Elevation of Ap4A level in bacteria is associated with increased sensitivity to heat and oxidative stress, reduced antibiotic tolerance, and decreased pathogenicity. ApaH is the major Ap4A hydrolase in gamma- and betaproteobacteria and has been recently proposed as a novel target to weaken the bacterial resistance to antibiotics. Here, we identified the orphan YqeK protein family (COG1713) as a highly efficient Ap4A hydrolase family, with members distributed in a consistent group of bacterial species that lack the ApaH enzyme. Among them are the pathogens Staphylococcus aureus, Streptococcus pneumoniae, and Mycoplasma pneumoniae. By identifying the player contributing to Ap4A homeostasis in these bacteria, we disclose a novel target to develop innovative antibacterial strategies.
KEYWORDS: Ap4A hydrolase, adenosine tetraphosphate, dinucleoside polyphosphates, Gram-positive bacteria, nucleotides
ABSTRACT
Diadenosine tetraphosphate (Ap4A) is a dinucleotide found in both prokaryotes and eukaryotes. In bacteria, its cellular levels increase following exposure to various stress signals and stimuli, and its accumulation is generally correlated with increased sensitivity to a stressor(s), decreased pathogenicity, and enhanced antibiotic susceptibility. Ap4A is produced as a by-product of tRNA aminoacylation, and is cleaved to ADP molecules by hydrolases of the ApaH and Nudix families and/or by specific phosphorylases. Here, considering evidence that the recombinant protein YqeK from Staphylococcus aureus copurified with ADP, and aided by thermal shift and kinetic analyses, we identified the YqeK family of proteins (COG1713) as an unprecedented class of symmetrically cleaving Ap4A hydrolases. We validated the functional assignment by confirming the ability of YqeK to affect in vivo levels of Ap4A in B. subtilis. YqeK shows a catalytic efficiency toward Ap4A similar to that of the symmetrically cleaving Ap4A hydrolases of the known ApaH family, although it displays a distinct fold that is typical of proteins of the HD domain superfamily harboring a diiron cluster. Analysis of the available 3D structures of three members of the YqeK family provided hints to the mode of substrate binding. Phylogenetic analysis revealed the occurrence of YqeK proteins in a consistent group of Gram-positive bacteria that lack ApaH enzymes. Comparative genomics highlighted that yqeK and apaH genes share a similar genomic context, where they are frequently found in operons involved in integrated responses to stress signals.
IMPORTANCE Elevation of Ap4A level in bacteria is associated with increased sensitivity to heat and oxidative stress, reduced antibiotic tolerance, and decreased pathogenicity. ApaH is the major Ap4A hydrolase in gamma- and betaproteobacteria and has been recently proposed as a novel target to weaken the bacterial resistance to antibiotics. Here, we identified the orphan YqeK protein family (COG1713) as a highly efficient Ap4A hydrolase family, with members distributed in a consistent group of bacterial species that lack the ApaH enzyme. Among them are the pathogens Staphylococcus aureus, Streptococcus pneumoniae, and Mycoplasma pneumoniae. By identifying the player contributing to Ap4A homeostasis in these bacteria, we disclose a novel target to develop innovative antibacterial strategies.
INTRODUCTION
Ap4A is a ubiquitous metabolite whose physiological function is still a matter of debate. Unknown is whether it represents simply a “damage metabolite” or is instead an “alarmone” with stress-signaling functions, or even a modulator of the stress response (1, 2). It is formed by a side reaction of aminoacyl-tRNA synthetase that, in the absence of tRNA, catalyzes the transfer of the AMP moiety of the bound aminoacyl adenylate to an ATP molecule (3). In mammalian and bacterial cells, Ap4A is present at a low micromolar concentration under normal conditions, but these levels rise significantly under various stress conditions, including heat, antibiotics, nutritional, and oxidative stresses (4–6). Accumulation of Ap4A is pleiotropic, resulting in loss of motility and defects in catabolite repression, increased sensitivity to heat, oxidative stress, and aminoglycoside antibiotics, along with impairment of biofilm formation and inhibition of sporulation under starvation conditions (4, 5, 7–9). The mechanism of action of Ap4A is still under investigation. Recent evidence showing that elevation of Np4A levels is associated with a novel Np4 capping of E. coli transcripts, leading to longer lifetimes, suggests that Ap4A might be involved in gene expression regulation (2). Ap4A homeostasis is maintained by the activity of hydrolases and/or phosphorylases, depending on the organism (10). Ap4A hydrolases belong to the ApaH (EC 3.6.1.41) and Nudix (EC 3.6.1.17) families. Hydrolases of the ApaH family catalyze the symmetrical cleavage of Ap4A, yielding two ADP molecules (11). They are widely distributed among gamma- and betaproteobacteria. Loss of ApaH function results in a marked increase in Ap4A levels, with consequent enhanced sensitivity to stressful conditions and decreased pathogenicity (8, 9, 12). Interestingly, the recent finding that apaH deletion in P. aeruginosa caused a significant increase in the bacterium sensitivity to kanamycin identified ApaH as a novel target for enhancing the killing potency of aminoglycosides (4). Ap4A hydrolases of the Nudix family (EC 3.6.1.17) asymmetrically hydrolyze Ap4A to ATP and AMP and are present both in eukaryotes and bacteria (13, 14). Notably, in pathogenic bacteria, both ApaH and Nudix hydrolase activities are essential for the intracellular invasion of the host (12). Finally, an Ap4A phosphorylase (EC 2.7.7.53) that converts Ap4A into ADP and ATP in the presence of phosphate can be found in lower eukaryotes (15) and some bacteria (16), including M. tuberculosis (17), where its absence is shown to cause a constant stress response that impairs bacterial growth and proliferation (18). In this work, based on biochemical studies and bioinformatic analyses, we identified the Staphylococcus aureus YqeK protein as the member of a novel bacterial family of symmetrically cleaving Ap4A hydrolases, distinct from the ApaH family, and conserved in a consistent group of bacterial species lacking the ApaH enzyme.
RESULTS
Biochemical characterization of S. aureus YqeK.
