Abstract
Background
Salmonella causes intracellular infections in humans. Besides quinolones, third generation cephalosporins are first line drugs used for salmonellosis therapy. An unresolved anomaly of this practice involves high relapse rates associated to quinolone- or cephalosporin-susceptible Salmonella isolates in patients that are discharged clinically following initial recovery. Reduced drug accessibility to intracellular locations has been hypothesized to impair pathogen eradication although supporting evidence is lacking in vivo. Here, we uncover a novel penicillin-binding protein as the first Salmonella factor likely contributing to relapse following beta-lactam, mainly ceftriaxone, therapy.
Methods
We used Salmonella enterica serovar Typhimurium mutants lacking the alternative penicillin-binding proteins PBP2SAL or PBP3SAL. Affinity of PBP2SAL and PBP3SAL for beta-lactam antibiotics was tested. Relapse after ceftriaxone therapy was analysed in the murine typhoid model.
Findings
S. Typhimurium does not express PBP2SAL or PBP3SAL in the Mueller-Hinton medium used for susceptibility testing. The pathogen produces these PBPs in response to acidic pH and nutrient limitation, conditions found in phagosomes of mammalian cells. PBP3SAL has low affinity for beta-lactams, even at acidic pH. In vitro susceptibility to ceftriaxone at low pH is strongly reduced. S. Typhimurium lacking PBP3SAL was unable to cause relapse in mice following ceftriaxone therapy.
Interpretation
The reduced capacity of ceftriaxone to clear S. Typhimurium in vivo is favoured by a switch in beta-lactam targets. This switch, involving production of the less-susceptible PBP3SAL, remains invisible for standard procedures used in clinical therapy. We conclude that eradication of salmonellosis will be possible only upon targeting of PBP3SAL with novel drugs.
Keywords: Salmonella enterica, Relapse, Penicillin-binding protein, Ceftriaxone, Intracellular, Phagosome
Research in context.
Evidence before this study
In an extensive search of literature about relapses and antibiotic resistance published up to March 2020, the available information refers to widespread resistance to third generation cephalosporins in typhoid and non-typhoid S. enterica serovars, and randomized controlled trials in which failure of the antibiotic therapy and relapse rates with the original isolate were estimated. The most representative original and review articles are cited in this study. Importantly, in early studies of 2002 involving randomized controlled trials using third generation cephalosporins in typhoid fever patients, principally ceftriaxone and cefixime, the rates of treatment failure were estimated 5 to 10 per cent, with relapse rates that were 3 to 6 per cent. In another study of 2016 performed in Nepal and involving uncomplicated enteric fever, third generation cephalosporins were associated with slow clinical improvement and high relapse burden. In a 2016 study in Japan, the relapse rate with intravenous ceftriaxone exceeded 10%. In a review published in 2011, relapse rates reaching up to 17% are reported in diverse randomized clinical trials involving typhoid fever patients. Relapses caused by drug-susceptible isolates have also been reported in cases of combined therapies involving quinolones and cephalosporins. Insufficient effect of beta-lactam antibiotics in vivo was attributed to low intracellular concentrations, although these observations are restricted to in vitro infection assays based on tissue cultured mammalian cells.
Added value of this study
Salmonella is recognized as one of the main bacterial pathogens causing asymptomatic and chronic infections in humans and livestock. Clinical evidence also associates drug-susceptible Salmonella isolates to high relapse rates in human patients that were clinically discharged after an antibiotic treatment that resulted in remission of symptoms. These relapses are attributed to a reduced antibiotic penetration into the intracellular locations where the pathogen resides, although no evidence has yet formally obtained in vivo for such potential cause. To our knowledge, our study provides the first clue explaining this clinical anomaly for a Salmonella enzyme that is involved in cell wall metabolism -the penicillin binding protein PBP3SAL- promoting pathogen persistence in host tissues after antibiotic chemotherapy. Targeting of this alternative PBP3SAL is therefore needed to successfully treat salmonellosis patients and eradicate this important pathogen.
Implications of all the available evidence
The data obtained in the assays involving binding of beta-lactams to PBP3SAL indicate that this enzyme, which replaces PBP3 in intracellular Salmonella, has low affinity for ceftriaxone, resulting in reduced in vitro susceptibility and a significant increase in relapse after therapy in an animal model. These findings suggest that modifications of current cephalosporins to better inhibit PBP3SAL might have consequences in the success in treatment of human infections.
Alt-text: Unlabelled box
1. Introduction
Salmonella enterica has an enormous impact in human health causing high morbidity and mortality [1,2]. The evidence accumulated in the last decades has demonstrated that S. enterica infections progress intracellularly with bacteria located inside epithelial cells and phagocytes [2]. Intracellularity has marked the evolution of S. enterica as pathogen following the acquisition of novel genes that enabled the bacterium to invade mammalian cells and to survive and proliferate inside host cells [3, 4].
Most intracellular infections impose a barrier for effective antimicrobial chemotherapy due to the poor penetration of commonly used antibiotics [5]. In vitro assays show that beta-lactams do not accumulate enough in intracellular locations to reach bactericidal effects [6]. Treatment failure of intracellular infections might also relate to other factors like: adaptive variations in the physiology of intracellular bacteria [2]; exposure of the pathogen to heterogeneous host stresses in distinct cell types leading to emergence of persistence phenotypes [7]; and, inhomogeneous distribution of the drug in intracellular locations [8]. These factors are thought to contribute the lack of effectiveness following antibiotic therapy in relapses associated to drug-susceptible isolates.
