Abstract
Cancer Informatics for Cancer Centers (CI4CC) is a grassroots, nonprofit 501c3 organization intended to provide a focused national forum for engagement of senior cancer informatics leaders, primarily aimed at academic cancer centers anywhere in the world but with a special emphasis on the 70 National Cancer Institute–funded cancer centers. Although each of the participating cancer centers is structured differently, and leaders’ titles vary, we know firsthand there are similarities in both the issues we face and the solutions we achieve. As a consortium, we have initiated a dedicated listserv, an open-initiatives program, and targeted biannual face-to-face meetings. These meetings are a place to review our priorities and initiatives, providing a forum for discussion of the strategic and pragmatic issues we, as informatics leaders, individually face at our respective institutions and cancer centers. Here we provide a brief history of the CI4CC organization and meeting highlights from the latest CI4CC meeting that took place in Napa, California from October 14-16, 2019. The focus of this meeting was “intersections between informatics, data science, and population science.” We conclude with a discussion on “hot topics” on the horizon for cancer informatics.
INTRODUCTION
Brief History of Cancer Informatics for Cancer Centers
Cancer Informatics for Cancer Centers (CI4CC) is a grassroots, nonprofit 501c3 organization intended to provide a focused national forum for engagement of senior cancer informatics leaders, primarily aimed at academic cancer centers anywhere in the world but with a special emphasis on the 70 National Cancer Institute (NCI)–funded cancer centers (https://www.ci4cc.org). CI4CC started as an attempt to maintain the sense of community and comradery that the NCI cancer Biomedical Informatics Grid—caBIG—program had developed, as that program was winding down in 2012. We held our first meeting in Dallas, Texas, in February of 2013. That meeting, attended by representatives of approximately 30 of the NCI-designated cancer centers, was testing the waters to see if such an event was needed and if the cancer informatics community was receptive and willing to participate. At that meeting, we had all the cancer informatics cores describe their structure, funding methodology (typically direct charge or effort on grants), and strategy for embedding informatics (and “data science”) in their cancer center. On the basis of the positive feedback from that initial meeting, we started meeting twice annually, with the next meeting in November of 2013 in the Bay Area of California. A preponderance of our meetings have been held in California, with only the meetings in Dallas, Washington DC, Park City, New Orleans, and Maui outside of California. Every meeting has had a different focus, most meetings have had two chairs, and many meetings have included aspects of precision oncology and how we can support precision oncology and learning health systems for cancer more effectively using informatics, data science, and machine learning (ML) approaches. It is important to foster diversity, with the goal of welcoming all members of the cancer informatics community and including them in CI4CC. CI4CC has been fortunate to have featured presentations from two NCI Directors, Drs Harold Varmus and Ned Sharpless. We have a “directors circle,” featuring cancer center directors, deputy directors, and NCI directors who have spoken at one or more CI4CC events (http://www.ci4cc.org/events/directors-circle). In addition to precision medicine, we have had meeting topics ranging from how informatics, data science, and ML can be applied to research problems in population health, pediatrics, cancer clinical trials, cancer registries, molecular tumor boards, and cancer surveillance.
THE FALL 2019 CI4CC MEETING
The biannual face-to-face conferences often focus on a particular theme, providing an opportunity for members of the informatics community to interact with leaders in related scientific disciplines to foster multidisciplinary approaches to cancer research and care delivery. Although we do not explicitly focus on team science, the ability of each of our member centers to successfully solve problems in any of these areas requires building interdisciplinary teams and using team science approaches.
CONTEXT
Key Objective
How best do we share ideas and best practices in the cancer center community in the informatics domain? Through the Cancer Informatics for Cancer Centers (CI4CC) organization, we enable cancer centers to explore and learn from each other.
Knowledge Generated
Cancer informatics is an ever-growing and diverse field. We focused the Fall 2019 meeting on the integration of data science and informatics with population science. Topics covered included: collection of cancer risk factor information and outcomes through digital tools, data science approaches for advancing insights from cohort and other large-scale population-based studies, data linkages for catchment area research, strategic approaches to creating and maintaining electronic data warehouses for the advancement of cancer research and care delivery (including common data models), leveraging artificial intelligence for observational research, and evolving Cancer Center Support Grant cores to support optimal data access, integration, and analysis.
Relevance
Through the Fall CI4CC meeting we were able to highlight scientific advances and ongoing efforts to better understand cancer etiology, identifying new approaches for cancer prevention and early detection, improving outcomes for patients with cancer, and enhancing cancer care delivery throughout the community.
