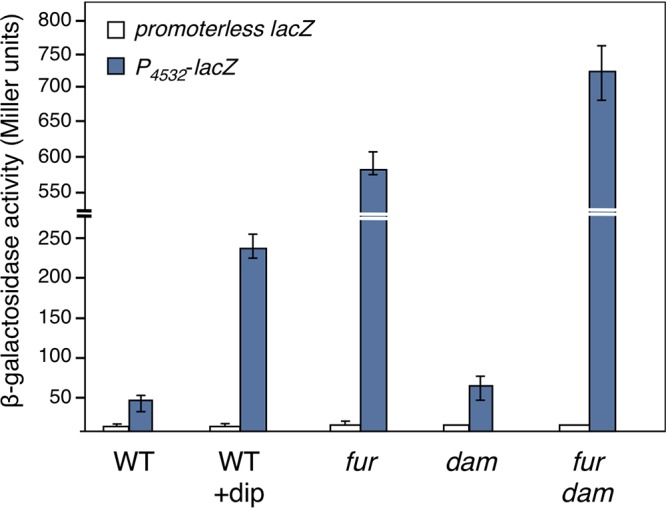
The 4532 promoter is under the control of iron levels, Fur, and Dam. β-Galactosidase activity (in Miller units) of a promoterless lacZ fusion and of the P4532-lacZ reporter fusion at OD600 = 0.8 in the WT EAEC 17-2 strain after a 30-min treatment with 2,2’-dipyridyl (+dip; 100 μM) or in the isogenic fur, dam, and fur-dam mutants.
Fur binds to the P4532 promoter and limits access to RNA polymerase.
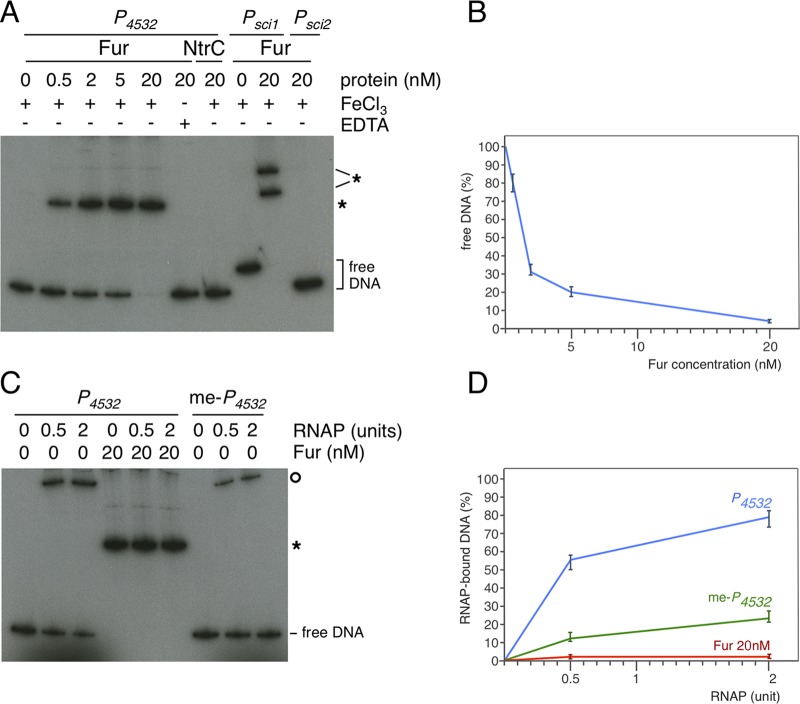
To test whether Fur binds the EC042_4532 promoter region in vitro, the purified E. coli Fur protein and the radiolabeled P4532 570-bp fragment were used for electrophoretic mobility shift assays (EMSA). As controls and as previously published (62), Fur bound to the sci1 promoter, yielding two bands due to the presence of two Fur boxes, but did not retard the Fur-independent sci2 promoter (Fig. 4A, lanes 8 to 10). Fur also shifted the P4532 fragment in the presence of iron, its corepressor (Fig. 4A, lanes 1 to 5; Fig. 4B). This shift was strictly dependent on metal-bound Fur, as no band retardation could be observed when the fragment and the purified regulator were incubated in the presence of the metal chelator EDTA (Fig. 4A, lane 6). In contrast, control experiments showed that the σ54 enhancer binding protein NtrC did not bind the P4532 fragment (Fig. 4A, lane 7). From these data, we conclude that Fur binds to the P4532 promoter in vitro, likely to the putative Fur-32 box.
FIG 4.
Fur binds to the 4532 promoter and prevents access to RNA polymerase in vitro. (A) Electrophoretic mobility shift assay of the EC042_4532 promoter (P4532) with the indicated concentration of Fur in the presence of FeCl3 or in the presence of EDTA or using a purified NtrC transcriptional activator. Controls include Fur shift assays of the Fur-dependent sci1 promoter or of the Fur-independent sci2 promoter. DNA-Fur complexes are indicated by stars. (B) Densitometry analysis of Fur binding on the P4532 fragment, represented as free P4532 DNA as a function of Fur concentration. (C) Electrophoretic mobility shift assay of the unmethylated (P4532) or methylated (me-P4532) EC042_4532 promoter with the indicated concentration of σ70-RNAP alone or in the presence of 20 nM Fur. DNA-Fur and DNA-RNAP complexes are indicated by the star and circle, respectively. (D) Densitometry analysis of RNAP binding on the unmethylated (blue curve), methylated (green curve), or Fur-bound unmethylated (red curve) P4532 fragment, represented as RNAP-bound DNA as a function of RNAP concentration.
