In bacterial populations, quorum sensing (QS) systems participate in the regulation of specialization processes and regulate collective behaviors that mediate interactions and allow survival of the species. In Gram-positive bacteria, QS systems of the RRNPP family (Rgg, Rap, NprR, PlcR, and PrgX) consist of intracellular receptors and their cognate signaling peptides. Two of these receptors, Rap and NprR, have regained attention in Bacillus subtilis and the Bacillus cereus group.
KEYWORDS: quorum sensing, Bacillus, Rap proteins, NprR, multifunctionality, redundancy
ABSTRACT
In bacterial populations, quorum sensing (QS) systems participate in the regulation of specialization processes and regulate collective behaviors that mediate interactions and allow survival of the species. In Gram-positive bacteria, QS systems of the RRNPP family (Rgg, Rap, NprR, PlcR, and PrgX) consist of intracellular receptors and their cognate signaling peptides. Two of these receptors, Rap and NprR, have regained attention in Bacillus subtilis and the Bacillus cereus group. Some Rap proteins, such as RapH and Rap60, are multifunctional and/or redundant in function, linking the specialization processes of sporulation and competence, as well as global expression changes in the transition phase in B. subtilis. NprR, an evolutionary intermediate between Rap and RRNPP transcriptional activators, is a bifunctional regulator that modulates sporulation initiation and activates nutrient scavenging genes. In this review, we discuss how these receptors switch between functions and connect distinct signaling pathways. Based on structural evidence, we propose that RapH and Rap60 should be considered moonlighting proteins. Additionally, we analyze an evolutionary and ecological perspective to understand the multifunctionality and functional redundancy of these regulators in both Bacillus spp. and non-Bacillus Firmicutes. Understanding the mechanistic, structural, ecological, and evolutionary basis for the multifunctionality and redundancy of these QS systems is a key step for achieving the development of innovative technologies for health and agriculture.
INTRODUCTION
The genus Bacillus comprises a group of Gram-positive, spore-forming, and rod-shaped bacteria found in every habitat (1–3). In the dawn of the genomic era, the sequencing of the genome of Bacillus subtilis 168 allowed new insights into the genetic basis governing the lifestyle of this species, including the molecular mechanisms controlling the regulation of cell differentiation, specialization, and biofilm formation (4). The information generated for B. subtilis can be extrapolated, with caution, to other Bacillus species, such as those of the Bacillus cereus group (5), a bacterial group of particular biotechnological and clinical interest.
Several microbial specialized behaviors are modulated by quorum sensing (QS). QS mechanisms are those of bacterial communication (6, 7) that depend on the synthesis and secretion of small signaling molecules which bind to specific protein receptors that perform the regulatory functions (8, 9). By regulating gene expression directly or indirectly, QS receptors and their cognate signaling molecules mediate bacterial group behavior and regulate functions at the population level (10, 11). Some of the QS-controlled bacterial functions are bioluminescence, virulence, sporulation, and genetic competence, all of which are called “collective traits” because they are only effective when carried out by large bacterial populations (10). Cell specialization processes in bacteria are of high evolutionary relevance, as they give place to phenotypically heterogeneous, while genotypically clonal, populations, differentiated in a manner homologous to that of phylogenetically advanced animal organs or plant tissues (12–14).
Gram-positive bacteria use mainly peptides as signaling molecules and possess complex systems of membrane-bound sensor kinase or cytoplasmic receptors (15). The RRNPP proteins (Rgg, Rap, NprR, PlcR, and PrgX) comprise a family of QS cytoplasmic receptors widespread in the phylum Firmicutes, specifically in the orders Bacillales and Lactobacillales and in the class Clostridia (16). RRNPP QS systems share an ancestor (17) and similar mechanisms of activation (9, 16, 18, 19) consisting of the synthesis, secretion, processing, and reinternalization of a signaling peptide that is then detected by the receptor. However, each receptor regulates different functions. In our 2010 review (18), we depicted the general characteristics of these QS systems and perspectives for basic research, and we discussed their biotechnological potential. Ten years later, many studies have been published on the structural features, functions, physiological roles, and relevance to microbe-host interactions of this protein family. The results continue to show that these QS systems are of high biotechnological and clinical relevance in both Bacillus and non-Bacillus Firmicutes.
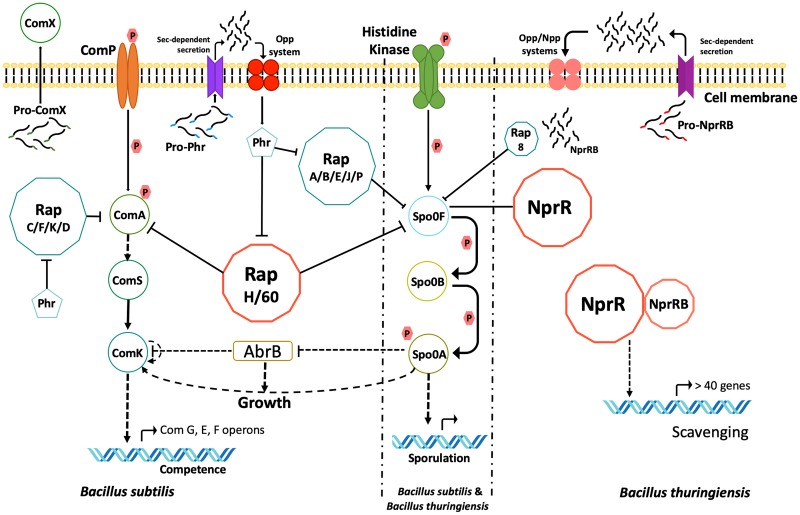
Two RRNPP receptors, Rap and NprR, have regained recent attention in B. subtilis and the B. cereus group. On one hand, several Rap paralogs with diverse functions, some multifunctional and/or redundant in function, are found in B. subtilis (20, 21). On the other hand, NprR, while found as a single copy in B. cereus group bacteria, is a bifunctional QS regulator. NprR uses two different structural domains to independently regulate sporulation initiation and activate the transcription of genes coding for nutrient scavenging enzymes (22–24). These two QS receptors integrate several signaling pathways in B. subtilis, Bacillus thuringiensis (Fig. 1), and probably other soil bacilli.
FIG 1.
Sporulation, competence, and metabolic scavenging are regulated and interconnected by Rap-Phr and NprR-NprRB in Bacillus species. Sensor histidine kinases and ComP are activated by autophosphorylation. ComA∼P activates early competence operons and ComK, which activates the late competence operons. Pro-Phr and Pro-NprRB are exported, processed, and reimported as mature signaling peptides. RapA, RapB, RapE, RapJ, RapP, RapH, and Rap60 dephosphorylate Spo0F∼P, decreasing intracellular Spo0A∼P and inhibiting sporulation onset. RapC, RapD, RapF, RapK, RapH, and Rap60 prevent competence development, through the inhibition of ComA binding to DNA. Phr peptides inhibit the activity of each cognate Rap. NprR binds to Spo0F and modulates (positively or negatively) sporulation initiation in B. thuringiensis. The NprR-NprRB complex regulates the transcription of >41 genes, some of which are involved in nutrient scavenging. Arrows indicate positive regulation, and blunt-end lines indicate negative regulation. Dotted lines indicate transcriptional regulation, and solid lines indicate protein-protein interactions (28 and 41).
Multiple studies have shown the genetic and structural basis for the functions of Rap and NprR receptors in the physiology of Bacillus species (22–28). However, the physiological and ecological implications of the interconnected functions of these receptors have not been fully addressed and need novel experimental approaches. Here, we analyze the conformations adopted by Rap and NprR receptors for switching functions and explore how these proteins integrate different signaling pathways. We also discuss the evolutionary and ecological implications of the multifunctionality and redundancy of these RRNPP family regulators; finally, we show a survey of similar Rap- and NprR-like regulators in other non-Bacillus Firmicutes.