The epidemiology of S. enterica in recent years shows an alarming increase in antibiotic resistance in both typhoid and non-typhoid serovars [9, 10]. Resistance is frequently reported to first choice antibiotics used to treat human salmonellosis such as quinolones and third generation cephalosporins. These drugs replaced chloramphenicol, trimethoprim-sulphamethoxazole and ampicillin due to the emergence of resistance in mobile genetic elements and, in the case of chloramphenicol, to undesirable haematologic side effects [11, 12]. In addition, worrisome relapse rates are also repeatedly reported in salmonellosis patients discharged clinically after remission of symptoms with third generation cephalosporin therapies [13]. Thus, in randomized controlled trials of third generation cephalosporins in typhoid fever, principally ceftriaxone and cefixime, the rates of treatment failure were 5 to 10 percent, with relapse rates that were 3 to 6 percent [14]. In other studies of uncomplicated enteric fever, third generation cephalosporins are generally associated with slow clinical improvement and high relapse burden [15]. In some series, the relapse rate with intravenous ceftriaxone, a first-choice cephalosporin, exceeded 10 percent [16]. The causes sustaining these high relapse rates remain unknown.
In this study, we characterize in serovar Typhimurium the production in intracellular bacteria of two pathogen-specific penicillin binding proteins (PBPs), natural targets of beta-lactam antibiotics. These alternative PBPs are expressed by the pathogen inside the mammalian cell and bind current beta-lactams with low affinity. Here, we demonstrate that these two factors, involving niche-dependant expression and reduced antibiotic binding, contribute to the failure of ceftriaxone treatment in terms of pathogen eradication.
2. Materials and methods
2.1. Bacterial strains, media and growth conditions
Bacterial strains and plasmids used are listed in Table S1. All S. enterica serovar Typhimurium (S. Typhimurium) strains used are derivates of virulent wild-type strain SV5015, an His+ prototroph of strain SL1344 [17]. Strains were grown in Luria-Bertani (LB) broth, composed of 1% (w/v) casein peptone, 0.5% (w/v) yeast extract and, 0.5% (w/v) sodium chloride; or, phosphate-carbon-nitrogen (PCN) minimal medium [18]. When necessary, pH was buffered with 80 mM MES [2-(N-morpholino) ethanesulfonic acid]. The composition of PCN medium is: 4 mM Tricine [N-[Tris(hydroxymethyl) methyl]glycine], 0.1 mM FeCl3, 376 µM K2SO4, 50 mM NaCl, 15 mM NH4Cl, 1 mM MgSO4, 1 µM CaCl2, 0.4% (w/v) glucose, 0.4 mM inorganic phosphate (Pi), and micronutrients [18]. The 80 mM MES solution was adjusted to the desired pH value with NaOH and used to buffer the medium. To grow strains harbouring plasmids, the media were supplemented with 100 µg/mL ampicillin or, 10 µg/mL chloramphenicol. For kanamycin, the antibiotic was used at 30 µg/mL or 60 µg/mL in neutral (7.0) or acidic (4.6) pH, respectively.
2.2. Construction of S. Typhimurium mutants lacking PBPs
To delete the native copy of mrdA encoding PBP2, a KanR cassette was used. This cassette contained upstream and downstream regions (50 bp) of mrdA including some codons of the gene due to short distance between mrdA and the flanking gene mrdB. The KanR cassette was amplified from pKD13 template plasmid [19] using primers KO STPBP2 FW/ KO STPBP2 RV (Table S2). Gene replacement was done using Lambda Red-mediated recombination [20]. After electroporation, Super Optimal Broth with Catabolite repression (SOC) medium buffered at pH 4.6 with MES was added and cells were grown for 3 h. Recombinants were selected on LB agar pH 4.6 supplemented with 60 µg/mL kanamycin. Primers pbp2-flanking FW and pbp2-flanking RV (Table S2) were used to confirm the gene deletion. The construction was verified by sequencing. Deletion of the SL1344_1845 gene, encoding PBP2SAL, was done with the primers KO PBP2* FW/ KO PBP2* RV (Table S2) using essentially similar inactivation procedure as for mrdA but selecting recombinants on LB agar pH 7.4 with 60 µg/mL kanamycin. Primers FL-2* Fw and FL-2* Rv (Table S2) were used to confirm gene deletion. S. Typhimurium mutants lacking PBP3 or PBP3SAL, have been published elsewhere [21].
2.3. Plasmid constructions involving PBP expression for Bollicin-650 binding assays
mrdA (PBP2) and SL1344_1845 (PBP2SAL) genes were cloned in the expression vector pFUS-Para [22]. The mrdA and SL1344_1845 genes were amplified using primers fwSpeI-PBP2/revSpe-PBP2 and primers fwSpeI-PBP2*/revSpeI-PBP2* (Table S2), respectively, digested with SpeI and ligated into pFUS-Para [21]. The final constructs were sequenced to ensure no undesired mutation. To construct pAC::PBP2SAL and pAC-HIS-::PBP2SAL, SpeI-SpeI fragments containing their respective coding sequences were ligated into pAC-Plac and pAC-6xHIS-Plac, respectively. To construct pAC-HIS::PBP2, the coding sequence of mrdA (SpeI-SpeI fragment) was ligated also into pAC-6xHIS-Plac. Generation of plasmids expressing PBP3-6xHIS and PBP3SAL-6xHIS variants has been previously described [21].