For the Fall 2019 conference, we focused on the integration of data science and informatics with population science, highlighting scientific advances and ongoing efforts to better understand cancer etiology, identifying new approaches for cancer prevention and early detection, improving outcomes for patients with cancer, and enhancing cancer care delivery throughout the community. Data collection and analysis have continuously evolved within the context of population science studies, increasingly incorporating digital tools for assessment of exposures and health behaviors. Furthermore, as we seek to unravel the complexity of cancer, novel informatics and data science approaches are required to integrate and analyze data across multiple biologic scales, including genomics, pathology, and radiology-based images and clinical and patient-reported outcomes. Given the complexity and amount of data currently available on patients with cancer and populations at risk for cancer, further convergence of data and population sciences is required to effectively advance data capture, integration, and analysis, with the common goal of reducing the cancer disease burden in the population. The Fall 2019 CI4CC conference covered topics such as collection of cancer risk factor information and outcomes through digital tools, data science approaches for advancing insights from cohort and other large-scale population-based studies, data linkages for catchment area research, strategic approaches to creating and maintaining electronic data warehouses for the advancement of cancer research and care delivery (including common data models), leveraging artificial intelligence (AI) for observational research, and evolving Cancer Center Support Grant (CCSG) cores to support optimal data access, integration, and analysis. Below we provide meeting highlights from the CI4CC meeting that took place in Napa, California from October 14-16, 2019.
HIGHLIGHTS FROM THE FALL 2019 CI4CC MEETING
Day 1
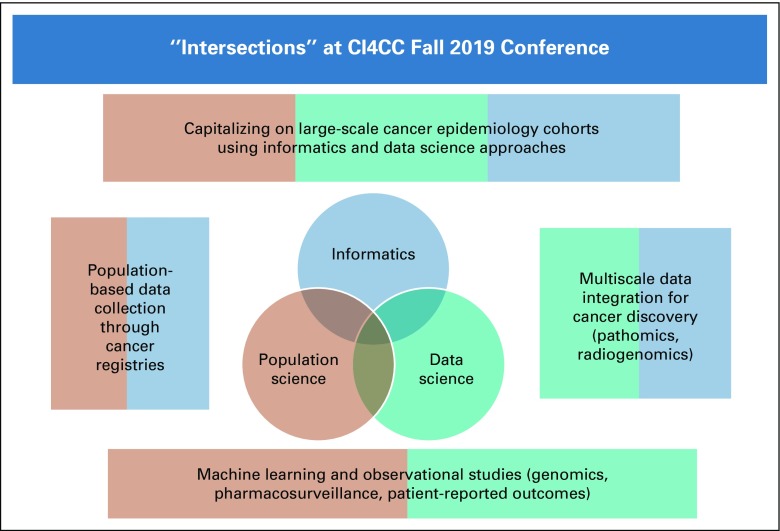
Drs Jill Barnholtz-Sloan (Case Western Reserve University School of Medicine and University Hospitals of Cleveland) and Dana Rollison (Moffitt Cancer Center), the conference co-chairs, set the stage for the conference by providing an overview of how data science, informatics, and population science intersect in a variety of areas of cancer research (Fig 1). Many large-scale cancer epidemiology cohort studies are recognizing the importance of leveraging state-of-the-art informatics and data science approaches to enhance data collection and promote data sharing. Informatics and data science approaches are also being used to model cancer biology across scales, including genomics, pathology, and radiology. Population-based cancer registries are using informatics approaches to augment information available in the registry with rich data available in other publicly available datasets. Finally, ML techniques are beginning to be leveraged by population-based datasets and cohort studies. The sessions of the conference focused on each of these intersections, and selected highlights from each session are described below.
FIG 1.
Intersections between population science, data science, and informatics highlighted at the Cancer Informatics for Cancer Centers Fall 2019 conference.
Population-based data collection for cancer prevention and care delivery.
This first session highlighted large-scale, population-based initiatives that leverage informatics tools to understand patterns in cancer incidence, treatment, survival, and related factors. Ms Lisa Mirel from the National Center for Health Statistics (NCHS) Data Linkage Program described how information collected through the NCHS population-based health surveys, such as the National Health and Interview Survey (NHIS) and the National Health and Nutrition Examination Survey (NHANES), are linked to vital and administrative data from other sources (National Death Index, Centers for Medicare & Medicaid Services enrollment and claims data, Department of Housing and Urban Development data on housing assistance), and discussed the informatics-related methodologies that enable use of the linked files (https://www.cdc.gov/nchs/data-linkage/index.htm). This program is designed to maximize the scientific value of the Center’s population-based surveys and enable researchers to examine the factors that influence disability, chronic disease, health care utilization, morbidity, and mortality. The linked files expand the analytic potential of both the survey and administrative data, enabling analyses that would not be possible with either data source alone. For example, linked files have been used to address a variety of cancer-related research topics, including the association between muscle-strengthening physical activity and cancer mortality, as well as the association between folate intake and biomarkers and cancer risk.1,2 The Data Linkage Program releases two types of public-use files: (1) public-use linked mortality files, containing a limited set of variables; and (2) feasibility files, designed to help interested researchers determine the maximum available sample sizes and assess the feasibility of analyses using the restricted-use linked files. To protect confidentiality of survey participants, all the other linked data files are restricted use and can be accessed only through the NCHS Research Data Centers (NCHS Research Data Center: https://www.cdc.gov/rdc/index.htm).