Rap PROTEINS: A REGULATORY REPERTOIRE FOR LINKING CELL DIFFERENTIATION PROCESSES
B. subtilis 168 has 11 QS-Rap proteins paralogs encoded in the chromosome (RapA through RapK) (reviewed in reference 16). Other Rap proteins encoded in plasmids (RapP, Rap40, Rap50, Rap60, and LS20) have been described in different strains (29–32). These Rap proteins modulate at least three signal transduction pathways, sporulation, integrative and conjugative transfer elements (ICEs), and competence (16, 29–31) (Fig. 1). Sporulation allows bacteria to enhance survival under adverse environmental conditions (33). Horizontal transfer of ICEs enables bacteria to acquire new genes and functions and therefore plays a role in bacterial evolution (34). Genetic competence facilitates uptake of external high-molecular-weight DNA (35), driving genomic diversity and bacterial adaptation to diverse conditions.
In B. subtilis, eight Rap paralogs modulate the Spo0A phosphorelay, a signal transduction pathway that integrates multiple intracellular and extracellular signals (36) and controls various downstream specialization processes, including sporulation initiation. Kinases A to E autophosphorylate in response to stress signals or environmental cues and transfer the phosphoryl group to Spo0F, which in turn is transferred to Spo0B, and finally to the transcriptional regulator Spo0A (37, 38) (see the Spo0A pathway in Fig. 1). Phosphorylated Spo0A (Spo0A∼P) regulates the transcription of different sets of genes as its cellular concentration increases (39). RapA, RapB, RapE, RapJ, RapP, RapH, and Rap60 dephosphorylate phosphorylated Spo0F (Spo0F∼P), preventing the flow of phosphate to Spo0A (36, 40–45); in the quorum state, each Rap is inactivated by the binding of its cognate signaling peptide Phr (28). Therefore, these seven Rap proteins are largely redundant in their function, and understanding the partial contribution of each to the overall modulation of sporulation initiation is complex (41, 46). The expression patterns of each rap gene, as well as the affinity of each Rap for Spo0F and its signaling peptide, play a role in defining the levels of Spo0A∼P and, therefore, which genes are activated and repressed (Fig. 1).
In addition to sporulation, some Rap paralogs regulate genetic competence in B. subtilis. The regulatory circuit for competence in B. subtilis comprises the membrane-bound sensor kinase ComP, its signaling peptide ComX, and ComA, the activator of early competence genes (47–49) (see the ComX-ComP-ComA pathway in Fig. 1). ComX triggers autophosphorylation of ComP and the subsequent phosphorylation of ComA; finally, phosphorylated ComA (ComA∼P) activates the transcription of 89 genes (47, 50), including comS (51), which encodes a protein that induces the synthesis of ComK (52). ComK in turn regulates the expression of late competence operons encoding the protein machinery needed for uptake, processing, and transcription of foreign DNA (52–54). RapC, RapD, RapF, RapK, RapG, RapH, and Rap60 are antiactivators of competence (41, 55–60) because they impair the transcriptional activator function of ComA (Fig. 1). Hence, RapH and Rap60 are two bifunctional regulators that connect sporulation and competence. Moreover, Rap60 also dephosphorylates KinA (41), further reducing the phosphate flux through the Spo0A phosphorelay.
RapA and RapE, both of which inhibit the phosphorelay at the Spo0F level, also connect sporulation and genetic competence, although in an indirect way. The genes rapA and rapE are included among the genes activated by the competence activator ComA∼P (36, 61). In this way, the activation of competence also causes the inhibition of sporulation through these two Rap paralogs (60).
All Rap proteins that target the Spo0A phosphorelay also cause dramatic global effects in the cells because Spo0A∼P represses AbrB, the master regulator of genes expressed during the transition between vegetative growth and the onset of stationary phase (62) (Fig. 1). When AbrB levels decrease below a critical concentration threshold, the repressive effect upon transition-state genes is lifted (63). Since AbrB is essential for cell survival, the intracellular levels of AbrB are tightly controlled. First, the expression of abrB is autoregulated, as AbrB represses the transcription of the abrB gene. Second, high levels of Spo0A∼P also repress abrB expression (62, 64). Third, AbrB is inactivated by the product of the gene abbA, which is activated at the transcriptional level by Spo0A∼P (65).
As in case of AbrB, the cell concentration of Spo0A is strictly controlled. The spo0A gene is transcribed from two different promoters, one is active during vegetative growth (Pv) and the other during sporulation (Ps). The Pv promoter is recognized by σA RNA polymerase and repressed by Spo0A∼P. On the other hand, the Ps promoter is recognized by σH RNA polymerase and induced by Spo0A∼P (66, 67). Furthermore, the repression of abrB by Spo0A∼P results in the derepression of σH transcription. In consequence, σH increases the expression of spo0A as well as Spo0A∼P levels under the appropriate conditions. Transcription of phr genes is also switched on by σH, increasing their cellular concentration and allowing the inhibition of Rap proteins (40, 63, 64).
In short, Rap proteins directly and indirectly affect the activities of the key transcription factors Spo0A, ComA, and AbrB. Bifunctional RapH and Rap60 interconnect the regulatory circuits that govern global expression patterns and drive differentiation. This robust regulatory repertoire aids the cell in efficient specialization, adaptation, and survival in the environment (4, 68).
Rap PROTEINS ADOPT DISTINCT CONFORMATIONS TO SWITCH THEIR FUNCTIONAL ACTIVITIES
A special group of bifunctional proteins are the moonlighting proteins, which employ nonconventional mechanisms to switch between functions due to their capacity for adopting different conformations, upon binding to different ligands (69). The different functions of a moonlighting protein are carried out by the same domain, and as such, multifunctionality is not the consequence of domain fusions (70). Furthermore, one region in a structural domain may bind to the same partner in different conformations or bind to unrelated partners. After binding, the protein undergoes structural reorganization, which is crucial for its functions (69, 71).
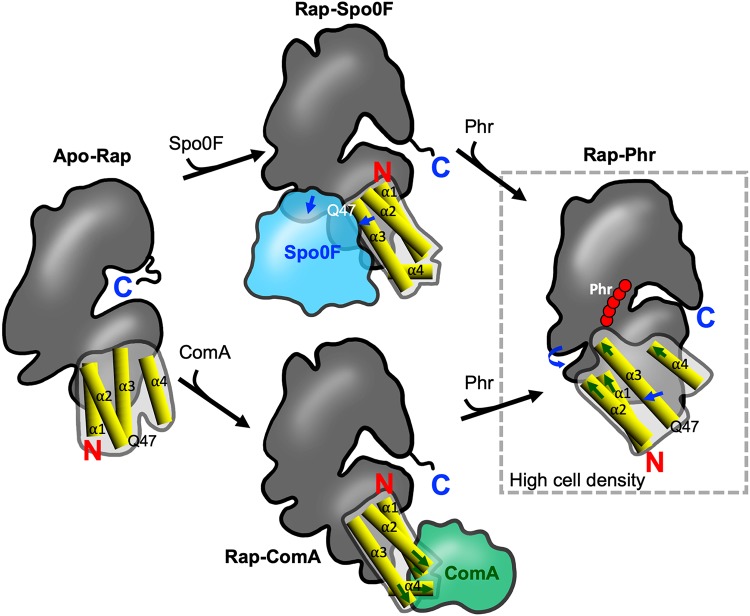
The Rap protein structure comprises an N-terminal domain, consisting of a three-helix bundle (3HB; α1, α2, and α3), and a superhelical C-terminal tetratricopeptide repeat-containing domain (TPR) helix-turn-helix (HTH) fold, connected by a flexible helix-containing linker (α4) (27, 43) (Fig. 2). The 3HB is essential for phosphatase activity (43) and for the interaction with proteins like ComA and Spo0F (72). The conserved residue (Q47 or E47) that catalyzes the dephosphorylation of Spo0F∼P is located in the α3 helix (Fig. 2). As previously mentioned, Rap60 and RapH regulate the Spo0A phosphorelay by dephosphorylating this phosphotransferase (29, 41, 43). Likewise, both RapH and Rap60 modulate competence by binding to ComA and inhibiting the DNA-binding activity. Rap60 forms a ternary complex, Rap60-ComA-DNA, which could block the access of the transcriptional machinery or inhibit the interaction of the RNA polymerase with DNA (41). In contrast, RapH binds to the DNA-binding domain of ComA, disrupting ComA dimerization and interfering with ComA binding to DNA (60). Hence, RapH and Rap60 are bifunctonal proteins that carry out two molecular mechanisms by the same domain; one of these functions (dephosphorylation of Spo0F∼P) implies a chemical reaction, while modulation of competence does not. Although genetic competence and sporulation are both signaling pathways, they are unrelated processes.
