Abstract
Background
Metastatic prostate cancer is a clonally heterogeneous disease state characterized by progressive somatic perturbations. The aim of this study was to identify cell free DNA- (cfDNA-) based alterations and their associations with outcomes in progressive metastatic prostate cancer.
Methods
In this longitudinal prospective cohort study plasma cfDNA/circulating tumor DNA (ctDNA) was analyzed before, during, and after androgen deprivation therapy (ADT) in 4 independent patient groups ranging from untreated metastatic hormone sensitive prostate cancer (mHSPC) to metastatic castrate resistant prostate cancer (mCRPC). Next generation sequencing was performed on ctDNA and germline DNA to characterize alterations and associations with clinical outcomes were determined for each group.
Findings
cfDNA yields were different in progressive mHSPC and mCRPC states (P < .001). In mHSPC, a higher than median ctDNA fraction was predictive of shorter time to ADT failure (HR, 2.29 [95% CI, 1.13–4.65]; Log-Rank P = .02). cfDNA, ctDNA taken with volume of metastatic disease in mHSPC and with alkaline phosphatase levels prognosticated survival better than clinical factors alone in mHSPC and mCRPC states (Log Rank P = 0.03). ctDNA-based AR, APC mutations were increased in mCRPC compared to mHSPC (P < ·05).TP53 mutations, RB1 loss, and AR gene amplifications correlated with poorer survival in mCRPC. Mutations in multiple DNA repair genes (ATM, BRCA1, BRCA2, CHEK2) were associated with time to ADT treatment failure and survival in mHSPC.
Interpretation
ctDNA fraction can further refine clinical prognostic factors in metastatic prostate cancer. Somatic ctDNA alterations have potential prognostic, predictive, and therapeutic implications in metastatic prostate cancer management.
Funding
Several funding sources have supported this study. A full list is provided in the Acknowledgments. No funding was received from Predicine, Inc. during the conduct of the study.
Keywords: Circulating tumor DNA, Metastatic prostate cancer, Genomic alterations
Research in context.
Evidence before this study
We consulted literature published after 2012, as before this time, genomic sequencing of tissue metastases or liquid biopsies were not reported for prostate cancer. Somatic mutational variations have been extensively reported in metastatic castrate resistant prostate cancer (mCRPC) since 2015 in tissue and in liquid biopsies. However, studies have yet to elucidate comprehensively the evolution in the continuum of the genomic landscape from metastatic hormone sensitive prostate cancer (mHSPC) to clinical and radiographically progressive mCRPC states, and studies have not explored its impact on clinical outcomes.
Added value of this study
This study provides an atlas of the somatic changes observed in each of the progressive metastatic prostate cancer states, starting with untreated mHSPC to treated mHSPC and mCRPC states. We were able to uncover the underlying clonal evolution in the continuum of these progressive metastatic states, which has not been previously published. Because clinical follow-up was obtained, we were also able to comprehensively profile the clinical relevance of the structural DNA-based variations. Until now, the selection of patients for treatments additional to androgen deprivation therapy has been guided using clinical prognostic factors (Gleason Score, the presence of visceral metastases, and/or the number of skeletal metastases).
Implications of all the available evidence
Our results provide insights based on the circulatory and measurable tumor genome and the presence or absence of key genomic perturbations. We observed that circulating tumor DNA yields is a key prognostic factor in each state of metastatic progression; this further refines currently recognized clinical prognostic factors. This finding may potentially assist in selecting patients for the intensification of upfront systemic treatment. Similarly, we identified specific genes in which alterations were associated with clinical outcomes, which may also have a potential for therapeutic impact.
Alt-text: Unlabelled box
1. Introduction
Prostate cancer is a significant public health concern. According to the GLOBOCAN 2018 database, there are 1·26 million cases of prostate cancer reported each year globally and over 375 000 associated deaths [1,2]. Recent advancements in the treatment of metastatic hormone-sensitive (mHSPC) and metastatic castrate-resistant (mCRPC) prostate cancer states include androgen receptor pathway inhibitors, cytotoxic chemotherapy, radioisotope treatment with radium-223, ex vivo purged autologous cellular immunotherapy with sipuleucel-T [3], and the poly (ADP-ribose) polymerase gamma inhibitor olaparib, which has received FDA breakthrough designation [4]. Several of these drug options have demonstrated clinical benefit when combined with androgen deprivation therapy (ADT) to treat patients with mHSPC [5], [6], [7], [8], [9], non-mCRPC, and high-risk locally advanced nonmetastatic disease [8,10]. However, drug choices are currently not selected on the basis of individual genomic variant profiling in any state of progressive disease.
The genomic landscape of solid metastatic tumors in the castrate-resistant state harbor a high degree of stage-specific somatic alterations and tumor heterogeneity compared to localized prostate cancer [11], [12], [13]. Stage-specific somatic alterations in individual patient tumors may aid in developing predictive, therapeutic, and prognostic molecular biomarkers, allowing for targeted drug–pairing choices with greater precision and therapeutic gain. As the genomic landscapes of mHSPC leading to mCRPC have not been well described, challenges remain for drug-targeted mutational profiling in patients with advanced prostate cancer. Our group previously reported the mutational landscape of solid metastases in patients with treatment-naive mCRPC before abiraterone acetate/prednisone drug therapy [11]. We successfully sequenced tumor nucleic acids from solid metastases, including bone, but obtaining a high yield and purity of nucleic acids from skeletal metastases before and after treatments was particularly challenging [14]. Additionally, the invasiveness of performing metastatic solid tissue biopsies for sequencing is not feasible in routine clinical practice. It is also impractical to perform metastatic solid tissue serial biopsies that capture an evolving genomic landscape to monitor minimal residual disease or identify pharmacodynamic markers before and after drug initiation under treatment selection pressure.
