The formation of ribosome dimers during periods of dormancy is widespread among bacteria. Dimerization is typically mediated by a single protein, hibernation-promoting factor (HPF). Bacteria lacking HPF exhibit strong defects in viability and pathogenesis and, in some species, extreme loss of rRNA. The mechanistic basis of these phenotypes has not been determined. Here, we report that HPF from the Gram-positive bacterium Bacillus subtilis preserves ribosomes by preventing the loss of essential ribosomal proteins at the dimer interface. This protection may explain phenotypes associated with the loss of HPF, since ribosome protection would aid survival during nutrient limitation and impart a strong selective advantage when the bacterial cell rapidly reinitiates growth in the presence of sufficient nutrients.
KEYWORDS: stationary phase, translation
ABSTRACT
When nutrients become scarce, bacteria can enter an extended state of quiescence. A major challenge of this state is how to preserve ribosomes for the return to favorable conditions. Here, we show that the ribosome dimerization protein hibernation-promoting factor (HPF) functions to protect essential ribosomal proteins. Ribosomes isolated from strains lacking HPF (Δhpf) or encoding a mutant allele of HPF that binds the ribosome but does not mediate dimerization were substantially depleted of the small subunit proteins S2 and S3. Strikingly, these proteins are located directly at the ribosome dimer interface. We used single-particle cryo-electron microscopy (cryo-EM) to further characterize these ribosomes and observed that a high percentage of ribosomes were missing S2, S3, or both. These data support a model in which the ribosome dimerization activity of HPF evolved to protect labile proteins that are essential for ribosome function. HPF is almost universally conserved in bacteria, and HPF deletions in diverse species exhibit decreased viability during starvation. Our data provide mechanistic insight into this phenotype and establish a mechanism for how HPF protects ribosomes during quiescence.
IMPORTANCE The formation of ribosome dimers during periods of dormancy is widespread among bacteria. Dimerization is typically mediated by a single protein, hibernation-promoting factor (HPF). Bacteria lacking HPF exhibit strong defects in viability and pathogenesis and, in some species, extreme loss of rRNA. The mechanistic basis of these phenotypes has not been determined. Here, we report that HPF from the Gram-positive bacterium Bacillus subtilis preserves ribosomes by preventing the loss of essential ribosomal proteins at the dimer interface. This protection may explain phenotypes associated with the loss of HPF, since ribosome protection would aid survival during nutrient limitation and impart a strong selective advantage when the bacterial cell rapidly reinitiates growth in the presence of sufficient nutrients.
INTRODUCTION
The majority of bacteria exist in a quiescent state and live in nutrient-limited conditions where the conservation of resources is crucial for long-term survival (1). Protein synthesis consumes more than half of the energy in the cell during active growth (2); thus, when nutrients become scarce, it must be tightly controlled (3). Protein synthesis is catalyzed by the ribosome, a large macromolecular complex (molecular weight of ∼2.6 million daltons) that constitutes >25% of the mass of the cell (4, 5). The production of new ribosomes is the most energy-intensive process in the cell (6); therefore, a mechanism to preserve active ribosomes imparts a strong selective advantage during stress and nutrient limitation.
Ribosomes in quiescent cells ranging from bacteria (3, 7, 8) to mammalian cells (9, 10) form dimers. However, the functional consequence of dimerization remains mysterious. In bacteria, ribosome dimerization is mediated by hibernation-promoting factor (HPF). Two monomers of HPF bind two 70S ribosomes and interact at their C termini to form a 100S ribosome dimer (11). Most bacteria encode an HPF homolog, and mutants lacking HPF exhibit pleiotropic phenotypes, including decreased viability during extended stationary phase (12), increased antibiotic sensitivity (13), decreased virulence (14), and a reduction in protein synthesis and growth after nutrient limitation (15, 16). Ribosome dimerization may inhibit protein synthesis (17, 18), and bacteria lacking HPF have more polysomes, which is indicative of increased protein synthesis (19). However, phenotypes associated with an HPF deletion may also be explained by ribosome instability, as rRNA is degraded in some mutants lacking HPF (15, 20, 21).
We investigated how HPF contributes to the physiology of quiescence in B. subtilis, which contains a well-characterized HPF homolog that dimerizes ribosomes during stationary phase (11, 16). Ribosomes harvested from stationary-phase cells lacking HPF were severely attenuated in protein synthesis. A mutant form of HPF that binds the ribosome but does not induce dimer formation also failed to preserve the translational activity of stationary-phase ribosomes. Using mass spectrometry to characterize these ribosomes, we found that two essential proteins of the small ribosomal subunit located at the dimer interface were significantly depleted in both HPF-deficient mutants. Cryo-EM analysis of ribosomes harvested from stationary-phase cells lacking HPF demonstrated that a substantial population of ribosomes was missing one or more essential proteins of the small subunit. Thus, our results support a model in which dimerization mediated by HPF preserves ribosomes for when conditions become favorable.
RESULTS
Ribosomes harvested from stationary-phase Δhpf cells are inactive.