FIG 2.
Schematic representation of the distinct conformations adopted by Rap proteins to switch functions. Shown are Apo-Rap (left), Rap-Spo0F (top), Rap-ComA (bottom), and Rap-Phr (right). The C-terminal TPR domain is shown as a dark-gray outline. The N-terminal domain, composed of α-helixes 1, 2, and 3 (3-helix bundle), and the linker region (α4) are shown as a light-gray outline, and each helix is detailed as yellow cylinders. Sites in the ComA-binding surface of Rap are indicated with green arrows; sites in the Spo0F-binding surface are indicated with blue arrows. Q47 indicates the position of the glutamine catalytic residue for phosphatase activity. C, carboxyl terminus; N, amino terminus. This schematic representation is based on the structures of Rap conformations from reference 44.
As a consequence, all of the data suggest that RapH and Rap60 are moonlighting proteins, in spite of not being included in the MoonProt database (http://www.moonlightingproteins.org/). The inclusion of a protein in the MoonProt database requires biochemical, biophysical, mutagenic, and other data supporting the presence of multiple physiologically relevant functions performed by a single polypeptide chain; additionally, these published data should be validated by the database creators (73). In the case of some Rap proteins, structural, biochemical, and mutagenesis data are available; however, they are not sufficient to confirm if RapH and Rap60 are moonlighting proteins, and more focused experiments should be performed.
The two Rap proteins (RapH and Rap60) must adopt different conformations upon binding to different ligands (Fig. 2). Detailed conformational changes in Rap proteins have been studied through the crystalized structures of Apo-RapI, RapHA-Spo0FA, RapF-ComAC, and RapJ-PhrC (44). Combining structural analysis with genetic and biochemical studies, major conformational changes have been described that may explain the bifunctionality of candidate moonlighting Rap proteins. The structure of Rap proteins undergoes radical conformational changes when they bind to their signaling peptide Phr or to target proteins (Spo0F or ComA) (Fig. 2). The structural domains 3HB and TPR are organized as separate structures in Apo-Rap and the Rap-Spo0F and Rap-ComA complexes (44) (Fig. 2). In the complex RapF-ComA, the N-terminal 3HB domain and the linker region form the ComA binding surface (27) (Fig. 2), and the phosphatase catalytic residue Q47 is buried. On the other hand, the complex RapH-Spo0F is a heterotetramer of two RapH and two Spo0F proteins, and each Spo0F coordinates a Mg2+ ion. Both the N-terminal 3HB and C-terminal TPR domains of RapH interact with Spo0F. Most of the 3HB residues that interact with Spo0F, including the Q47 residue that is essential for phosphatase activity, are located in the α3 helix of RapH (43) (Fig. 2).
In the Rap-Phr complex, the protein becomes a single domain made of nine TPR-like folds (28, 44); the entire N-terminal region (3HB and the linker) turns over and packs against the TPR domain to merge with the C terminus, forming an extended TPR domain (44) (Fig. 2). This separates and moves the Rap residues that form the Spo0F binding surface on both 3HB and TPR domains to opposite sides so they cannot simultaneously interact with Spo0F (44) (Fig. 2). The conformational changes in RapH induced by PhrH also hide the residues necessary for ComA binding (44) (Fig. 2). It is possible that the structure of Rap60 is modified in a similar way when binding its Phr and protein targets. However, structural, genetic, and biochemical studies of the Rap60-Phr, Rap60-Spo0F, and Rap60-ComA complexes are required to understand all of the detailed changes that occur as a consequence of these interactions.
NprR, AN EVOLUTIONARY INTERMEDIATE BETWEEN Rap PROTEINS AND RRNPP TRANSCRIPTIONAL REGULATORS
NprR is a bifunctional QS protein that is found across the B. cereus group and absent in B. subtilis. NprR functions as a transcriptional regulator and modulates the Spo0A phosphorelay (23) (Fig. 1, right). Hence, this is a very special QS receptor that shares characteristics from both Rap proteins and the transcriptional regulators of the RRNPP family. For this reason, it is generally regarded as an evolutionary intermediate in this group of QS regulators (17).
The transcriptional regulator function of NprR is carried out by the N-terminal HTH DNA-binding domain (23), whereas modulation of the Spo0A phosphorelay probably takes place in the 3HB domain, as it happens in Rap proteins (43). In fact, NprR directly binds to Spo0F in vitro, similar to Rap proteins, and the HTH domain of NprR is not required for modulating the phosphorelay (23). Perchat et al. (17) proposed that NprR shifts from a Rap-like phosphatase to a transcriptional activator, and the NprRB (or NprX) signaling peptide acts as the switch. Based on these evidences, they proposed that NprR is a moonlighting quorum sensor. However, the bifunctionality of NprR depends on two different structural domains (23); therefore, it does not fit one of the criteria for moonlighting proteins, that the different functions must be carried out by the same domain (http://www.moonlightingproteins.org/) (70).
The X-ray crystal structure of full-length NprR (either in the apo form or with its ligands) is not available; the crystal structure has been solved only for the truncated ΔHTH NprR-NprRB mutant. Additionally, the structure of full-length NprR in solution has been characterized (22, 23, 74). Likewise, some biochemical and genetic function studies gave insights into how NprR carries out both functions. Some data indicate that apo-NprR forms dimers, tetramers, and hexamers in solution (23), while other authors indicate that both the full-length and the truncated NprR proteins form only dimers (74). However, there is consensus that (i) NprR forms a tetramer when it binds to the signaling peptide and (ii) only the NprR-NprRB complex binds to DNA (22, 23, 74) (Fig. 1, right). On the other hand, the full-length NprR binds to Spo0F, forming a heterotetramer consisting of two NprR proteins and two Spo0F molecules (23).
The proposed model for the molecular mechanism of NprR as a transcriptional regulator involves the binding of the signaling peptide to apo-NprR. This causes the shift in conformations from an inactive apo dimer to an active tetrameric complex. As consequence, the NprR-NprRB complex recognizes its target DNA sequences (22, 74) (Fig. 1).
Although it is clear that NprR modulates sporulation initiation by binding to Spo0F, some experimental results are contradictory. In one hand, some data suggest that it is a Rap-like protein that negatively modulates the phosphorelay by dephosphorylating Spo0F∼P (24); however, those in vitro results showed that KinA-dependent phosphorylation of Spo0F is reduced in the presence of NprR (24) but not that NprR dephosphorylates Spo0F∼P. Direct demonstration of Spo0F dephosphorylation has been shown for some Rap proteins from B. subtilis (36, 43, 44). In contrast, we have suggested that the molecular mechanisms of Rap proteins and NprR on the phosphorelay may be opposed, as indicated by several accumulated evidence, as follows. (i) NprR appears to be a positive regulator of the Spo0A phosphorelay, as sporulation is delayed in B. thuringiensis ΔnprR mutants. This is supported by the decrease in the expression of the early sporulation genes spoIIA and spoIIIG, which are directly activated by Spo0A, in the ΔnprR mutant (23). In contrast, sporulation and expression of early sporulation genes in Δrap mutants are increased compared to those in the corresponding wild-type (WT) strain (43, 75). (ii) NprR seems to play a positive role in sporulation initiation in Bacillus anthracis (76). (iii) The catalytic residue conserved in all Rap proteins that dephosphorylate Spo0F (Q47 or E47) is absent in NprR (23, 43). (iv) Finally, the signaling peptide NprRB is unable to disrupt the Spo0F-NprR complex in vitro (22), which suggests that the peptide does not act as the switch for exchanging functions. In contrast, PhrA breaks the RapA-Spo0F complex (77), releasing its inhibitory function on the phosphorelay. In short, although different models have been proposed for the bifunctionality of NprR, the molecular mechanisms are still unclear, and these mechanisms are a question that needs to be addressed.