Dr Marta Induni from the Cancer Registry of Greater California (CRGC) spoke about informatics advances facilitating rapid reporting of pathology reports in a standard format, in compliance with California’s new law requiring pathologists to report cancer cases within 2 weeks (AB 2325; (https://leginfo.legislature.ca.gov/faces/billNavClient.xhtml?bill_id=201520160AB2325). This accelerated and structured reporting allows a central cancer registry to become more relevant to patients with active disease and can facilitate near–real-time use of cancer data to benefit the quality of life (QoL) and improve outcomes for patients with cancer in its catchment. Several use cases were presented from CRGC, including a proposed alert system for pathologists who do not follow best practices, standards of care, or standard operating procedures. The College of American Pathologists encourages structured reporting of required data elements for the purpose of interoperability, data capture, and data exchange (https://documents.cap.org/protocols/dSynoptic_Report_DefinitionAndExamples_v4.0.pdf). Furthermore, this rapid reporting to centralized registries could facilitate the matching of eligible patients to clinical trials soon after initial diagnosis.
Dr Elizabeth Shenkman from the University of Florida focused her talk on novel approaches for leveraging real-world data points from electronic health records, health care claims, patient report, and other sources to generate evidence about cancer-related outcomes for diverse patient populations, including those that may be under-represented in biomedical research.3 Such approaches for cancer control include: (1) incorporation and linking of contextual data, such as census tract and environmental information, to real-world health data; and (2) use of diverse computational and statistical modeling techniques.4 The OneFlorida Clinical Research Consortium was created to form an enduring infrastructure for cohort discovery, pragmatic clinical trials, and observational studies.5 A hallmark of OneFlorida is its centralized Data Trust, which contains linked electronic health record, tumor registry, claims, and geospatial data for > 14 million Floridians. Currently, 24 cancer-related studies are being conducted in OneFlorida (examples are: https://maps.cancer.gov/overview/DCCPSGrants/abstract.jsp?applId=9816491&term=CA234030; and https://www.pcori.org/research-results/2019/natural-language-processing-connect-social-determinants-and-clinical-factors). This statewide approach to cancer prevention can be used as a model for other states or large regions of the country.
Novel ML/AI approaches to observational studies.
This second session focused on state-of-the-art data science approaches to the analysis of observational data, with speakers presenting examples in genomics, pharmacosurveillance, and patient-reported outcomes (PROs), the latter of which was the focus of the presentation by Dr Amrita Basu from the University of California, San Francisco. PROs may be used to measure QoL, a topic of increasing importance to women diagnosed with breast cancer who often experience some form of drug-related toxicity, psychosocial distress, and subsequent impairments in their QoL. Impairments in QoL can interfere with treatment adherence and engagement in health-promoting behaviors, whereas effective management of symptoms during treatment has been associated with improved QoL, adherence, and increased survival. Dr Basu discussed the development of the neoadjuvant Clinical Benefit Index, a novel longitudinal approach to assessing QoL in breast oncology that provides a single numerical index of QoL and clinical efficacy, and went on to describe the impact of age, stage, and educational status on QoL in a separate study of patients with breast cancer enrolled in the California Athena Breast Cancer Study, where impaired QoL in one domain was associated with more severe symptoms in another. These insights may lead to strategies to prevent or delay symptom onset or interference, promote intervention at the earliest detectable onset of symptoms, and result in supportive care and treatment adaptations.
Paul fearn memorial lecture and poster session.