YqeK from S. aureus (SaYqeK) was expressed in E. coli and purified to homogeneity. A molecular weight of about 23 kDa was estimated upon SDS-PAGE analysis (Fig. 1A). Gel filtration experiments showed a native molecular weight of about 44 kDa, which is consistent with a dimeric structure of the protein (Fig. 1B). During the purification of the protein, we noticed that the UV spectrum of the fraction eluted from the Ni-nitrilotriacetic acid (Ni-NTA) column had a shape and a A260/A280 ratio that indicated a mixture of protein and nucleotides (Fig. 1C) (19). Conversely, the spectrum of the protein after gel filtration was typical of a pure protein, with a A260/A280 ratio very close to 0.6 (20) (Fig. 1C). This suggested that an unknown nucleotide(s), likely bound to YqeK during its expression in E. coli, could have been released during the gel filtration chromatography. To identify such a ligand(s), we denatured the protein obtained from the affinity chromatography either by acidic treatment or by heating and performed a reversed-phase high-performance liquid chromatography (RP-HPLC) analysis on the deproteinized samples. In both cases, one single peak was observed, whose identity as genuine ADP was confirmed by coelution with an ADP standard (Fig. 1D).
Fig. 1.
Biochemical characterization of SaYqeK. (A) SDS-PAGE analysis of recombinant SaYqeK purification. M, molecular weight standards; a, crude extract; b, Ni-NTA pool. (B) Gel filtration chromatography of Ni-NTA pool (solid line) and standard proteins (dotted line). (C) UV spectral analysis of the Ni-NTA pool (solid line) and gel filtration pool (dashed line). (D) HPLC analysis of the acid-soluble fraction of the Ni-NTA pool (red) and ADP standard (blue). (E) SaYqeK ligand screening by thermal shift analysis. Ligands were tested at 5 μM concentration. For ease of viewing, bars are colored with different shades of gray. (F) HPLC profile at 260 nm of the reaction mixture of SaYqeK (50 μg/ml) at 0 and 20 min of incubation in the presence of 0.2 mM Ap4A. A standard ADP profile is also shown (std).
The finding that an ADP molecule copurified with YqeK, together with the observation that molecules of GDP and dGDP have been found in the active sites of the protein’s available 3D structures (PDB codes: 2O08 and 2OGI), prompted us to screen as possible ligands a series of metabolites comprising ADP and GDP in their moieties. To this end, we used a thermal shift assay as a preliminary screening test. Several compounds were tested, and those affecting the thermal stability of the protein are shown in Fig. 1E. Among them, ADP, Ap5A, Ap4A, Ap3A, Ap4U, and Ap4G were the most effective. Also, GDP and Gp4G increased the melting temperature of the protein, although to a lesser extent, whereas ATP, GTP, and the corresponding deoxyribonucleotides were the least effective. No effect was exerted by Ap2A, AMP or GMP, pyrimidine nucleotides, NAD or nicotinate adenine dinucleotide (NaAD), ADP- or GDP-linked sugars (like ADP-ribose, ADP-mannose, ADP-glucose, or GDP-glucose), cyclic AMP or GMP, or cyclic di-GMP or cyclic di-AMP. Likewise, no thermal shift was observed in the presence of phosphate, pyrophosphate, or phosphoribosyl pyrophosphate.
Prompted by these results, we investigated whether YqeK was able to use the candidate ligands as substrates. As a first screening, we incubated the enzyme at 0.05 mg/ml with 0.2 mM ligand and analyzed the reaction mixture by RP-HPLC after a 10 min incubation at 37°C. While GDP, ADP, GTP, ATP, and the corresponding deoxyribonucleotides were not consumed, Ap3A, Ap4A, and Ap5A entirely disappeared, being converted to ADP plus AMP, ADP, and ADP plus ATP, respectively. Likewise, efficient hydrolysis of Ap4U, Ap4G, and Gp4G was observed, always releasing ADP or GDP as one of the products. Hydrolysis of Ap4A is shown in Fig. 1F. By assaying YqeK activity under initial velocity conditions, we found that the activity with Ap3A was only 5% of that measured with Ap4A. Therefore, Ap3A was not considered further. The results of the kinetic analysis performed with the tested nucleotides are shown in Table 1. The enzyme exhibits a marked preference for Ap4A, which is mainly exerted at the substrate affinity level. Indeed, the catalytic efficiency is at least twice that determined for the other dinucleoside tetraphosphates tested, despite a lower kcat value. Analysis of product inhibition revealed that the enzyme is not inhibited by the ADP product. Based on all the above results, we proposed a functional assignment of SaYqeK as a symmetrically cleaving Ap4A hydrolase.
TABLE 1.
SaYqeK kinetic constants for preferred substrates
| Substrate | Km (μM ± SD) | kcat (s−1 ± SD) | Kcat/km (s−1 μM−1) |
|---|---|---|---|
| Ap4A | 3.2 ± 0.3 | 107 ± 2 | 34 |
| Ap5A | 21.5 ± 2.5 | 32 ± 1 | 1.5 |
| Gp4G | 68.0 ± 22 | 1,002 ± 186 | 15 |
| Ap4G | 19.7 ± 4.0 | 210 ± 16 | 11 |
| Ap4U | 17.6 ± 2.3 | 324 ± 12 | 18 |
As outlined below, the available crystal structures of YqeK bacterial orthologs show the presence of a diiron cluster in the putative active site, indicating that the enzymatic activity might be iron dependent. Indeed, we found that an overnight preincubation of YqeK with 50 mM EDTA at room temperature fully inactivated the enzyme, provided that 50 mM EDTA was also present in the assay mixture, whereas a 25% inhibition was measured without the preincubation step. These results confirm that YqeK is metal dependent and suggest that the metal-binding site is poorly accessible to chelators. As we found that the recombinant enzyme was active in the absence of added metal ions in the reaction mixtures, we hypothesized that the metal was taken up from the host during protein synthesis.
In vivo functional activity of yqeK gene.