Ribosome dimerization mediated by HPF occurs during stationary phase in many bacteria (22), including B. subtilis (see Fig. S1 in the supplemental material). Phenotypes of B. subtilis Δhpf strains include a reduction in protein synthesis and a longer lag phase after nutrient limitation (15, 16). Since these phenotypes suggest that Δhpf strains may have impaired translation capacity, we hypothesized that HPF is necessary to preserve ribosomes during stationary phase. To investigate this, we harvested ribosomes from wild-type (JDB1772) and Δhpf (JDB4221) strains in stationary phase (optical density [OD] of ∼7.0; corresponding to time point number 5) (Fig. S1A) and compared their activity by monitoring the production of a reporter protein by a PURExpress in vitro translation system containing these ribosomes (23, 24). Ribosomes harvested from stationary-phase wild-type cultures produced protein within 10 min, similar to ribosomes harvested from an exponential-phase culture (Fig. 1). The stationary-phase ribosomes were present mainly as dimers upon addition to the reaction (Fig. S1). This experiment, therefore, suggests that the 100S dimer can be disassembled into free subunits that can then initiate translation in vitro, indicating that stoichiometric amounts of HPF can be removed from the ribosome in a highly purified translation system. This result is consistent with the observation that ribosome dimers are resolved within minutes after entry into fresh media (8).
FIG 1.
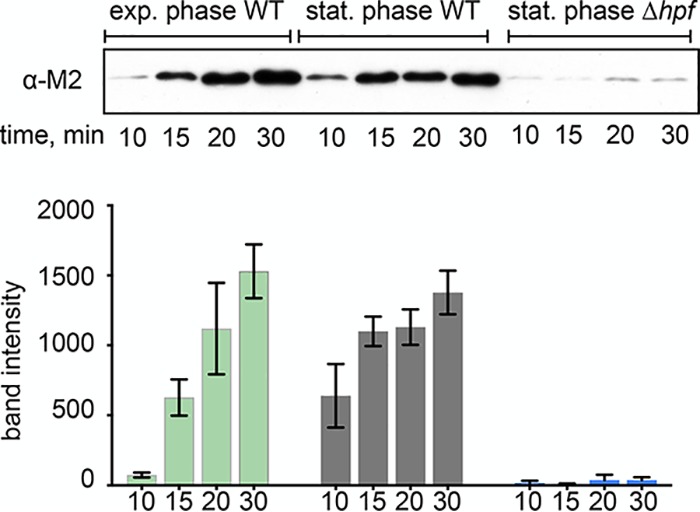
Stationary-phase ribosomes harvested from HPF null strains are inactive. Ribosomes harvested from exponential-phase or stationary-phase wild-type cells (JDB1772) or from stationary-phase Δhpf (JDB4221) cells were assayed in a purified ribosome-free in vitro translation system. Sedimentation profiles of wild-type stationary-phase ribosomes showed that these ribosomes were present mainly as dimers. The production of an M2-tagged protein encoded by an added template was monitored by Western blot with an anti-M2 antibody (top). Reactions proceeded for 10, 15, 20, or 30 min at 37°C. The bar graph shows band intensity from 3 independent translation experiments. Error bars indicate standard deviation.
In contrast, ribosomes from a stationary-phase culture of a Δhpf strain were almost entirely inactive. Protein produced by Δhpf ribosomes was barely detectable even after 30 min of incubation, and Δhpf ribosomes exhibited a 97% ± 2% reduction in protein synthesis compared with the wild-type stationary-phase ribosomes (Fig. 1). These data suggest that HPF is required for maintaining active ribosomes.
The dimerization function of HPF is required for maintaining active ribosomes.
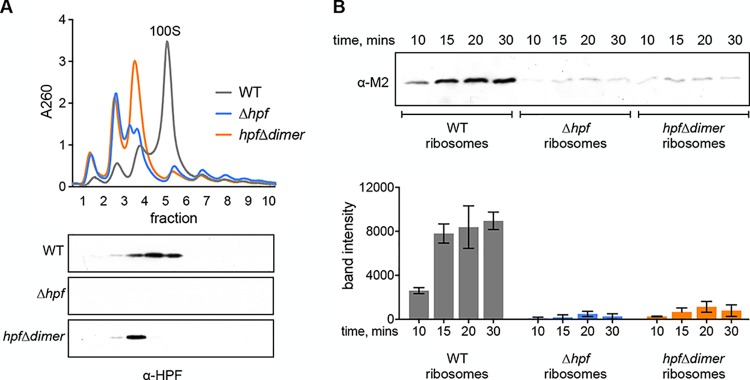
The ribosome dimerization activity of HPF proteins is broadly conserved (22), but the functional consequences of dimerization are not clear. B. subtilis strains encoding a defective HPF that can bind the ribosome but cannot form ribosome dimers are equally defective in recovery from stationary phase as a strain lacking HPF (11). Since ribosomes harvested from the HPF null strain were almost completely inactive in vitro, we hypothesized that the dimerization activity of HPF is required to maintain active ribosomes in the stationary phase. We tested this by constructing a strain expressing HPFΔdimer, an HPF mutant that binds the ribosome but is unable to form dimers. Comigration of this mutant protein, which encodes an M2 tag at the C terminus, with the 70S peak indicates that it binds the ribosome but fails to mediate dimerization (Fig. 2A). We purified ribosomes from stationary-phase cultures (optical density at 600 nm [OD600] of ∼7.0) of this strain (hpfΔdimer), as well as from wild-type and Δhpf strains. Ribosomes harvested from the hpfΔdimer strain were almost as inactive as the HPF null strain (Fig. 2B) and exhibited a 91% ± 7% reduction in protein synthesis at 30 min compared with the wild type. Thus, ribosome dimerization, and not just ribosome binding, is necessary for preserving active ribosomes.
