Abstract
The metagenomic datasets of the microbial DNA from tropical bed bugs (Cimex hemipterus) after feeding on human blood were presented. Next-generation sequencing of the community DNA was carried out on an Illumina Miseq platform and the raw fastq files were analyzed using QIIME (version 1.9.1). The metagenome of three samples comprised of 108,198 sequences representing 44,646,263 bps with a mean length of 412.63 bps. The sequence data is accessible at the NCBI SRA under the bioproject number PRJNA600667. Community analysis showed Proteobacteria was the most abundance (more than 99%) microbial community that present in the guts of fully fed tropical bed bugs.
Keywords: Cimex hemipterus, Metagenome, Microbial DNA, Proteobacteria
Specifications table
| Subject | Microbiology |
| Specific subject area | Metagenomic study on the microbial community in the guts of Cimex hemipterus |
| Type of data | Figures, table and 16S rDNA Illumina sequence |
| How data were acquired | 16S v3-v4 amplicon metagenomics sequencing followed by community metagenome analysis. |
| Data format | Raw fastq files |
| Parameters for data collection | Laboratory strain tropical bed bugs after feeding on human blood |
| Description of data collection | The microbial DNA was extracted from the crushed guts of fully fed tropical bed bugs using HiYield™ Genomic DNA isolation kit (Real Biotech Corporation, Taiwan). 16S v3-v4 amplicon metagenomics sequencing was carried in Illumina MiSeq platform. |
| Data source location | Visual inspection was conducted with the aids of flashlight and the bed bugs were collected using forceps from cushion seat at the waiting area in Kuala Lumpur International Airport (KLIA) (25 Feb 2014) at the coordinates of 2.7456 N 101.7072 E. |
| Data accessibility | Repository name: NCBI SRA |
| Data identification number: PRJNA600667 | |
| Direct URL to data: | |
| https://www.ncbi.nlm.nih.gov/bioproject/PRJNA600667 |
Value of the data
-
•
The metagenomics data provide full taxonomic profiles of the microbial diversity and abundance in the guts of fully fed Cimex hemipterus.
-
•
The data also provides an initial picture of the functional capabilities of the gut microbial community of C. hemipterus.
-
•
The data is important for profiling, annotation or pathway reconstruction in understanding the metabolic process within the guts of C. hemipterus performed by microorganisms for forensic and research purposes
-
•
Provide information regarding the microbial community that may associate in blood meal digestion or pathogen defense of C. hemipterus
-
•
Provide possibility in recovery of novel biocatalysts from metagenomic data
-
•
Provide information on the bacterial species and functional groups that play important role for the host's success, offers scope for future studies to develop new pest management approaches that exploit novel targets of chemical, genetic and biological control for C. hemipterus and other insect pests.
1. Data description
The Illumina Miseq sequencer produce 32,816, 43,211, 32,171 sequences with average read length of 413.50, 411.61, 413.13 from samples BB1, BB2 and BB3 respectively (Table 1).
Table 1.
Number of sequences, base pairs and average length of the sequences from each sample.
