Dear Editor,
Tuberculosis is one of the most important global concerns, as it is considered as the second leading cause of death due to the infectious diseases after HIV. According to the WHO reports in 2020, it is estimated that around 10 million people have affected to TB and 1.5 million deaths occurred due to that in 2019 [1,2].
Iran is also one of the countries in the Middle-East region with a population of about 82 million; incidence of TB in Iran is estimated at about 11,000 people. Tuberculosis control and monitoring in Iran is very important due to its proximity to the countries such as Azerbaijan and Armenia (both high MDR-TB) and Afghanistan and Pakistan (with high-TB burden) [1], [2], [3]. One of the most important strategies in designing the regional programs for the control and prevention of tuberculosis is the identification and tracking of community circulating Mycobacterium tuberculosis (Mtb) strains by the molecular epidemiology techniques [3,4].
According to the literature review, the most common Mtb families in Iran are Beijing (Beijing / W), Haarlem (HaarlemI / HaarlemII and New-1), Delhi/Cas, East-Asian Indian, while TUR, Ural, LAM and bovis are less separable [5,6].
Evidence has been suggested in recent years about the relationship of drug resistance and some Mtb lineages [5], [6], [7]. Thus, this study was performed aiming to determine the frequency of dominant lineages in Iran to evaluate the possible relationship of each of the Beijing, Delhi/Cas, Haarlem and EAI lineages with the first-line drug resistance, especially isoniazid, rifampin and MDR-TB (Multi-drug resistant TB).
In order to fulfil the study, we attempted to investigate all the required studies of the Iranian population by searching PubMed, Scopus, Embase, Cochrane library, Google Scholar and Iranian databases Iranmedex, SID and ISC by March 2020. The search was done with the MeSH keywords including “Mycobacterium tuberculosis”, “TB”, “drug-resistance”, and “genotyping”. The inclusion criteria included: 1- Studies involving the prevalence of our genotypes in susceptible isolates and drug resistance, and 2- Studies in which genotyping was determined by the standard techniques of Spoligotyping, MIRU-VNTR, IS6110 RFLP, and Whole genome sequencing (WGS); and case reports, repetitive sample studies with, and nonclinical studies were included. Finally, the probable relation between the infection and each of the Beijing, Delhi/Cas, Haarlem and EAI genotypes with first-line drug resistance was measured by the Odds Ratio (ORs) with 95% confidence intervals and the Egger weighted regression method was also determined [8].
Based on our investigation, 28 original articles including Persian and English articles were included in the present quantitative analysis (Table 1). In the present study, 6671 Mtb strain data were evaluated. The studies were limited to 2006–2019 and were conducted in the areas including Tehran (National Research Institute of Tuberculosis and Lung Disease), Tabriz, Golestan, Kermanshah and Mashhad.
Table 1.
Characteristics of included studies.