FIG 2.
The dimerization function of HPF is required for maintaining active ribosomes. Ribosomes were harvested from the wild type (JDB1772), a Δhpf strain (JDB4221), or a strain with a mutant HPF that can bind the ribosome but does not form ribosome dimers (hpfΔdimer strain [JDB4227]) grown to stationary phase. (A) Sucrose density gradients of ribosomes. Fractions collected from gradients were probed with a polyclonal antibody raised against HPF. (B) Western blot with quantification from independent experiments showing translational activity of ribosomes harvested from wild-type, Δhpf, and hpfΔdimer strains. Error bars represent standard deviation.
Stationary-phase ribosomes from strains lacking functional HPF are depleted for essential proteins.
Ribosomes isolated from strains lacking a functional HPF (Δhpf and hpfΔdimer) are defective in translation in vitro (Fig. 1 and 2B). rRNA was examined on an agarose gel and appeared intact in all strains (see Fig. S2 in the supplemental material), suggesting that the ribosomal protein composition may be different from wild-type ribosomes. To identify differences, we analyzed ribosomes by mass spectrometry (Fig. 3A; Table 1). S2 and S3, two essential proteins of the small subunit, were significantly depleted in ribosomes derived from both strains relative to the wild type (Fig. 3A; Table 1). A Western blot using an antibody to S2 of whole-cell lysates of either wild-type or Δhpf cells revealed that S2 levels are similar in both strains (Fig. S2). Thus, since S2 levels were lower in ribosomes harvested from HPF mutants compared with the wild type but not in whole-cell extracts, it is likely that S2 is free in the cytoplasm.
FIG 3.
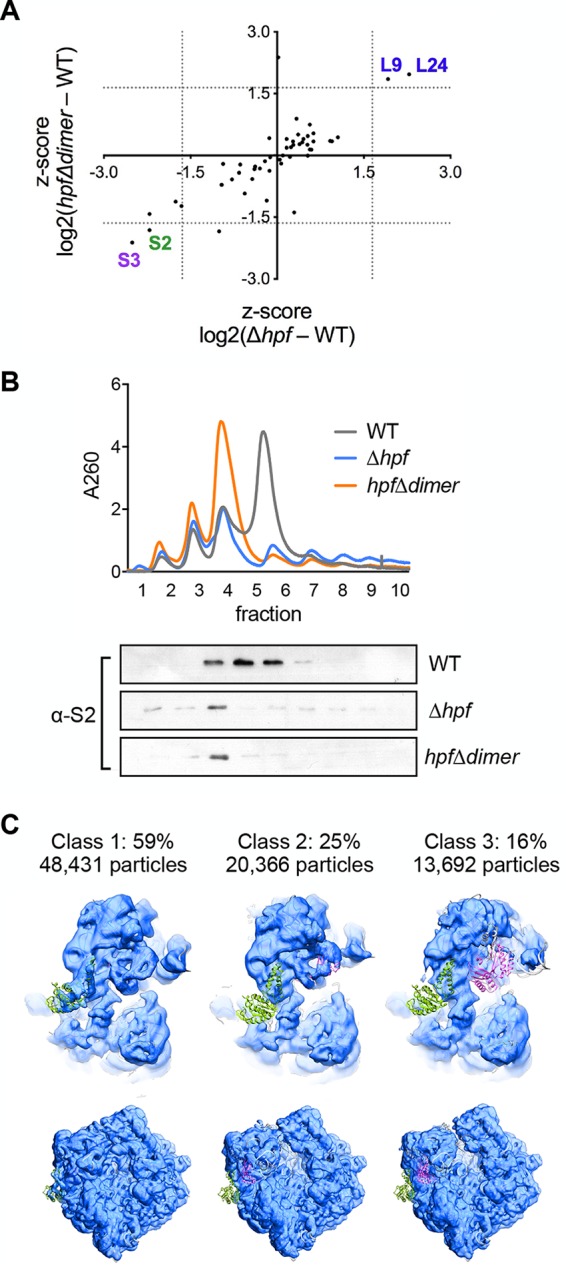
Ribosomes from strains lacking functional HPF are depleted for S2 and S3. Ribosomes were harvested from wild-type (JDB1772) cells or a Δhpf (JDB4221) or hpfΔdimer (JDB4227) strain in stationary phase and analyzed by mass spectrometry and by Western blot. (A) Plot of the difference in levels of ribosomal proteins between the wild type and each HPF mutant. Proteins in the bottom left quadrant are depleted in both mutants compared with the wild type. Proteins in the top right quadrant are enriched in both mutants compared with the wild type. Dotted lines indicate a Z-score value of 1.645. (B, top) Sucrose density gradients of ribosomes purified from each strain. (Bottom) Gradient fractions were probed with a polyclonal antibody raised against purified S2 protein. (C) Cryo-EM analysis of ribosomes harvested from the Δhpf strain in stationary phase. The small subunit model from the structure of the 70S ribosome (PDB code 4V4A) (ribbon diagram) was rigid-body fitted into the maps of three different classes of the Δhpf ribosome (blue). Proteins S2 (green) and S3 (purple) are indicated. Two views are shown, namely, a zoomed-in view of the small subunit (top) and the complete 70S particle (bottom).