Day 1 wrapped up with the Paul Fearn Memorial Lecture and Poster Session. Dr Paul Fearn was a respected leader in cancer informatics. He contributed to the field not only through his vast technical expertise but also through a powerful combination of creative strategy, innovative approaches, insightful empathy, and a passion for training the next generation of informatics professionals. He provided steadfast guidance and support not only to his team but also to many he encountered, promoting innovation and openness among colleagues across fields. The CI4CC community aimed to honor Dr Fearn’s legacy through the Inaugural Paul Fearn Memorial Lecture and Poster Session. Nine CI4CC investigators presented their work as part of these events in honor of Dr Fearn, including Dr Jack DiGiovanna from Seven Bridges, who discussed leveraging cloud-based analysis ecosystems to support training the next generation of data scientists. The National Institutes of Health (NIH) Strategic Plan for Data Science includes expanding the national research workforce and collaboration across disciplines as key objectives (https://datascience.nih.gov/strategicplan). However, training biomedical scientists with diverse backgrounds on essential, complex, and iterative data science techniques is challenging. Once data scientists have the appropriate theoretical background, applying it to real-world research questions has an additional learning curve. For example, there are multiple hurdles associated with accessing controlled datasets, exploring them, and understanding them sufficiently to investigate relevant questions. A final obstacle is adequate “hands-on” training with scientific analysis software. To tackle these challenges, the Seven Bridges team leveraged three data ecosystems: the NCI Cancer Research Data Common (CRDC) Cancer Genomics Cloud,6 the NHLBI BioData Catalyst, and the NIH Common Fund Gabriella Miller Kids First DRC’s Cavatica (https://kidsfirstdrc.org/). Diverse trainees were successfully connected to actionable data in collaborative, powerful, and cost-efficient environments. Representative examples span high school students visualizing gene expression levels to graduate student geneticists learning to code association studies. Leveraging cloud-based analysis ecosystems could provide an important component of training the national research workforce.
Day 2
The second day continued the conference theme by starting with presentations and a panel discussion focused on ongoing large-scale epidemiologic cohort studies that are leveraging data science and informatics to advance the generation of new knowledge. These studies are essential components of the cancer research enterprise, providing ideal settings for assessing how real-world data affect cancer risks and outcomes.
Cancer cohorts and other large-scale population studies: the importance of data science and informatics.
Dr Montserrat Garcia-Closas from the NCI described NCI-Connect, a new, prospective cohort study being conducted by investigators in the NCI Division of Cancer Epidemiology and Genetics (DCEG) in collaboration with integrated health care systems in the United States. The primary aim is to build a comprehensive research resource using new technologies and methods for the scientific community to study cancer etiology, precursor to tumor transformation, cancer risk prediction, early detection of cancer, and second cancers and cancer survivorship. To this end, the cohort is designed to enroll up to 200,000 adults free of cancer for long-term follow-up with serial data and biospecimen collections, following rigorous epidemiologic principles. Data will be collected by multiple mechanisms, including questionnaires, electronic health care records (EHRs), medical images, mobile and wearable technologies, and data linkages to resources such as cancer and mortality registries and environmental-monitoring data. Biospecimens will be used to generate data on biomarkers of susceptibility, internal dose, and early biologic effects using targeted and -omic technologies. NCI-Connect is being designed using an information technology system’s architecture to support a mobile app for participant engagement and data collection to supporting big data analytics by scientists, all while facilitating reproducible dissemination in the public domain. This infrastructure will enable Epidemiology Data Commons (the colocation of data assets and code in the cloud) to facilitate following of the Findable, Accessible, Interoperable, Reusable (FAIR) principles for scientific data management and stewardship.7 To achieve these goals, DCEG is developing a serverless cloud execution model facilitated by resources available through the NIH Science and Technology Research Infrastructure for Discovery, Experimentation, and Sustainability—STRIDES—program (https://datascience.nih.gov/strides). The key design motivations for this data system are: (1) interoperability: decoupling of the data layer from the application layer by stateless application programming interfaces (APIs); (2) user-centric governance at the service (API call) layer level: this model is a far safer than “security at the perimeter,” and maximizes collaborative use and data analysis, because usages are associated with the identity and scope of the user making the API call to the data service; (3) delivery of cloud-based, consumer-facing Web apps: the API ecosystem acts as a “marketplace” for pluggable components. The investigators expect that a broader cohort commons infrastructure will eventually emerge, maybe under the aggregation of other resources, such as the NIH All of Us program.
Dr Alexander Borowsky from University of California, Davis, provided an overview of the NIH All of Us Program, which aims to build an ambitious and unprecedented longitudinal cohort of 1 million persons, representative of the population of the United States,8 including demographic subgroups that are underrepresented in current and past biomedical research, defined by race, ethnicity, age, sex, gender identity, sexual orientation, disability status, access to health care, income, education level, and/or geographic location.9 Participants agree to provide access to their medical records, to complete survey questionnaires, and to provide blood (or saliva) and urine samples. Participants are provided permanent access through a password-protected web portal and smart phone app to review details of their own data, to refresh or add to questionnaire data, to link wearable fitness tracker data, and to see aggregated information about the overall cohort. To achieve the goals of the study, multiple critical informatics systems with connections were developed.10 The participant portal serves as the initial enrollment tool, serving video descriptions of the purpose and specific consent elements of the program, as well as handling the informed consent documentation with e-signature. This portal was designed to recognize the state of residence/enrollment of the participant to serve state-specific information and documents to comply with state as well as federal health information and research consent regulations. Enrollment on the portal is monitored by the system with location-based information transmitted to the appropriate health care provider partners or “direct volunteer” enrollment sites. After enrollment and collection of biospecimens, the participant electronic medical record data are mapped to a common data model Observational Medical Outcomes Partnership (OMOP) and securely transmitted quarterly to the central database.