| First author | Publication year | City | Patients | MTB strains | MTB genotypes distribution | Typing method | Ref | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beijing | CAS/Delhi | Haarlem | East African-Indian | |||||||||||
| DR | Total | DR | Total | DR | Total | DR | Total | |||||||
| Ramazanzadeh | 2006 | NRITLD | 345 | 195 | 195 | 195 | 0 | 0 | 0 | 0 | 0 | 0 | Spoligotyping | 9 |
| Farnia | 2006 | NRITLD | 3812 | 1074 | 12 | 12 | 11 | 11 | 44 | 44 | 10 | 10 | Spoligotyping | 10 |
| Farnia | 2007 | NRITLD | NA | 31 | 0 | 0 | 2 | 2 | 13 | 13 | 14 | 14 | Spoligotyping | 11 |
| Amirmozafari | 2007 | NRITLD | NA | 439 | 27 | 68 | 0 | 38 | 0 | 149 | 0 | 31 | Spoligotyping | 12 |
| Farnia | 2008 | NRITLD | NA | 258 | 1 | 12 | 0 | 49 | 8 | 20 | 0 | 58 | Spoligotyping | 13 |
| Dousstdar | 2008 | NRITLD | NA | 30 | 9 | 9 | 12 | 12 | 7 | 7 | 1 | 2 | Spoligotyping | 14 |
| Doustdar | 2008 | NRITLD | NA | 50 | 15 | 15 | 0 | 0 | 10 | 10 | 0 | 0 | IS6110 RFLP | 15 |
| Doustdar | 2009 | NRITLD | NA | 34 | 7 | 7 | 10 | 10 | 5 | 5 | 8 | 8 | IS6110 RFLP | 16 |
| Rohani | 2009 | Mashhad | NA | 113 | 5 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | Spoligotyping | 17 |
| Tajeddin | 2009 | NRITLD | NA | 238 | 9 | 13 | 21 | 76 | 10 | 66 | 13 | 55 | Spoligotyping | 18 |
| Velayati | 2009 | NRITLD | NA | 146 | 5 | 5 | 4 | 4 | 9 | 9 | 5 | 5 | MIRU-VNTR | 19 |
| Ahmadi | 2009 | NRITLD | 238 | 238 | 9 | 13 | 21 | 66 | 10 | 66 | 13 | 55 | Spoligotyping | 20 |
| Mozafari | 2010 | NRITLD | NA | 105 | 7 | 20 | NA | 22 | NA | 2 | 0 | 0 | Spoligotyping | 21 |
| Merza | 2010 | NRITLD | 3812 | 1074 | 6 | 12 | NA | 181 | NA | 44 | NA | 227 | Spoligotyping | 22 |
| Jafarian | 2012 | NRITLD | NA | 96 | 1 | 3 | 7 | 7 | 15 | 36 | 0 | 0 | MIRU-VNTR | 23 |
| Haeili | 2013 | NRITLD | NA | 291 | 3 | 3 | 5 | 70 | 0 | 3 | 0 | 4 | Spoligotyping | 24 |
| Sharifipour | 2014 | NRITLD | NA | 190 | 5 | 11 | 8 | 37 | 8 | 72 | 0 | 0 | Spoligotyping | 25 |
| Varahram | 2014 | NRITLD | 151 | 151 | 10 | 14 | 4 | 23 | 12 | 31 | 14 | 42 | Spoligotyping | 26 |
| Kardan Yamchi | 2015 | NRITLD | NA | 31 | 8 | 8 | 2 | 2 | 1 | 1 | 1 | 1 | Spoligotyping | 27 |
| Mohajeri | 2016 | Kermanshah | 523 | 146 | 3 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | Multiplex PCR | 28 |
| Rezaei | 2016 | NRITLD | NA | 20 | 3 | 3 | 3 | 3 | 10 | 10 | 0 | 0 | Spoligotyping | 29 |
| Khanipour | 2016 | NRITLD | NA | 723 | 9 | 9 | 1 | 1 | 11 | 11 | Spoligotyping | 30 | ||
| Khosravi | 2017 | NRITLD | NA | 88 | 9 | 9 | 0 | 0 | 3 | 3 | 0 | 0 | IS6110 RFLP | 31 |
| Mansoori | 2018 | Golestan | 11,807 | 164 | 5 | 19 | 3 | 31 | 0 | 2 | 0 | 0 | MIRU-VNTR | 32 |
| Zaniani | 2018 | Isfahan | 33 | 18 | 7 | 7 | 4 | 4 | 0 | 0 | 1 | 1 | MIRU-VNTR | 33 |
| Vaziri | 2019 | NRITLD | 13,892 | 606 | 14 | 14 | 7 | 7 | 0 | 0 | 0 | 0 | WGS | 34 |
| Kazemian | 2018 | NRITLD | NA | 34 | 11 | 11 | 4 | 4 | 0 | 0 | 1 | 1 | Spoligotyping | SID |
| Afaghi-Gharamaleki | 2019 | Tabriz | 125 | 115 | 3 | 7 | 0 | 1 | 0 | 0 | 0 | 0 | MIRU-VNTR | 35 |
NRITLD: National Research Institute of tuberculosis and lung disease; WGS: whole genome sequencing.