Because of its participation in the Spo0A phosphorelay and being a transcriptional regulator of many genes involved in nutrient scavenging (Fig. 1, bottom right), NprR probably plays a role in other specialization processes and collective functions, e.g., biofilm formation, virulence, spreading, etc. Hence, new hypotheses should be tested in order to understand the physiological and ecological relevance of this bifunctional regulator in the B. cereus group.
EVOLUTION AND ECOLOGY OF MULTIFUNCTIONALITY AND REDUNDANCY OF NprR AND Rap PARALOGS
Multiple signaling pathways in bacteria have undergone evolutionary expansion and diversification, increasing their complexity and resulting in overlapping and interconnected functions. This is the case of the RRNPP family regulators; understanding the evolution of these proteins in the context of their ecology facilitates our perception of how Bacillus species respond to the environment.
In B. subtilis, Rap-Phr evolution by duplication and horizontal gene transfer (78) has resulted in multiple paralog systems working in parallel, i.e., with no cross talk between pathways and with redundant functions for inhibiting the functions of either Spo0F or ComA. Although the maintenance of multiple Rap receptors with the same function could represent an energetic burden with no immediate gain of novel functions, it is known to provide advantages in social settings, e.g., in a population or a community. The selection of redundant Rap proteins has been driven by a facultative cheating mechanism, where variants with additional Rap-Phr systems are able to exploit their ancestral strain with fewer QS systems (21). Exploitation refers to the capability of individuals with additional Rap proteins to inhibit the activation of genes related to the synthesis of extracellular compounds (such as biofilm matrix components, surfactants, and extracellular enzymes) while using the compounds produced by ancestral cells. This cheating behavior should cease at high cell density when the corresponding novel Phr is accumulated, internalized, and recognized by the Rap protein, antagonizing its repressive function. As a result, the population with additional Rap-Phr systems increases its relative fitness at low relative abundance (with respect to the ancestral population) and cooperates at higher abundance (quorum state). This mechanism seems to be possible only when the newly acquired QS receptor has a dominant repression upon its target function in the absence of the novel signaling peptide. This is the case of redundant Rap-Phr systems, in which a single Rap protein can inhibit the functions of its target response regulator or transcriptional activator, even when additional redundant Rap proteins are not expressed or are bound to their corresponding Phr. These mechanisms have allowed Rap-Phr systems to evolve in complex signaling circuits composed of multiple paralog systems in each species (21, 78).
Multifunctionality in Rap proteins did not require the acquisition of new domains. The loss of pathway specificity in novel Rap-Phr systems led to diversification (78) and therefore to the acquisition of new functions. The evolutionary advantages of multifunctionality may be based on the optimization of energetic costs. Because QS systems are independent signaling pathways, it is likely that redundant Rap paralogs are not all active at the same time; instead, some of them could be activated only under certain conditions.
B. subtilis is considered the Gram-positive model to study the molecular mechanisms controlling the regulation of cell differentiation in spite of the genomic and ecological differences with other species. It is well known that the Spo0A phosphorelay is conserved between B. subtilis and the B. cereus group (79), and it maintains at least one common function in both groups, which is the modulation of sporulation initiation. However, the mechanisms by which the phosphorelay is regulated are not conserved. First, although several Rap protein paralogs are encoded in both B. subtilis and the B. cereus group genomes, they have diversified independently after speciation, with no known examples of horizontal transfer between the two groups; therefore, no Rap homolog is shared between these groups. That is, all Rap proteins from B. subtilis share a common ancestor, while all Rap proteins for the B. cereus group share a different common ancestor (23, 78, 80); second, NprR, which targets Spo0F in the B. cereus group, is not found in B. subtilis. Additionally, not all of the cell differentiation processes from B. subtilis are shared in the B. cereus group species; for example, homologs of the ComP-ComA two-component system are largely absent in species from the B. cereus group (79). Hence, it is difficult to generate feasible predictions about the molecular mechanisms, structural basis of molecular interactions, ecology, and evolution of Rap proteins and other elements regulating differentiation in the B. cereus group based on what is known about B. subtilis.
Recent reports showed that some divergent Rap proteins from the B. cereus group keep the Spo0F-binding activity (75, 81, 82). Hence, it has been proposed that the ancestral Rap receptor of the Rap proteins from B. subtilis and B. cereus targeted Spo0F, and later, the ability to control other regulators was gained, e.g., ComA or DegU in B. subtilis (78, 83). We propose that the capability to target multiple response regulators is directed by the ecology of each species. Consequently, ecological features (referring to habitats and niches) of each bacterial group should be considered separately. B. subtilis is a rhizospheric bacterium (84), and Rap proteins in this species regulate features related to root colonization, including interactions with plants and microbes, biofilm formation, sporulation, and competence (68, 85, 86). In contrast, B. thuringiensis is a soil-inhabiting, insect-pathogenic, and saprophytic bacterium (87) that may use its Rap repertoire to regulate virulence, sporulation, Cry toxin production, nutrient scavenging, and dispersal. Accordingly, we recently found that the lab strain B. thuringiensis Bt8741 has expanded the regulation of extracellular proteases to six out of the eight Rap proteins encoded in its genome (80). This contrasts acutely with only one Rap protein involved in the regulation of extracellular proteases in B. subtilis (83).
The ecological roles of RRNPP proteins, especially Rap proteins and NprR, are not fully understood. However, some efforts have been recently made to assess the impact of proteins that target the Spo0A phosphorelay in more realistic, ecologically relevant settings, in contrast to classic, unrealistic experiments in liquid medium. For example, coinoculation experiments using B. subtilis strains showed that RapP is involved in maintaining a low production of surfactin to avoid subsequent cheating in swarms of B. subtilis (30). Also, in B. thuringiensis, the loss of NprR causes only slight delays in sporulation initiation in liquid medium (23), but this protein is essential for necrotrophism and sporulation in insect hosts (88); therefore, this QS system may provide a fitness advantage to the bacterial population in nature. Another example is Rap8 from B. thuringiensis HD73; rap8- or phr8-deficient mutants in flask cultures show levels of sporulation similar to that of the WT strain; however, sporulation is decreased ≈5-fold in Δphr8 mutants in insect cadavers (82). Although the decrease in sporulation is slight and spores are still detectable, it could represent a disadvantage for spore dissemination after the necrotrophic phase. Future studies should be aimed to explain how the presence of multifunctional and redundant Rap proteins, and the bifunctional NprR, is advantageous in nature for the B. cereus group bacteria to overcome such a genetic burden.
PERSPECTIVES: BIFUNCTIONAL RRNPP RECEPTORS IN THE B. CEREUS GROUP AND NON-BACILLUS FIRMICUTES
Different signaling pathways are integrated via the repertoire of Rap proteins in B. subtilis, and because multiple Rap paralogs are found in all strains from the B. cereus group (78, 81, 89, 90), they could serve the same physiological function in these bacteria. Although Rap proteins from B. cereus group are largely understudied, it is known that RapBXA0205 and BA3790 from B. anthracis strain A2012, Rap8 from B. thuringiensis HD73, and RapF and RapK (also known as Rap8 and Rap7, respectively) from B. thuringiensis Bt407 participate in the Spo0A phosphorelay (75, 81, 82).