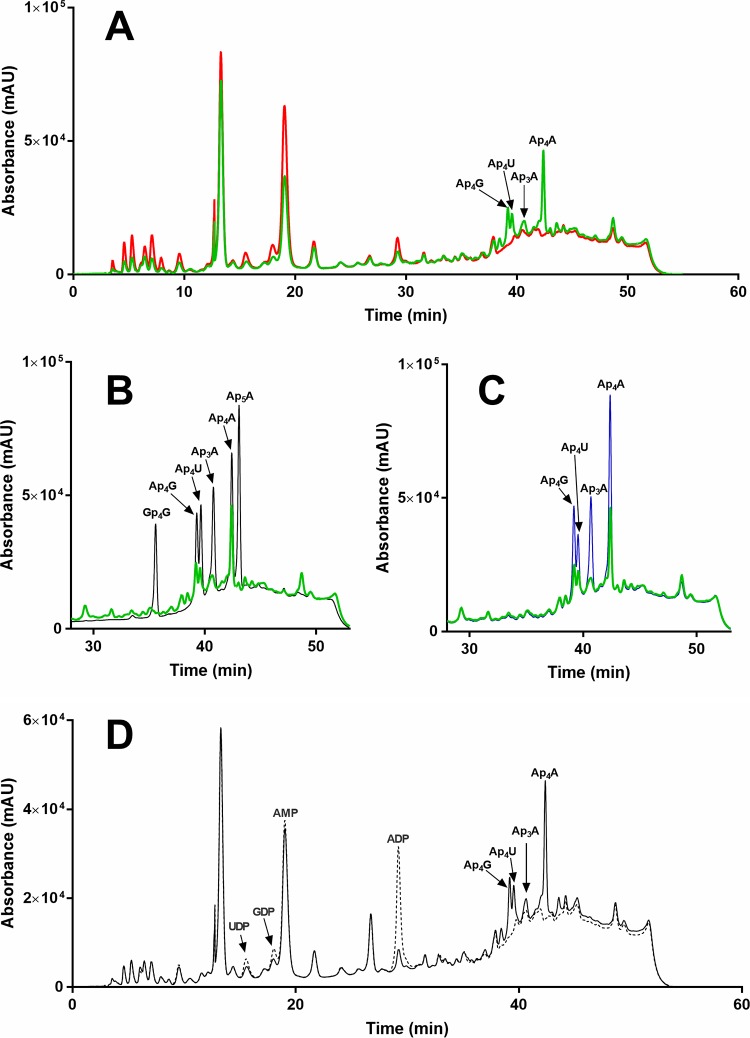
To validate our functional assignment, we grew B. subtilis wild type and the strain lacking the yqeK gene (GenBank BSU25630, Uniprot ID P54456) to compare the in vivo levels of the dinucleotides that are hydrolyzed in vitro by recombinant YqeK. No difference in the growth rate of wild-type and mutant cells was observed. As shown in Fig. 2A, the HPLC analysis of the mutant revealed several peaks that were not detectable in the wild type. Among them, we identified Ap4G, Ap4U, Ap3A, and Ap4A based on the finding that the corresponding chromatographic peaks (i) coeluted with standard molecules (Fig. 2B), (ii) increased in spiked samples (Fig. 2C), and (iii) disappeared after incubation with an excess of SaYqeK (Fig. 2D). Notably, a concomitant increase of UDP, GDP, AMP, and ADP, i.e., the expected products of YqeK-catalyzed reactions, was observed (Fig. 2D). These results indicate that YqeK affects the intracellular levels of these dinucleotides.
Fig. 2.
In vivo validation of YqeK function. HPLC absorbance profiles at 260 nm of ethanol-boiled extracts from wild type B. subtilis (red) and the ΔyqeK mutant (green) (A), mutant extract (green) and nucleotide standards (black) (B), mutant extract (green) and nucleotide-spiked extract (blue) (C), or mutant extract incubated in the absence (solid line) and in the presence (dotted line) of SaYqeK enzyme (D).
Phylogenetic distribution of yqeK and apaH.
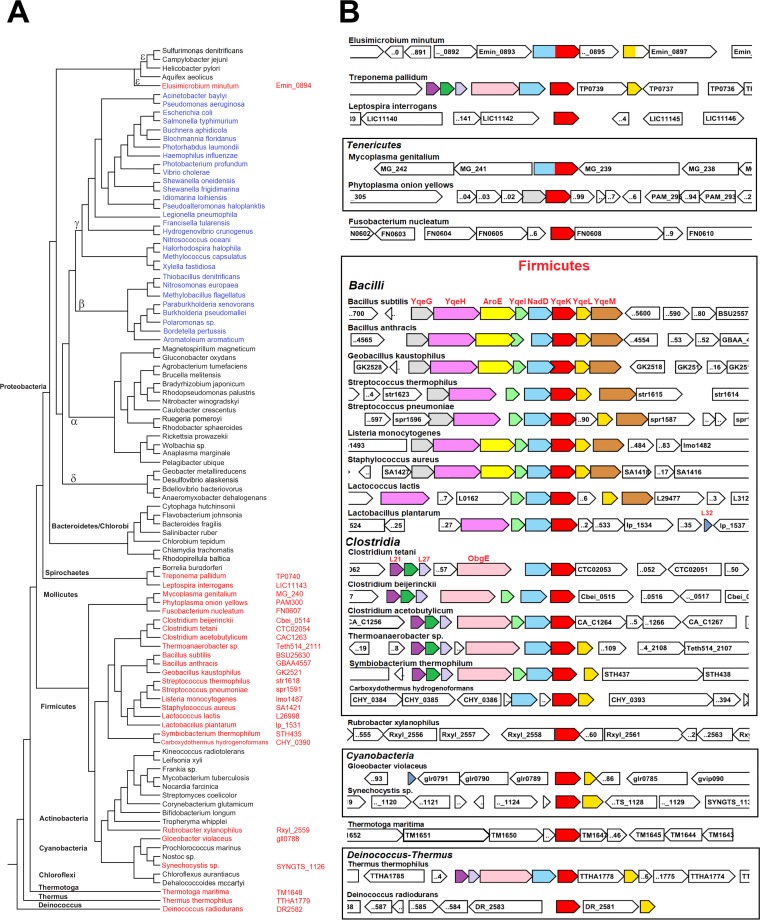
As current literature suggests that Ap4A hydrolase symmetrical cleavage activity belongs to the ApaH family, we next explored the distribution of yqeK and apaH genes using comparative genomics. Using SayqeK as the query, we performed a homology search analysis against a selection of one hundred bacterial genomes that maximized the diversity within the eubacterial kingdom. As a result, we observed an overall presence of YqeK in Gram-positive bacteria. All analyzed Firmicutes, including the class of Mollicutes, invariably possess the yqeK gene (Fig. 3A). Outside Firmicutes, yqeK is found in the Thermotoga and Thermus-Deinococcus groups, a few species of Cyanobacteria and Spirochaetes, and in an isolated case of Actinobacteria (Rubrobacter xylanophilus) (Fig. 3A). In contrast, apaH is consistently distributed in Gram-negative bacteria of the classes Beta- and Gammaproteobacteria, and no instances of yqeK and apaH genomic cooccurrence have been detected (Fig. 3A). This observation, along with our biochemical and in vivo functional characterization, supports the notion that YqeK represents the symmetrically cleaving Ap4A hydrolase in Firmicutes.