Dr James Lacey Jr from City of Hope shared lessons learned from modernizing an existing epidemiology cohort study to be compatible with the NCI’s CRDC and the principles of FAIR data. Since 2015, the California Teachers Study (CTS; www.calteachersstudy.org/for-researchers), which began in 1995-1996 and has followed > 133,000 women continuously since, has replaced its legacy methods with a data warehouse, a secure remote desktop, and standardized processes configured for population sciences. Data visualizations, reusable workflows, and analytic tools provide scalability, and the environment meets the technical requirements of the CRDC. As the CTS has deployed these tools, a consistent challenge has emerged: how can cohorts like the CTS present their vast data in ways that enable users to perform the complex, individualized, and unpredictable analyses that epitomize epidemiologic research? The CTS includes > 5,000 columns of patient-reported data, 30,000 cancers and 30,000 deaths, and 500,000 hospitalization end points, but most projects make their “go v no-go” decisions on the basis of only a handful of data points. Epidemiology data commons need to make all of their data available and provide user-friendly query and analytic tools that allow users to define as many combinations as possible and capitalize on the complexity that makes cancer cohorts valuable.
Multiscale data integration for cancer discovery.
The second session on Day 2 focused on the complexities of analyzing data across scales, including genomics, pathomics, and radiomics. As our understanding of cancer biology deepens within each of these scales, development of quantitative approaches for integrating information across scales will be increasingly important for convergence research.
Dr Joel Saltz from Stonybrook University discussed the evolution of his team’s work to develop digital pathology ML/AI whole-slide image methodologies.11 Their focus has been on developing methods for using routinely collected biopsy and excision specimens to generate pathophysiological ground truth for large-scale clinical investigations to improve clinical workflows and thereby affect patient care. The methodologies encompass ML/deep learning methods, software for viewing whole-slide images and annotations, and data management methods for digital pathology analytic algorithm results.12 They coined the term “pathomics” to describe digital pathology information; this includes segmented cell nuclei, maps of tumors and infiltrating lymphocytes, and features directly extracted from digital pathology images using AI algorithms.13
Dr Jennifer Permuth from Moffitt Cancer Center discussed approaches to analyzing radiologic images and genomics (“radiogenomics”) in the context of pancreatic cancer (PaCa) and its precursors. Intraductal papillary mucinous neoplasms (IPMNs) are the most common cystic precursors to PaCa, the only solid malignancy with a 5-year relative survival rate < 10%.14,15 To date, existing imaging modalities and molecular markers cannot reliably distinguish low/moderate grade (benign) IPMNs that merit surveillance from high-grade/invasive (malignant) IPMNs that warrant surgical resection, posing a great clinical challenge.14 By leveraging expertise in population, clinical, and data science and informatics and multi-institutional infrastructure known as the Florida Pancreas Collaborative,16 the team seeks to discover a combined quantitative imaging and biomarker approach that is noninvasive and has added value in predicting IPMN pathology beyond that provided by standard radiologic and clinical characteristics. The team will build on their preliminary studies to evaluate underexplored categories of quantitative “radiomic” features extracted from preoperative computed tomography scans,17,18 along with a circulating plasma microRNA blood test that they have developed,18 and generate prototype clinical decision-making models (nomograms) to predict malignant IPMN pathology. They will also evaluate the relationship between radiomic features and biologic processes characterized by microRNA and/or mucin expression that underlie IPMN tumor development and/or progression to glean diagnostic and prognostic information. This line of translational research has potential to foster clinically actionable information that may be used to rapidly and cost effectively personalize care for individuals with IPMNs and reduce PaCa burden.
Dr Alex Bui from the University of California, Los Angeles discussed challenges and opportunities for data integration against the backdrop of the increasing volume and variety of biomedical and clinical big data, such as -omics, imaging, EHRs, and mHealth. The potential of using these data with new computational methods like ML to advance our understanding of cancer and its treatment is driving significant excitement. But harnessing this information requires facing two challenges: combining such data to support comprehensive analyses, and ensuring that the data used are appropriate to inform real-world applications. First, often each type of data is analyzed in isolation, thereby missing an opportunity to view the disease as a phenomenon seen across spatial scales (eg, from the molecular to the person) and time (eg, from screening and diagnosis through to treatment and survivorship). For example, crossover areas like radiogenomics frequently consider features separately (ie, radiomics, genomics) before combining them, when joint analyses may proffer different results. Increased computational power and novel algorithms for automated knowledge graph construction can help make these connections over large observational datasets. Second, integration is itself insufficient, as the nature and quality of the data used in downstream analyses must be considered. Indeed, the (clinical) applicability of a model developed using ML is contingent on its ability to work with real-world data, given inherent noise, missing data, and bias. Moreover, as the environment in which an ML model is deployed changes (eg, new technologies or therapies), it may become outdated. Infrastructure for continual data curation and performance re-evaluation of data-driven analyses are thus necessary, and how these models evolve can guide future insights about cancer outcomes and research directions.