Fur repression is usually caused by preventing access of the RNA polymerase (RNAP) to the promoter. We hypothesized that such a mechanism might be likely at promoter P4532, as the putative Fur-32 box overlaps with the −10 RNAP-binding element (Fig. 2B). We therefore tested whether σ70-RNAP holoenzyme binds to the P4532 promoter and whether Fur influences σ70-RNAP binding. Figure 4C shows that the σ70-RNAP complex binds to the P4532 promoter (Fig. 4C, lanes 1 to 3) and that preincubation of the P4532 fragment with Fur prevents binding of the σ70-RNAP, demonstrating that Fur and RNAP compete for binding on P4532 (Fig. 4C, lanes 4 to 6; Fig. 4D).
Dam methylation at the GATC-32 site decreases RNAP binding to the P4532 promoter.
To gain insight on the contribution of Dam to the regulation of EC042_4532, we measured the β-galactosidase activity of the P4532-lacZ translational fusion in dam and fur-dam EAEC strains. Deletion of dam did not cause a significant variation in the activity of the promoter fusion compared to its parental wild-type strain (Fig. 3). In contrast, the activity of the promoter fusion in the fur-dam strain increased ∼16-fold compared to the wild-type strain, and ∼1.4-fold compared to the fur mutant. These results show that Dam and Fur have additive negative effects on regulation at the P4532 promoter and that the contribution of Dam is masked in the presence of Fur. Based on these results, we hypothesized that GATC-32 methylation affects RNAP binding. A Dam-methylated P4532 fragment was subjected to EMSA with the reconstituted σ70-RNAP complex. As shown in Fig. 4C and D, σ70-RNAP binding was diminished on the methylated P4532 fragment.
Fur-Dam competition at the P4532 promoter.
The observation that the Dam effect was masked by Fur in vivo raised the idea that, similar to the Psci1 situation, Fur binding to the Fur-32 box prevents Dam methylation of the GATC-32 site. To test this hypothesis, in vitro and in vivo assays were conducted.
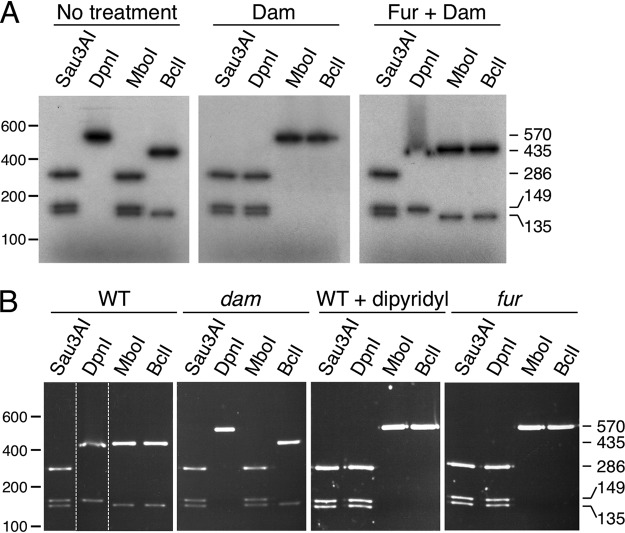
(i) Fur binding at the P4532 promoter prevents GATC-32 methylation in vitro. To test the impact of Fur binding on GATC-32 methylation in vitro, we added purified Dam methylase to radiolabeled P4532 fragments preincubated or not preincubated with purified Fur protein. The P4532 fragments were then used for enzymatic digestion using enzymes that cleave GATC motifs (Fig. S2). We used the fortuitous fact that the GATC-32 site is part of a larger palindromic sequence, TGATCA, which is the target for BclI, a restriction enzyme that is sensitive to Dam methylation (Fig. S2). In addition to GATC-32, the P4532 fragment contains a GATC site at position 149 (GATC149) that does not overlap a Fur box (Fig. S2). Figure 5A shows that, as expected, incubation with the Dam methylase caused methylation of the GATC sites as P4532 is cleaved into three fragments when incubated with DpnI, an enzyme that specifically recognizes methylated GATC motifs. In agreement with this result, P4532 was resistant to MboI and BclI, two enzymes that are sensitive to GATC adenine methylation (Fig. 5A, middle panel). When the P4532 fragment was preincubated with Fur, only the GATC149 site was digested by DpnI. In contrast, only the GATC-32 site was digested by MboI or BclI (Fig. 5A, right panel). These experiments demonstrate that in the presence of Fur, GATC149 is methylated whereas GATC-32 is not, suggesting that Fur protects GATC-32 methylation by steric occlusion.
FIG 5.