Fig. 3.
Phylogenomics of yqeK. (A) Phylogenetic distribution of YqeK (red) and ApaH (blue) in bacteria. Locus tags of YqeK orthologs are next to the species name. (B) Genomic context of the yqeK gene. Genes are colored according to the locus organization in B. subtilis and Clostridium tetani. The gene abbreviations are explained in the text.
Genomic neighborhood of yqeK.
Comparative genome analysis shows that in Firmicutes yqeK is always adjacent to nadD, a key gene in the NAD biosynthetic pathway, with the only exception being Carboxydothermus hydrogenoformans (Fig. 3B). Such genetic proximity is also found outside the Firmicutes, although less frequently. Nonetheless, yqeK, when present, invariably cooccurs with nadD in the genome. In addition, the two genes are fused in Mycoplasma genitalium and Elusimicrobium minutum, thus reinforcing their putative functional association (Fig. 3B). To further investigate such functional association, we asked whether NaAD, the product of the NadD-catalyzed reaction, and NAD, the ultimate product of the NadD-driven pathway, might be substrates or inhibitors of the Ap4A hydrolase activity of YqeK. However, YqeK proved to be insensitive to both dinucleotides. Likewise, NadD activity was not affected by Ap4A. In addition, NadD activity did not change in the presence of YqeK, and YqeK activity was insensitive to the presence of NadD. These results appear to rule out direct cross talk between the two enzymes.
Another strong association is with yqeL, encoding the ribosomal silencing factor RsfS (formerly Iojap), which is often next to yqeK and even fused to it in some cases (as in Clostridium sp. strain SY8519 and Eubacterium nodatum [not shown]). Other closely neighboring genes, often belonging to the same operon as in Bacilli, include yqeG, yqeH, aroE, yqeI, and yqeM (Fig. 3B). The yqeG gene encodes a 5′-nucleotidase of broad specificity with an as yet uncertain physiological role. In B. subtilis, YqeG is necessary for normal colony formation on solid medium and is induced by oxidative stress (21). The yqeH gene encodes an essential GTPase of the Era/Obg family involved in ribosome 30S assembly (22). In Clostridia, Thermus thermophilus, and Treponema pallidum, the obgE gene replaces yqeH in the yqeK-containing operons. Notably, ObgE is another essential GTPase that has been recently proposed as a (p)ppGpp binding target with a role in the stringent response to amino acid starvation and in other downstream effects of (p)ppGpp metabolism (23). The aroE gene, found in the majority of Bacilli, encodes NAD(P)-dependent shikimate dehydrogenase, an indispensable enzyme committed to the biosynthesis of aromatic amino acids (24). YqeI, present in nearly all Firmicutes, is a putative rRNA-binding protein, required for the proper assembly of 50S and 30S ribosome subunits (25). The last member of the yqeK-containing operon is yqeM (smtA in E. coli), a gene typically found in Bacilli and shown to be a dispensable, putative S-adenosylmethionine-dependent methyltransferase overexpressed under biofilm conditions in S. aureus (26).
YqeK structural analysis.
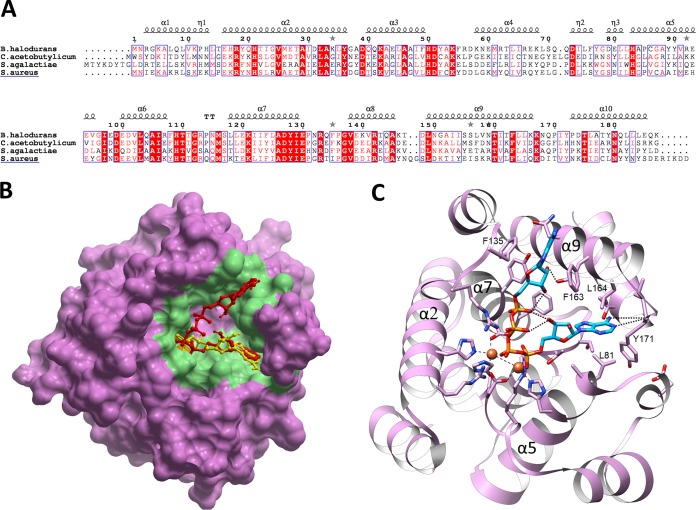
YqeK is a member of the largely uncharacterized HD domain superfamily of enzymes (27). This superfamily comprises phosphohydrolases with nucleotidase or phosphodiesterase activity and oxygenases that catalyze the O2-dependent cleavage of C-C or C-P bonds (28). HD-domain proteins are metal dependent, and enzymes with one, two, or three metal-binding sites have been characterized. YqeK belongs to a subfamily identified as COG1713 (a predicted HD-superfamily hydrolase involved in NAD metabolism) that features a diiron catalytic cluster, as seen in the three crystal structures of YqeK orthologs from Streptococcus agalactiae (PDB ID, 2OGI), Bacillus halodurans (PDB ID, 2O08), and Clostridium acetobutylicum (PDB ID, 3CCG), solved by the Joint Center for Structural Genomics. Of note, the S. agalactiae and B. halodurans YqeK structures have dGDP and GDP molecules bound to the dimetal centers, whereas a free phosphate is present in the C. acetobutylicum protein. By using the structure of B. halodurans YqeK, which shares the highest sequence identity (42%) with SaYqek (Fig. 4A), we employed ICM Pocket Finder to predict the shape of the putative active site. The prediction was for an extended, V-shaped cleft (with an area of 633 Å2 and a volume of 651 Å3) half occupied by the GDP molecule, with the dinuclear cluster located at its vertex (Fig. 4B). Molecular docking of Ap4A revealed that one of the two ADP moieties of Ap4A ligand overlaps with the natural nucleotide found in the ligand-bound structure (Fig. 4B). The adenine ring of this ADP moiety might be stabilized by two H-bonds with the Tyr171 backbone amide and further coordinated by van der Waals contacts with Leu81 and Leu164 (Fig. 4C). Notably, this conformation also fits with the docking pose of the ADP stand-alone ligand (not shown), suggesting that this moiety of Ap4A leaves the active site last. Indeed, in our docking simulation, the other ADP moiety occupies the less-buried arm of the V-shaped binding cleft, with the adenine slightly protruding outward and only partially stabilized by Phe135 and Phe163 (Fig. 4C). In agreement with our gel filtration experiment, all the available YqeK structures share a dimeric assembly with an average interface area of ∼650 Å2. Analysis by the PISA server classifies the dimeric structures in complex with the nucleotides as stable, in contrast with the unstable phosphate-bound complex, suggesting that the dimer formation is favored by the ligand binding.