While bifunctionality has been well studied for NprR in B. thuringiensis, the multifunctionality of Rap proteins in the B. cereus group has not been established at the molecular level. However, all Rap paralogs retain their multidomain architecture, which indicates that they establish different interactions with proteins and peptides (oligomerization, binding to signaling peptides or response regulators). Furthermore, Spo0F-binding Rap proteins are likely involved in the regulation of multiple functions, as the Spo0A phosphorelay is a signal integration pathway that modulates many differentiation programs (28). Another level of complexity is the presence of a gene coding for a putative precursor of a Phr signaling peptide in most of the rap paralogs of the B. cereus group (78, 80, 81), which indicates that they retain a QS capability that directs their functions.
Based on these evidences, we recently studied eight Rap paralogs encoded in the genome of B. thuringiensis Bt8741 (89) and found that they indeed constitute a multifunctional and redundant regulatory repertoire (80); five paralogs participate in more than one of the collective functions studied, and all of those functions are regulated by more than one Rap paralog. Notably, our data suggested that RapC, RapF2, and RapLike link the regulation of colony spread and the production of extracellular proteases, while RapC and RapLike also participate in the regulation of sporulation. One pending aspect for understanding the multifunctionality and redundancy of Rap proteins from the B. cereus group is the identification of the target response regulators (other than Spo0F) and transcriptional activators that mediate these functions (80).
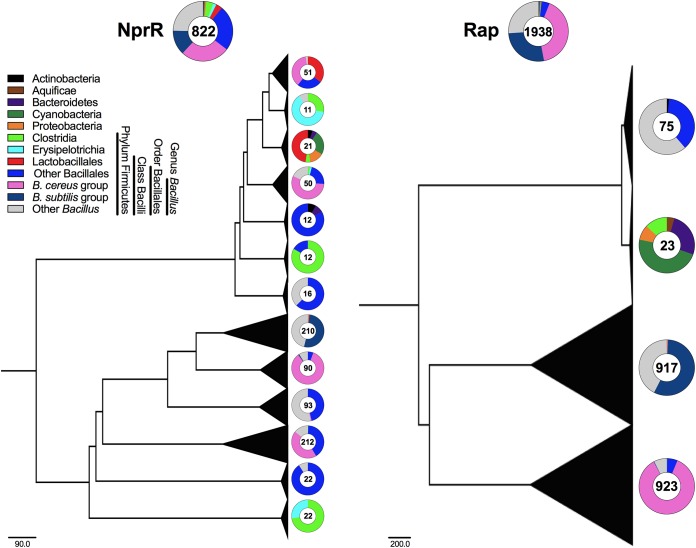
RRNPP-like proteins that mediate overlapping functions, or with multiple functions, are likely also found outside the Bacillus genus. A recent review showed the diversity of regulators with structural similarity to the RRNPP family (16). Although homologs of NprR are only found in genomes from the B. cereus group (22) (see Files S1 and S2 in the supplemental material), phylogenetic reconstruction of sequences with structural similarity to NprR shows that proteins with conserved NprR-like domains are found across the Bacillus genus (including the B. subtilis group), other non-Bacillus Bacillales, bacteria from the order Lactobacillales, members of the classes Clostridia and Erysipelotrichia, and a few representatives from the phyla Actinobacteria, Bacteroidetes, Cyanobacteria, and Proteobacteria (Fig. 3). On the other hand, Rap proteins are found across bacteria from the order Bacillales (B. cereus group, B. subtilis group, other Bacillus spp., and non-Bacillus spp.) and a few representatives are also found in the phyla Bacteroidetes, Cyanobacteria, and Proteobacteria. Additionally, Rap-like proteins (with structural similarity to Rap proteins) are found in the class Clostridia and the phyla Actinobacteria, Aquificae, Bacteroidetes, Cyanobacteria, and Proteobacteria (Fig. 3 and Files S1 and S2). The broad presence of RRNPP-like regulators across Firmicutes implies that many proteins with similar structures, molecular mechanisms, and functions will likely be described, and the family will continue to grow rapidly (19). For this reason, the current acronym RRNPP will no longer be suitable for grouping these receptors. We propose that the protein family should be renamed the TCQR (TPR-containing, cytoplasmic, quorum-sensing receptor) family, considering the most important characteristics that these proteins share.
FIG 3.
Maximum-likelihood phylogenetic trees showing the distributions of NprR- and Rap-like proteins across the domain Bacteria. The sequence data set was obtained from a previous work (16), and sequences from unknown organisms were excluded (File S1). Triangles indicate collapsed branches, and the triangle area is proportional to the number of sequences collapsed. The total number of sequences and their percentage are shown on top of each tree; the number of sequences and their percentages in each taxon are shown on the right of each collapsed branch. Scale bars indicate the number of substitutions per site.
The presence of paralogs with a degree of redundancy in their functions, as well as bifunctionality in other TCQR receptors, has been described in non-Bacillus Firmicutes. Rgg receptors and small hydrophobic peptide (SHP) signaling peptides are widely studied systems that were more recently included in the protein family. Rgg-SHP systems are found across low-G+C Gram-positive bacteria (except for Clostridiaceae) but have been broadly studied in streptococci (reviewed in references 91 and 92). Rgg paralogs control functions such as virulence (RpoB), enzymatic functions, formation of capsule and biofilms (Rgg2 and Rgg3), and natural competence (ComR). Interestingly, there are cases where several paralogs have been identified in a single genome; e.g., up to 7 paralogs have been identified in Streptococcus thermophilus (93), and Streptococcus pyogenes codes for 4 paralogs which have been thoroughly studied (94). Although the functions of Rgg paralogs have been regarded as nonredundant (95), Rgg2 and Rgg3 have overlapping mechanisms. While Rgg2 is a transcriptional activator, Rgg3 is a repressor, and they are highly coordinated through the activation SHP2 and SHP3 by Rgg2. Additionally, they bind to common DNA regions with antagonizing effects on the expression of genes that control biofilm biogenesis (96). This mechanism ensures that expression is repressed by Rgg3 under peptide-limiting conditions and activated by Rgg2 in a quorum state (97). Notably, the presence of Rgg regulators and their involvement in the activation of virulence are common to other Streptococcus species (98), as well as other Firmicutes of clinical relevance, such as Pneumococcus spp. (99).
Another bifunctional QS regulator that has been recently studied is RstA from Clostridioides difficile (100, 101). RstA is now described as a RRNPP (TCQR) family regulator, as it contains the canonic architecture consisting of an N-terminal DNA-binding domain, followed by an Spo0F-like binding domain (although Spo0F is not conserved in C. difficile) and a C-terminal TPR-containing domain. This regulator appears to be activated by a signaling molecule, but a cognate signaling peptide or its gene remain elusive. Similar to the functions of NprR, RstA positively modulates the early stages of sporulation by a DNA-binding independent mechanism, probably through protein-protein interactions with an unknown target response regulator, and it is also a transcriptional regulator that directly and indirectly represses the expression of TcdA and TcdB, two major toxins of this pathogen (100, 101). The structural and evolutionary bases of redundancy and multifunctionality have not been fully addressed for these TCQR regulators from non-Bacillus Firmicutes, but they illustrate how these phenomena are widely spread in the bacterial world and may be highly relevant in the context of human pathogenesis.
QS studies have had an impact in human health through novel disease diagnosis strategies and antimicrobial agents (102), and we expect they will soon have an impact in agriculture, as many research efforts on microbiology are aiming to decrease the harmful impacts of this human activity on the environment. However, some QS systems show unexpected complexity that includes multifunctionality and redundancy in their functions, which hinders the development of technologies based on these molecular circuits. Understanding the mechanistic, structural, ecological, and evolutionary basis for multifunctionality and redundancy of these QS systems is a key step for achieving the development of innovative technologies.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by CONACYT (Mexico) grant 267837 to M.D.L.T. A fellowship from CONACYT was given to A.V.-F. and G.G.