2.4. Bocillin-650 binding assays to detect PBPs
Subcellular fractions containing bacterial membranes were prepared from 100 mL cultures of E. coli strains SP4500 [mrdA (ts)] [23] or RP41 [ftsI(ts)] [23] expressing PBP2/PBP2SAL or PBP3/PBP3SAL, respectively, from a lac inducible promoter. At OD600 = 0.3, 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce expression of the PBP of interest. After 2 h induction, bacteria were pelleted by centrifugation (4400 x g, 10 min, 4 °C), washed in 0.1 M phosphate buffer adjusted to pH 7.4, 5.8 or 4.6 and frozen at -80 °C. Preparation of lysates and binding assays with Bocillin-650 were done as previously described [21].
2.5. DNA techniques
All primers used in this study are listed in Table S2. PCR was performed using Q5 polymerase (New England Biolabs) according to manufacture instructions. PCR fragments were purified using the Speedtools PCR Clean-Up kit (Biotools).
2.6. Immunoblot analyses
The following antibodies were used as primary antibodies: mouse monoclonal anti-Flag (1:5000; Sigma), mouse monoclonal anti-6xHis (1:2500), rabbit polyclonal anti-PBP2 (1:1000; lab collection), rabbit polyclonal anti-PBP3 (1:1000, lab collection) [21]. Goat polyclonal anti-mouse (1:20,000) or anti-rabbit IgG (1:20,000) conjugated to horseradish peroxidase (Bio-Rad) were used as secondary antibodies. Proteins were separated by 4-20% precasted polyacrylamide gels, 7% tricine gels, or 6% acrylamide gels.
2.7. Synthesis of PBPs during a shift from pH 7⋅4 to 4⋅6
S. Typhimurium strain MD5064 (PBP2SAL::3xFlag PBP3SAL::3xFlag) (Table S1) was grown overnight at 37 °C in PCN pH 7.4 medium. Cultures were prepared to a starting optical density at 600 nm (OD600) of 0.05 in PCN pH 4.6 medium. Bacteria were collected at different times for protein analysis by western blot, as described [21].
2.8. Synthesis of PBPs in macrophage and mouse infections
The murine macrophage line RAW 264.7 (Cell Lines Service) was cultured in Dulbecco's modified Eagle´s medium (DMEM) with 5% (v/v) foetal bovine serum (FBS) and 4 mM glutamine at 37 °C in a 5% CO2 atmosphere. RAW 264.7 macrophages were seeded in p150 plates and pre-cultured for 24 h before infection in the presence of 2.5 ng/mL interferon-γ. For infection, culture medium was replaced by fresh medium without interferon-γ and macrophages exposed to S. Typhimurium strain MD5064 (PBP2SAL::3xFlag PBP3SAL::3xFlag) (Table S1) at a multiplicity of infection (MOI) of 10:1 during 10 min. Macrophages were washed with phosphate-buffered saline (PBS) supplemented with 0.5 mM MgCl2 and 0.9 mM CaCl2. Extracellular non-phagocytosed bacteria were killed by incubation in medium containing gentamicin (100 µg/mL for 1 h, followed by 10 µg/mL for the remaining incubation). To determine levels of PBPs produced by intracellular bacteria the infected macrophages were lysed at 2 and 8 hpi, as described [21]. Production of PBPs in bacteria colonizing spleen of BALB/c mice was monitored with S. Typhimurium strain MD5064 (PBP2SAL::3xFlag PBP3SAL::3xFlag), as described [21].
2.9. Spot dilution and antibiotic susceptibility assays
Bacterial viability was monitored after spotting 5 µL of 10-fold serially dilutions of the cultures onto agar plates containing PCN medium at pH 7.4 or 4.6, with or without antibiotic added. Antibiotic susceptibility was also tested using MIC strips (MIC Test Strip; Liofilchem) in PCN agar plates containing 0.01% yeast extract (w/v) at neutral or acid pH.
2.10. Relapse experiments
BALB/c mice were challenged intraperitoneally with a dose of ~5 × 103 cfu of S. Typhimurium wild-type strain SV5015 or mutant derivates lacking PBPs of interest (PBP2, PBP3, PBP2SAL or PBP3SAL) (Table S1). One group of infected mice was left non-treated. In the second infected group, ceftriaxone was administrated orally at a dose of 400 mg/kg per 24 h for three days. This antibiotic therapy controlled infection (no symptoms after 3 days) although all mice of this group showed relapse after 7 days of therapy discontinuation. Mice were sacrificed at 3 days (group with no antibiotic therapy) and 10 days (group showing relapse after antibiotic therapy) for bacterial counts in spleen and liver, as previously described [21].
2.11. Ethical regulations
Animal experiments were performed in accordance with the guidelines of the European Commission for the handling of laboratory animals (directive 2010/63/EU) and approved by the Environment Council (Consejería de Medio Ambiente) of the Regional Government of Madrid, under license PROEX 110/19.