Fig. 4.
YqeK structural analysis. (A) Structure-based sequence alignment of SaYqeK and structurally characterized YqeK proteins. α and 310 helices (η labels) are shown as spirals on top of the alignment according to the structure of B. halodurans YqeK (PDB 2O08). TT represents sharp turns. Identical amino acids are indicated by a solid red background, while similar amino acids are boxed. UNIPROT IDs for the sequences used are as follows: Q9KD90, Bacillus halodurans; Q97JL1, Clostridium acetobutylicum; Q8DY32, Streptococcus agalactiae; A0A0Y9ZU75, Staphylococcus aureus. (B) Ap4A (red) docking pose in putative binding site (colored in green) of B. halodurans YqeK. Ap4A is superposed with cocrystallized GDP ligand (yellow). (C) Close-up view of B. halodurans YqeK active site with modeled Ap4A. Residues that are likely to interact with the dinucleoside tetraphosphate are displayed. Iron ions are shown as orange spheres, and the side chains of the coordinating residues are shown in stick format and colored by atom type.
DISCUSSION
Despite great advances in the functional annotation of genes since the publication of the first fully sequenced genome, a significant fraction of bacterial genes still lack functional characterization (29). Among them is the gene yqeK, which is conserved in most Gram-positive species. In this work, we identified it as an NpnN hydrolase gene, highly specific for Ap4A. Among known NpnN hydrolases, YqeK mostly resembles the ApaH enzyme, which is the major Ap4A hydrolase in proteobacteria. Both enzymes are active toward several NpnN nucleotides but exhibit the highest specificity toward Ap4A, which is symmetrically cleaved into ADP molecules. The efficiency of YqeK in hydrolyzing Ap4A is very similar to that exhibited by ApaH (30), and YqeK significantly affects the in vivo intracellular level of the dinucleotide in B. subtilis, as reported for ApaH in gammaproteobacteria (7, 12). Deletion of the yqeK gene in B. subtilis also increased the content of Ap4G and Ap4U. The in vivo occurrence of these dinucleotides has been well documented in E. coli as the result of the activity of aminoacyl-tRNA transferases (31). To our knowledge, this is the first report showing their occurrence in B. subtilis. Despite the described similarities, ApaH and YqeK belong to different families, i.e., the serine/threonine phosphatase family (32) and the HD domain superfamily (27), respectively, which is suggestive of a different catalytic mechanism. Our phylogenetic analysis revealed that the two enzymes occur in a mutually exclusive manner, with YqeK present in all bacterial species that lack ApaH. Furthermore, both genes play a role in the stress response, as evidenced by gene deletion studies. In particular, E. coli apaH mutants show impaired growth and change in morphology under starvation conditions or heat stress (1), suggesting that the enzyme is essential for the cell to adapt to stress. Similarly, the deletion of yqeK in B. subtilis impairs the bacterium’s capability to form biofilm, which is a typical adaptation to environmental cues, as cells living in biofilms are better adapted to survive periods of environmental stress (33). apaH and yqeK also share similarities in genomic context. In E. coli, apaH is part of an operon comprising the gene ksgA, encoding an rRNA modification enzyme involved in 16S rRNA maturation and ribosome biogenesis, and is essential for the fidelity of translation during antibiotic stress (34, 35). Similarly, yqeK is often found associated with genes encoding proteins involved in ribosome assembly/maturation (yqeH, yqeG, and yqeI) and the control of protein translation under nutrient shortage (yqeL). Notably, in B. subtilis these genes together with yqeK form a large operon, also comprising genes encoding metabolic enzymes like shikimate dehydrogenase (aroE) and NadD (nadD) that catalyze key steps in the biosynthesis of aromatic amino acids and the coenzyme NAD, respectively. Expression of this operon is constitutive, but increases during germination and under conditions of oxidative stress (36), indicating that it might represent a complex and integrated response to stress signals. Consistent with this, and with the findings that shortage of aromatic amino acids is a hallmark of starvation (37) and that coping with stress-induced events requires the activity of several NAD-consuming enzymes (38), the upregulation of aroE and nadD might ensure amino acid and NAD replenishment, respectively. In general, the stress response requires cellular reprogramming at metabolic, transcriptional, and translational levels. Thus, a yqeK-containing operon might control the coordination of multiple responses at these various levels. Although other biological substrates could exist for YqeK, our finding that the enzyme efficiently hydrolyzes Ap4A both in vitro and in vivo indicates that maintenance of Ap4A levels should be coordinated with NAD and amino acids synthesis, as well as with the control of translation.
Evidence is accumulating that targeting of Ap4A hydrolase might present a promising approach to weakening the bacterial response to stress signals (including antibiotics) and to decreasing pathogenicity. In this view, the functional annotation of YqeK as a novel Ap4A hydrolase in a group of Gram-positive pathogenic bacteria discloses a novel target for antibacterial strategies.
MATERIALS AND METHODS
Preparation of recombinant protein.