The studied Mtb strains included two groups of susceptible and drug-resistant TB, with drug-resistant TB strains being resistant to the antibiotics such as isoniazid, rifampin, streptomycin, ethambutol and pyrazinamide.
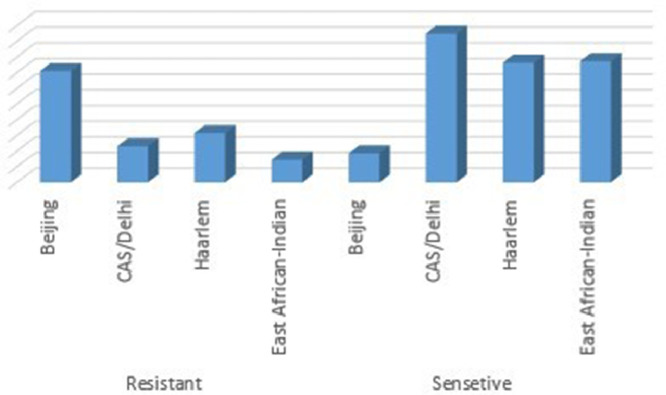
In general, the frequency of Beijing (22.5%), Delhi/Cas (66.7/66.7%), Haarlem (264/60.2) and EAI (22.3/514.5%) lineages were evaluated, while the distribution of lineages in first-line drug resistant strains were evaluated as Beijing (76.2%/398%), Delhi/Cas (19 / 129.5%), Haarlem (29/17.1%) and EAI (16/81%). The distribution of Mtb lineages between susceptible and drug-resistant TB is demonstrated in Fig. 1.
Fig. 1.
Distribution of Iranian Predominant Mycobacterium tuberculosis lineages between susceptible and drug-resistant Mtb strains.
Based on statistical analysis by our random effects model, a significant relationship was observed between the infection with Mtb Beijing strains and drug resistance (ORs: 1.54 (with 95% CIs); 0.7–3.5; Q = 35.94; I2 = 61.04; P = 0.02). However, no significant relationship was observed between the infection with Delhi/Cas families (0.17; 0.11–0.22; Q = 131.4; I2 = 84.0; P = 0.00), Harrlem (0.18; 0.1–0.2; Q = 126.5; I2 = 84.9; EAI (0.15; 0.0–0.2; Q = 70.3; I2 = 81.5; P = 0.00) and drug resistance. Moreover, there was a significant relationship between first line drugs with Mtb Beijing strains and resistance to isoniazid and rifampin (2.19; 1.0–4.2; Q = 77.6; I2 = 88.4; P = 0.027 and 1.60; 0.75–3.41; Q = 59.1; I2 = 89.84; P = 0.2 respectively). Thus, according to the present meta-analysis results, the Delhi/Cas, Haarlem, and Beijing families were the three most common lineages in the tuberculosis patients in Iran, respectively, while Beijing was evaluated with the most significant difference amongst DR-TB strains. We also found a significant relationship between the infection with Beijing Mtb strains and resistance to isoniazid and rifampin in the Iranian patients.