Fur protects GATC-32 from methylation in vitro and in vivo. (A) A radiolabeled PCR product corresponding to the 570-bp P4532 fragment was digested by the restriction enzymes indicated on top. Left panel, untreated PCR product; middle panel, PCR product treated with the Dam methylase; right panel, PCR product incubated with purified Fur (20 nM) prior to Dam methylation. Molecular weight markers (in base pairs) are indicated on the left. The sizes of the digestion products (in base pairs) are indicated on the right. See Fig. S2 in the supplemental material for positions of restriction sites and sizes of expected DNA fragments. (B) The P4532 promoters isolated from pGE573 vectors carrying the P4532-lacZ fusion purified from the EAEC wild-type strain (WT) or its isogenic dam or fur mutant strain or from the WT strain treated with 2,2’-dipyridyl were digested by the restriction enzymes indicated on top. Molecular weight markers (in base pairs) are indicated on the left. The sizes of the digestion products (in base pairs) are indicated on the right. The dashed lines indicate reorganization of the lanes from the same gel. See Fig. S2 for positions of restriction sites and sizes of expected DNA fragments.
(ii) Fur binding at the P4532 promoter prevents GATC-32 methylation in vivo. The methylation status of the P4532 GATC sites was then tested in vivo. The pGE573 plasmid bearing the P4532-lacZ fusion was extracted from various genetic backgrounds, the EcoRI-BamHI fragment comprising the P4532 promoter was purified and the methylation state of GATC-32 was assessed by restriction. In the WT strain grown in LB medium, the MboI and BclI enzymes cleaved GATC-32 (Fig. 5B, left panel), revealing that this site is unmethylated. The absence of methylation is likely due to the presence of Fur bound to the Fur box overlapping with GATC-32, as GATC-32 was methylated in the fur isogenic background (Fig. 5B, right panel) or when WT cells were grown in the presence of the 2,2’-dipyridyl iron chelator (Fig. 5B, third panel from left).
Taken together, the results of the in vitro and in vivo Dam methylation assays demonstrate that Fur binding on the Fur-32 box prevents access of the Dam methylase to the GATC-32 site in iron-rich conditions. In contrast, Fur repression is relieved in iron limiting conditions and the GATC-32 site is then methylated.
(iii) GATC-32 Dam methylation decreases the affinity of Fur for the P4532 promoter. The observation that the GATC-32 site is methylated once Fur repression is relieved raised the question of whether methylation of the GATC-32 motif interferes with Fur binding. We therefore performed mobility shift assays with Fur using the P4532 fragment, methylated by Dam in vitro. Figure 6 shows that methylation of GATC-32 caused a significant decrease in the affinity of Fur for the P4532 promoter.
FIG 6.
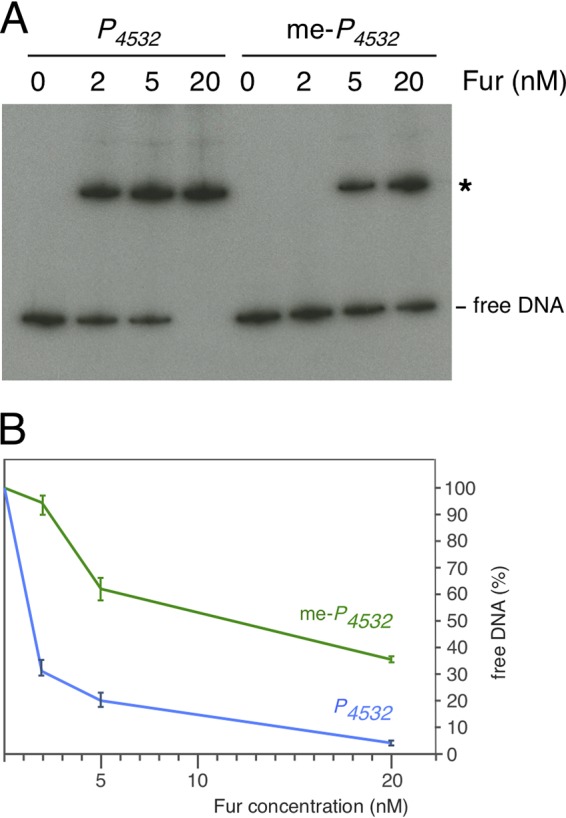
GATC-32 methylation influences Fur binding on P4532. (A) Electrophoretic mobility shift assay of the unmethylated (P4532) or methylated (me-P4532) P4532 fragment with the indicated concentration of purified Fur. (B) Densitometry analysis of Fur binding on the unmethylated or methylated P4532 fragment, represented as free P4532 DNA as a function of Fur concentration.