Plasmid pOHypY.SA.4, carrying Staphylococcus aureus yqek under the control of the T7 promoter and encoding a protein with an N-terminal His tag, was kindly provided by Andrei L. Osterman (Sanford Burnham Prebys Medical Discovery Institute, San Diego, CA). The construct was used to transform BL21(DE3) cells for protein expression. Cells were grown at 37°C in Luria Bertani medium supplemented with 0.1 mg/ml ampicillin. After reaching an optical density at 600 nm (OD600) of 0.6, expression was induced with 1 mM isopropyl-β-d-thiogalactoside. After 4 h of induction, cells were harvested by centrifugation at 5,000 × g for 10 min. A pellet from 250 ml of culture was resuspended in 13 ml of lysis buffer (50 mM HEPES [pH 7.5], 0.3 M NaCl) containing 1 mM phenylmethanesulfonyl fluoride and proteases inhibitors. The recombinant protein was purified to homogeneity by nickel affinity chromatography. Briefly, the cell suspension was passed twice through a French press (18,000 lb/in2) and centrifuged at 20,000 × g for 25 min at 4°C. The supernatant was loaded onto a 1 ml Ni-NTA resin (GE Healthcare) equilibrated with lysis buffer. After a washing with 30 mM imidazole in lysis buffer, elution was performed with a linear gradient from 30 to 350 mM imidazole. Fractions containing the recombinant protein were pooled and applied to a Superose 12 HR 10/30 column (GE Healthcare) eluted with lysis buffer. About 6 mg of pure protein was obtained from a starting volume of 250 ml of culture.
Thermal shift assays.
The assay was performed according to reference 39. The mixtures contained 1 μM SaYqeK, 5 μM ligand, 10× Sypro Orange (Invitrogen), 40 mM HEPES (pH 7.5), and 0.25 M NaCl in a final volume of 25 μl. They were heated from 40 to 99°C with a heating rate of 0.5°C every 5 s in a real-time PCR device (Rotor-Gene 3000, Corbett Life Science). The fluorescence intensity was measured with excitation and emission wavelengths of 470 and 585 nm, respectively.
Enzyme activity assays.
For YqeK activity, reaction mixtures contained 40 mM HEPES (pH 7.5), 0.25 M NaCl, and appropriate amounts of substrate and YqeK protein in 100 μl volume. For kinetic analyses, 1 to 2 ng protein was used, and the substrates ranged from 1 μM to 160 μM. Incubation was carried out at 37°C for an appropriate incubation time ensuring both a linear change of product formation with time and a substrate consumption of less than 5%. Reactions were stopped with 0.6 M HClO4, and after 10 min on ice, the samples were centrifuged for 1 min at 12,000 × g. The supernatants were neutralized with 0.8 M K2CO3 and injected into an HPLC system. Nucleotide separation was performed as described in reference 40. Control mixtures in the absence of YqeK were always processed in parallel. Km and kcat values were calculated by fitting initial rates to a standard Michaelis-Menten model using the software Prism 6 (GraphPad). The effect of NAD, NaAD, and ADP on the Ap4A hydrolase activity of YqeK was assayed in the presence of 80 μM Ap4A and 0.25 mM effector.
Pure recombinant NadD from B. anthracis was prepared as described in (41). NadD reaction mixtures contained 50 mM HEPES (pH 7.5), 10 mM MgCl2, 0.1 mM nicotinate mononucleotide (NaMN), 0.1 mM ATP, and 0.05 mU of enzyme in a final volume of 50 μl, in the presence and absence of 80 μM Ap4A. Incubation was carried out at 37°C for 20 min, and product formation was analyzed by HPLC as described above. The effect of the NadD protein on the Ap4A hydrolase activity and vice versa was tested by assaying YqeK or NadD in the presence of the other enzyme at molar ratios ranging from 1:1 to 1:10.
Deletion mutant analysis.
Bacillus subtilis subsp. subtilis 168 (BGSCID: 1A1) and ΔyqeK knockout mutant (BGSCID: BKE25630) (42) were obtained from the Bacillus Genetic Stock Center (OH). Cells were grown to mid-log phase in Luria-Bertani medium and harvested by centrifugation for 10 min at 5,000 × g. Pellets were washed twice with 0.9% NaCl and stored at –80°C. For the extraction of the nucleotides, pellets deriving from 15 ml of culture were resuspended in 3 ml of boiling buffered ethanol (75% ethanol in 10 mM HEPES, pH 7.1) and suspensions were incubated at 84°C for 3 min (43). After cooling on ice for 3 min, samples were centrifuged for 5 min at 20,000 × g and supernatants were dried in a Speed-vac concentrator before resuspension in 0.75 ml double-distilled water. Samples were acidified to pH 5.0 by addition of 1 μl of 0.4 M HClO4 and nucleotides were analyzed by HPLC as reported previously (40). The column was maintained at 25°C in order to separate Ap4G from Ap4U. Spiked samples were prepared by adding standard nucleotides in the samples before HPLC injection.
To evaluate the effect of YqeK activity on the nucleotide extract prepared from the mutant, 100 μl of extract was incubated with 0.5 μg recombinant SaYqeK in 50 mM HEPES, pH 7.5, and 0.3 M NaCl, in a final volume of 100 μl. A control mixture in the absence of YqeK was processed in parallel. After a 15-min incubation at 37°C, samples were analyzed by HPLC as described above.
Comparative genome analysis and bioinformatics tools.
Orthologs of YqeK and ApaH were identified by PSI-BLAST (44) searches (E value cutoff, e−20). We used a representative set of 100 bacterial genomes as the organismal search set (Genomic Encyclopedia of Bacteria and Archaea project) (45). Physical clustering and gene fusions were analyzed with the microbial genomic context viewer (46) and STRING 11.0 (47). Structure-based multiple alignment was constructed using PROMALS3D (48) and rendered with Espript 3.0 (49). Visualization, comparison of protein structures, and molecular docking were performed with Molsoft ICM 3.8. We used Pocket Finder (50) in ICM for automatic detection of ligand-binding sites in the high resolution (1.9 Å) crystal structure of B. halodurans YqeK (PDB entry 2O08). One of the identified pockets, encompassing the cocrystal ligand and the metal dinuclear center, has been used for docking simulations. Ligands were fully flexible, and the receptor was set as rigid during all docking studies.
ACKNOWLEDGMENTS
We kindly thank Andrei L. Osterman and Oleg V. Kurnasov (Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA) for supplying the plasmid construct encoding Staphylococcus aureus YqeK.
This work was supported by grants from the Polytechnic University of Marche and partly from PRIN 2017CBNCYT to N.R.