We report the results of a prospective longitudinal cohort study, in which the individual genomic variants of patients with 4 different metastatic prostate cancer states, starting with treatment-naive mHSPC and leading to mCRPC, were profiled using plasma circulating tumor DNA (ctDNA) sequencing. We aimed to characterize tumor-associated copy number alterations (CNAs), single-nucleotide variations (SNVs), and commonly observed rearrangements in independent cohorts of patients with metastatic prostate cancer before, during, and at the time of ADT failure, defined both on the basis of biochemical relapse and clinical/radiographic progression criteria. We also used a second independent castrate-resistant prostate cancer cohort to validate ctDNA-based genomic variants. In doing so, we have comprehensively profiled the genomic landscape across the spectrum of metastatic prostate cancer and have deciphered key clinical and molecular insights into this heterogeneous disease.
2. Materials and methods
2.1. Patient enrollment and sample collection
Patients with metastatic prostate cancer were prospectively enrolled between September 2009 and March 2014 in a single tertiary-level cancer center (Mayo Clinic, Rochester, MN) after obtaining written informed consent for serial blood collections, as previously described [15], [16], [17], [18]. A second cohort of clinical mCRPC patients was enrolled from Monash University (Australia). Methods for circulating free DNA (cfDNA) and germline DNA (gDNA) extraction are detailed in Supplementary Methods.
2.2. Library preparation, capture, and sequencing
Five to thirty nanograms of extracted cfDNA and 40 ng per unique patient sample of fragmented gDNA were subjected to library construction, including end-repair dA-tailing and adapter ligation. Ligated library fragments with appropriate adapters were amplified via PCR. The amplified DNA libraries were then checked using Bioanalyzer 2100, and samples with yields > 700 ng were proceeded to hybrid capture. Details of library capture, hybridization, and the 120-gene PredicineLDT panel are provided in Supplementary Methods.
2.3. Analyses of next generation sequencing data from cfDNA
Data were analyzed using the Predicine in-house–developed analysis pipeline, which starts from the raw sequencing database call files (·BCL) and outputs the final mutation calls. Briefly, the pipeline first does an adapter trim, barcode checking, and correction. Cleaned paired FASTQ files were aligned to human reference genome build hg19 using Burrows-Wheeler Aligner [19]. Consensus bam files are then derived by merging paired-end reads originated from the same molecules (based on mapping location and unique molecular identifiers) as single strand fragments. Single strand fragments from the same double strand DNA molecules were further merged as double stranded. By using error suppression method described in [20], both sequencing and PCR errors were mostly corrected during this process. Candidate variants consisting of point mutations and small insertions and deletions were identified by comparing with local variant background [20] (defined based on plasma samples from health donors and historical data). Variants were further filtered by log-odds (LOD) threshold [21], base quality and mapping quality thresholds, repeat regions and other quality metrics. Detailed variant filtering and analysis of sequencing data from gDNA are provided under “Supplementary Methods”.
2.4. CNV estimation methods
We first calculated the on-target unique fragment coverage on the basis of consensus sequence from BAM files; the fragment was also corrected for GC bias. The GC-adjusted unique fragment was then compared against corresponding coverage from a group of normal reference samples to estimate the significance of the copy number variant. Amplification or deletion of a gene copy number with an absolute z-score > 2·58 (the inverse of the cumulative distribution function of a Gaussian distribution at 99·5% confidence level) were deemed as true events. Methods used in detection of DNA rearrangements are elaborated in Supplementary Methods.
2.5. ctDNA fraction calculation
ctDNA fraction was estimated on the basis of the allele fractions of autosomal somatic mutations, as described previously [22]. Briefly, the mutant allele fraction (MAF) and the ctDNA fraction were related, as MAF=(ctDNA*1)/([1-ctDNA]*2+ctDNA*1); therefore, ctDNA=2/([1/MAF]+1). Somatic mutations in genes with a detectable copy number amplification were omitted from ctDNA fraction estimates. ctDNA fractions were then estimated as the product of ctDNA fraction and total cfDNA yield in the corresponding sample following a normalization by plasma volume.
2.6. Plasma tumor mutational burden calculation
Plasma tumor mutational burden (pTMB) was defined as the number of somatic coding SNVs, including synonymous and nonsynonymous variants, detected in the plasma samples after removing germline single-nucleotide polymorphisms. The full 240 KB coding regions covered by the PredicineLDT panel (Predicine Inc., Hayward, CA, USA) were used to calculate pTMB. Only samples with the maximum somatic allelic frequency (MSAF) ≥0.5% were analyzed for pTMB. The 0.5% of MSAF was selected according to the assay performance metrics (shown in the Supplementary Figure S1) and previous publication [1].
2.7. Statistical analysis methods
Wilcoxon signed rank test was used to evaluate statistical differences of cfDNA yield/ctDNA fraction between patients with different states of progressive mHSPC and mCRPC due to skewed distribution of DNA yields. Paired Wilcoxon tests were performed to examine differences of cfDNA/ctDNA and pTMB between matched untreated mHSPC patients and 3-month post-ADT mHSPC patients and between untreated mHSPC and mCRPC patients. Fisher Exact tests were used to calculate Odds Ratios and estimate the significance of differences of the genomic alterations among different disease states. Kruskal Wallis tests were performed to compare the statistical differences of ctDNA, cfDNA, TMB among patients in the different states.