Jorge Peralta provided assistance with the design of Fig. 2. We thank Gabriel del Rio for his useful comments.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.La Duc MT, Satomi M, Venkateswaran K. 2004. Bacillus odysseyi sp. nov., a round-spore-forming Bacillus isolated from the Mars Odyssey spacecraft. Int J Syst Evol Microbiol 54:195–201. doi: 10.1099/ijs.0.02747-0. [DOI] [PubMed] [Google Scholar]
- 2.Hong HA, To E, Fakhry S, Baccigalupi L, Ricca E, Cutting SM. 2009. Defining the natural habitat of Bacillus spore-formers. Res Microbiol 160:375–379. doi: 10.1016/j.resmic.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Heyndrickx M. 2011. The importance of endospore-forming bacteria originating from soil for contamination of industrial food processing. Appl Environ Soil Sci 2011:561975. doi: 10.1155/2011/561975. [DOI] [Google Scholar]
- 4.López D, Kolter R. 2010. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev 34:134–149. doi: 10.1111/j.1574-6976.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- 5.Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, Chu L, Mazur M, Goltsman E, Larsen N, D'Souza M, Walunas T, Grechkin Y, Pusch G, Haselkorn R, Fonstein M, Ehrlich SD, Overbeek R, Kyrpides N. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87–91. doi: 10.1038/nature01582. [DOI] [PubMed] [Google Scholar]
- 6.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 7.Monnet V, Juillard V, Gardan R. 2016. Peptide conversations in Gram-positive bacteria. Crit Rev Microbiol 42:339–351. doi: 10.3109/1040841X.2014.948804. [DOI] [PubMed] [Google Scholar]
- 8.Ng W-L, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Do H, Kumaraswami M. 2016. Structural mechanisms of peptide recognition and allosteric modulation of gene regulation by the RRNPP family of quorum-sensing regulators. J Mol Biol 428:2793–2804. doi: 10.1016/j.jmb.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papenfort K, Bassler BL. 2016. Quorum sensing signal–response systems in Gram-negative bacteria. Nat Rev Microbiol 14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams P, Cámara M. 2009. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol 12:182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Veening J-W, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 13.Veening JW, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW. 2008. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci U S A 105:4393–4398. doi: 10.1073/pnas.0700463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro JA. 1998. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol 52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 15.Rutherford ST, Bassler BL. 2012. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neiditch MB, Capodagli GC, Prehna G, Federle MJ. 2017. Genetic and structural analyses of RRNPP intercellular peptide signaling of Gram-positive bacteria. Annu Rev Genet 51:311–333. doi: 10.1146/annurev-genet-120116-023507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perchat S, Talagas A, Zouhir S, Poncet S, Bouillaut L, Nessler S, Lereclus D. 2016. NprR, a moonlighting quorum sensor shifting from a phosphatase activity to a transcriptional activator. Microb Cell 3:573–575. doi: 10.15698/mic2016.11.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocha-Estrada J, Aceves-Diez AE, Guarneros G, De La Torre M. 2010. The RNPP family of quorum-sensing proteins in Gram-positive bacteria. Appl Microbiol Biotechnol 87:913–923. doi: 10.1007/s00253-010-2651-y. [DOI] [PubMed] [Google Scholar]
- 19.Perez-Pascual D, Monnet V, Gardan R. 2016. Bacterial cell-cell communication in the host via RRNPP peptide-binding regulators. Front Microbiol 7:706. doi: 10.3389/fmicb.2016.00706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reizer J, Reizer A, Perego M, Saier MH. 1997. Characterization of a family of bacterial response regulator aspartyl-phosphate (Rap) phosphatases. Microb Comp Genomics 2:103–111. doi: 10.1089/omi.1.1997.2.103. [DOI] [PubMed] [Google Scholar]
- 21.Even-Tov E, Omer Bendori S, Valastyan J, Ke X, Pollak S, Bareia T, Ben-Zion I, Bassler BL, Eldar A. 2016. Social evolution selects for redundancy in bacterial quorum sensing. PLoS Biol 14:e1002386. doi: 10.1371/journal.pbio.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabrera R, Rodríguez-Romero A, Guarneros G, de la Torre M. 2016. New insights into the interaction between the quorum-sensing receptor NprR and its DNA target, or the response regulator Spo0F. FEBS Lett 590:3243–3253. doi: 10.1002/1873-3468.12371. [DOI] [PubMed] [Google Scholar]
- 23.Cabrera R, Rocha J, Flores V, Vázquez-Moreno L, Guarneros G, Olmedo G, Rodríguez-Romero A, de la Torre M. 2014. Regulation of sporulation initiation by NprR and its signaling peptide NprRB: molecular recognition and conformational changes. Appl Microbiol Biotechnol 98:9399–9412. doi: 10.1007/s00253-014-6094-8. [DOI] [PubMed] [Google Scholar]
- 24.Perchat S, Talagas A, Poncet S, Lazar N, de la Sierra-Gallay IL, Gohar M, Lereclus D, Nessler S. 2016. How quorum sensing connects sporulation to necrotrophism in Bacillus thuringiensis. PLoS Pathog 12:e1005779. doi: 10.1371/journal.ppat.1005779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubois T, Perchat S, Verplaetse E, Gominet M, Lemy C, Aumont-Nicaise M, Grenha R, Nessler S, Lereclus D. 2013. Activity of the Bacillus thuringiensis NprR-NprX cell-cell communication system is co-ordinated to the physiological stage through a complex transcriptional regulation. Mol Microbiol 88:48–63. doi: 10.1111/mmi.12168. [DOI] [PubMed] [Google Scholar]
- 26.Perchat S, Dubois T, Zouhir S, Gominet M, Poncet S, Lemy C, Aumont-Nicaise M, Deutscher J, Gohar M, Nessler S, Lereclus D. 2011. A cell-cell communication system regulates protease production during sporulation in bacteria of the Bacillus cereus group. Mol Microbiol 82:619–633. doi: 10.1111/j.1365-2958.2011.07839.x. [DOI] [PubMed] [Google Scholar]
- 27.Baker MD, Neiditch MB. 2011. Structural basis of response regulator inhibition by a bacterial anti-activator protein. PLoS Biol 9:e1001226. doi: 10.1371/journal.pbio.1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perego M. 2013. Forty years in the making: understanding the molecular mechanism of peptide regulation in bacterial development. PLoS Biol 11:e1001516. doi: 10.1371/journal.pbio.1001516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koetje EJ, Hajdo-Milasinovic A, Kiewiet R, Bron S, Tjalsma H, Bron S. 2003. A plasmid-borne Rap-Phr system of Bacillus subtilis can mediate cell-density controlled production of extracellular proteases. Microbiology 149:19–28. doi: 10.1099/mic.0.25737-0. [DOI] [PubMed] [Google Scholar]
- 30.Lyons NA, Kolter R. 2018. A single mutation in rapP induces cheating to prevent cheating in Bacillus subtilis by minimizing public good production. Commun Biol 1:133–145. doi: 10.1038/s42003-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rösch TC, Graumann PL. 2015. Induction of plasmid conjugation in Bacillus subtilis is bistable and driven by a direct interaction of a Rap/Phr quorum-sensing system with a master repressor. J Biol Chem 290:20221–20232. doi: 10.1074/jbc.M115.664110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meijer WJJ, Wisman GBA, Terpstra P, Thorsted PB, Thomas CM, Holsappel S, Venema G, Bron S. 1998. Rolling-circle plasmids from Bacillus subtilis: complete nucleotide sequences and analyses of genes of pTA1015, pTA1040, pTA1050 and pTA1060, and comparisons with related plasmids from Gram-positive bacteria. FEMS Microbiol Rev 21:337–368. doi: 10.1111/j.1574-6976.1998.tb00357.x. [DOI] [PubMed] [Google Scholar]
- 33.Nicholson WL. 2002. Roles of Bacillus endospores in the environment. Cell Mol Life Sci 59:410–416. doi: 10.1007/s00018-002-8433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellanger X, Roberts AP, Morel C, Choulet F, Pavlovic G, Mullany P, Decaris B, Guédon G. 2009. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J Bacteriol 191:2764–2775. doi: 10.1128/JB.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dubnau D. 1991. The regulation of genetic competence in Bacillus subtilis. Mol Microbiol 5:11–18. doi: 10.1111/j.1365-2958.1991.tb01820.x. [DOI] [PubMed] [Google Scholar]