TABLE 1.
Proteins most enriched in wild-type or HPF-deficient ribosomesa
| Gene name by category | Protein | Log2(Δhpf-wt)b | Log2(hpfΔdimer-wt)c |
|---|---|---|---|
| Most depleted in HPF-deficient mutants | |||
| hpf | Hibernation-promoting factor (HPF) | −2.64 ± 0.21 | −0.83 |
| fusA | Elongation factor G | −0.75 ± 0.07 | −0.52 |
| rpsC | 30S ribosomal protein S3 | −0.61 ± 0.18 | −0.37 |
| rpsB | 30S ribosomal protein S2 | −0.53 ± 0.13 | −0.31 |
| Most enriched in HPF-deficient mutants | |||
| rbfA | Ribosome biogenesis factor RbfA | 1.15 ± 0.01 | 0.57 |
| rsfS | Ribosome silencing factor RsfS | 0.83 ± 0.08 | 0.84 |
| rimM | Ribosome maturation factor RimM | 0.71 ± 0.21 | 0.48 |
| rimP | Ribosome maturation factor RimP | 0.65 ± 0.28 | 0.48 |
Values were taken from Data Set S1 in the supplemental material. The highest-ranking ribosomal proteins or known ribosome-interacting proteins with either positive or negative values are listed.
The average from two biological replicates.
Reports values from one sample.
To confirm that S2 is missing from the ribosomes, we used a polyclonal antibody raised against B. subtilis S2 and probed fractions collected from ribosome gradients of wild-type and the two HPF-deficient mutant strains. In ribosomes isolated from both mutant strains, S2 levels were significantly reduced, confirming the results of the mass spectrometry (Fig. 3B). Thus, the loss of S2 and/or S3 may be sufficient to explain the defect of ribosomes isolated from cells lacking a functional HPF.
Cryo-EM reveals classes of ribosomes lacking S2 and S3.
Since both S2 and S3 are essential for translation, the loss of only one of these proteins would be sufficient to inactivate the ribosome. We hypothesized that ribosomal subpopulations lacking only one of these proteins, or both, exist. To test this, we used single-particle cryo-EM to further analyze the composition of different classes of ribosomes in the population. The Δhpf strain was grown to stationary phase, and the 70S ribosomal peak was isolated from a sucrose gradient and analyzed by cryo-EM. We observed three abundant classes of 70S ribosomes after classification and refinement. To determine which ribosomal proteins were absent, we rigid-body fitted the model of a small subunit of the Escherichia coli 70S ribosome (PDB code 4V4A) (25) into cryo-EM maps for each of these classes (Fig. 3C). The first class, comprising 59% of the particles, had prominent density in the regions of both S2 and S3. A second class of particles (25%) lacked density in the region of S2, and the third class (16%) lacked density in the region of both S2 and S3 even at higher thresholds.
DISCUSSION
We find that B. subtilis HPF contributes to ribosome preservation by protecting essential proteins found on the small ribosomal subunit at the dimer interface. In strains that lack HPF or express an HPF mutant unable to mediate ribosome dimerization, ribosomal proteins S2 and S3 were significantly depleted from stationary-phase ribosomes (Fig. 3A and B). Cryo-electron microscopy further confirmed that a significant population of ribosomes harvested from a Δhpf strain in stationary phase were missing S2, S3, or both (Fig. 3C). Thus, dimerization is a mechanism to protect essential ribosomal proteins susceptible to loss and/or degradation (Fig. 4).
FIG 4.
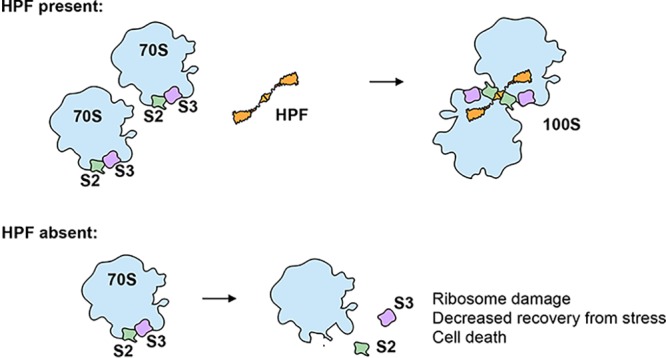
Model for HPF-mediated protection of ribosomes in B. subtilis. HPF binds two 70S ribosomes to form a 100S dimer. S2 and S3 proteins at the dimer interface (green and purple) are protected and occluded from the solvent surface. If HPF is not functional, S2 and S3 are exposed and may not remain bound to the ribosome.