The Beijing tuberculosis family originated from Beijing, China; it was first introduced by Soolingen et al. (1995) as a predominant tuberculosis genotype of East Asia but has worldwide distribution and global distribution today [37,38]. The incidence of Beijing genotypes in recent years has been estimated at between 50% (East Asian countries) to 8.9% (Western countries) [6,38]. According to the review of the literature, the Beijing genotype is one of the predominant tuberculosis family in the Middle-East countries, especially in Iran [5,6,39]. Based on our analysis, the Beijing genotypes were the third most common genotype in Iran. Numerous studies have been published in recent years on the relationship between the Beijing TB genotypes and drug resistance [39], [40], [41]. Based on WGS (whole genome sequencing) data, the presence of multiple SNPs in the katG, rpoB, embB, rpsL, rrs, eis, gyrA and gyrB genes compared to other genotypes could be effective in increasing MIC relative to the anti-tuberculosis drugs [42]. There have been several studies, so far, on the relationship of the infection with Beijing genotypes and drug resistance. However, some other studies contradict this phenomenon [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45]. According to Ramazanzadeh et al. (2014) about 80% of drug resistant Mtb isolates from the Iranian patients belonged to the Beijing genotype. Also, in the study by Tarashi et al. (2017), amongst the MDR-TB isolates of Iranian patients the frequency of Beijing and Haarlem genotypes were 19.3% and 18.7%, respectively [5,6]. We also showed in this study that the Beijing genotype was the third most common genotype in the Iranian tuberculosis (TB) patients, and the infection with the Beijing family had significant relationship with the resistance to isoniazid and rifampin. Farnia et al. (2006) showed that Haarlem I and Beijing genotypes are the most common TB lineages of the Afghan population residing in Iran, who are affected to MDR-TB [10].
Other similar studies have also found that the Beijing genotype has been isolated from a significant population of TB patients in Iran neighbouring countries, especially Afghanistan and Pakistan. Hence, in order to prevent the spread of Mtb Beijing family infection, it seems that designing the guidelines has a significant impact on reducing the DR -TB cases in the Iranian population [5,6,39].
Ethical
Ethical Statement is not applicable for this manuscript.
References
- 1.Keikha M., Esfahani B.N. The relationship between tuberculosis and lung cancer. Adv Biomed Res. 2018;7:8–11. doi: 10.4103/abr.abr_182_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . Global tuberculosis report 2019. World Health Organization; 2020. [Google Scholar]
- 3.Zaniani F.R., Moghim S., Mirhendi H., Safaei H.G., Fazeli H., Salehi M. Genetic lineages of mycobacterium tuberculosis isolates in Isfahan, Iran. Curr Microbiol. 2017;74(1):14–21. doi: 10.1007/s00284-016-1145-2. [DOI] [PubMed] [Google Scholar]
- 4.Supply P., Allix C., Lesjean S., Cardoso-Oelemann M., Rüsch-Gerdes S., Willery E. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44(12):4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarashi S., Fateh A., Jamnani F.R., Siadat S.D., Vaziri F. Prevalence of Beijing and Haarlem genotypes among multidrug-resistant Mycobacterium tuberculosis in Iran: systematic review and meta-analysis. Tuberculosis. 2017;107:31–37. doi: 10.1016/j.tube.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Ramazanzadeh R., Sayhemiri K. Prevalence of Beijing family in Mycobacterium tuberculosis in world population: systematic review and meta-analysis. Int J Mycobacteriol. 2014;3(1):41–45. doi: 10.1016/j.ijmyco.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Garzon-Chavez D., Zurita J., Mora-Pinargote C., Franco-Sotomayor G., Leon-Benitez M., Granda-Pardo J.C., Trueba G., Garcia-Bereguiain M.A., de Waard J.H. Prevalence, drug resistance, and genotypic diversity of the Mycobacterium tuberculosis Beijing Family in Ecuador. Microbial Drug Resistance. 2019;25(6):931–937. doi: 10.1089/mdr.2018.0429. [DOI] [PubMed] [Google Scholar]