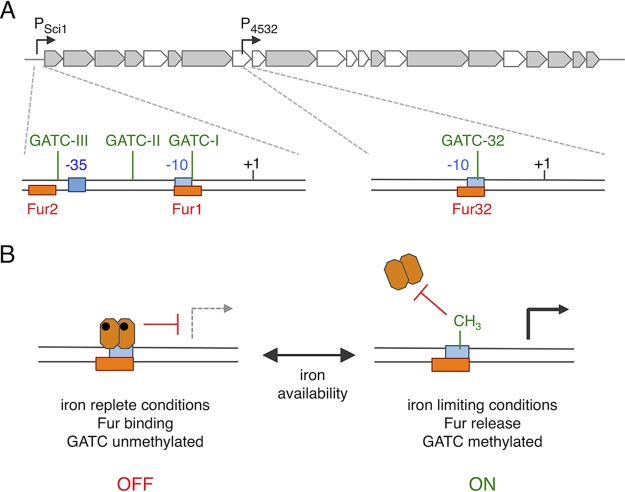
To summarize, we report in this study the presence of an internal promoter within the sci1 T6SS gene cluster of enteroaggregative E. coli. The presence of internal promoters that serve as transcriptional restarts or that are necessary for ensuring proper stoichiometric production is common in large gene clusters. It has been well documented for gene clusters encoding amino acid synthesis pathways such as histidine, tryptophan, threonine, or branched-chain amino acids (63–67). More recently, an internal promoter within the gene cluster encoding the ESX-3 type VII secretion system has been identified in Mycobacterium smegmatis (68). Here, we show that this internal promoter, P4532, is under the control of a regulatory mechanism similar to that controlling the main promoter (Fig. 7). Expression from the P4532 promoter is repressed by the Fur protein, which binds to a Fur box overlapping with the −10 transcriptional element. In addition, a GATC site, GATC-32, which is a target of the Dam methylase, overlaps with the Fur-binding box. In iron-rich conditions, Fur binding to the promoter prevents methylation of this motif. However, during iron starvation, Fur removal allows methylation of the GATC-32 site and the methylation decreases the affinity of Fur for its binding box. Therefore, Fur controls the switch between on and off expression, whereas Dam methylation stabilizes the on phase (Fig. 7). This mechanism is therefore similar to that previously reported for the sci1 main promoter (62). However, differences can be noticed. First, the level of methylation and the activity of the Dam methylase might be slightly different on the main and the internal promoters, as the sequences flanking the GATC motifs have different AT content. Indeed, sequences flanking Dam sites have been previously shown to modulate the catalytic activity or the processivity of Dam (69). Second, an ∼13-fold derepression of the internal promoter is observed in the absence of Fur, while a >25-fold derepression was observed for the main promoter (62). These results are consistent with the lower degree of consensus for the Fur-32 box compared to the Fur box overlapping with the −10 element of the main promoter (Fig. 2C) and with the potential cooperativity of the two Fur-binding boxes at the main promoter (62).
FIG 7.
Schematic representation of sci1 gene cluster regulation. (A) The sci1 T6SS gene cluster is represented on top with the location of the main (PSci1) and internal (P4532) promoters. Expanded genetic architectures of these promoters are shown below: +1, transcriptional start; −10 and −35 transcriptional elements (blue); Fur binding box (orange); Dam methylation GATC site (green). (B) Model of regulation of the sci1 main and internal promoters by Fur and Dam. In iron-replete conditions (left), a Fur dimer (hexagons) complexed to iron (dots) is bound to the Fur box, preventing methylation of the GATC site and access by RNA polymerase. Expression from the promoter is repressed (off state). In iron-limiting conditions (right), Fur is released from the promoter, allowing GATC methylation by Dam and binding of RNA polymerase. Expression from the promoter is turned on (on state).
The role of the Dam methylase in transcriptional gene regulation is well documented. In addition to its role in mismatch repair and replication initiation, Dam is involved in epigenetic control of the expression of many genes, including genes encoding type III secretion systems, adhesins, or fimbriae or those involved in lipopolysaccharide modifications (for reviews, see references 70, to ,72). GATC sites can be found in intergenic regions, and in some cases these sites overlap transcriptional elements such as the −10 element (73). Hence Dam-dependent methylation may directly impact transcription. However, in most cases, GATC sites found in promoter regions do not overlap transcriptional elements, but rather overlap regulator-binding boxes. In these cases, the methylation status may control binding of the regulator, and reciprocally, regulator binding may prevent methylation of certain GATC sites. Several studies have reported competition between Dam-dependent methylation and regulator fixation, such as the OxyR repressor at the agn43 promoter, or the Lrp repressor at the pap operon promoter (74–76). In general, competition between methylation and regulator binding results in the transition between off and on expression phases (72).
In conclusion, the sci1 gene cluster is subjected to Fur-Dam regulation, and a transcriptional restart occurs after the eighth gene of the operon. Further experiments will be necessary to define whether this restart is necessary because transcription of the mRNA from the initial promoter stops before the last gene, or because the distal part of the operon requires additional copies of mRNA for proper stoichiometry.
MATERIALS AND METHODS
Bacterial strains, plasmids, medium, and growth conditions.
E. coli K-12 strain DH5α was used for all cloning procedures. The EAEC strains used in this study are all derivatives of strain 17-2 and have been previously described (62). The plasmid-borne P4532-lacZ fusion was engineered by ligating a blunt-end 570-bp fragment encompassing the 4532 promoter (corresponding to bases –450 to +120, respective to the EC042_4532 transcriptional start site [nucleotides 4892656 to 4893121], amplified from EAEC 17-2 chromosomal DNA using oligonucleotides 5′-CGCACCATGATCGTCTCTGTATCGC and 5′-CTGAAACGAACTGCTCATGGCTCTCTC) into the SmaI-linearized pGE573, a vector that carries a promoterless lacZ gene (77). In this construct, the lacZ gene is under the control of the P4532 promoter. Proper insertion, orientation, and sequence of the fragment into the pGE-P4532 plasmid were verified by restriction, PCR, and DNA sequencing (MWG). E. coli cells were routinely grown in Luria broth (LB) or Sci1-inducing medium (SIM; M9 minimal medium supplemented with glycerol 0.25%, vitamin B1 200 μg · ml−1, Casamino Acids 40 μg · ml−1, MgCl2 2 mM, CaCl2 0.1 mM, and LB [10% vol/vol] [62]) supplemented with antibiotics when necessary (kanamycin 50 μg · ml−1, ampicillin 100 μg · ml−1 for K-12, or 200 μg · ml−1 for EAEC).