Ribosomes isolated from stationary-phase cells lacking HPF are not active in vitro (Fig. 1). This finding may explain why B. subtilis strains lacking HPF exhibit decreased recovery from stationary phase. Interestingly, a B. subtilis strain expressing a mutant form of HPF that can bind the ribosome but cannot mediate dimer formation also exhibits this phenotype (11, 16). We tested a strain expressing a similar mutant protein (the hpfΔdimer strain) and found that ribosomes harvested from this strain were nearly as inactive as ribosomes from the HPF null strain. We hypothesized that ribosomes harvested from either the Δhpf or the hpfΔdimer strain might be defective in similar ways. Therefore, we characterized ribosomes from both mutants by mass spectrometry (Table 1), and we found that they deviated significantly from wild-type ribosomes in their protein composition (Fig. 3A). In particular, both mutants were significantly depleted for S2 and S3, essential proteins of the small ribosomal subunit that are located directly at the interface between the two ribosomes of the dimer. The protection of S2 is consistent with the presence of numerous direct contacts between HPF and S2 (11); thus, in free 70S or 30S complexes, S2 is much more exposed than in the 100S dimer configuration. S3 is near the dimer interface and participates in dimer formation in E. coli but not in B. subtilis (11, 18). However, ribosome dimers are flexible, and as there is some movement of the 70S monomers relative to one another, S3 may also be protected by dimer formation (11, 26).
Why are S2 and S3 specifically in need of protection during stationary phase? S2 and S3 are the last proteins to be added to the ribosome during assembly and, thus, may easily dissociate from the ribosome (27, 28). As much as 17% of S2 has been detected free from the ribosome in the cytosol (29). Loss of S2/S3 may further destabilize the ribosome because ribosomes not participating in translation are especially susceptible to degradation due to exposure of the rRNA when the 30S and 50S subunits are separated (30). Cryo-EM analysis resolved specific populations of ribosomes with defects in the small subunit. Our analysis revealed three classes of 70S ribosomes following classification and refinement. Two of these classes, comprising 40% of these particles, were missing either S2 or S3. Since both of these proteins are essential for translation, these data indicate that a substantial portion of the ribosomes in the Δhpf strain would be inactive.
HPF is essential for preserving 16S or 23S rRNA in E. coli, Pseudomonas aeruginosa, and Mycobacterium smegmatis (15, 20, 21), but significant degradation of rRNA has not been reported in B. subtilis and was not observed in this study (Fig. S2). Could the loss of S2 and S3 be the first step in the degradation of ribosomes in these species? Loss of S2 and S3 and the subsequent failure to translate and form active 70S ribosomes could contribute to further destabilization of the ribosome and eventual loss of rRNA. Thus, the defective small subunit reported here may be a degradation intermediate on the way to further loss of rRNA.
Proteins most enriched in the HPF-deficient strains include factors associated with immature ribosomes (Table 1). One of these proteins is the ribosome maturation factor RbfA that binds the 30S subunit in a position that overlaps with the A- and P-sites similar to HPF (31) and prevents initiation by a 30S subunit that has not fully matured (32, 33). The ribosome maturation factors L24, RimM, and RimP were also enriched in the HPF-deficient ribosomes. Mutations in E. coli RimM lead to the loss of both S2 and S3 from immature 30S subunits (34). Similarly, ribosomes isolated from E. coli strains lacking RimP are depleted for S2, S3, and S21 (35). Since S2 and S3 are late-binding proteins, the association of these factors with ribosomes isolated from Δhpf strains further indicates that the 30S is structurally similar to an immature small subunit. L24 is involved in the maturation of the 50S subunit and is not required for the activity of the large subunit in peptide bond formation (36). The large subunit protein L9 was also enriched in the mutant ribosomes. L9 is not essential in B. subtilis (37) but has recently been shown to contribute to translation fidelity by preventing compaction during ribosome collisions (38). Another protein more abundant in both mutant ribosomes is the ribosome silencing factor RsfS that binds the ribosome via ribosomal protein L14 (39), interferes with subunit joining, and modestly inhibits protein synthesis (40). However, deletion of rsfS did not rescue the inactive phenotype of Δhpf ribosomes (see Fig. S4 in the supplemental material), indicating that increased RsfS binding to Δhpf and hpfΔdimer ribosomes is not sufficient to explain their inactivity.
Given that ribosome synthesis is the single most energy-consuming process in cells, preventing ribosome degradation, even when protein synthesis levels are low, is evolutionarily favorable (6). A readily available, excess store of ribosomes under nutrient limitation would facilitate rapid resumption of growth when nutrients become available (41). The ability to transition to maximal protein synthesis as quickly as possible imparts a clear selective advantage for cells with higher ribosomal content (42, 43). Consistent with this interpretation, stationary-phase B. subtilis cells lacking HPF (16) or expressing a mutant HPF unable to dimerize ribosomes exhibit both decreased viability during stationary phase and an increased lag phase following transfer to fresh media (11, 16). The long-term protection of excess ribosomes depends specifically on the ability of the cell to store ribosomes in the form of dimers, a mechanism that may have evolved to guard against the loss of essential proteins of the small subunit.