Survival analyses was used to evaluate associations of feature variables with ADT failure and overall survival (OS). ADT failure was defined as the time from a patient initiating continuous ADT therapy until biochemical relapse with 2 successive serum PSA readings above the nadir, with the second reading taken at least 4 weeks apart. A patient was right censored if the patient did not have PSA relapse at the time of the last follow-up or at the cutoff date of the analysis. Similarly, overall survival was calculated as the difference between the date of study enrollment to the date of death or to the last follow-up for alive patients (right censored) at the time of the cutoff date of the analysis. For a given feature variable, Cox proportional hazards regression and the Log-rank test were used in univariate and multivariate analyses to assess the significance of its association with ADT failure or OS. Gleason score, ctDNA fraction, and metastatic volume (for time to ADT failure)/alkaline phosphatase levels (for OS) were included in the multivariate analysis as co-variables. To dichotomize the patient cohorts, the median cutoff was used for ctDNA fraction and alkaline phosphatase levels. A high Gleason score patient was defined as a patient with Gleason scores greater than 7, and a low Gleason score was defined as scores less than or equal to 7. The definition of metastatic volume was based on the CHAARTED trial [6]. Scaled Schoenfeld residuals and deviance residuals with time were examined to ensure the validity of the Cox regression assumptions. All tests of statistical significance were two-tailed, with a significance set at P ≤ ·05. To account for multiple hypothesis testing, adjusted P values by the Benjamini and Hochberg procedure were also reported [23].
All statistical analyses were performed using R version 3·5·3 [24], and all tests of statistical significance were two-tailed with a significance set at P ≤ ·05.
2.8. Role of the funding source
This study was based on a non-commercial collaboration between academic institutions. No funding was received from Predicine, Inc. during the conduct of the study. The discussion to submit the paper for publication was made by MK, AA, JU, and SJ; the final decision was made by MK.
3. Results
Between September 2009 and January 2013, 303 metastatic prostate cancer patients were prospectively enrolled at the Mayo Clinic and followed until death. A second biobank of metastatic prostate cancer patients at Monash University recruited prospective patients between 2016 and 2018. Metastatic prostate cancer patients in both biobanks with adequate plasma volume from whole blood for cfDNA extraction were included, and for the larger Mayo Clinic cohort, which had adequate longitudinal follow-up, the cutoff date for all analyses was September 1, 2018. The workflow for the final 250 (82%) patients with analyzable cfDNA sequencing data with clinical outcomes is shown in Supplementary Figure S1. Individual deidentified patient-level results can be found in the Supplementary Raw Data.
The study cohort was divided into 4 independent groups. The first group, “Untreated mHSPC,” included mHSPC patients whose first sample collection was performed before ADT initiation. Several, but not all, patients in this group had a second serial blood sample collection after 3 months of ADT; these serially collected patients were labeled the “3-month mHSPC” subgroup. The second group, “mHSPC on ADT,” included patients who were enrolled at the time of the first sample collection while undergoing continuous ADT. These patients had a biochemical response confirmed by stable to lower serum prostate-specific antigen (PSA) levels compared to pre-ADT initiation levels. The third independent group, “Biochemical progressive mCRPC,” included patients with biochemical progression on ADT (defined as serially rising PSA levels above a previous PSA nadir) and castrate testosterone levels at the time of first blood sample collection and before a secondary hormonal maneuver or any additional new drug was administered for progression. No evidence of radiographic progression was observed in these patients. The fourth group, “Clinical mCRPC,” consisted of mCRPC patients with both biochemical failure (rising serial PSA levels above a previous nadir) and appearance of new radiographic metastases, with the sample collection performed before any new drug addition for progressive disease. The demographic data of all patients enrolled in each group are detailed in Table 1 and Supplementary Table S1.
Table 1.
Clinical characteristics of patients in different states of metastatic prostate cancer progression.
| Untreated mHSPC group (n = 99) |
mHSPC on ADT group | Biochemical progressive mCRPC group | Clinical mCPRC group | ||
|---|---|---|---|---|---|
| Entire group, n = 97 | 3-month mHSPC subgroup, n = 29 | ||||
| Total patients, no. | 97 | 37 | 40 | 81 | 85 |
| Patients with analyzable NGS data (N) | 73 | 29 | 33 | 75 | 69 |
| Age in years at the time specimen collection, median (range) | 66 (45–90) | 70 (45–90) | 71 (50–89) | 70 (52–87) | 72 (50–92) |
| PSA at time of sample collection, median ng/ml (IQR) | |||||
| First sample | 6.35 (1.0–28.3) | 11.20 (4.0–24.5) | 0.53 (0.0–5.6) | 2.8 (0.6–8.5) | 16.7 (4.5–60.5) |
| Second sample | N/A | 0.94 (0–30.6) | N/A | N/A | N/A |
| ALP at the time of sample collection, median (range) | 80 (39–485) | 77 (46–255) | 82 (42–670) | 78 (34–392) | 99.5 (44–605) |