| Sample | Sequences | Bases (bp) | Average Length (bp) |
|---|---|---|---|
| BB1 | 32,816 | 13,569,406 | 413.50 |
| BB2 | 43,211 | 17,786,156 | 411.61 |
| BB3 | 32,171 | 13,290,701 | 413.13 |
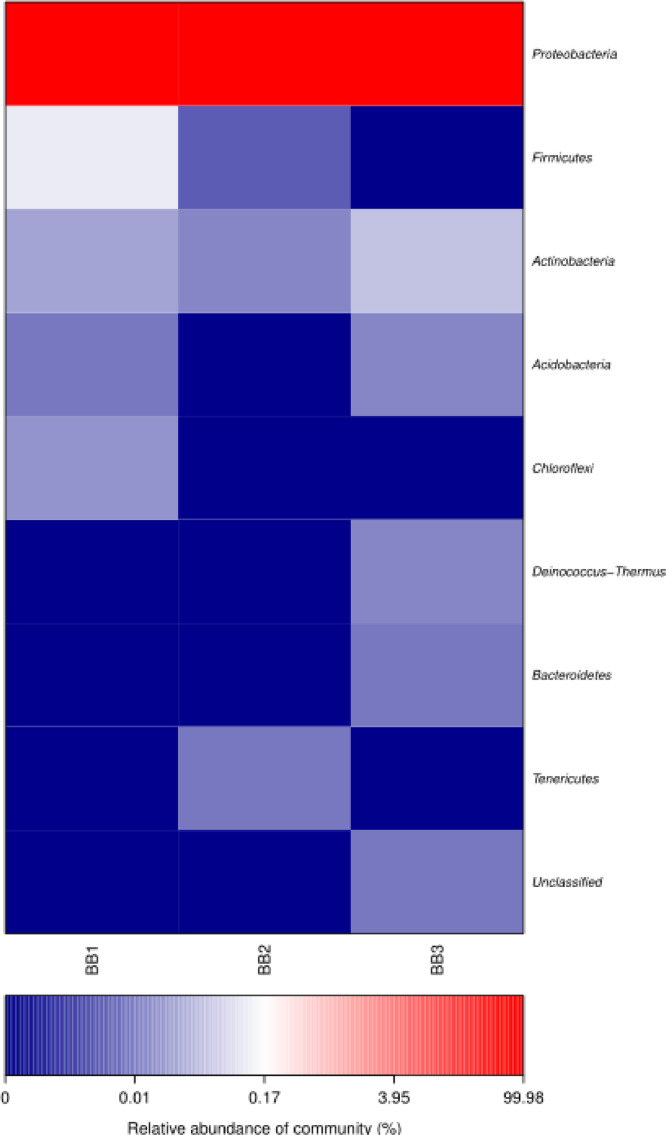
The community analysis showed that more than 99% of sequences of the three samples were assigned to two families within the Proteobacteria: Anaplasmataceae and the Enterobacteriaceae. Two genera including Wolbachia and Pectobacterium comprised the majority of these families. Less than 1% of the reads from 7 phyla of bacteria including Firmicutes, Actinobacteria, Acidobacteria, Chloroflexi, Deinococcus-Thermus, Bacteroidetes, Tenericutes and unclassified reads (Fig. 1).
Fig. 1.
Heatmap analysis which showed the relative abundances of microbial community in C. hemipterus individuals (Samples BB1, BB2 & BB3).
2. Experimental design, material and methods
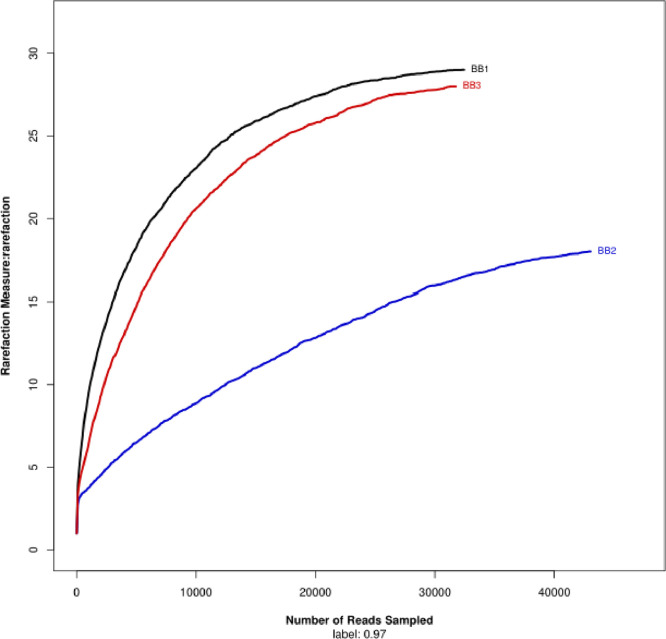
The microbial DNA was extracted from guts of fully fed tropical bed bugs (C. hemipterus) with three replications (BB1, BB2 and BB3) using HiYield™ Genomic DNA isolation kit (Real Biotech Corporation, Taiwan) following the manufacturer's protocol. 16S v3-v4 amplicon metagenomics sequencing was completed via the Illumina MiSeq platform according to the standard protocol. The sequences were analysed using QIIME (version 1.9.1) [1]. Low quality reads with average quality score <20 were trimmed with Trimmomatic [2] software and trimmed reads with lengths shorter than 50 bp were discarded. Paired-reads were merged became single read using FLASH (Fast Length Adjustment of Short reads) [3] based on overlapped relationship. Sequences that overlap longer than 10 bps were assembled while reads that could not be assembled were discarded. The merged reads were used to Operational Taxonomic Unit (OTU) clustering with 97% similarity cut-off using UPARSE [4] software while chimeric sequences were detected using UCHIME [5] software and were removed from the analyses. The taxonomy of the 16S rRNA gene sequences were analyzed using RDP Classifier [6] against the SILVA 16S rRNA database [7] with confidence threshold of 0.7. The sequence coverage of each sample (BB1, BB2 and BB3) was evaluated by rarefaction analysis (Fig. 2) using mothur [8] and R [9] software. The detailed protocol was available on dx.doi.org/10.17504/protocols.io.bc9giz3w.
Fig. 2.