2.12. Statistical analysis
We analysed data with GraphPad Prism, version 8.0, software (GraphPad Inc. San Diego, CA). In the mice experiments, total bacterial numbers per organ were normalized to the median of the group infected with wild-type bacteria. Significant differences amongst these normalized values for each of the groups challenged with the different bacterial mutants (WT, ΔPBP2, ΔPBP3, ΔPBP2SAL, ΔPBP3SAL) were determined by Kruskal-Wallis multiple comparison tests, using the group of mice challenged with wild-type bacteria as control. Significance was established at P values ≤ 0.05.
3. Results
3.1. S. Typhimurium replaces PBP2 and PBP3 by the alternative PBP2SAL and PBP3SAL in infection conditions
The clinical bases accounting for therapy failure in cases of salmonellosis associated to drug-susceptible isolates remain unknown [11, 14]. Third generation cephalosporins, commonly used to treat salmonellosis, target penicillin-binding proteins (PBPs) involved in cell wall metabolism. We reasoned that S. enterica, as intracellular pathogen, could persist during the initial antibiotic therapy by modifying expression of alternative PBPs in the intracellular niche of mammalian cells. The first insight supporting this hypothesis came with the comparison of genome sequence from S. enterica serovar Typhimurium reference strain SL1344 (genome sequence NC_016810.1) and that of the non-pathogenic Escherichia coli K-12 strain MG1655 (genome sequence NC_000913.3). This comparative analysis showed the presence in the pathogen of several genes encoding enzymes involved in peptidoglycan metabolism that are absent in E. coli. amongst these are PBP3SAL, which S. Typhimurium uses to replicate inside mammalian cells [21,24], and a second pathogen-specific PBP that we named PBP2SAL due to its high homology to the shape-maintaining PBP2 (63% identity at the amino acid level) (Fig. S1). BLAST analyses demonstrated that the genes of S. Typhimurium strain SL1344 encoding PBP2SAL (SL1344_1845) and PBP3SAL (SL1344_1765) are conserved in all species, subspecies, and serovars of the Salmonella genus. Importantly, in addition to PBP2SAL and PBP3SAL, all members of the Salmonella genus have functional the "classic" PBP2 and PBP3, which are targets of beta-lactams and are responsible for peptidoglycan synthesis during cell elongation and cell division, respectively [25].
Similar to PBP3SAL, whose production is stimulated in intracellular S. Typhimurium [21], we reasoned that PBP2SAL could be expressed selectively by bacteria located in the acidic phagosome of mammalian cells. S. Typhimurium produces PBP2SAL exclusively in acidic pH (Fig. 1a), a feature reminiscent of what it is observed for PBP3SAL [21]. We also noted that as the nutrient rich media (Mueller-Hinton, LB) was prepared progressively more acidic, the bacteria produced the complete repertories of PBPs involved in cell elongation and division: PBP2 vs PBP2SAL and PBP3 vs PBP3SAL (Fig. 1a). However, in contrast to such behaviour in nutrient rich medium, we noticed that bacteria growing in a minimal medium with limited amount of nutrients expressed preferentially PBP2SAL and PBP3SAL, compared to PBP2 and PBP3 (Fig. 1b). PBP2SAL and PBP3SAL also prevailed over PBP2 and PBP3 in infection conditions, as denoted in intracellular bacteria obtained from infected macrophages or, in bacteria isolated from spleen of infected mice (Fig. 1c-d). Altogether, these data demonstrated that PBP2SAL and PBP3SAL, which are not produced by S. Typhimurium in Mueller-Hinton medium at neutral pH (Fig. 1a), are however the main PBPs that sustain pathogen growth in the host.
Fig. 1.
S. Typhimurium produces the pathogen-specific PBP2SAL and PBP3SAL in acidic pH and during infection. (a) Levels of PBP2, PBP2SAL, PBP3, and PBP3SAL determined by western blot in bacteria grown in the indicated media and pH values. For these assays, bacteria expressing functional 3x-flag-tagged PBP2SAL and PBP3SAL, were used; (b) replacement of PBP2/PBP3 by PBP2SAL/PBP3SAL occurring in minimal medium PCN adjusted to pH 4.6. Note that such replacement is favoured at the lowest pH value tested; (c) production of PBP2SAL/PBP3SAL prevail over that of PBP2/PBP3 in intracellular bacteria; (d) PBP2SAL and PBP3SAL replace PBP2 and PBP3 in bacteria colonizing the spleen of susceptible mice used in a murine typhoid model. The assays were repeated in at least two independent biological replicates.
3.2. PBP3SAL has low affinity for beta-lactams, even in acidic pH
Our initial premise considered that S. Typhimurium could modify PBP expression inside the mammalian cells. Our findings indeed unveiled major changes consistent with the idea of a “replacement” of functional PBPs involving main beta-lactam targets (Fig. 1). Although it remains unknown the exact location of surviving bacteria in the period that follows antimicrobial therapy and that precedes relapses, it has been shown for S. Typhimurium in the murine model that bacteria surviving antibiotic therapy are in a non-growing physiological state that is induced “intracellularly” and, that allow them to regrow in the absence of antibiotic pressure [26], [27], [28]. How PBPs contribute to reach this intracellular persistence state is unknown. We hypothesized that the novel PBP2SAL and/or PBP3SAL produced by intracellular bacteria could have less affinity for known beta-lactams and, as consequence, contribute to survival inside the infected host cell despite exposure to the drug along the antibiotic therapy.