| Metastatic Volume, no. | |||||
| Low | 47 | 20 | NA | NA | NA |
| High | 22 | 7 | NA | NA | NA |
| Unknown | 4 | 2 | NA | NA | NA |
| Median time from ADT initiation for mHSPC stage to biochemically progress to CRPC stage, mo (range) | 14.98 (0–78.5) | 17.9 (7.33–57.8) | 15.93 (1.2–130.43) | 15.70 (0–211.67) | 16.77 (3.2–184.8) |
| Median follow up time from date of HSPC specimen collection to last follow up, mo (range) | 94.53 (0.03–107.3) | 97.07 (25.37–107.3) | 85.17 (0–104.73) | N/A | N/A |
| Median time from ADT initiation to failure, mo (range) | 14.99 (0–78.5) | 16.77 (7.33–57.8) | 15.93 (1.2–130.43) | ||
| Median time for mHSPC patients from initiation of ADT to death/last follow up, mo (range) | 71.2 (1.13–115.1) | 75.8 (25.37–106.93) | 63 (0.33–141.33) | N/A | N/A |
| Median follow-up time from date of mCRPC specimen collection to last follow up, mo (range) | N/A | N/A | N/A | 74.7 (1.53–102.77) | 94.67 (1.6–94.67) |
| Median time to death/last follow-up for mCRPC patients, mo (range) | N/A | N/A | N/A | 43.33 (0–102.77) | 25.93 (1.6–94.67) |
| Patients progressed on ADT, no. | 34 | 16 | 23 | N/A | N/A |
| Patients dead upon follow-up, no. | 38 | 15 | 20 | 58 | 62 |
All demographic data mentioned above only relate to NGS-analyzable patients. Disease progression in subjects diagnosed and treated for localized stage disease was defined by the treating physician on the basis of either serial PSA rise on 2 separate occasions after achieving a posttreatment nadir PSA value or appearance of new radiological disease or with the initiation of a new cancer-specific intervention, whichever came first. ADT initiation for the hormone-sensitive stage was defined by the treating physician and included either serial PSA rise on 2 separate occasions after achieving a nadir PSA value or appearance of new radiological disease or with the initiation of a new cancer specific intervention, whichever came first. Biochemical progression to mCRPC stage during ADT for mHSPC was defined by the treating physician and included either serial PSA rise on 2 separate occasions after achieving a nadir PSA value during ongoing continuous ADT or the initiation of a secondary hormonal maneuver whichever came first. Clinical progression to CRPC stage during ADT for the hormone-sensitive stage was defined as the first appearance of new radiological disease or the initiation of a new cancer-specific intervention, whichever came first during ongoing continuous ADT.
Abbreviations: ADT, androgen deprivation therapy; ALP, alkaline phosphatase; IQR, interquartile range (25–75); mCRPC, metastatic castrate-resistant prostate cancer; mHSPC, metastatic hormone-sensitive prostate cancer; mo, months; N/A, not applicable; NGS, next generation sequencing; PSA, prostate-specific antigen.
3.1. cfDNA, ctDNA, pTMB distributions in progressive metastatic prostate cancer states
We calculated the cfDNA yield and ctDNA fraction and the number of variants in the coding regions of the genes covered by the panel (denoted as pTMB) for all patients in the 4 groups and compared the overall group and intergroup-wise distributions for differences (Table 2). The per group distributions and comparisons between them are also shown in Supplementary Figure S2 (cfDNA), Fig. 1A (ctDNA), and Fig. 1B (pTMB). cfDNA yield/ctDNA fraction and pTMB levels were significantly greater in the mCRPC groups than in the mHSPC groups (P < ·001, Kruskal–Wallis test). There were no differences in cfDNA yield/ctDNA fraction or in the pTMB levels between untreated mHSPC and mHSPC on ADT groups.
Table 2.
cfDNA, ctDNA, and mutational load tumor profile across metastatic prostate cancer states.
| Untreated mHSPC group n = 73 | Serial 3-month mHSPC group (n = 29/73) | mHSPC on ADT group (n = 33) | Biochemical Progressive CRPC group (n = 75) | Clinical mCRPC group (n = 69) | |
|---|---|---|---|---|---|
| Median cfDNA, ng/ml (range) | 4.32 (1.58–212.88) | 4.2 (1.78–10.67) | 3.7 (1.6–103.56) | 12.26 (3.87–84.71) | 17.97 (5.54–678.86) |
| Group comparison of cfDNA yields, type (P value) | Untreated vs 3-month ADT treated (P = .22) (N = 29) |
3-month post ADT vs continuous ADT (P = .83) | Untreated mHSPC vs biochemical CRPC relapse (P < .001) | Untreated mHSPC vs clinical CRPC (P < .001) | |
| Median ctDNA fraction (range) | 0.01(0.006–0.63) | 0.009 (0.006–0.23) | 0.01(0.006–0.92) | 0.01(0.006–0.76) | 0.04(0.006–0.75) |
| Group comparison of ctDNA fraction, type (P value) | Untreated vs 3-month ADT treated (P = .02) (N = 29) |
3-month post ADT vs continuous ADT (P = .79) | Untreated mHSPC vs biochemical CRPC relapse (P < .001) | Untreated mHSPC vs clinical CRPC (P < .001) | |
| pTMB variantsa, no. | 3 (1–28) | 4 (1–12) | 3 (1–13) | 5 (1–41) | 5 (1–28) |
| pTMB-based group comparisons, type (P value) | Untreated vs 3-month ADT treated (P = .18) (N = 29) |
3-month post ADT vs continuous ADT (P = .47) | Untreated mHSPC vs biochemical CRPC relapse (P < .001) | Untreated mHSPC vs clinical CRPC (P < .001) | |
| pTMB-based group comparisons for mHSPC state vs. mCRPC, P value | NA | NA | NA | P < .001 | |
Number of variants in the coding region covered by the panel.
Abbreviations: ADT, androgen deprivation therapy; cfDNA, circulating free DNA; ctDNA, circulating tumor DNA; CRPC, castrate-resistant prostate cancer; mCRPC, metastatic castrate-resistant prostate cancer; mHSPC, metastatic hormone-sensitive prostate cancer; NA, not applicable; pTMB, plasma tumor mutational burden.
Fig. 1.