REFERENCES
- 1.Despotovic D, Brandis A, Savidor A, Levin Y, Fumagalli L, Tawfik DS. 2017. Diadenosine tetraphosphate (Ap4A)—an E. coli alarmone or a damage metabolite? FEBS J 284:2194–2215. doi: 10.1111/febs.14113. [DOI] [PubMed] [Google Scholar]
- 2.Luciano DJ, Levenson-Palmer R, Belasco JG. 2019. Stresses that raise Np4A levels induce protective nucleoside tetraphosphate capping of bacterial RNA. Mol Cell 75:957–966. doi: 10.1016/j.molcel.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goerlich O, Foeckler R, Holler E. 1982. Mechanism of synthesis of adenosine(5’)tetraphospho(5’)adenosine (AppppA) by aminoacyl-tRNA synthetases. Eur J Biochem 126:135–142. doi: 10.1111/j.1432-1033.1982.tb06757.x. [DOI] [PubMed] [Google Scholar]
- 4.Ji X, Zou J, Peng H, Stolle AS, Xie R, Zhang H, Peng B, Mekalanos JJ, Zheng J. 2019. Alarmone Ap4A is elevated by aminoglycoside antibiotics and enhances their bactericidal activity. Proc Natl Acad Sci U S A 116:9578–9585. doi: 10.1073/pnas.1822026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura Y, Tanaka C, Sasaki K, Sasaki M. 2017. High concentrations of intracellular Ap4A and/or Ap5A in developing Myxococcus xanthus cells inhibit sporulation. Microbiology 163:86–93. doi: 10.1099/mic.0.000403. [DOI] [PubMed] [Google Scholar]
- 6.Lee PC, Bochner BR, Ames BN. 1983. AppppA, heat-shock stress, and cell oxidation. Proc Natl Acad Sci U S A 80:7496–7500. doi: 10.1073/pnas.80.24.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farr SB, Arnosti DN, Chamberlin MJ, Ames BN. 1989. An apaH mutation causes AppppA to accumulate and affects motility and catabolite repression in Escherichia coli. Proc Natl Acad Sci U S A 86:5010–5014. doi: 10.1073/pnas.86.13.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnstone DB, Farr SB. 1991. AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40. EMBO J 10:3897–3904. doi: 10.1002/j.1460-2075.1991.tb04959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monds RD, Newell PD, Wagner JC, Schwartzman JA, Lu W, Rabinowitz JD, O'Toole GA. 2010. Di-adenosine tetraphosphate (Ap4A) metabolism impacts biofilm formation by Pseudomonas fluorescens via modulation of c-di-GMP-dependent pathways. J Bacteriol 192:3011–3023. doi: 10.1128/JB.01571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guranowski A. 2000. Specific and nonspecific enzymes involved in the catabolism of mononucleoside and dinucleoside polyphosphates. Pharmacol Ther 87:117–139. doi: 10.1016/s0163-7258(00)00046-2. [DOI] [PubMed] [Google Scholar]
- 11.Guranowski A, Jakubowski H, Holler E. 1983. Catabolism of diadenosine 5’,5‴-P1,P4-tetraphosphate in procaryotes. Purification and properties of diadenosine 5’,5‴-P1,P4-tetraphosphate (symmetrical) pyrophosphohydrolase from Escherichia coli K12. J Biol Chem 258:14784–14789. [PubMed] [Google Scholar]
- 12.Ismail TM, Hart CA, McLennan AG. 2003. Regulation of dinucleoside polyphosphate pools by the YgdP and ApaH hydrolases is essential for the ability of Salmonella enterica serovar typhimurium to invade cultured mammalian cells. J Biol Chem 278:32602–32607. doi: 10.1074/jbc.M305994200. [DOI] [PubMed] [Google Scholar]
- 13.Abdelghany HM, Gasmi L, Cartwright JL, Bailey S, Rafferty JB, McLennan AG. 2001. Cloning, characterisation and crystallisation of a diadenosine 5′,5‴-P1,P4-tetraphosphate pyrophosphohydrolase from Caenorhabditis elegans. Biochim Biophys Acta Prot Struc Mol Enz 1550:27–36. doi: 10.1016/S0167-4838(01)00263-1. [DOI] [PubMed] [Google Scholar]
- 14.Cartwright JL, Britton P, Minnick MF, McLennan AG. 1999. The IalA invasion gene of Bartonella bacilliformis encodes a (de)nucleoside polyphosphate hydrolase of the MutT motif family and has homologs in other invasive bacteria. Biochem Biophys Res Commun 256:474–479. doi: 10.1006/bbrc.1999.0354. [DOI] [PubMed] [Google Scholar]
- 15.Plateau P, Fromant M, Schmitter JM, Blanquet S. 1990. Catabolism of bis(5′-nucleosidyl) tetraphosphates in Saccharomyces cerevisiae. J Bacteriol 172:6892–6899. doi: 10.1128/jb.172.12.6892-6899.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLennan AG, Mayers E, Adams DG. 1996. Anabaena flos-aquae and other cyanobacteria possess diadenosine 5′,5‴-P1,P4-tetraphosphate (Ap4A) phosphorylase activity. Biochem J 320:795–800. doi: 10.1042/bj3200795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori S, Shibayama K, Wachino J, Arakawa Y. 2011. Structural insights into the novel diadenosine 5′,5‴-P(1),P(4)-tetraphosphate phosphorylase from Mycobacterium tuberculosis H37Rv. J Mol Biol 410:93–104. doi: 10.1016/j.jmb.2011.04.059. [DOI] [PubMed] [Google Scholar]
- 18.Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 19.Porterfield JZ, Zlotnick A. 2010. A simple and general method for determining the protein and nucleic acid content of viruses by UV absorbance. Virology 407:281–288. doi: 10.1016/j.virol.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldfarb AR, Saidel LJ. 1951. Ultraviolet absorption spectra of proteins. Science 114:156–157. doi: 10.1126/science.114.2954.156. [DOI] [PubMed] [Google Scholar]
- 21.Terakawa A, Natsume A, Okada A, Nishihata S, Kuse J, Tanaka K, Takenaka S, Ishikawa S, Yoshida KI. 2016. Bacillus subtilis 5’-nucleotidases with various functions and substrate specificities. BMC Microbiol 16:249. doi: 10.1186/s12866-016-0866-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morimoto T, Loh PC, Hirai T, Asai K, Kobayashi K, Moriya S, Ogasawara N. 2002. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 148:3539–3552. doi: 10.1099/00221287-148-11-3539. [DOI] [PubMed] [Google Scholar]
- 23.Persky NS, Ferullo DJ, Cooper DL, Moore HR, Lovett ST. 2009. The ObgE/CgtA GTPase influences the stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 73:253–266. doi: 10.1111/j.1365-2958.2009.06767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parish T, Stoker NG. 2002. The common aromatic amino acid biosynthesis pathway is essential in Mycobacterium tuberculosis. Microbiology 148:3069–3077. doi: 10.1099/00221287-148-10-3069. [DOI] [PubMed] [Google Scholar]