Rarefaction curve showing the species richness of the microbiome of C. hemipterus individuals. Samples BB1 (black curve), BB2 (blue curve) and BB3 (red curve) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Acknowledgments
Acknowledgement
This work was funded by the Fundamental Research Grant (FRGS) (203/PBIOLOGI/6711681).
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., Bai Y., Bisanz J.E., Bittinger K., Brejnrod A., Brislawn C.J., Brown C.T., Callahan B.J., Caraballo-Rodríguez A.M., Chase J., Cope E.K., Da Silva R., Diener C., Dorrestein P.C., Douglas G.M., Durall D.M., Duvallet C., Edwardson C.F., Ernst M., Estaki M., Fouquier J., Gauglitz J.M., Gibbons S.M., Gibson D.L., Gonzalez A., Gorlick K., Guo J., Hillmann B., Holmes S., Holste H., Huttenhower C., Huttley G.A., Janssen S., Jarmusch A.K., Jiang L., Kaehler B.D., Kang K.B., Keefe C.R., Keim P., Kelley S.T., Knights D., Koester I., Kosciolek T., Kreps J., Langille M.G.I., Lee J., Ley R., Liu Y.X., Loftfield E., Lozupone C., Maher M., Marotz C., Martin B.D., McDonald D., McIver L.J., Melnik A.V., Metcalf J.L., Morgan S.C., Morton J.T., Naimey A.T., Navas-Molina J.A., Nothias L.F., Orchanian S.B., Pearson T., Peoples S.L., Petras D., Preuss M.L., Pruesse E., Rasmussen L.B., Rivers A., Robeson M.S., Rosenthal P., Segata N., Shaffer M., Shiffer A., Sinha R., Song S.J., Spear J.R., Swafford A.D., Thompson L.R., Torres P.J., Trinh P., Tripathi A., Turnbaugh P.J., Ul-Hasan S., van der Hooft J.J.J., Vargas F., Vázquez-Baeza Y., Vogtmann E., von Hippel M., Walters W., Wan Y., Wang M., Warren J., Weber K.C., Williamson C.H.D., Willis A.D., Xu Z.Z., Zaneveld J.R., Zhang Y., Zhu Q., Knight R., Caporaso J.G. Reproducible, interactive, scalable and extensible microbiome data science using Qiime 2. Nat. Biotech. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magoč T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10(10):996. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 5.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., Glöckner F.O. SILVA: a comprehensive online resource for quality checked and aligned ribosomal rna sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., Sahl J.W., Stres B., Thallinger G.G., van Horn D.J., Weber C.F. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Team R.C. R: a language and environment for statistical computing. dim (ca533) 2019;1(1358):34. [Google Scholar]