RNA purification.
EAEC total RNAs were extracted using the PureYield RNA midiprep system (Promega) from 8 × 109 cells grown in SIM and harvested in exponential growth phase (optical density at λ = 600 nm [OD600] ∼0.8). RNAs were eluted with 1 ml of water, cleared with DNase I (Ambion), and precipitated overnight at – 80°C by ammonium sulfate-ethanol procedures. The RNA pellet was washed and resuspended in 45 μl of nuclease-free water. RNA quality and integrity were tested on agarose gels and by the absorbance ratio at λ = 260/280 nm. The absence of DNA contamination was further tested by PCR using 35 cycles of amplification. Quantifications gave an average RNA concentration of 70 μg · ml−1. Total RNAs were then subjected to reverse transcription-PCR (Access RT-PCR; Promega) or transcriptional +1 mapping (5′ RACE; Invitrogen).
Reverse transcription-PCR.
The reverse transcription and PCR assays were performed with the one-tube procedure, using the Access RT-PCR system (Promega), with 200 ng of total RNA and oligonucleotides allowing amplification of 550- to 750-bp regions overlapping the two contiguous genes (Fig. 1A) (primer sequences available upon request), following the supplier’s guidelines. Briefly, both reverse transcriptase and Tfl Taq polymerase were added in each tube. The reverse transcription was carried out for 45 min at 45°C, and, after inactivation of the reverse transcriptase at 94°C for 5 min, a 30-cycle PCR was performed (denaturation at 94°C for 30 s, annealing at 55°C for 40 s, and amplification at 68°C for 50 s). As negative controls to test for DNA contamination, RT-PCRs were also performed in the absence of reverse transcriptase. As positive controls, the regions overlapping the two contiguous genes were amplified from 30 ng of genomic DNA.
5′ RACE assay.
Total RNAs (80 μg · ml−1) were subjected to transcriptional +1 mapping using the 5′ RACE system (Invitrogen).
β-Galactosidase assays.
β-Galactosidase activity was measured by the method of Miller (78) on whole cells harvested at an OD600 of 0.8. Reported values represent the average from technical triplicates from three independent biological cultures, and standard deviations are shown on the graphs.
Protein purification.
The Fur and NtrC proteins were purified as described previously (62, 79). The σ70-saturated RNAP holoenzyme was purchased from USB Corp. The Dam methylase and restriction enzymes were obtained from New England BioLabs and used as recommended by the manufacturer.
Electrophoretic mobility gel shift assay and Dam methylation assays.
DNA radiolabeling, EMSA, Fur/RNAP competition EMSA, and in vivo and in vitro Dam methylation assays have been performed as previously described (62).
Supplementary Material
ACKNOWLEDGMENTS
We thank Emmanuelle Bouveret and Mireille Ansaldi for sharing strains, plasmids, and protocols; Laure Journet and the members of the Cascales, Lloubès, Bouveret, and Sturgis research groups for insightful discussions; and Isabelle Bringer, Annick Brun, and Olivier Uderso for technical assistance.
This work was supported by grants from the Agence Nationale de la Recherche to E.C. (ANR-10-JCJC-1303-03 and ANR-14-CE14-0006-02). Work in the E.C. laboratory is supported by the CNRS, the Aix-Marseille Université, the Fondation pour la Recherche Médicale (DEQ20180339165), and the Fondation Bettencourt-Schueller. Y.R.B. was a recipient of a doctoral fellowship from the French Ministry of Research.