MATERIALS AND METHODS
Strains and medium.
Strains were derived from B. subtilis 168 trpC2 and grown in LB. For detailed information on strain construction, see the supplemental material.
Sucrose gradient density centrifugation.
Cells were lysed using a Fast Prep machine in ribosome gradient buffer (20 mM Tris-acetate [pH4°C 7.4], 60 mM ammonium chloride, 7.5 mM magnesium acetate, 6 mM β-mercaptoethanol, and 0.5 mM EDTA). Lysate was cleared of debris by centrifugation at 4°C at 20,000 relative centrifugal force (rcf) for 20 min. Ribosomes were purified by sucrose cushion (37.7% sucrose in ribosome gradient buffer). A260 for lysates or purified ribosomes was determined by Nanodrop, and 100 μl of normalized lysate or ribosomes (as indicated in figure legends) was loaded on a 10% to 40% sucrose gradient in ribosome gradient buffer. Gradients were run for 3 hours at 30,000 rpm in a Beckman SW41 rotor. Samples were collected with a BioComp gradient station and a BioComp TRiAX flow cell monitoring continuous absorbance at 260 nm.
In vitro translation.
Ribosomes for in vitro translation were harvested by loading cleared cell lysate on a 1.5-ml buffered sucrose cushion (20 mM Tris-acetate [pH4°C 7.4], 100 mM ammonium chloride, 10 mM magnesium acetate, 0.5 mM EDTA, 6 mM β-mercaptoethanol, and 37.7% sucrose). Ribosomes were pelleted in a Beckman TLA100.3 rotor at 85,000 rpm for 2 hours. Pellets were washed and resuspended in ribosome gradient buffer by gentle agitation at 4°C. Ribosomes were added to 100 nM to the PURExpress ribosome-free kit (New England BioLabs) (24). A template encoding an M2-tagged peptide was added to the reaction which was then resolved by SDS-PAGE and probed with an anti-M2 antibody (Sigma). Further details are described in the supplemental material.
Mass spectrometry.
Protein sample processing and protein quantification by mass spectrometry was performed as described previously, with minor modifications (44, 45). Raw data were analyzed with MaxQuant software version 1.6.0.16 (46) using a UniProt Bacillus subtilis 168 database (proteome UP000001570; downloaded March 2019).
Cryo-electron microscopy sample preparation and data acquisition.
Ribosomes harvested from the Δhpf strain were resolved on a 10% to 40% sucrose gradient as described. The 70S peak was isolated, exchanged into ribosome buffer without sucrose, and concentrated with a Corning 100K cutoff polyethersulfone (PES) filter to 200 nM. The sample was applied to plasma-treated holey carbon grids and vitrified in liquid ethane using a FEI Vitrobot system. Images were automatically acquired using Leginon/Appion (47, 48) on a Titan Krios microscope (ThermoFisher Scientific) operated at 300 keV and equipped with a K2 direct electron detector (Gatan) and a postcolumn energy filter (operated at 20-eV slit width). Movies were recorded in counting mode for 8 s (20 ms/frame), with a total dose on a specimen level of 56.7 e-/Å2. Defocus range was set to vary between 0.7 and 2.0 μm. Raw frames were aligned and dose-weighted using MotionCor2 (49). Contrast transfer function (CTF) estimation was done with CTFFIND4 (50).
Cryo-electron microscopy data processing and heterogeneity analysis.
Particle picking was done using template-based picking (FindEM) in Appion (51) using templates generated from manually picked particles. Two-dimensional (2D) classification, ab initio reconstruction, and heterorefinement steps, as well as final refinements were done in cryoSPARC2 (52) using default settings and three classes (see Fig. S3 in the supplemental material). Consensus refinement and masked classification were done with RELION3 (53, 54). Final maps are submitted to the Electron Microscopy Data Bank (EMDB) with the following accession codes: EMD-21467, EMD-21478, EMD-21479, and EMD-21480.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of our lab for helpful discussions and comments on the manuscript. We thank Sandro Pereira for constructing the Δhpf (JDB4221) strain. We thank Luke Berchowitz for generously providing access to his ribosome gradient analyzer.