To test this hypothesis involving low affinity for current drugs, we performed antibiotic binding assays using a fluorescent-labelled beta-lactam, Bocillin-650, commonly used to detect PBPs [29]. We expressed S. Typhimurium PBP2SAL and PBP3SAL in E. coli strains bearing endogenous PBP2 and PBP3 labile variants that do not bind Bocillin-650. In this manner, we were confident that the Bocillin-650 signal detected at the expected molecular weight corresponded to PBP2SAL or, PBP3SAL. Antibiotic binding was performed at three different pH values (7.4, 5.8, and 4.6) with membrane material prepared from the corresponding recombinant strains overproducing PBP2SAL or, PBP3SAL. For comparison, we also produced in E. coli the fully functional PBP2 or PBP3 from S. Typhimurium. These Bocillin-650 binding assays showed that PBP2SAL binds efficiently the beta-lactam at acidic pH (5.8 and 4.6), although at neutral pH the affinity was lower than that displayed by PBP2 (Fig. 2). Remarkably, PBP3SAL did not bind Bocillin-650 at neutral pH with poor binding capacity even at the acidic pH (Fig. 2), at which this enzyme is functional in vivo [21]. This observation prompted us to classify PBP3SAL as a specialized enzyme that plays an essential role in intracellular S. Typhimurium persistence and has low affinity to known beta-lactams currently used in clinics.
Fig. 2.
PBP3SAL has low affinity for binding of Bocillin-650, a fluorescent reagent that detects PBPs. The binding was performed at the indicated pH values. The upper panels refer to levels of the respective PBPs, the lower panels to the signal obtained with the fluorescent Bocillin-650. Note that PBP3SAL binds Bocillin-650 with low efficiency at all pH values tested, despite acidic pH the optimal for its function. The assays were repeated in three independent biological replicates.
3.3. PBP3SAL expression decreases beta-lactam susceptibility in S.Typhimurium
To further determine whether PBP2SAL or PBP3SAL could alter beta-bactam susceptibility in S. Typhimurium, we generated mutants lacking each of the four PBPs under study: PBP2, PBP2SAL, PBP3, or, PBP3SAL (see Materials and methods). As we previously reported, the S. Typhimurium mutant lacking PBP3 is unable to grow at neutral pH since PBP3SAL is only active at acid pH [21]. These four mutants were then tested for antimicrobial susceptibility to ceftriaxone, a drug of choice to treat salmonellosis that binds with high affinity to PBP3 [30]. The production of PBP3SAL at pH 4.6 resulted in a pronounced decrease in ceftriaxone susceptibility (Fig. 3a). Thus, viability assays revealed that the mutant lacking PBP3SAL was four-log more sensitive to ceftriaxone when plated on low-pH plates containing 5 µg/mL of the drug (Fig. 3a). E-test assays further confirmed that the presence of PBP3SAL in acid pH also reduces cephalothin, cefuroxime, ceftazidime, and aztreonam susceptibility (Fig. 3b-c). Certainly, the effect of reduction in ceftriaxone susceptibility in conditions mimicking the intra-phagosomal environment, might be enhanced by the relatively low intracellular active concentrations of the drug, facilitating pathogen persistence in the host, and, as consequence, the relapse.
Fig. 3.
PBP3SAL decreases ceftriaxone susceptibility at acidic pH. (a) Bacterial viability was tested in the indicated strains in PCN minimal medium at pH values 7.4 and 4.6 in the presence/absence of ceftriaxone 5 µg/mL. While the antibiotic drastically reduces viability at neutral pH (condition of PBP3 production), it only affects bacterial viability in acidic pH in the presence of PBP3SAL. (b) MIC values obtained by the E-test assay for different beta-lactams that bind with high affinity to PBP3: aztreonam (ATM), cefotaxime (CTX), ceftazidime (CAZ), cefuroxime (CXM) and cephalotin (KF). (c) Representative E-test assays showing a marked increase in the MIC value to aztreonam (ATM) in response to acid pH (4.6) that is further enhanced in the ΔPBP3 mutant, expressing only PBP3SAL. The assays were repeated in three independent biological replicates.