Day 2 of the Fall 2019 meeting concluded with a panel discussion on data integration and availability through CCSG cores. Dr Jeanine Genkinger from Columbia University discussed that different models exist for cancer center shared resources that are designed to develop and support studies to address and promote clinically relevant and innovative research. Starting in 2014, the DataBase Shared Resource (DBSR) at the Columbia University Herbert Irving Comprehensive Cancer Center was tasked to develop, integrate, and maintain a centralized, cost-effective, and well-characterized research database. As such, the DBSR has: (1) integrated all prior adult solid tumor registries into one standardized protocol, (2) created a retrospective cohort of patients through linkage of data from EHRs across multiple platforms and data warehouses (ie, New York-Presbyterian Hospital [NYPH] Tumor Registry) with existing residual tissue samples, and (3) developed and maintained a prospective cohort implemented through a standardized protocol with universal consent of individuals with cancer or at risk for cancer, collecting biospecimens and epidemiologic data, and linking to data from the EHR/NYPH Tumor Registry and residual tissue samples. To date, DBSR integrated 24 clinical registries into one overarching research database. Because of highly qualified and bilingual recruiters, DBSR has met its accrual targets and enrolled > 4,700 individuals, from whom DBSR has collected > 2,400 biospecimens and > 2,600 questionnaires. To foster research, the DBSR has approved and fulfilled > 120 data and biospecimen requests and supported > 10 ancillary studies through efficient enrollment and data extraction. Through this, the DBSR has supported several federally funded grants and peer-reviewed publications.
Dr Wenjin (Jim) Zheng from the University of Texas Health Science Center at Houston discussed his vision for proactive, data-driven cancer informatics support, whereby data analysts in the informatics core can take a lead role with data analysis and mining to make discoveries or generate hypotheses and work together with experimentalists and clinician to drive cancer research. A successful proactive informatics core needs several critical components. First, informaticians should have broad knowledge of cancer research so that they can initiate scientifically sound projects or work with experimentalists and clinicians to do so.20 Second, sufficient financial support should be in place so informaticians can afford the high-risk and time-consuming nature of exploratory projects. Third, there should be open-minded experimental or clinical collaborators who are willing to embrace data-driven research. Last but not least, there should be a robust computing infrastructure that allows quick, prototypical, and exploratory analysis of a large amount of data to generate sufficient results to initiate new projects or to make significant contributions to existing projects. Satisfying these conditions can ensure a cancer informatics core support that is highly productive and impactful.
Dr Travis Gerke from Moffitt Cancer Center discussed the Collaborative Data Services Core (CDSC), which provides access to the Center’s robust data assets. Discrete data on > 570,000 patients are available through an enterprise-wide data warehouse, which spans clinical, administrative, patient-reported, biospecimen, and molecular domains. CDSC provides three primary services: (1) study design consultations, which feature cohort identification and assess feasibility; (2) provisioning of patient data from source systems, often complemented by a manual medical record abstraction service for information not available in discrete format; and (3) individual or small-group training on self-service querying tools and best practices. A high volume of service requests (400-500 annually) are fulfilled on an hourly chargeback system by a team of 8 data scientists, 6 database abstractors, and 6 operational team members.
Day 3
Day 3 featured a Cancer Center Directors Keynote Lecture from Dr Cornelia Ulrich, Executive Director of the Comprehensive Cancer Center at Huntsman Cancer Institute (HCI). HCI is home to a unique multilevel data environment that enables its researchers to leverage intersections between informatics, data science, and population science. Its Research Informatics shared resource offers robust software solutions that allow our researchers to collect and mine data from local, state, and national sources, such as the Utah Cancer Registry, Utah Population Database, tumor registries, Oncology Research Information Exchange Network (ORIEN), ColoCare, and Flatiron. It uses state-of-the-art capabilities, including natural language processing.21 HCI researchers are implementing novel strategies for identifying and managing individuals with hereditary cancer through the EHR to facilitate translation of genetic discoveries to the clinic and population. The Utah Population Database (UPDB) is a unique and vital tool for conducting population-based studies, gene discovery, and health services research. Data in the UPDB comprise medical, public, health, and demographic data for 11 million individuals across the span of decades. The UPDB has been instrumental in the discovery of several key genetic discoveries in cancer, such as BRCA 122 and CDKN2A.23 In addition, UPDB has been used for research in cancer survivorship and disease risk quantification and prevention.24,25 The UPDB is also linked to the Utah Cancer Registry, which is part of the NCI’s SEER program. On a national scale, HCI has partnered with the ORIEN Network and Flatiron. HCI has used its partnership with Flatiron to link with data on patient-reported outcomes, assessed in their clinics at regular intervals, and have used these patient-reported outcomes as clinical predictors for advanced cancers. HCI leverages a unique multilevel data environment and capabilities in state-of-the-art data science to advance cancer research and translation of discoveries for maximum impact to the clinic and population.