- 25.Gagarinova A, Stewart G, Samanfar B, Phanse S, White CA, Aoki H, Deineko V, Beloglazova N, Yakunin AF, Golshani A, Brown ED, Babu M, Emili A. 2016. Systematic genetic Screens reveal the dynamic global functional organization of the bacterial translation machinery. Cell Rep 17:904–916. doi: 10.1016/j.celrep.2016.09.040. [DOI] [PubMed] [Google Scholar]
- 26.Resch A, Rosenstein R, Nerz C, Gotz F. 2005. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol 71:2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aravind L, Koonin EV. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci 23:469–472. doi: 10.1016/S0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 28.Worsdorfer B, Lingaraju M, Yennawar NH, Boal AK, Krebs C, Bollinger JM Jr, Pandelia ME. 2013. Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases. Proc Natl Acad Sci U S A 110:18874–18879. doi: 10.1073/pnas.1315927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarboe LR. 2018. Improving the success and impact of the metabolic engineering design, build, test, learn cycle by addressing proteins of unknown function. Curr Opin Biotechnol 53:93–98. doi: 10.1016/j.copbio.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 30.Plateau P, Fromant M, Brevet A, Gesquiere A, Blanquet S. 1985. Catabolism of bis(5′-nucleosidyl) oligophosphates in Escherichia coli: metal requirements and substrate specificity of homogeneous diadenosine-5′,5‴-P1,P4-tetraphosphate pyrophosphohydrolase. Biochemistry 24:914–922. doi: 10.1021/bi00325a016. [DOI] [PubMed] [Google Scholar]
- 31.Brevet A, Chen J, Leveque F, Plateau P, Blanquet S. 1989. In vivo synthesis of adenylylated bis(5’-nucleosidyl) tetraphosphates (Ap4N) by Escherichia coli aminoacyl-tRNA synthetases. Proc Natl Acad Sci U S A 86:8275–8279. doi: 10.1073/pnas.86.21.8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang QH, Hu WX, Gao W, Bi RC. 2006. Crystal structure of the diadenosine tetraphosphate hydrolase from Shigella flexneri 2a. Proteins 65:1032–1035. doi: 10.1002/prot.21106. [DOI] [PubMed] [Google Scholar]
- 33.Branda SS, Gonzalez-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. 2004. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol 186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Connolly K, Rife JP, Culver G. 2008. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol Microbiol 70:1062–1075. doi: 10.1111/j.1365-2958.2008.06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blanchin-Roland S, Blanquet S, Schmitter JM, Fayat G. 1986. The gene for Escherichia coli diadenosine tetraphosphatase is located immediately clockwise to folA and forms an operon with ksgA. Mol Gen Genet 205:515–522. doi: 10.1007/bf00338091. [DOI] [PubMed] [Google Scholar]
- 36.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Härtig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RAT, Nannapaneni P, Noone D, Pohl S, Rinn B, Rügheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, Bessières P, Noirot P. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 37.Ely B, Pittard J. 1979. Aromatic amino acid biosynthesis: regulation of shikimate kinase in Escherichia coli K-12. J Bacteriol 138:933–943. doi: 10.1128/JB.138.3.933-943.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorci L, Ruggieri S, Raffaelli N. 2014. NAD homeostasis in the bacterial response to DNA/RNA damage. DNA Repair (Amst)) 23:17–26. doi: 10.1016/j.dnarep.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 39.Lo MC, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, Ellestad G. 2004. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem 332:153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 40.Mori V, Amici A, Mazzola F, Di Stefano M, Conforti L, Magni G, Ruggieri S, Raffaelli N, Orsomando G. 2014. Metabolic profiling of alternative NAD biosynthetic routes in mouse tissues. PLoS One 9:e113939. doi: 10.1371/journal.pone.0113939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorci L, Pan Y, Eyobo Y, Rodionova I, Huang N, Kurnasov O, Zhong S, MacKerell AD Jr, Zhang H, Osterman AL. 2009. Targeting NAD biosynthesis in bacterial pathogens: structure-based development of inhibitors of nicotinate mononucleotide adenylyltransferase NadD. Chem Biol 16:849–861. doi: 10.1016/j.chembiol.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koo BM, Kritikos G, Farelli JD, Todor H, Tong K, Kimsey H, Wapinski I, Galardini M, Cabal A, Peters JM, Hachmann AB, Rudner DZ, Allen KN, Typas A, Gross CA. 2017. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst 4:291–305.e7. doi: 10.1016/j.cels.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loret MO, Pedersen L, Francois J. 2007. Revised procedures for yeast metabolites extraction: application to a glucose pulse to carbon-limited yeast cultures, which reveals a transient activation of the purine salvage pathway. Yeast 24:47–60. doi: 10.1002/yea.1435. [DOI] [PubMed] [Google Scholar]
- 44.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu M, Eisen JA. 2008. A simple, fast, and accurate method of phylogenomic inference. Genome Biol 9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Overmars L, Kerkhoven R, Siezen RJ, Francke C. 2013. MGcV: the microbial genomic context viewer for comparative genome analysis. BMC Genomics 14:209. doi: 10.1186/1471-2164-14-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pei J, Kim BH, Grishin NV. 2008. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gouet P, Robert X, Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.An J, Totrov M, Abagyan R. 2005. Pocketome via comprehensive identification and classification of ligand binding envelopes. Mol Cell Proteomics 4:752–761. doi: 10.1074/mcp.M400159-MCP200. [DOI] [PubMed] [Google Scholar]