- 8.Lee J. Odds ratio or relative risk for cross-sectional data? Int J Epidemiol. 1994;23(1):201–203. doi: 10.1093/ije/23.1.201. [DOI] [PubMed] [Google Scholar]
- 9.Ramazanzadeh R., Farnia P., Amirmozafari N., Ghazi F., Ghadertotonchi Z., Kamran J. Comparison between molecular epidemiology, geographical regions and drug resistance in Mycobacterium tuberculosis strains isolated from Iranian and Afghan patients. Chemotherapy. 2006;52(6):316–320. doi: 10.1159/000095971. [DOI] [PubMed] [Google Scholar]
- 10.Farnia P., Masjedi M.R., Mirsaeidi M., Mohammadi F., Vincent V., Bahadori M., Velayati A.A. Prevalence of Haarlem I and Beijing types of Mycobacterium tuberculosis strains in Iranian and Afghan MDR-TB patients. J Infect. 2006;53(5):331–336. doi: 10.1016/j.jinf.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 11.Farnia P., Masjedi M.R., Nasiri B., Mirsaedi M., Sorooch S., Kazeampour M., Velayati A.A. Instability of IS6110 patterns in multidrug-resistant strains of Mycobacterium tuberculosis. Epidemiol Infect. 2007;135(2):346–352. doi: 10.1017/S0950268806006790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amirmozafari N., Ramezanzadeh R., Farnia P., Ghazi F. The frequency of Beijing genotype of Mycobacterium tuberculosis isolated from tuberculosis patients. Razi J Med Sci. 2006;13(52):7–17. [Google Scholar]
- 13.Farnia P., Masjedi M.R., Varahram M., Mirsaeidi M., Ahmadi M., Khazampour M. The recent-transmission of Mycobacterium tuberculosis strains among Iranian and Afghan relapse cases: a DNA-fingerprinting using RFLP and spoligotyping. BMC Infect Dis. 2008;8(1):109. doi: 10.1186/1471-2334-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doustdar F., Khosravi A.D., Farnia P., BAHREHMAND A., Masjedi M.R., Velayati A.A. Mutations in rpoB gene and genotypes of rifampin resistant Mycobacterium tuberculosis isolates in Iran. Tanaffos. 2008;7(2):11–17. [Google Scholar]
- 15.Doustdar F., Khosravi A.D., Farnia P., Masjedi M.R., Velayati A.A. Molecular analysis of isoniazid resistance in different genotypes of Mycobacterium tuberculosis isolates from Iran. Microbial Drug Resistance. 2008;14(4):273–279. doi: 10.1089/mdr.2008.0842. [DOI] [PubMed] [Google Scholar]
- 16.Doustdar F., Khosravi A.D., Farnia P. Mycobacterium tuberculosis genotypic diversity in pyrazinamide-resistant isolates of Iran. Microbial Drug Resistance. 2009;15(4):251–256. doi: 10.1089/mdr.2009.0066. [DOI] [PubMed] [Google Scholar]
- 17.Rohani M., Farnia P., Nasab M.N., Moniri R., Torfeh M., Amiri M.M. Beijing genotype and other predominant Mycobacterium tuberculosis spoligotypes observed in Mashhad city, Iran. Indian J Med Microbiol. 2009;27(4):306. doi: 10.4103/0255-0857.55441. [DOI] [PubMed] [Google Scholar]
- 18.Tajeddin E., Farnia P., Kargar M., Noroozi J., Ahmadi M., Kazempour M., Masjedi M.R., Velayati A.A. Identification of Mycobacterium tuberculosis Beijing genotype using three different molecular methods. Koomesh. 2009;11(1) [Google Scholar]
- 19.Velayati A.A., Masjedi M.R., Farnia P., Tabarsi P., Ghanavi J., ZiaZarifi A.H. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest. 2009;136(2):420–425. doi: 10.1378/chest.08-2427. [DOI] [PubMed] [Google Scholar]
- 20.Ahmadi M.O., Farnia P., Tajedin E., Tabarsi P., Baghaei P.A., Masjedi M.R., Velayati A. Mycobacterium Tuberculosis Complex Strains Identification with Spoligotyping Method in Patients Attending to Masih Daneshvari Hospital. J Adv Med Biomed Res. 2009 Aug 10;17(67):23–32. [Google Scholar]