TOPICS ON THE HORIZON FOR CANCER INFORMATICS
The focus on intersections between population science, data science, and informatics highlighted several topics requiring further discussion and dialogue at future CI4CC meetings, including harmonization of data across existing common data models, linkage of data across initiatives in a way that preserves de-identification yet facilitates the assessment of overlap between data sets, and challenges integrating information back into the EHR to enhance clinical decisions. More broadly, cancer informatics encompasses a wide array of topics around data capture, data organization, data integration, data visualization, and data interpretation. Corresponding topics on the horizon for cancer informatics that the CI4CC organization will be highlighting at future meetings and through their initiatives are: AI and ML in oncology, involvement with the ASCO efforts around CancerLinQ and Minimal Common Oncology Data Elements (mCODE), precision medicine and learning health platforms, cancer data sharing, data interoperability (with a particular focus on the link between -omic data and the EMR), digital health innovation, and NCI CCSG–focused shared informatics resources.
ACKNOWLEDGMENT
We thank all participants in Cancer Informatics for Cancer Centers (CI4CC). We also thank Tim and Sarah Rose for all of their CI4CC organizational help with meeting plans and execution. The NCI-Connect for Cancer Prevention cohort thanks Dr Jonas Almeida, Chief Data Scientist in the Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute, for his contributions to the CI4CC meeting report as a lead of the IT and data systems architecture for Connect, as well as the contributions of the Connect study team at DCEG and participating health care centers (https://dceg.cancer.gov/research/who-we-study/cohorts/connect).
AUTHOR CONTRIBUTIONS
Conception and design: Jill S. Barnholtz-Sloan, Dana E. Rollison, Alexander D. Borowsky, Jack DiGiovanna, Montserrat Garcia-Closas, Jeanine M. Genkinger, Travis Gerke, Marta Induni, Lisa Mirel, Jennifer B. Permuth, Joel Saltz, Elizabeth A. Shenkman, Cornelia M. Ulrich, W. Jim Zheng, Sorena Nadaf, Warren A. Kibbe
Financial support: Alexander D. Borowsky, Sorena Nadaf
Administrative support: Jill S. Barnholtz-Sloan, Sorena Nadaf
Provision of study material or patients: Alexander D. Borowsky, Marta Induni, Elizabeth A. Shenkman, Sorena Nadaf
Collection and assembly of data: Jill S. Barnholtz-Sloan, Dana E. Rollison, Alexander D. Borowsky, Alex Bui, James V. Lacey Jr, Elizabeth A. Shenkman
Data analysis and interpretation: Dana E. Rollison, Amrita Basu, James V. Lacey Jr, Elizabeth A. Shenkman
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Dana E. Rollison
Travel, Accommodations, Expenses: Caserta Analytics
Amrita Basu
Employment: Leidos Health
Travel, Accommodations, Expenses: Leidos Health
Alexander D. Borowsky
Research Funding: Danaher (Inst)
Travel, Accommodations, Expenses: Agendia
Open Payments Link: https://openpaymentsdata.cms.gov/physician/1310389/summary
Jack DiGiovanna
Employment: Biogen (I)
Stock and Other Ownership Interests: Biogen (I)
Jennifer B. Permuth
Expert Testimony: Shook, Hardy, and Bacon Legal Group
No other potential conflicts of interest were reported.