H.A.F. was supported by NIH F32GM122266; M.J. was supported by R35GM128802; and J.D. was supported by R01 GM114213 and R21AI135427 and is a Burroughs-Wellcome Investigator in the Pathogenesis of Infection Disease. Some of this work was performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Rittershaus ES, Baek SH, Sassetti CM. 2013. The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe 13:643–651. doi: 10.1016/j.chom.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tempest DW, Neijssel OM. 1984. The status of YATP and maintenance energy as biologically interpretable phenomena. Annu Rev Microbiol 38:459–486. doi: 10.1146/annurev.mi.38.100184.002331. [DOI] [PubMed] [Google Scholar]
- 3.Wilson DN, Nierhaus KH. 2007. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol 42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 4.Voorhees RM, Ramakrishnan V. 2013. Structural basis of the translational elongation cycle. Annu Rev Biochem 82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 5.Green R, Noller HF. 1997. Ribosomes and translation. Annu Rev Biochem 66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 6.Maitra A, Dill KA. 2015. Bacterial growth laws reflect the evolutionary importance of energy efficiency. Proc Natl Acad Sci U S A 112:406–411. doi: 10.1073/pnas.1421138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wada A, Yamazaki Y, Fujita N, Ishihama A. 1990. Structure and probable genetic location of a “ribosome modulation factor” associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc Natl Acad Sci U S A 87:2657–2661. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maki Y, Yoshida H, Wada A. 2000. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells 5:965–974. doi: 10.1046/j.1365-2443.2000.00389.x. [DOI] [PubMed] [Google Scholar]
- 9.Krokowski D, Gaccioli F, Majumder M, Mullins MR, Yuan CL, Papadopoulou B, Merrick WC, Komar AA, Taylor D, Hatzoglou M. 2011. Characterization of hibernating ribosomes in mammalian cells. Cell Cycle 10:2691–2702. doi: 10.4161/cc.10.16.16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franken LE, Oostergetel GT, Pijning T, Puri P, Arkhipova V, Boekema EJ, Poolman B, Guskov A. 2017. A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy. Nat Commun 8:722. doi: 10.1038/s41467-017-00718-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckert B, Abdelshahid M, Schafer H, Steinchen W, Arenz S, Berninghausen O, Beckmann R, Bange G, Turgay K, Wilson DN. 2017. Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J 36:2061–2072. doi: 10.15252/embj.201696189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basu A, Yap M-N. 2016. Ribosome hibernation factor promotes Staphylococcal survival and differentially represses translation. Nucleic Acids Res 44:4881–4893. doi: 10.1093/nar/gkw180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKay SL, Portnoy DA. 2015. Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob Agents Chemother 59:6992–6999. doi: 10.1128/AAC.01532-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kline BC, McKay SL, Tang WW, Portnoy DA. 2015. The Listeria monocytogenes hibernation-promoting factor is required for the formation of 100S ribosomes, optimal fitness, and pathogenesis. J Bacteriol 197:581–591. doi: 10.1128/JB.02223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akiyama T, Williamson KS, Schaefer R, Pratt S, Chang CB, Franklin MJ. 2017. Resuscitation of Pseudomonas aeruginosa from dormancy requires hibernation promoting factor (PA4463) for ribosome preservation. Proc Natl Acad Sci U S A 114:3204–3209. doi: 10.1073/pnas.1700695114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akanuma G, Kazo Y, Tagami K, Hiraoka H, Yano K, Suzuki S, Hanai R, Nanamiya H, Kato-Yamada Y, Kawamura F. 2016. Ribosome dimerization is essential for the efficient regrowth of Bacillus subtilis. Microbiology 162:448–458. doi: 10.1099/mic.0.000234. [DOI] [PubMed] [Google Scholar]
- 17.Trösch R, Willmund F. 2019. The conserved theme of ribosome hibernation: from bacteria to chloroplasts of plants. Biol Chem 400:879–893. doi: 10.1515/hsz-2018-0436. [DOI] [PubMed] [Google Scholar]
- 18.Kato T, Yoshida H, Miyata T, Maki Y, Wada A, Namba K. 2010. Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure 18:719–724. doi: 10.1016/j.str.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Hood RD, Higgins SA, Flamholz A, Nichols RJ, Savage DF. 2016. The stringent response regulates adaptation to darkness in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci U S A 113:E4867–E4876. doi: 10.1073/pnas.1524915113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niven GW. 2004. Ribosome modulation factor protects Escherichia coli during heat stress, but this may not be dependent on ribosome dimerisation. Arch Microbiol 182:60–66. doi: 10.1007/s00203-004-0698-9. [DOI] [PubMed] [Google Scholar]
- 21.Trauner A, Lougheed KE, Bennett MH, Hingley-Wilson SM, Williams HD. 2012. The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. J Biol Chem 287:24053–24063. doi: 10.1074/jbc.M112.364851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prossliner T, Winther KS, Sørensen MA, Gerdes K. 2018. Ribosome hibernation. Annu Rev Genet 52:321–348. doi: 10.1146/annurev-genet-120215-035130. [DOI] [PubMed] [Google Scholar]