- 21.Mozafari M., Farnia P., Jafarian M., Razavi Deligani M., Kazempour M., Masjedi M., Velayati A.A. Comparison of Mycobacterium tuberculosis Beijing genotype with other Mycobacterium tuberculosis strains Using MIRU-VNTR method. ISMJ. 2012;15(1):1–2. [Google Scholar]
- 22.Merza M.A., Farnia P., Salih A.M., Masjedi M.R., Velayati A.A. The most predominant spoligopatterns of Mycobacterium tuberculosis isolates among Iranian, Afghan-immigrant, Pakistani and Turkish tuberculosis patients: a comparative analysis. Chemotherapy. 2010;56(3):248–257. doi: 10.1159/000316846. [DOI] [PubMed] [Google Scholar]
- 23.Jafarian M., Farnia P., Kargar M., Aghalimerza M., Ramazanzadeh R., Ahmadi M. Study of genetic diversity of M.tuberculosis strains isolated by MIRU-VNTR technique in Masih Daneshvari Hospital, Tehran. SJKU. 2010;14(4):29–39. [Google Scholar]
- 24.Haeili M., Darban‐Sarokhalil D., Fooladi A.A., Javadpour S., Hashemi A., Siavoshi F. Spoligotyping and drug resistance patterns of Mycobacterium tuberculosis isolates from five provinces of Iran. Microbiologyopen. 2013;2(6):988–996. doi: 10.1002/mbo3.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharifipour E., Nasiri M., Farnia P., Mozafari M., Irani S. Evaluation of molecular diversity of Mycobacterium tuberculosis strains by polymorphisms in RD Regions. J Mycobac Dis. 2014;4(153) 2161-1068. [Google Scholar]
- 26.Varahram M., Farnia P., Nasiri M.J., Karahrudi M.A., Dizagie M.K., Velayati A.A. Association of Mycobacterium tuberculosis lineages with IFN-γ and TNF-α gene polymorphisms among pulmonary tuberculosis patient. Mediterr J Hematol Infect Dis. 2014;6(1) doi: 10.4084/MJHID.2014.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamchi J.K., Haeili M., Feyisa S.G., Kazemian H., Shahraki A.H., Zahednamazi F., Fooladi A.A., Feizabadi M.M. Evaluation of efflux pump gene expression among drug susceptible and drug resistant strains of Mycobacterium tuberculosis from Iran. Infect Genet Evol. 2015;36:23–26. doi: 10.1016/j.meegid.2015.08.036. [DOI] [PubMed] [Google Scholar]
- 28.Mohajeri P., Moradi S., Atashi S., Farahani A. Mycobacterium tuberculosis Beijing genotype in western Iran: distribution and drug resistance. J Clin Diagnostic Res: JCDR. 2016;10(10):DC05. doi: 10.7860/JCDR/2016/20893.8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rezaei F., Haeili M., Mohajeri P., Shahraki A.H., Fooladi A.A., Zahednamazi F., Feizabadi M.M. Frequency of mutational changes in the embB among the ethambutol-resistant strains of Mycobacterium tuberculosis in Iran. J Infect Dev Countries. 2016;10(04):363–368. doi: 10.3855/jidc.7215. [DOI] [PubMed] [Google Scholar]
- 30.Khanipour S., Ebrahimzadeh N., Masoumi M., Sakhaei F., Alinezhad F., Safarpour E. Haarlem 3 is the predominant genotype family in multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis in the capital of Iran: a 5-year survey. J Glob Antimicrob Resist. 2016;5:7–10. doi: 10.1016/j.jgar.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Khosravi A.D., Shahraki A.H., Dezfuli S.K., Hashemzadeh M., Goodarzi H., Mohajeri P. Genetic diversity of multidrug-resistant Mycobacterium tuberculosis strains isolated from tuberculosis patients in Iran using MIRU-VNTR technique. Kaohsiung J Med Sci. 2017;33(11):550–557. doi: 10.1016/j.kjms.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansoori N., Yaseri M., Vaziri F., Douraghi M. Genetic diversity of Mycobacterium tuberculosis complex isolates circulating in an area with high tuberculosis incidence: using 24-locus MIRU-VNTR method. Tuberculosis. 2018;112:89–97. doi: 10.1016/j.tube.