Author contributions. Y.R.B. and E.C. conceived the study and designed the experiments. Y.R.B. performed all in vivo and in vitro experiments, with the help of C.S.B. for RNA analyses. Y.R.B. and E.C. analyzed the data. E.C. wrote the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.West SA, Griffin AS, Gardner A. 2007. Evolutionary explanations for cooperation. Curr Biol 17:661–672. doi: 10.1016/j.cub.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Blango MG, Mulvey MA. 2009. Bacterial landlines: contact-dependent signaling in bacterial populations. Curr Opin Microbiol 12:177–181. doi: 10.1016/j.mib.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strassmann JE, Gilbert OM, Queller DC. 2011. Kin discrimination and cooperation in microbes. Annu Rev Microbiol 65:349–367. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- 4.Cornforth DM, Foster KR. 2013. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol 11:285–293. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 5.Aussel L, Beuzón CR, Cascales E. 2016. Meeting report: adaptation and communication of bacterial pathogens. Virulence 7:481–490. doi: 10.1080/21505594.2016.1152441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rakoff-Nahoum S, Foster KR, Comstock LE. 2016. The evolution of cooperation within the gut microbiota. Nature 533:255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chassaing B, Cascales E. 2018. Antibacterial weapons: targeted destruction in the microbiota. Trends Microbiol 26:329–338. doi: 10.1016/j.tim.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 8.García-Bayona L, Comstock LE. 2018. Bacterial antagonism in host-associated microbial communities. Science 361:eaat2456. doi: 10.1126/science.aat2456. [DOI] [PubMed] [Google Scholar]
- 9.Granato ET, Meiller-Legrand TA, Foster KR. 2019. The evolution and ecology of bacterial warfare. Curr Biol 29:521–537. doi: 10.1016/j.cub.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 10.Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, Riley M, Slatin S, Cavard D. 2007. Colicin biology. Microbiol Mol Biol Rev 71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruhe ZC, Low DA, Hayes CS. 2013. Bacterial contact-dependent growth inhibition. Trends Microbiol 21:230–237. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coulthurst S. 2019. The Type VI secretion system: a versatile bacterial weapon. Microbiology 165:503–515. doi: 10.1099/mic.0.000789. [DOI] [PubMed] [Google Scholar]
- 13.Ho BT, Dong TG, Mekalanos JJ. 2014. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zoued A, Brunet YR, Durand E, Aschtgen MS, Logger L, Douzi B, Journet L, Cambillau C, Cascales E. 2014. Architecture and assembly of the Type VI secretion system. Biochim Biophys Acta 1843:1664–1673. doi: 10.1016/j.bbamcr.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Basler M. 2015. Type VI secretion system: secretion by a contractile nanomachine. Philos Trans R Soc B 370:20150021. doi: 10.1098/rstb.2015.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cascales E. 2017. Microbiology: and Amoebophilus invented the machine gun! Curr Biol 27:1170–1173. doi: 10.1016/j.cub.2017.09.025. [DOI] [PubMed] [Google Scholar]
- 17.Brackmann M, Nazarov S, Wang J, Basler M. 2017. Using force to punch holes: mechanics of contractile nanomachines. Trends Cell Biol 27:623–632. doi: 10.1016/j.tcb.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Taylor NMI, van Raaij MJ, Leiman PG. 2018. Contractile injection systems of bacteriophages and related systems. Mol Microbiol 108:6–15. doi: 10.1111/mmi.13921. [DOI] [PubMed] [Google Scholar]
- 19.Cherrak Y, Flaugnatti N, Durand E, Journet L, Cascales E. 2019. Structure and activity of the type VI secretion system. Microbiol Spectr 7. doi: 10.1128/microbiolspec.PSIB-0031-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudryashev M, Wang RY, Brackmann M, Scherer S, Maier T, Baker D, DiMaio F, Stahlberg H, Egelman EH, Basler M. 2015. Structure of the type VI secretion system contractile sheath. Cell 160:952–962. doi: 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Brackmann M, Castaño-Díez D, Kudryashev M, Goldie KN, Maier T, Stahlberg H, Basler M. 2017. Cryo-EM structure of the extended type VI secretion system sheath-tube complex. Nat Microbiol 2:1507–1512. doi: 10.1038/s41564-017-0020-7. [DOI] [PubMed] [Google Scholar]
- 22.Aschtgen MS, Gavioli M, Dessen A, Lloubès R, Cascales E. 2010. The SciZ protein anchors the enteroaggregative Escherichia coli Type VI secretion system to the cell wall. Mol Microbiol 75:886–899. doi: 10.1111/j.1365-2958.2009.07028.x. [DOI] [PubMed] [Google Scholar]
- 23.Zoued A, Durand E, Bebeacua C, Brunet YR, Douzi B, Cambillau C, Cascales E, Journet L. 2013. TssK is a trimeric cytoplasmic protein interacting with components of both phage-like and membrane anchoring complexes of the type VI secretion system. J Biol Chem 288:27031–27041. doi: 10.1074/jbc.M113.499772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.English G, Byron O, Cianfanelli FR, Prescott AR, Coulthurst SJ. 2014. Biochemical analysis of TssK, a core component of the bacterial Type VI secretion system, reveals distinct oligomeric states of TssK and identifies a TssK-TssFG subcomplex. Biochem J 461:291–304. doi: 10.1042/BJ20131426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunet YR, Zoued A, Boyer F, Douzi B, Cascales E. 2015. The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet 11:e1005545. doi: 10.1371/journal.pgen.1005545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durand E, Nguyen VS, Zoued A, Logger L, Péhau-Arnaudet G, Aschtgen MS, Spinelli S, Desmyter A, Bardiaux B, Dujeancourt A, Roussel A, Cambillau C, Cascales E, Fronzes R. 2015. Biogenesis and structure of a type VI secretion membrane core complex. Nature 523:555–560. doi: 10.1038/nature14667. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen VS, Logger L, Spinelli S, Legrand P, Huyen Pham TT, Nhung Trinh TT, Cherrak Y, Zoued A, Desmyter A, Durand E, Roussel A, Kellenberger C, Cascales E, Cambillau C. 2017. Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex. Nat Microbiol 2:17103. doi: 10.1038/nmicrobiol.2017.103. [DOI] [PubMed] [Google Scholar]