REFERENCES
- 1. Hu J, Juan W, Sahyoun NR: Intake and biomarkers of folate and risk of cancer morbidity in older adults, NHANES 1999-2002 with Medicare Linkage. PloS One 11:e0148697, 2016. [DOI] [PMC free article] [PubMed]
- 2.Siahpush M, Farazi PA, Wang H, et al. Muscle-strengthening physical activity is associated with cancer mortality: Results from the 1998-2011 National Health Interview Surveys, National Death Index record linkage. Cancer Causes Control. 2019;30:663–670. doi: 10.1007/s10552-019-01169-z. [DOI] [PubMed] [Google Scholar]
- 3. Stewart M, Norden AD, Dreyer N, et al: An exploratory analysis of real-world end points for assessing outcomes among immunotherapy-treated patients with advanced non-small-cell lung cancer. JCO Clin Cancer Inform 10.1200/CCI.18.00155. [DOI] [PMC free article] [PubMed]
- 4. Madhavan G, Phelps CE, Rouse WB, et al: Vision for a systems architecture to integrate and transform population health. Proc Natl Acad Sci USA 115:12595-12602, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shenkman E, Hurt M, Hogan W, et al: OneFlorida Clinical Research Consortium: Linking a clinical and translational science institute with a community-based distributive medical education model. Acad Med 93:451-455, 2018. [DOI] [PMC free article] [PubMed]
- 6. doi: 10.1158/0008-5472.CAN-17-0387. Lau JW, Lehnert E, Sethi A, et al: The Cancer Genomics Cloud: Collaborative, reproducible, and democratized-a new paradigm in large-scale computational research. Cancer Res 77:e3-e6, 2017 [Erratum: Cancer Res 78:5179, 2018] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al: The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3:160018, 2016. [DOI] [PMC free article] [PubMed]
- 8.All of Us Research Program Investigators. Denny JC, Rutter JL, et al. The “All of Us” research program. N Engl J Med. 2019;381:668–676. doi: 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khoury MJ, Evans JP: A public health perspective on a national precision medicine cohort: Balancing long-term knowledge generation with early health benefit. JAMA 313:2117-2118, 2015. [DOI] [PMC free article] [PubMed]
- 10. Ohno-Machado L, Agha Z, Bell DS, et al: pSCANNER: Patient-centered Scalable National Network for Effectiveness Research. J Am Med Inform Assoc 21:621-626, 2014. [DOI] [PMC free article] [PubMed]
- 11.Gupta R, Kurc T, Sharma A, et al. The emergence of pathomics. Curr Pathobiol Rep. 2019;7:73–84. [Google Scholar]
- 12. Saltz J, Sharma A, Iyer G, et al: A containerized software system for generation, management, and exploration of features from whole slide tissue images. Cancer Res 77:e79-e82, 2017. [DOI] [PMC free article] [PubMed]
- 13. Saltz J, Gupta R, Hou L, et al: Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep 23:181-193.e7, 2018. [DOI] [PMC free article] [PubMed]
- 14. Matthaei H, Schulick RD, Hruban RH, et al: Cystic precursors to invasive pancreatic cancer. Nat Rev Gastroenterol Hepatol 8:141-150, 2011. [DOI] [PMC free article] [PubMed]
- 15. American Cancer Society: Cancer Facts and Figures. Atlanta, GA, American Cancer Society, 2019. [Google Scholar]
- 16.Permuth JB, Trevino J, Merchant N, et al. Partnering to advance early detection and prevention efforts for pancreatic cancer: The Florida Pancreas Collaborative. Future Oncol. 2016;12:997–1000. doi: 10.2217/fon-2016-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Permuth-Wey J, Chen DT, Fulp WJ, et al: Plasma microRNAs as novel biomarkers for patients with intraductal papillary mucinous neoplasms of the pancreas. Cancer Prev Res (Phila) 8:826-834, 2015. [DOI] [PMC free article] [PubMed]
- 18. Permuth JB, Choi J, Balarunathan Y, et al: Combining radiomic features with a miRNA classifier may improve prediction of malignant pathology for pancreatic intraductal papillary mucinous neoplasms. Oncotarget 7:85785-85797, 2016. [DOI] [PMC free article] [PubMed]
- 19.Zhu L, Zheng WJ. Informatics, data science, and artificial intelligence. JAMA. 2018;320:1103–1104. doi: 10.1001/jama.2018.8211. [DOI] [PubMed] [Google Scholar]
- 20.Chang J. Core services: Reward bioinformaticians. Nature. 2015;520:151–152. doi: 10.1038/520151a. [DOI] [PubMed] [Google Scholar]
- 21. Mowery DL, Kawamoto K, Bradshaw R, et al: Determining onset for familial breast and colorectal cancer from family history comments in the electronic health record. AMIA Jt Summits Transl Sci Proc 2019:173-181, 2019. [PMC free article] [PubMed]
- 22.Futreal PA, Liu Q, Shattuck-Eidens D, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 23.Kamb A, Gruis NA, Weaver-Feldhaus J, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 24. Soisson S, Ganz PA, Gaffney D, et al: Long-term cardiovascular outcomes among endometrial cancer survivors in a large, population-based cohort study. J Natl Cancer Inst 110:1342-1351, 2018. [DOI] [PMC free article] [PubMed]
- 25.Samadder NJ, Curtin K, Tuohy TM, et al. Characteristics of missed or interval colorectal cancer and patient survival: A population-based study. Gastroenterology. 2014;146:950–960. doi: 10.1053/j.gastro.2014.01.013. [DOI] [PubMed] [Google Scholar]