- 36.Perego M, Hanstein C, Welsh KM, Djavakhishvili T, Glaser P, Hoch JA. 1994. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell 79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 37.Burbulys D, Trach KA, Hoch JA. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552. doi: 10.1016/0092-8674(91)90238-T. [DOI] [PubMed] [Google Scholar]
- 38.Higgins D, Dworkin J. 2012. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujita M, González-Pastor JE, Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol 187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perego M. 1997. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc Natl Acad Sci U S A 94:8612–8617. doi: 10.1073/pnas.94.16.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boguslawski KM, Hill PA, Griffith KL. 2015. Novel mechanisms of controlling the activities of the transcription factors Spo0A and ComA by the plasmid-encoded quorum sensing regulators Rap60-Phr60 in Bacillus subtilis. Mol Microbiol 96:325–348. doi: 10.1111/mmi.12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang M, Grau R, Perego M. 2000. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J Bacteriol 182:303–310. doi: 10.1128/jb.182.2.303-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parashar V, Mirouze N, Dubnau DA, Neiditch MB. 2011. Structural basis of response regulator dephosphorylation by Rap phosphatases. PLoS Biol 9:e1000589. doi: 10.1371/journal.pbio.1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parashar V, Jeffrey PD, Neiditch MB. 2013. Conformational change-induced repeat domain expansion regulates Rap phosphatase quorum-sensing signal receptors. PLoS Biol 11:e1001512. doi: 10.1371/journal.pbio.1001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parashar V, Konkol MA, Kearns DB, Neiditch MB. 2013. A plasmid-encoded phosphatase regulates Bacillus subtilis biofilm architecture, sporulation, and genetic competence. J Bacteriol 195:2437–2448. doi: 10.1128/JB.02030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Omer Bendori S, Pollak S, Hizi D, Eldar A. 2015. The RapP-PhrP quorum-sensing system of Bacillus subtilis strain NCIB3610 affects biofilm formation through multiple targets, due to an atypical signal-insensitive allele of RapP. J Bacteriol 197:592–602. doi: 10.1128/JB.02382-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magnuson R, Solomon J, Grossman AD. 1994. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell 77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 48.Roggiani M, Dubnau D. 1993. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J Bacteriol 175:3182–3187. doi: 10.1128/jb.175.10.3182-3187.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinrauch Y, Penchev R, Dubnau E, Smith I, Dubnau D. 1990. A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev 4:860–872. doi: 10.1101/gad.4.5.860. [DOI] [PubMed] [Google Scholar]
- 50.Comella N, Grossman AD. 2005. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum‐sensing transcription factor ComA in Bacillus subtilis. Mol Microbiol 57:1159–1174. doi: 10.1111/j.1365-2958.2005.04749.x. [DOI] [PubMed] [Google Scholar]
- 51.Ogura M, Yamaguchi H, Yoshida K, Fujita Y, Tanaka T. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res 29:3804–3813. doi: 10.1093/nar/29.18.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hahn J, Luttinger A, Dubnau D. 1996. Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis. Mol Microbiol 21:763–775. doi: 10.1046/j.1365-2958.1996.371407.x. [DOI] [PubMed] [Google Scholar]
- 53.van Sinderen D, Luttinger A, Kong L, Dubnau D, Venema G, Hamoen L. 1995. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol Microbiol 15:455–462. doi: 10.1111/j.1365-2958.1995.tb02259.x. [DOI] [PubMed] [Google Scholar]
- 54.Dubnau D. 1997. Binding and transport of transforming DNA by Bacillus subtilis: the role of type-IV pilin-like proteins–a review. Gene 192:191–198. doi: 10.1016/s0378-1119(96)00804-9. [DOI] [PubMed] [Google Scholar]
- 55.Auchtung JM, Lee CA, Grossman AD. 2006. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J Bacteriol 188:5273–5285. doi: 10.1128/JB.00300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogura M, Fujita Y. 2007. Bacillus subtilis rapD, a direct target of transcription repression by RghR, negatively regulates srfA expression. FEMS Microbiol Lett 268:73–80. doi: 10.1111/j.1574-6968.2006.00559.x. [DOI] [PubMed] [Google Scholar]
- 57.Solomon JM, Lazazzera BA, Grossman AD. 1996. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev 10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 58.Bongiorni C, Ishikawa S, Stephenson S, Ogasawara N, Perego M. 2005. Synergistic regulation of competence development in Bacillus subtilis by two Rap-Phr systems. J Bacteriol 187:4353–4361. doi: 10.1128/JB.187.13.4353-4361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayashi K, Kensuke T, Kobayashi K, Ogasawara N, Ogura M. 2006. Bacillus subtilis RghR (YvaN) represses rapG and rapH, which encode inhibitors of expression of the srfA operon. Mol Microbiol 59:1714–1729. doi: 10.1111/j.1365-2958.2006.05059.x. [DOI] [PubMed] [Google Scholar]
- 60.Smits WK, Bongiorni C, Veening J-W, Hamoen LW, Kuipers OP, Perego M. 2007. Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis. Mol Microbiol 65:103–120. doi: 10.1111/j.1365-2958.2007.05776.x. [DOI] [PubMed] [Google Scholar]
- 61.Jiang M, Shao W, Perego M, Hoch JA. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol 38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- 62.Perego M, Spiegelman GB, Hoch JA. 1988. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol 2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 63.Robertson JB, Gocht M, Marahiel MA, Zuber P. 1989. AbrB, a regulator of gene expression in Bacillus, interacts with the transcription initiation regions of a sporulation gene and an antibiotic biosynthesis gene. Proc Natl Acad Sci U S A 86:8457–8461. doi: 10.1073/pnas.86.21.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strauch M, Webb V, Spiegelman G, Hoch JA. 1990. The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci U S A 87:1801–1805. doi: 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Banse AV, Chastanet A, Rahn-Lee L, Hobbs EC, Losick R. 2008. Parallel pathways of repression and antirepression governing the transition to stationary phase in Bacillus subtilis. Proc Natl Acad Sci U S A 105:15547–15552. doi: 10.1073/pnas.0805203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chibazakura T, Kawamura F, Takahashi H. 1991. Differential regulation of spo0A transcription in Bacillus subtilis: glucose represses promoter switching at the initiation of sporulation. J Bacteriol 173:2625–2632. doi: 10.1128/jb.173.8.2625-2632.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strauch MA, Trach KA, Day J, Hoch JA. 1992. Spo0A activates and represses its own synthesis by binding at its dual promoters. Biochimie 74:619–626. doi: 10.1016/0300-9084(92)90133-y. [DOI] [PubMed] [Google Scholar]
- 68.Mielich-Süss B, Lopez D. 2015. Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ Microbiol 17:555–565. doi: 10.1111/1462-2920.12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tompa P, Szász C, Buday L. 2005. Structural disorder throws new light on moonlighting. Trends Biochem Sci 30:484–489. doi: 10.1016/j.tibs.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 70.Mani M, Chen C, Amblee V, Liu H, Mathur T, Zwicke G, Zabad S, Patel B, Thakkar J, Jeffery CJ. 2015. MoonProt: a database for proteins that are known to moonlight. Nucleic Acids Res 43:D277–D282. doi: 10.1093/nar/gku954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Widjaja M, Harvey KL, Hagemann L, Berry IJ, Jarocki VM, Raymond BBA, Tacchi JL, Gründel A, Steele JR, Padula MP, Charles IG, Dumke R, Djordjevic SP. 2017. Elongation factor Tu is a multifunctional and processed moonlighting protein. Sci Rep 7:11227. doi: 10.1038/s41598-017-10644-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Core L, Perego M. 2003. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol Microbiol 49:1509–1522. doi: 10.1046/j.1365-2958.2003.03659.x. [DOI] [PubMed] [Google Scholar]