- 28.Cherrak Y, Rapisarda C, Pellarin R, Bouvier G, Bardiaux B, Allain F, Malosse C, Rey M, Chamot-Rooke J, Cascales E, Fronzes R, Durand E. 2018. Biogenesis and structure of a type VI secretion baseplate. Nat Microbiol 3:1404–1416. doi: 10.1038/s41564-018-0260-1. [DOI] [PubMed] [Google Scholar]
- 29.Rapisarda C, Cherrak Y, Kooger R, Schmidt V, Pellarin R, Logger L, Cascales E, Pilhofer M, Durand E, Fronzes R. 2019. In situ and high-resolution cryo-EM structure of a bacterial type VI secretion system membrane complex. EMBO J 38:e100886. doi: 10.15252/embj.2018100886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunet YR, Espinosa L, Harchouni S, Mignot T, Cascales E. 2013. Imaging type VI secretion-mediated bacterial killing. Cell Rep 3:36–41. doi: 10.1016/j.celrep.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 32.Kapitein N, Bönemann G, Pietrosiuk A, Seyffer F, Hausser I, Locker JK, Mogk A. 2013. ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol Microbiol 87:1013–1028. doi: 10.1111/mmi.12147. [DOI] [PubMed] [Google Scholar]
- 33.Cascales E, Cambillau C. 2012. Structural biology of type VI secretion systems. Philos Trans R Soc Lond B Biol Sci 367:1102–1111. doi: 10.1098/rstb.2011.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeRoux M, De Leon JA, Kuwada NJ, Russell AB, Pinto-Santini D, Hood RD, Agnello DM, Robertson SM, Wiggins PA, Mougous JD. 2012. Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proc Natl Acad Sci U S A 109:19804–19809. doi: 10.1073/pnas.1213963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basler M, Ho BT, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, Catalano CE, Gonen T, Mougous JD. 2013. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell 51:584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. 2013. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durand E, Cambillau C, Cascales E, Journet L. 2014. VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol 22:498–507. doi: 10.1016/j.tim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 39.Whitney JC, Beck CM, Goo YA, Russell AB, Harding BN, De Leon JA, Cunningham DA, Tran BQ, Low DA, Goodlett DR, Hayes CS, Mougous JD. 2014. Genetically distinct pathways guide effector export through the type VI secretion system. Mol Microbiol 92:529–542. doi: 10.1111/mmi.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alcoforado Diniz J, Coulthurst SJ. 2015. Intraspecies competition in Serratia marcescens is mediated by Type VI-secreted Rhs effectors and a conserved effector-associated accessory protein. J Bacteriol 197:2350–2360. doi: 10.1128/JB.00199-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alcoforado Diniz J, Liu YC, Coulthurst SJ. 2015. Molecular weaponry: diverse effectors delivered by the Type VI secretion system. Cell Microbiol 17:1742–1751. doi: 10.1111/cmi.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unterweger D, Kostiuk B, Ötjengerdes R, Wilton A, Diaz-Satizabal L, Pukatzki S. 2015. Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J 34:2198–2210. doi: 10.15252/embj.201591163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitney JC, Quentin D, Sawai S, LeRoux M, Harding BN, Ledvina HE, Tran BQ, Robinson H, Goo YA, Goodlett DR, Raunser S, Mougous JD. 2015. An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell 163:607–619. doi: 10.1016/j.cell.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flaugnatti N, Le TT, Canaan S, Aschtgen MS, Nguyen VS, Blangy S, Kellenberger C, Roussel A, Cambillau C, Cascales E, Journet L. 2016. A phospholipase A(1) antibacterial Type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol Microbiol 99:1099–1118. doi: 10.1111/mmi.13292. [DOI] [PubMed] [Google Scholar]
- 45.Unterweger D, Kostiuk B, Pukatzki S. 2017. Adaptor proteins of type VI secretion system effectors. Trends Microbiol 25:8–10. doi: 10.1016/j.tim.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Coyne MJ, Comstock LE. 2019. Type VI secretion systems and the gut microbiota. Microbiol Spectr 7. doi: 10.1128/microbiolspec.PSIB-0009-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu Y, Waldor MK, Mekalanos JJ. 2013. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14:652–663. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bachmann V, Kostiuk B, Unterweger D, Diaz-Satizabal L, Ogg S, Pukatzki S. 2015. Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Negl Trop Dis 9:e0004031. doi: 10.1371/journal.pntd.0004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wexler AG, Bao Y, Whitney JC, Bobay LM, Xavier JB, Schofield WB, Barry NA, Russell AB, Tran BQ, Goo YA, Goodlett DR, Ochman H, Mougous JD, Goodman AL. 2016. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci U S A 113:3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, Durand E, Journet L, Cascales E, Monack DM. 2016. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A 113:E5044–5051. doi: 10.1073/pnas.1608858113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson MC, Vonaesch P, Saffarian A, Marteyn BS, Sansonetti PJ. 2017. Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe 21:769–776. doi: 10.1016/j.chom.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 52.Bernard CS, Brunet YR, Gueguen E, Cascales E. 2010. Nooks and crannies in type VI secretion regulation. J Bacteriol 192:3850–3860. doi: 10.1128/JB.00370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leung KY, Siame BA, Snowball H, Mok YK. 2011. Type VI secretion regulation: crosstalk and intracellular communication. Curr Opin Microbiol 14:9–15. doi: 10.1016/j.mib.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 54.Silverman JM, Brunet YR, Cascales E, Mougous JD. 2012. Structure and regulation of the type VI secretion system. Annu Rev Microbiol 66:453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyata ST, Bachmann V, Pukatzki S. 2013. Type VI secretion system regulation as a consequence of evolutionary pressure. J Med Microbiol 62:663–676. doi: 10.1099/jmm.0.053983-0. [DOI] [PubMed] [Google Scholar]