A. Distribution of ctDNA fractions across metastatic groups with significant differences in yield observed across the 4 independent groups of patients (P < . 001, Kruskal–Wallis test). B. Distribution of plasma-based tumor mutation burden across groups. C. Distribution of cfDNA yields based on metastatic volume in the untreated hormone-sensitive group and serum alkaline phosphatase (ALP) in mCRPC states. Samples are dichotomized into low and high groups based on the median value of cfDNA yields (median: 9.6 ng/mL) and ALP levels (Median: 83 IU/L), respectively. Percentage of samples with different ctDNA fractions are shown in different colors for each description. D. Combined analysis of ctDNA fraction and metastatic volume for the prediction of ADT failure in mHSPC patients. High and low ctDNA fractions are defined based on the third quartile of ctDNA fraction across the samples. E. Overall survival in the mHSPC group based on the combined analysis of volume of metastatic disease with ctDNA fraction in mHSPC patients. F. Combined analysis of ctDNA fraction and serum ALP levels of overall survival in mCRPC patients.
A median cfDNA yield cutoff value of 9·6 ng/mL was used for all study samples based on which the ctDNA fraction distribution was determined (top panel of Fig. 1C). The ECOG 3805 CHAARTED trial's [6] definition of high- and low- volume metastatic disease was used to stratify high vs low metastatic volume in the untreated metastatic hormone-sensitive group. Fig. 1C (middle panel) shows the distribution of ctDNA fractions in high- and low- volume metastatic disease. The lower panel in Fig. 1C shows ctDNA fraction distributions above and below the median serum alkaline phosphatase (ALP) levels (median, 83 IU/L), a known prognostic factor for survival in castration-resistant state [25].
The impact of metastatic volume on cfDNA yield/ctDNA fraction and pTMB is shown in Supplementary Figures S3 (A-C) and detailed in the Supplementary Results. Change in nucleic acid yields under the effect of ADT after 3 months of treatment in 29 paired mHSPC samples is detailed in Table 2, Supplementary Figures S4, and the Supplementary Results with an observed decrease in ctDNA fraction. Supplementary Figure S5 shows no significant changes in cfDNA yield/ctDNA fraction after prolonged ADT exposure compared to samples analyzed after 3 months of ADT.
3.2. ctDNA/cfDNA yields and clinical outcomes
To determine the impact of cfDNA yield/ctDNA fraction and pTMB levels on clinical outcomes, we evaluated the predictive value of these variables for ADT efficacy in patients in the untreated mHSPC group using ADT failure time and assessed their prognostic value for OS in patients in mHSPC and mCRPC states (Supplementary Results). Supplementary Figure S6 summarizes the prognostic and predictive (to ADT) value of cfDNA yield/ctDNA fraction/pTMB in mHSPC state.
As anticipated, patients with high-volume metastatic disease in the untreated mHSPC group had the shortest OS (Supplementary Figure 7SA); however, metastatic volume was not predictive of the duration of ADT failure (Supplementary Figure 7SB). We further explored the combined effect of cfDNA yield/ctDNA fraction and metastatic disease volume on survival. Untreated mHSPC patients with high-volume metastatic disease and high-yield cfDNA/ctDNA had the shortest OS, and patients with low-volume metastatic disease and low cfDNA yield/ctDNA fraction had the longest OS. Interestingly, not all patients with high-volume metastatic disease had poor OS, as a group of mHSPC patients with high-volume metastatic disease and low nucleic acid yields had intermediate OS similar to those of patients with low-volume metastatic disease and high cfDNA yield/ctDNA fraction. Fig. 1D shows the Kaplan-Meier plots for OS based on volume status and ctDNA fraction in the untreated mHSPC group. Fig. 1E shows the combined effect of ctDNA fraction and metastatic disease volume on ADT failure rates; patients with high-volume metastases and high ctDNA fraction exhibited the shortest time to ADT failure.
The prognostic value of ALP levels on OS, a known clinical prognostic factor in mCRPC, was determined (Supplementary Figure S8) and nucleic acid yield/fraction–based prognosis evaluated (Supplementary Figure S9). The combined effect of ALP and nucleic acid yield/fraction on OS for all mCRPC patients is shown in Fig. 1F for ctDNA and Supplementary Figure S10 for cfDNA, and detailed results are presented in the Supplementary Results. Results of nucleic acid–based prognosis on OS in the subset of patients with clinical mCRPC alone is shown in Supplementary Figures S11A (cfDNA), S11B (ctDNA), S11C (pTMB), S12A (serum ALP), and S12B and S12C (combined nucleic acid yield/fraction with ALP).