- 73.Chen C, Zabad S, Liu H, Wang W, Jeffery C. 2018. MoonProt 2.0: an expansion and update of the moonlighting proteins database. Nucleic Acids Res 46:D640–D644. doi: 10.1093/nar/gkx1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zouhir S, Perchat S, Nicaise M, Perez J, Guimaraes B, Lereclus D, Nessler S. 2013. Peptide-binding dependent conformational changes regulate the transcriptional activity of the quorum-sensor NprR. Nucleic Acids Res 41:7920–7933. doi: 10.1093/nar/gkt546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bongiorni C, Stoessel R, Shoemaker D, Perego M. 2006. Rap phosphatase of virulence plasmid pXO1 inhibits Bacillus anthracis sporulation. J Bacteriol 188:487–498. doi: 10.1128/JB.188.2.487-498.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang H, Sikavi C, Tran K, McGillivray SM, Nizet V, Yung M, Chang A, Miller JH. 2011. Papillation in Bacillus anthracis colonies: a tool for finding new mutators. Mol Microbiol 79:1276–1293. doi: 10.1111/j.1365-2958.2011.07519.x. [DOI] [PubMed] [Google Scholar]
- 77.Ishikawa S, Core L, Perego M. 2002. Biochemical characterization of aspartyl phosphate phosphatase interaction with a phosphorylated response regulator and its inhibition by a pentapeptide. J Biol Chem 277:483–489. doi: 10.1074/jbc.M201086200. [DOI] [PubMed] [Google Scholar]
- 78.Even-Tov E, Omer Bendori S, Pollak S, Eldar A. 2016. Transient duplication-dependent divergence and horizontal transfer underlie the evolutionary dynamics of bacterial cell-cell signaling. PLoS Biol 14:e2000330. doi: 10.1371/journal.pbio.2000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anderson I, Sorokin A, Kapatral V, Reznik G, Bhattacharya A, Mikhailova N, Burd H, Joukov V, Kaznadzey D, Walunas T, Souza MÕ, Larsen N, Pusch G, Liolios K, Grechkin Y, Lapidus A, Goltsman E, Chu L, Fonstein M, Ehrlich SD. 2005. Comparative genome analysis of Bacillus cereus group genomes with Bacillus subtilis. FEMS Microbiol Lett 250:175–184. doi: 10.1016/j.femsle.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 80.Gastélum G, de la Torre M, Rocha J. 2019. Rap protein paralogs of Bacillus thuringiensis: a multifunctional and redundant regulatory repertoire for the control of collective functions. J Bacteriol 202:e00747-19. doi: 10.1128/JB.00747-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cardoso PDF, Perchat S, Vilas-Boas LA, Lereclus D, Vilas-Bôas GT. 2019. Diversity of the Rap-Phr quorum-sensing systems in the Bacillus cereus group. Curr Genet 65:1367–1381. doi: 10.1007/s00294-019-00993-9. [DOI] [PubMed] [Google Scholar]
- 82.Fazion F, Perchat S, Buisson C, Vilas-Bôas G, Lereclus D. 2018. A plasmid-borne Rap-Phr system regulates sporulation of Bacillus thuringiensis in insect larvae. Environ Microbiol 20:145–155. doi: 10.1111/1462-2920.13946. [DOI] [PubMed] [Google Scholar]
- 83.Ogura M, Shimane K, Asai K, Ogasawara N, Tanaka T. 2003. Binding of response regulator DegU to the aprE promoter is inhibited by RapG, which is counteracted by extracellular PhrG in Bacillus subtilis. Mol Microbiol 49:1685–1697. doi: 10.1046/j.1365-2958.2003.03665.x. [DOI] [PubMed] [Google Scholar]
- 84.Earl AM, Losick R, Kolter R. 2008. Ecology and genomics of Bacillus subtilis. Trends Microbiol 16:269–275. doi: 10.1016/j.tim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schultz D, Wolynes PG, Ben JE, Onuchic JN. 2009. Deciding fate in adverse times: sporulation and competence in Bacillus subtilis. Proc Natl Acad Sci U S A 106:21027–21034. doi: 10.1073/pnas.0912185106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen Y, Cao S, Chai Y, Clardy J, Kolter R, Guo J, Losick R. 2012. A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol Microbiol 85:418–430. doi: 10.1111/j.1365-2958.2012.08109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Argôlo-Filho RC, Loguercio LL. 2013. Bacillus thuringiensis is an environmental pathogen and host-specificity has developed as an adaptation to human-generated ecological niches. Insects 5:62–91. doi: 10.3390/insects5010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dubois T, Faegri K, Perchat S, Lemy C, Buisson C, Nielsen-LeRoux C, Gohar M, Jacques P, Ramarao N, Kolstø A-B, Lereclus D. 2012. Necrotrophism is a quorum-sensing-regulated lifestyle in Bacillus thuringiensis. PLoS Pathog 8:e1002629. doi: 10.1371/journal.ppat.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Slamti L, Perchat S, Huillet E, Lereclus D. 2014. Quorum sensing in Bacillus thuringiensis is required for completion of a full infectious cycle in the insect. Toxins (Basel) 6:2239–2255. doi: 10.3390/toxins6082239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sheppard AE, Poehlein A, Rosenstiel P, Liesegang H, Schulenburg H. 2013. Complete genome sequence of Bacillus thuringiensis strain 407 Cry-. Genome Announc 1:e00158-12. doi: 10.1128/genomeA.00158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jimenez JC, Federle MJ. 2014. Quorum sensing in group A Streptococcus. Front Cell Infect Microbiol 4:127. doi: 10.3389/fcimb.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cook LC, Federle MJ. 2014. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev 38:473–492. doi: 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ibrahim M, Nicolas P, Bessieres P, Bolotin A, Monnet V, Gardan R. 2007. A genome-wide survey of short coding sequences in streptococci. Microbiology 153:3631–3644. doi: 10.1099/mic.0.2007/006205-0. [DOI] [PubMed] [Google Scholar]
- 94.Federle M. 2012. Pathogenic streptococci speak, but what are they saying? Virulence 3:92–94. doi: 10.4161/viru.3.1.18652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parashar V, Aggarwal C, Federle MJ, Neiditch MB. 2015. Rgg protein structure–function and inhibition by cyclic peptide compounds. Proc Natl Acad Sci U S A 112:5177–5182. doi: 10.1073/pnas.1500357112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. 2011. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog 7:e1002190. doi: 10.1371/journal.ppat.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.LaSarre B, Aggarwal C, Federle MJ. 2012. Antagonistic Rgg regulators mediate quorum sensing via competitive DNA binding in Streptococcus pyogenes. mBio 3:e00333-12. doi: 10.1128/mBio.00333-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xie Z, Meng K, Yang X, Liu J, Yu J, Zheng C, Cao W, Liu H. 2019. Identification of a quorum sensing system regulating capsule polysaccharide production and biofilm formation in Streptococcus zooepidemicus. Front Cell Infect Microbiol 9:121. doi: 10.3389/fcimb.2019.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhi X, Abdullah IT, Gazioglu O, Manzoor I, Shafeeq S, Kuipers OP, Hiller NL, Andrew PW, Yesilkaya H. 2018. Rgg-Shp regulators are important for pneumococcal colonization and invasion through their effect on mannose utilization and capsule synthesis. Sci Rep 8:6369. doi: 10.1038/s41598-018-24910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Edwards AN, Tamayo R, McBride SM. 2016. A novel regulator controls Clostridium difficile sporulation, motility and toxin production. Mol Microbiol 100:954–971. doi: 10.1111/mmi.13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Edwards AN, Anjuwon-Foster BR, McBride SM. 2019. RstA is a major regulator of Clostridioides difficile toxin production and motility. mBio 10:e01991-18. doi: 10.1128/mBio.01991-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yong Y-C, Zhong J-J. 2013. Impacts of quorum sensing on microbial metabolism and human health. Adv Biochem Eng Biotechnol 131:25–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.