- 23.Shimizu Y, Kanamori T, Ueda T. 2005. Protein synthesis by pure translation systems. Methods 36:299–304. doi: 10.1016/j.ymeth.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Tuckey C, Asahara H, Zhou Y, Chong S. 2014. Protein synthesis using a reconstituted cell-free system. Curr Protoc Mol Biol 108:16.31.1–16.31.22. doi: 10.1002/0471142727.mb1631s108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vila-Sanjurjo A, Ridgeway WK, Seymaner V, Zhang W, Santoso S, Yu K, Cate JH. 2003. X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli. Proc Natl Acad Sci U S A 100:8682–8687. doi: 10.1073/pnas.1133380100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matzov D, Aibara S, Basu A, Zimmerman E, Bashan A, Yap M-N, Amunts A, Yonath AE. 2017. The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus. Nat Commun 8:723. doi: 10.1038/s41467-017-00753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Traub P, Nomura M. 1968. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc Natl Acad Sci U S A 59:777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. 2010. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science 330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulbrich B, Nierhaus KH. 1975. Pools of ribosomal proteins in Escherichia coli. Studies on the exchange of proteins between pools and ribosomes. Eur J Biochem 57:49–54. doi: 10.1111/j.1432-1033.1975.tb02275.x. [DOI] [PubMed] [Google Scholar]
- 30.Deutscher MP. 2003. Degradation of stable RNA in bacteria. J Biol Chem 278:45041–45044. doi: 10.1074/jbc.R300031200. [DOI] [PubMed] [Google Scholar]
- 31.Nord S, Bhatt MJ, Tukenmez H, Farabaugh PJ, Wikstrom PM. 2015. Mutations of ribosomal protein S5 suppress a defect in late-30S ribosomal subunit biogenesis caused by lack of the RbfA biogenesis factor. RNA 21:1454–1468. doi: 10.1261/rna.051383.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma IM, Woodson SA. 2019. IF3 licenses newly made 30S subunits for translation during stress. bioRxiv 10.1101/655696. [DOI]
- 33.Goto S, Kato S, Kimura T, Muto A, Himeno H. 2011. RsgA releases RbfA from 30S ribosome during a late stage of ribosome biosynthesis. EMBO J 30:104–114. doi: 10.1038/emboj.2010.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Q, Goto S, Chen Y, Feng B, Xu Y, Muto A, Himeno H, Deng H, Lei J, Gao N. 2013. Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process. Nucleic Acids Res 41:2609–2620. doi: 10.1093/nar/gks1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR. 2014. A combined quantitative mass spectrometry and electron microscopy analysis of ribosomal 30S subunit assembly in E. coli. Elife 3:e04491. doi: 10.7554/eLife.04491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spillmann S, Nierhaus KH. 1978. The ribosomal protein L24 of Escherichia coli is an assembly protein. J Biol Chem 253:7047–7050. [PubMed] [Google Scholar]
- 37.Akanuma G, Nanamiya H, Natori Y, Yano K, Suzuki S, Omata S, Ishizuka M, Sekine Y, Kawamura F. 2012. Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation. J Bacteriol 194:6282–6291. doi: 10.1128/JB.01544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith AM, Costello MS, Kettring AH, Wingo RJ, Moore SD. 2019. Ribosome collisions alter frameshifting at translational reprogramming motifs in bacterial mRNAs. Proc Natl Acad Sci U S A 116:21769–21779. doi: 10.1073/pnas.1910613116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Sun Q, Jiang C, Yang K, Hung LW, Zhang J, Sacchettini JC. 2015. Structure of ribosomal silencing factor bound to Mycobacterium tuberculosis ribosome. Structure 23:1858–1865. doi: 10.1016/j.str.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Häuser R, Pech M, Kijek J, Yamamoto H, Titz B, Naeve F, Tovchigrechko A, Yamamoto K, Szaflarski W, Takeuchi N, Stellberger T, Diefenbacher ME, Nierhaus KH, Uetz P. 2012. RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet 8:e1002815. doi: 10.1371/journal.pgen.1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korem Kohanim Y, Levi D, Jona G, Towbin BD, Bren A, Alon U. 2018. A bacterial growth law out of steady state. Cell Rep 23:2891–2900. doi: 10.1016/j.celrep.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Mori M, Schink S, Erickson DW, Gerland U, Hwa T. 2017. Quantifying the benefit of a proteome reserve in fluctuating environments. Nat Commun 8:1225. doi: 10.1038/s41467-017-01242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Remigi P, Ferguson GC, McConnell E, De Monte S, Rogers DW, Rainey PB. 2019. Ribosome provisioning activates a bistable switch coupled to fast exit from stationary phase. Mol Biol Evol 36:1056–1070. doi: 10.1093/molbev/msz041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng Z, Otto GM, Powers EN, Keskin A, Mertins P, Carr SA, Jovanovic M, Brar GA. 2018. Pervasive, coordinated protein-level changes driven by transcript isoform switching during meiosis. Cell 172:910–923.e16. doi: 10.1016/j.cell.2018.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng Z, Mugler CF, Keskin A, Hodapp S, Chan LY, Weis K, Mertins P, Regev A, Jovanovic M, Brar GA. 2019. Small and large ribosomal subunit deficiencies lead to distinct gene expression signatures that reflect cellular growth rate. Mol Cell 73:36–47.e10. doi: 10.1016/j.molcel.2018.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 47.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. 2005. Automated molecular microscopy: the new Leginon system. J Struct Biol 151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 48.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B. 2009. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohou A, Grigorieff N. 2015. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol 192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roseman AM. 2004. FindEM—a fast, efficient program for automatic selection of particles from electron micrographs. J Struct Biol 145:91–99. doi: 10.1016/j.jsb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 52.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. 2017. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 53.Scheres SH. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH. 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.