3.3. CNAs, SNVs, and rearrangements in ctDNA
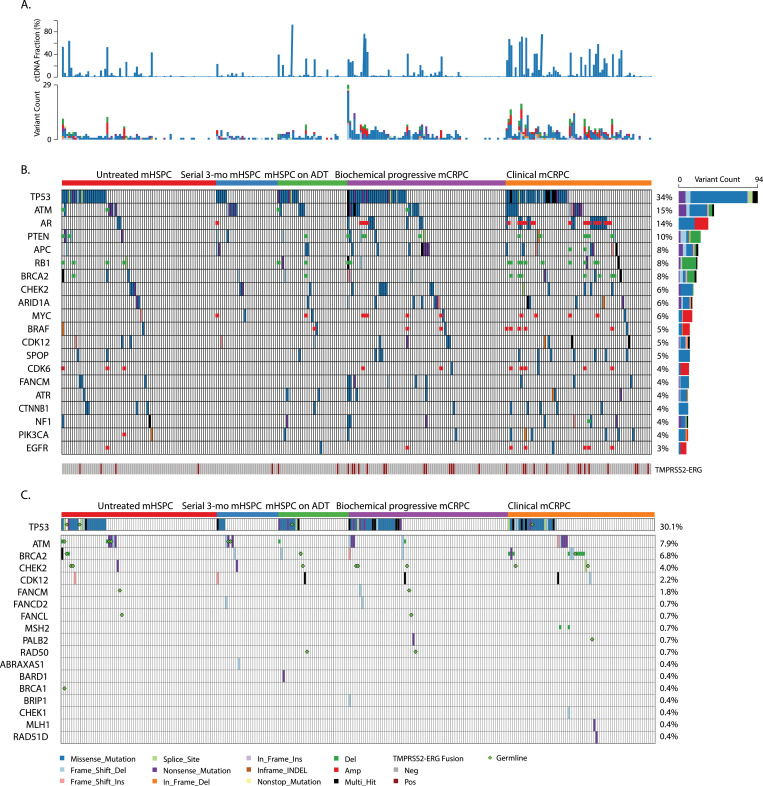
We characterized the spectrum of alterations in each of the 4 independent groups of mHSPC and mCRPC patients. A 120 gene panel was used to identify ctDNA-based CNAs, SNVs, and the frequency of TMPRSS2-ERG fusion. Fig. 2 shows the individual patient ctDNA fractions and variant counts (Fig. 2A) of all patients. Fig. 2B shows the overall distribution of plasma ctDNA somatic alterations in the top 26 genes across the spectrum of all 4 groups (n = 250) that were altered in ≥3% of the study patients using the targeted 120 gene panel. Fig. 2C represents deleterious/likely deleterious alterations detected in genes that were involved in DNA damage repair (DDR) pathways and includes copy number loss and germline pathogenic mutations. Supplementary Table S2 describes the number of patients in each metastatic group who had a genomic alteration of any kind and shows the intergroup comparisons that were performed. All 3 types of somatic alterations (SNVs, CNAs, and TMPRSS2-ERG fusions) were detected more frequently in mCRPC patients than in mHSPC patients. Within the mCRPC groups, a significantly higher proportion of clinical mCRPC group patients had somatic events compared to all other groups (Supplementary Table S2). To identify any effects that cfDNA yield/ctDNA fraction might have on detecting ctDNA-based events, we correlated these yields with event frequencies (r2 = 0·42 for cfDNA yield; r = 0·82 for ctDNA fraction) (Supplementary Figure S13). Additional descriptive details of alteration characteristics are provided in Supplementary Figure S14 and in the Supplementary Results.
Fig. 2.
A: Individual patient ctDNA fractions and variant counts across metastatic groups. B. Overall heatmap of individual somatic alterations observed in metastatic prostate cancer groups. C. Overall heatmap of deleterious/likely deleterious alterations detected in genes involved in DNA damage repair pathways. Copy number loss is shown as a blue box, and germline deleterious/likely deleterious mutations are marked with a green diamond shape.
Fig. 2B and C also shows TP53, AR, DDR pathway genes, cell cycle control and differentiation pathway genes, and well-known tumor suppressor genes to be among the most frequent genes within the top 20 genes with detectable somatic alterations. An increased number of genes in mCRPC patients (n = 33 [23%]) had detectable CNAs (amplifications/deletions) compared to mHSPC patients (n = 8 [8%]). AR gene amplification was the most common CNA and was largely detected in the mCRPC group patients. EGFR, MYC, BRAF, and CDK6 gene amplifications were detected in both mHSPC and mCRPC patients. RB1 deletion/loss was the most frequently detected deletion/loss, and 41/250 patients also had copy number loss of DNA repair pathway genes (Fig. 2C) in both the mHSPC (n = 14/106) and mCRPC groups (n = 27/144). An increased number of mCRPC patients (n = 33 [23%]) had detectable CNAs (amplifications/deletions) compared to mHSPC patients (n = 8 [8%]).
The overall frequency of ctDNA mutations, which were significantly increased in patients in the mCRPC groups compared to patients in the mHSPC groups (Fig. 3A), were observed in AR, APC, and KIT genes (P < ·05). AR hotspot mutations were detectable only in patients in the mCRPC groups. Fig. 3B shows that these mutations were in the ligand-binding domain of the receptor and indicates the number of patients with each mutation. Previously reported mutations T742L, T742C, V716M, T878A, L702H, H875Y and other novel hotspot AR mutations were among those detected in patients in the mCRPC groups [26], but we did not detect the previously reported F788L mutation in this cohort. Fig. 3C further shows the distribution of AR hotspot mutations across exon regions in mCRPC patients at the different levels of variant allelic frequency of detection. Fig. 3D shows the per-patient occurrence of detectable AR mutations, AR copy number gain, and individual patient-level ctDNA fractions in both mCRPC groups.
Fig. 3.
A. Alteration frequencies in key genes between mCRPC and mHSPC groups. B. Lollipop plot of AR somatic mutations detected in mHSPC and mCRPC patients. Known hotspot AR mutations are labeled with detailed amino acid changes. C. Distribution of AR hotspot mutations across exon regions in mCRPC patients. Each dot represents a patient, and the distinct colors indicate different levels of variant allelic frequency (VAF). D. Distribution of AR mutations and AR copy number gain along with matching ctDNA fractions in mCRPC patients detected with these alterations. Each colored bar represents an individual patient.
Co-occurrence of multiple somatic mutations in individual genes across the spectrum of metastatic groups was also observed in select patients, with a higher number detected in the mCRPC groups than in the mHSPC groups. In the clinical mCRPC and biochemical progressive mCRPC patients (11 [16%] and 3 [4%], respectively), cooccurrence of multiple somatic mutations was detected in TP53, APC, BRCA2, RBI1, CDK12, and ARID1A. In the untreated mHSPC patients (2 [3%]), multi-hits were detected in 2 genes: BRCA2 and NF1.