3.4. Production of PBP3SAL in the host contributes to S. Typhimurium relapse in the typhoid murine model
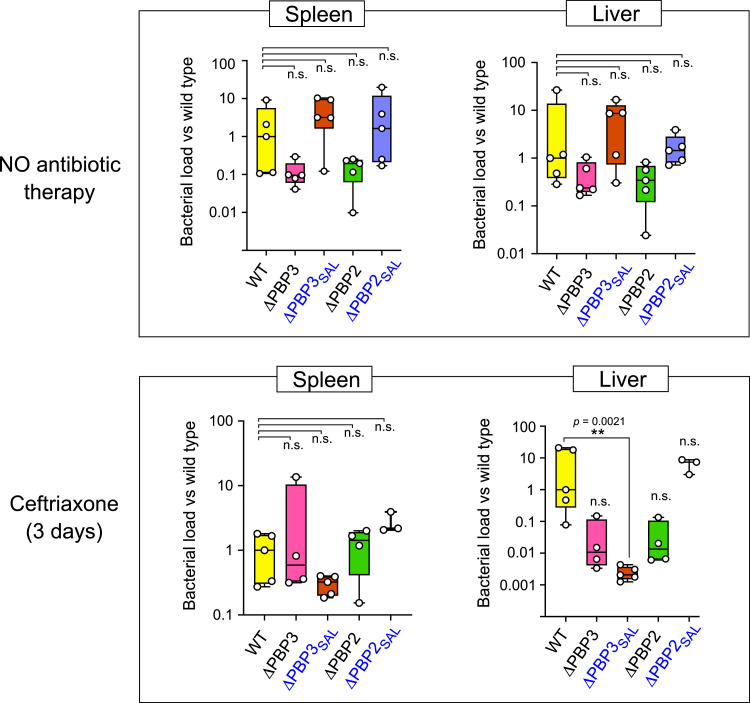
The possible contribution of alternative PBP2SAL and PBP3SAL to relapses after ceftriaxone therapy was further tested by using the murine typhoid model, in which serovar Typhimurium infection progresses as an invasive disease similar to the typhoid fever caused by serovar Typhi in humans [31]. Mice were challenged with the isogenic mutants lacking each of the PBPs of interest. Of the two groups of infected mice, one received ceftriaxone therapy for 3 days starting at 24 h after inoculation, which prevented appearance of symptoms. However, once the antibiotic therapy was discontinued, this group of mice showed relapse after seven days, with a significant bacterial burden in target organs such as liver and spleen (Fig. 4). The only exception was the mutant lacking PBP3SAL, which reached up to 3-logs less bacterial loads in the liver during relapse (P = 0.0021, Kruskal-Wallis multiple comparison test) (Fig. 4). These observations unveiled an unsuspected contribution of PBP3SAL to relapses associated to Salmonella invasive infections.
Fig. 4.
PBP3SAL allows S. Typhimurium to resist antibiotic (ceftriaxone) therapy and facilitate relapses. One group of susceptible mice (lower panel) was treated with ceftriaxone preventing appearance of symptoms. All members of this group showed relapse seven days after discontinuing ceftriaxone treatment. Note that colonization of the liver in the group of mice with relapse was much lower, 3-log difference and statistically significant, **, P = 0.0021, (Kruskal-Wallis multiple comparison test), in bacteria lacking PBP3SAL compared to the rest of strains used. No statistically significant differences were found in untreated mice (upper panels). n.s., not significant.
4. Discussion
Many human trials involving patients infected with Salmonella have reported unusual high rate of relapses caused by isolates that show in vitro susceptibility to the drug used in the initial therapy. These relapses occur weeks to months after remission of first symptoms, with recurrence of clinical disease that is usually milder than the initial illness. In the case of typhoid in humans, with an estimated annual global burden greater than 27 million, clinical relapse rate accounts for 5% to 20% of patients treated with antimicrobials, especially third generation cephalosporins [32]. This clinical anomaly manifests inability of the drug to effectively eradicate the pathogen. In many instances such treatment failure has been linked to inhomogeneous distribution of the drug in host tissues [8], or poor accessibility to intracellular locations where the pathogen resides [32, 33]. Reactivation and dissemination of dormant bacteria have been reported in some cases to occur from mesenteric and caecum draining lymph nodes [28, 34]. Intracellularity might also impair an effective immune response facilitating relapses and decreasing the effectivity of vaccination [35, 36]. However, pathogen factors that contribute to relapse have not yet been reported to date.
We show here that Salmonella replaces the main PBPs involved in cell elongation and division concomitantly to the adaptation to an intracellular lifestyle inside an acidic phagosome. This event definitively influences antibiotic susceptibility in a way that is undetectable by standard susceptibility testing. In these tests, Salmonella only expresses the known repertoire of PBPs for elongation and division used in extracellular conditions, PBP2 and PBP3, the main beta-lactams targets that are non-expressed by intracellular bacteria (Fig. 1). Noteworthy, the experiments performed in mice showed that the ΔPBP3SAL mutant causes disease (Fig. 4), despite the replacement of PBP3 by this PBP3SAL that was observed in wild-type bacteria (see Fig. 1d). A possibility explaining this phenotype is that the lack of PBP3SAL due to a genetic inactivation may alter the regulation that normally down-regulates PBP3 production in response to acid pH and limited nutrients (Fig. 1b). These PBPs are essential for cell division, so the lack of one of them may render constitutive the expression of the other, an idea supported by the growth exhibited by the ΔPBP3SAL mutant on minimal PCN medium plates at pH 4.6 without antibiotic (Fig. 2a). Note, however, that the reverse does not apply for PBP3SAL since the ΔPBP3 mutant is unable to growth at neutral pH (Fig. 2a).
The advantage for S. Typhimurium for acquiring PBP2SAL and PBP3SAL remains at present unknown, although we could speculate about a probable correlation between the production of these alternative PBPs and a reduction in the growth rate, required to successfully persist inside the mammalian cell. Growth control is essential to co-exist in the infected host and, the fact that PBP2SAL and PBP3SAL are exquisitely regulated when the growth conditions are not optimal like acidic pH and nutrient limitation, led us to support such hypothesis. In this context, it is also tempting to postulate a putative relation between the origin of these PBPs and competition with other microbiota producing antimicrobial compounds affecting mainly actively growing cells, which is the beta-lactams mechanism of action. Together with the acquisition of other functions allowing invasion and survival inside mammalian cells, like the specialized type III secretion systems encoded by pathogenicity islands, the growth control linked to the production of PBP2SAL and PBP3SAL might have culminated in the successful pathogen that Salmonella is, prone to cause persistent intracellular infections.