- 56.LeRoux M, Kirkpatrick RL, Montauti EI, Tran BQ, Peterson SB, Harding BN, Whitney JC, Russell AB, Traxler B, Goo YA, Goodlett DR, Wiggins PA, Mougous JD. 2015. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. Elife 4. doi: 10.7554/eLife.05701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.LeRoux M, Peterson SB, Mougous JD. 2015. Bacterial danger sensing. J Mol Biol 427:3744–3753. doi: 10.1016/j.jmb.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. 2007. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol 9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 59.Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. 2006. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol 61:1267–1282. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- 60.Journet L, Cascales E. 2016. The type VI secretion system in Escherichia coli and related species. EcoSalPlus 7:1–20. doi: 10.1128/ecosalplus.ESP-0009-2015. [DOI] [PubMed] [Google Scholar]
- 61.Morin N, Santiago AE, Ernst RK, Guillot SJ, Nataro JP. 2013. Characterization of the AggR regulon in enteroaggregative Escherichia coli. Infect Immun 81:122–132. doi: 10.1128/IAI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brunet YR, Bernard CS, Gavioli M, Lloubès R, Cascales E. 2011. An epigenetic switch involving overlapping fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet 7:e1002205. doi: 10.1371/journal.pgen.1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grisolia V, Carlomagno MS, Bruni CB. 1982. Cloning and expression of the distal portion of the histidine operon of Escherichia coli K-12. J Bacteriol 151:692–700. doi: 10.1128/JB.151.2.692-700.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grisolia V, Riccio A, Bruni CB. 1983. Structure and function of the internal promoter (hisBp) of the Escherichia coli K-12 histidine operon. J Bacteriol 155:1288–1296. doi: 10.1128/JB.155.3.1288-1296.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jackson EN, Yanofsky C. 1972. Internal promoter of the tryptophan operon of Escherichia coli is located in a structural gene. J Mol Biol 69:307–313. doi: 10.1016/0022-2836(72)90232-x. [DOI] [PubMed] [Google Scholar]
- 66.Saint Girons I, Margarita D. 1985. Evidence for an internal promoter in the Escherichia coli threonine operon. J Bacteriol 161:461–462. doi: 10.1128/JB.161.1.461-462.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wek RC, Hatfield GW. 1986. Examination of the internal promoter, PE, in the ilvGMEDA operon of E. coli K-12. Nucleic Acids Res 14:2763–2777. doi: 10.1093/nar/14.6.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maciag A, Piazza A, Riccardi G, Milano A. 2009. Transcriptional analysis of ESAT-6 cluster 3 in Mycobacterium smegmatis. BMC Microbiol 9:48. doi: 10.1186/1471-2180-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peterson SN, Reich NO. 2006. GATC flanking sequences regulate Dam activity: evidence for how Dam specificity may influence pap expression. J Mol Biol 355:459–472. doi: 10.1016/j.jmb.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 70.Wion D, Casadesús J. 2006. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol 4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Casadesús J, Low D. 2006. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev 70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sánchez-Romero MA, Casadesús J. 2020. The bacterial epigenome. Nat Rev Microbiol 18:7–20. doi: 10.1038/s41579-019-0286-2. [DOI] [PubMed] [Google Scholar]
- 73.Camacho EM, Serna A, Madrid C, Marqués S, Fernández R, de la Cruz F, Juárez A, Casadesús J. 2005. Regulation of finP transcription by DNA adenine methylation in the virulence plasmid of Salmonella enterica. J Bacteriol 187:5691–5699. doi: 10.1128/JB.187.16.5691-5699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haagmans W, van der Woude M. 2000. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol Microbiol 35:877–887. doi: 10.1046/j.1365-2958.2000.01762.x. [DOI] [PubMed] [Google Scholar]
- 75.Waldron DE, Owen P, Dorman CJ. 2002. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol Microbiol 44:509–520. doi: 10.1046/j.1365-2958.2002.02905.x. [DOI] [PubMed] [Google Scholar]
- 76.Peterson SN, Reich NO. 2008. Competitive Lrp and Dam assembly at the pap regulatory region: implications for mechanisms of epigenetic regulation. J Mol Biol 383:92–105. doi: 10.1016/j.jmb.2008.07.086. [DOI] [PubMed] [Google Scholar]
- 77.Eraso JM, Weinstock GM. 1992. Anaerobic control of colicin E1 production. J Bacteriol 174:5101–5109. doi: 10.1128/jb.174.15.5101-5109.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller J. 1972. Experiments in Molecular Genetics, p 352–355. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 79.Bernard CS, Brunet YR, Gavioli M, Lloubès R, Cascales E. 2011. Regulation of type VI secretion gene clusters by sigma54 and cognate enhancer binding proteins. J Bacteriol 193:2158–2167. doi: 10.1128/JB.00029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.