Of the 250 patients profiled for ctDNA alterations, we were able to successfully sequence the gDNA of 219 to probe for germline pathogenic variations in key genes. Pathogenic variants were detected in 20/219 patients, including 11/92 patients in the mHSPC and 9/127 in mCRPC groups. Nine patients had both germline and somatic ctDNA alterations in DDR genes. Fig. 2C shows patients with cooccurrence of pathogenic germline variants and somatic alterations in key genes throughout the spectrum of progression.
3.4. ctDNA clonal dynamic changes after ADT initiation in paired plasma of untreated mHSPC patients
Pharmacodynamic changes in ctDNA-based alterations were interrogated before and after 3 months of ADT in the 29 paired plasma collections of the untreated mHSPC group patients (n = 29; [40%]). Changes in PSA (Fig. 4A), ctDNA fractions (Fig. 4B) and specific clonal changes of the most frequently mutated genes before and after 3 months of ADT treatment in the untreated mHSPC group are shown in Fig. 4C. After 3 months of ADT, several patients demonstrated a PSA response but not a complete disappearance of plasma ctDNA fraction and/or detectable mutations (Fig. 4B). Appearance of new mutations after 3 months of ADT with PSA response was also observed (Fig. 4C: patients 1175, 1284, 1054, and 1073).
Fig. 4.
A. PSA changes after 3-months of ADT in untreated mHSPC paired patient samples. B. ctDNA fraction changes after 3-months of ADT in untreated mHSPC paired patient samples. C. ctDNA-based somatic alterations of top frequently mutated genes detected in 29 paired untreated mHSPC patients before and after 3 months of androgen deprivation therapy.
3.5. Comparison of ctDNA-based alterations in 2 independent clinical mCRPC cohorts
We compared our findings from the Mayo Clinic clinical mCRPC group (n = 69) to an independent second matched cohort of 34 clinical mCRPC patients from Monash Health and Chris O'Brien Lifehouse, Australia, who underwent plasma ctDNA profiling. The baseline demographic profile of the Australian clinical mCRPC cohort is provided in Supplementary Table S3. We observed similar frequencies of ctDNA-based perturbations between the Mayo Clinic and Australian cohorts in key altered genes: TP53, AR, BRCA2, and BRAF (Supplementary Figure S15). These data highlight common findings of genomic alterations in 2 independent racially matched cohorts (for example, near similar AR amplification frequency [23% vs. 20%]).
3.6. Plasma ctDNA alterations and clinical outcomes
The effect of plasma ctDNA-based alterations on clinical outcomes among the mHSPC and mCRPC groups was determined in the Mayo Clinic longitudinal cohort. Median time from date of specimen collection to death/last follow-up for each independent group in provided in Table 1. Several patients in each group were not detected to have alteration events (Supplementary Table S2). Among untreated mHSPC patients (n = 21 [29%]) without detectable alterations, the median OS was 86·5 months (95% CI, 49·27; not reached). Among untreated mHSPC patients (n = 53 [73%]) with detectable alterations, the median OS was 66·4 (range, 53·6; not reached) months (HR, 1·27 [95% CI, 0·62–2·64]; P = ·51) (Supplementary Figure S16A). Presence of detectable somatic events did not affect ADT failure rates (Supplementary Figure S16B). In comparison, in the mCRPC groups, patients not detected to have somatic events had a significantly longer OS than mCRPC patients with detected events (54·6 vs 30·93 months; HR, 1·84 [95% CI, 0·99–3·43]; Log-Rank P = .05) (Supplementary Figure S16C). We also explored MSI status and correlation with plasma TMB in mHSPC/mCRPC patients detected with MMR-deficiency mutations (and/or MSI-high status, hypermutation) and are reported under “Supplementary Results” and Supplementary Figure S17.
OS outcomes were also determined for the mHSPC and mCRPC groups on the basis of individual-gene and multiple-gene alterations after adjusting for known prognostic variables in both groups. In untreated mHSPC patients at the individual-gene level, alterations in TP53 and ATM were significantly associated with shorter OS. These alterations were not significant after adjusting for metastatic volume and Gleason Score. Collectively, untreated mHSPC patients with somatic alterations detected in multiple DNA repair genes (ATM, BRCA1, BRCA2, and CHEK2) had significantly short OS (HR, 4·0 [95% CI, 1·4–11·8]; P = 4·75 × 10–05) (Supplementary Table S4) and a shorter time to failure with ADT even after adjusting for clinical prognostic factors.
For the mCRPC groups, RB1 deletions had the most significant prognostic value for poor outcomes in multivariate analyses after adjusting for Gleason Score and ALP levels (HR, 4.2 [95% CI, 2.0–8.7]; P =1.5 × −04). Somatic perturbations detected in other genes in multivariable analyses that had prognostic significance included TP53, CDH1, CDK6 and BRCA2 (Supplementary Table S5). At the individual gene level, Fig. 5A shows RB1 copy number deletion to be associated with significantly worse OS in mCRPC patients. Similarly, Fig. 5B shows poor OS in mCRPC patients with AR copy number gain. mCRPC patients who harbored different TP53 mutations and an increasing number of mutations were also associated with poorer OS (Fig. 5C).
Fig. 5.
A. RB1 wild type vs copy number deletion and overall survival in mCRPC patients. B. AR copy number gain compared to wild type and overall survival in mCRPC patients. C. TP53 mutations vs wild type and overall survival in mCRPC patients.