Our study therefore indicates that, to improve current antimicrobial therapy aimed to control salmonellosis, we need novel drugs with increased affinity for PBP3SAL. Wild type bacteria exposed to such drugs would be unable to progress in the infection since the intracellular signals -acid pH and nutrient limitation- would trigger the substitution of PBP3 by PBP3SAL during the course of the adaptative program to the intracellular lifestyle. We also envision such novel drugs will not only reduce relapse rates but also chronic and asymptomatic infections involving dormant intracellular bacteria, major contributors of pathogen dissemination, both in humans and livestock.25 The activity of this novel PBP3SAL, which might evolved to ensure a long lasting residence of Salmonella inside mammalian cells, will certainly illuminate medical research to improve therapies to control this highly successful human pathogen.
Declarations of Competing Interest
The authors declare no conflict of interest. Dr. García-del Portillo, Dr. Pucciarelli, Dr. Castanheira, Dr. Cestero and, David López-Escarpa have a patent EP20382036 pending to Spanish Office of Patents, which is relevant to this work.
Acknowledgments
Acknowledgements
We thank José R. Penadés for suggestions and critical reading of the manuscript and José Luis Martínez for antibiotic reagents. D.L.E and J.J.C were supported by PhD fellowships from the ‘Programa de Formación de Personal Investigador’ (FPI) and ‘Programa Severo Ochoa’, respectively, funded by the Spanish Ministry of Science and Innovation.
Funding sources
This work was supported by grants BIO2016-77639-P and PCIN-2016–082 from the Spanish Ministry of Science and Innovation. The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to the data in the study and had final responsibility for the decision to submit for publication.
Author Contributions
SC, FB and FG-P contributed to the study design. SC, DL-E, MGP and JJC collected and analysed the data that were further discussed with FB and FG-P. SC, FB, MGP and FG-P interpreted the data. The first draft was written by FG-P, which was critically commented by FB and SC. All authors revised and approved the final version of the manuscript.
Footnotes
Funding. Spanish Ministry of Science and Innovation grants BIO2016-77,639-P and PCIN-2016–082.
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102771.
Appendix. Supplementary materials
References
- 1.Crump J.A., Luby S.P., Mintz E.D. The global burden of typhoid fever. Bull World Health Organ. 2004;82(5):346–353. [PMC free article] [PubMed] [Google Scholar]
- 2.Pucciarelli M.G., Garcia-Del Portillo F. Salmonella intracellular lifestyles and their impact on host-to-host transmission. Microbiol Spectr. 2017;5(4) doi: 10.1128/microbiolspec.mtbp-0009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ilyas B., Tsai C.N., Coombes B.K. Evolution of salmonella-host cell interactions through a dynamic bacterial genome. Front Cell Infect Microbiol. 2017;7:428. doi: 10.3389/fcimb.2017.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiyoshi H., Tiffany C.R., Bronner D.N., Baumler A.J. Typhoidal Salmonella serovars: ecological opportunity and the evolution of a new pathovar. FEMS Microbiol Rev. 2018;42(4):527–541. doi: 10.1093/femsre/fuy024. [DOI] [PubMed] [Google Scholar]
- 5.Abed N., Couvreur P. Nanocarriers for antibiotics: a promising solution to treat intracellular bacterial infections. Int J Antimicrob Agents. 2014;43(6):485–496. doi: 10.1016/j.ijantimicag.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Bongers S., Hellebrekers P., Leenen L.P.H., Koenderman L., Hietbrink F. Intracellular penetration and effects of antibiotics on Staphylococcus aureus inside human neutrophils: a comprehensive review. Antibiotics (Basel) 2019;8(2) doi: 10.3390/antibiotics8020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bumann D., Cunrath O. Heterogeneity of Salmonella-Host interactions in infected host tissues. Curr Opin Microbiol. 2017;39:57–63. doi: 10.1016/j.mib.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Bumann D., Fanous J., Li J., Goormaghtigh F. Antibiotic chemotherapy against heterogeneous pathogen populations in complex host tissues. F1000Res. 2019:8. doi: 10.12688/f1000research.19441.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDermott P.F., Zhao S., Tate H. Antimicrobial resistance in nontyphoidal Salmonella. Microbiol Spectr. 2018;6(4) doi: 10.1128/microbiolspec.arba-0014-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Britto C.D., Wong V.K., Dougan G., Pollard A.J. A systematic review of antimicrobial resistance in Salmonella enterica serovar typhi, the etiological agent of typhoid. PLoS Negl Trop Dis. 2018;12(10) doi: 10.1371/journal.pntd.0006779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler T. Treatment of typhoid fever in the 21st century: promises and shortcomings. Clin Microbiol Infect. 2011;17(7):959–963. doi: 10.1111/j.1469-0691.2011.03552.x. [DOI] [PubMed] [Google Scholar]
- 12.Soe G.B., Overturf G.D. Treatment of typhoid fever and other systemic salmonelloses with cefotaxime, ceftriaxone, cefoperazone, and other newer cephalosporins. Rev Infect Dis. 1987;9(4):719–736. doi: 10.1093/clinids/9.4.719. [DOI] [PubMed] [Google Scholar]
- 13.Dragsted U.B., Pedersen P. Relapse of multiresistant Salmonella typhi after combined therapy with ciprofloxacin and ceftriaxone. Clin Microbiol Infect. 2000;6(3):167–168. doi: 10.1046/j.1469-0691.2000.00036.x. [DOI] [PubMed] [Google Scholar]