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Zaniani F.R., Moghim S., Esfahani B.N. Genetic diversity of drug-resistant Mycobacterium tuberculosis isolates in Isfahan province of Iran. Adv Biomed Res. 2018:7. doi: 10.4103/2277-9175.225594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaziri F., Kohl T.A., Ghajavand H., Kamakoli M.K., Merker M., Hadifar S. Genetic diversity of multi-and extensively drug-resistant Mycobacterium tuberculosis isolates in the capital of Iran, revealed by whole-genome sequencing. J Clin Microbiol. 2019;57(1) doi: 10.1128/JCM.01477-18. e01477-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afaghi G.A., Moaddab S.R., Darbouy M., Ansarin K., Hanifian S. Genotypic diversity of resistant Mycobacterium tuberculosis strains isolated from tuberculosis patients in East Azerbaijan center by MIRU-VNTR. J Microbial World. 2009;11(4):320–331. [Google Scholar]
- 36.Van Soolingen D., Qian L., De Haas P.E., Douglas J.T., Traore H., Portaels F. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol. 1995;33(12):3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toungoussova O.S., Sandven P., Mariandyshev A.O., Nizovtseva N.I., Bjune G., Caugant D.A. Spread of drug-resistant Mycobacterium tuberculosis strains of the Beijing genotype in the Archangel Oblast, Russia. J Clin Microbiol. 2002;40(6):1930–1937. doi: 10.1128/JCM.40.6.1930-1937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanveer M., Hasan Z., Siddiqui A.R., Ali A., Kanji A., Ghebremicheal S. Genotyping and drug resistance patterns of M. tuberculosis strains in Pakistan. BMC Infect Dis. 2008;8(1):171. doi: 10.1186/1471-2334-8-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffner S., Sahebi L., Ansarin K., Sabour S., Mohajeri P. Mycobacterium tuberculosis of the Beijing genotype in Iran and the World Health Organization Eastern Mediterranean Region: a meta-analysis. Microbial Drug Resistance. 2018;24(6):693–698. doi: 10.1089/mdr.2017.0160. [DOI] [PubMed] [Google Scholar]
- 40.Sun H., Zhang C., Xiang L., Pi R., Guo Z., Zheng C., Li S., Zhao Y., Tang K., Luo M., Rastogi N. Characterization of mutations in streptomycin-resistant Mycobacterium tuberculosis isolates in Sichuan, China and the association between Beijing-lineage and dual-mutation in gidB. Tuberculosis. 2016;96:102–106. doi: 10.1016/j.tube.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Nhu N.T., Lan N.T., Phuong N.T., van V., Chau N., Farrar J., Caws M. Association of streptomycin resistance mutations with level of drug resistance and Mycobacterium tuberculosis genotypes. Int. J. Tuberculosis Lung Dis. 2012;16(4):527–531. doi: 10.5588/ijtld.11.0202. [DOI] [PubMed] [Google Scholar]
- 42.Alvarez N., Haft D., Hurtado U.A., Robledo J., Rouzaud F. Whole-genome sequence of a Beijing extensively drug-resistant Mycobacterium tuberculosis clinical isolate from Buenaventura, Colombia. Genome Announc. 2016;4(1) doi: 10.1128/genomeA.01549-15. e01549-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan L., Huang Y., Mi L.G., Li Y.X., Liu P.Z., Zhang J., Liang H.Y., Li F., Li H., Zhang S.Q., Li W.J. There is no correlation between sublineages and drug resistance of Mycobacterium tuberculosis Beijing/W lineage clinical isolates in Xinjiang, China. Epidemiol Infect. 2015;143(1):141–149. doi: 10.1017/S0950268814000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., Cao X., Li S., Wang H., Wei J., Liu P. Characterization of Mycobacterium tuberculosis isolates from Hebei, China: genotypes and drug susceptibility phenotypes. BMC Infect Dis. 2016;16(1):107. doi: 10.1186/s12879-016-1441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y., Jiang X., Li W., Zhang X., Wang W., Li C. The study on the association between Beijing genotype family and drug susceptibility phenotypes of Mycobacterium tuberculosis in Beijing. Sci Rep. 2017;7(1):1–7. doi: 10.1038/s41598-017-14119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]