- 14.Parry C.M., Hien T.T., Dougan G., White N.J., Farrar J.J. Typhoid fever. N Engl J Med. 2002;347(22):1770–1782. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- 15.Arjyal A., Basnyat B., Nhan H.T., Koirala S., Giri A., Joshi N. Gatifloxacin versus ceftriaxone for uncomplicated enteric fever in Nepal: an open-label, two-centre, randomised controlled trial. Lancet Infect Dis. 2016;16(5):535–545. doi: 10.1016/S1473-3099(15)00530-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matono T., Kato Y., Morita M., Izumiya H., Yamamoto K., Kutsuna S. Case series of imported enteric fever at a referral center in Tokyo, Japan: antibiotic susceptibility and risk factors for relapse. Am J Trop Med Hyg. 2016;95(1):19–25. doi: 10.4269/ajtmh.15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vivero A., Banos R.C., Mariscotti J.F., Oliveros J.C., Garcia-del Portillo F., Juarez A. Modulation of horizontally acquired genes by the HHA-YDGT proteins in Salmonella enterica serovar Typhimurium. J Bacteriol. 2008;190(3):1152–1156. doi: 10.1128/JB.01206-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deiwick J., Hensel M. Regulation of virulence genes by environmental signals in Salmonella typhimurium. Electrophoresis. 1999;20(4–5):813–817. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<813::AID-ELPS813>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 19.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellermeier C.D., Janakiraman A., Slauch J.M. Construction of targeted single copy lac fusions using lambda red and FLP-mediated site-specific recombination in bacteria. Gene. 2002;290(1–2):153–161. doi: 10.1016/s0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- 21.Castanheira S., Cestero J.J., Rico-Perez G., Garcia P., Cava F., Ayala J.A. A specialized peptidoglycan synthase promotes Salmonella cell division inside host cells. MBio. 2017;8(6) doi: 10.1128/mBio.01685-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobato-Marquez D., Moreno-Cordoba I., Figueroa V., Diaz-Orejas R., Garcia-del Portillo F. Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci Rep. 2015;5:9374. doi: 10.1038/srep09374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia del Portillo F., de Pedro M.A. Differential effect of mutational impairment of penicillin-binding proteins 1A and 1B on Escherichia coli strains harboring thermosensitive mutations in the cell division genes ftsA, ftsQ, ftsZ, and pbpB. J Bacteriol. 1990;172(10):5863–5870. doi: 10.1128/jb.172.10.5863-5870.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castanheira S., Cestero J.J., Garcia-del Portillo F., Pucciarelli M.G. Two distinct penicillin binding proteins promote cell division in different Salmonella lifestyles. Microb Cell. 2018;5(3):165–168. doi: 10.15698/mic2018.03.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spratt B.G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci U S A. 1975;72(8):2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helaine S., Cheverton A.M., Watson K.G., Faure L.M., Matthews S.A., Holden D.W. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343(6167):204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Claudi B., Sprote P., Chirkova A., Personnic N., Zankl J., Schurmann N. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell. 2014;158(4):722–733. doi: 10.1016/j.cell.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser P., Regoes R.R., Dolowschiak T., Wotzka S.Y., Lengefeld J., Slack E. Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment. PLoS Biol. 2014;12(2) doi: 10.1371/journal.pbio.1001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao G., Meier T.I., Kahl S.D., Gee K.R., Blaszczak L.C. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob Agents Chemother. 1999;43(5):1124–1128. doi: 10.1128/aac.43.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kocaoglu O., Carlson E.E. Profiling of beta-lactam selectivity for penicillin-binding proteins in Escherichia coli strain DC2. Antimicrob Agents Chemother. 2015;59(5):2785–2790. doi: 10.1128/AAC.04552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsolis R.M., Kingsley R.A., Townsend S.M., Ficht T.A., Adams L.G., Baumler A.J. Of mice, calves, and men Comparison of the mouse typhoid model with other Salmonella infections. Adv Exp Med Biol. 1999;473:261–274. [PubMed] [Google Scholar]
- 32.Ahmad K.A., Khan L.H., Roshan B., Bhutta Z.A. Factors associated with typhoid relapse in the era of multiple drug resistant strains. J Infect Dev Ctries. 2011;5(10):727–731. doi: 10.3855/jidc.1192. [DOI] [PubMed] [Google Scholar]
- 33.Brauner A., Fridman O., Gefen O., Balaban N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol. 2016;14(5):320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 34.Griffin A.J., Li L.X., Voedisch S., Pabst O., McSorley S.J. Dissemination of persistent intestinal bacteria via the mesenteric lymph nodes causes typhoid relapse. Infect Immun. 2011;79(4):1479–1488. doi: 10.1128/IAI.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffin A.J., McSorley S.J. Development of protective immunity to Salmonella, a mucosal pathogen with a systemic agenda. Mucosal Immunol. 2011;4(4):371–382. doi: 10.1038/mi.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abrahams G.L., Hensel M. Manipulating cellular transport and immune responses: dynamic interactions between intracellular Salmonella enterica and its host cells. Cell Microbiol. 2006;8(5):728–737. doi: 10.1111/j.1462-5822.2006.00706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.