4. Discussion
A comprehensive analysis of the genomic landscape of metastatic prostate cancer, from mHSPC to clinical and radiographically progressive mCRPC, and its impact on clinical outcomes has not been elucidated. In metastatic prostate cancer, somatic mutational variations have been extensively profiled in the castrate-resistant state [11,12,[27], [28], [29], [30]], and prognostic and predictive genomic perturbations identified. However, in practice, it is difficult to obtain tissue biopsies of progressive metastatic prostate cancer before ADT initiation, during response to ADT, and at the time of ADT initiation. At the same time, this uncharacterized and uncaptured underlying clonal evolution in the continuum of progressive metastatic states is likely to result in cumulative late-stage alterations. This longitudinal cohort study aimed to fill the gap in this continuum, from the untreated mHSPC state to the development of castration resistance, by profiling plasma ctDNA-based alterations in the coding regions of 120 key biology-specific cancer and prostate cancer genes. Because radiographic and clinical symptoms of mHSPC patients who progress on ADT typically follow biochemical failure (defined as rising serum PSA levels and often used in clinical practice for additional therapeutic interventions) by a median of 8 months, we separately delineated somatic genomic alterations in mCRPC patients experiencing biochemical and radiographic/clinical progressions [6]. Although our study was not designed to be a predictive and prognostic biomarker study, follow-up data obtained from this longitudinal cohort allowed for us to comprehensively profile the clinical relevance of the structural DNA-based variations observed in each of these metastatic groups with clinical outcomes.
We report several interesting findings in our study, which add practical value beyond currently used clinical factors. We observed that ctDNA fraction may serve as prognostic factors in mHSPC and mCRPC patients. mHSPC patients with low ctDNA fraction and low-volume metastatic disease status achieved better OS than patients with high-volume disease and/or high ctDNA fraction. These data if validated prospectively may serve in selecting patients for the intensification of upfront systemic treatment beyond known clinical factors to achieve optimal OS in patients with high-volume disease and/or high ctDNA yield. Until now, the selection of patients for treatments additional to ADT has been guided using non-genomic clinical factors such as Gleason Score, the presence of visceral metastases, and/or the number of skeletal metastases. With an increasing array of systemic treatment options available to mHSPC patients [5,31], these data potentially provide further insights into selection of initial therapy for metastatic prostate cancer based on the circulatory and measurable tumor genome and the presence or absence of key genomic perturbations.
Similarly, among mCRPC patients, we observed a poor prognosis subgroup of patients with high ctDNA fraction and high levels of serum ALP, which a known clinical poor prognostic marker. These results suggest that these patients could be the focus of clinical trials testing more aggressive combination therapies. Notably, in both mHSPC and mCRPC patients, pTMB levels were not strongly linked to clinical outcomes, suggesting that pTMB is not a useful biomarker in advanced prostate cancer. This finding is consistent with the low mutational burden observed in prostate cancer [32], particularly in comparison to malignancies such as lung cancer and melanoma [33].
We identified several genes in which alterations were associated with clinical outcomes in both mHSPC and mCRPC patients which have potential therapeutic impact. In mHSPC patients, perturbations in the TP53 and DDR genes (ATM + BRCA 1/2 ± CHEK2) were significantly associated with survival after adjusting for several clinical factors, although not for ADT treatment failure. In mCRPC state, alterations in TP53, RB1, BRCA2, CDK6, and CHD1 were linked to shorter rates of OS. It would appear that inactivation of tumor suppressor genes and selected DDR genes appears to confer worse outcomes in advanced prostate cancer. It is increasingly recognized that the loss of function of key tumor suppressor genes, including TP53, RB1, and PTEN, may be a poor prognostic marker for both mHSPC and mCRPC patients [30,34,35], and our ctDNA-based results largely support this finding. Although the prognostic value of DDR gene alterations for mCRPC patients is controversial [36,37], their prognostic significance for mHSPC patients is largely unknown; our findings suggest that patients with composite DDR alterations have poor rates of OS. This will require independent and prospective validation.
The emergence of somatic alterations in the mCRPC state in 2 independent cohorts (the biochemical progressive mCRPC group and the clinical mCRPC group), has not been consistently reported. Mayrhofer et al. [38] recently conducted a small study in which plasma-based genomes of hormone-naive and castrate-resistant patients were compared using ctDNA genome profiling. The authors reported ctDNA and ctDNA-based somatic variations in 6 hormone-sensitive, 23 hormone-naive, and 188 castrate-resistant patients, which were used to evaluate variations in cfDNA/ctDNA and genomic alterations from the hormone-naive to the castrate-resistant state. Given this study's limited number of patients and the distinctive subset of patients with biochemical and radiographic breakdown at the time of castration resistance, it is difficult to draw conclusions regarding clinical effects or to create an atlas of changes across the spectrum of progression. In our study of larger mCRPC cohorts, we observed qualitative and quantitative differences in genomic perturbations between these 2 mCRPC groups, with a greater accumulation of mutational alterations detected with radiographic progression than biochemical progression alone while on ADT. It is unclear whether these differences in genomic perturbations will have clinical implications. However, the emergence of cumulative and novel somatic perturbations at the time of ADT failure, which can be detected earlier in their course has value for impacting practice, as these alterations could be used to monitor minimally progressive disease as well as provide a potential therapeutic advantage for earlier interventions that affect clinical outcomes.
Another key application of ctDNA profiling in metastatic prostate cancer is the facilitation of personalized medicine therapeutics. Given the recently announced PROfound clinical trial results [39], in which olaparib was found to significantly improve radiographic progression-free survival in patients with BRCA 1/2 or ATM aberrant mCRPC, it is notable that we detected BRCA 1/2 or ATM perturbations in 16% of our mCRPC cohort. Use of a ctDNA assay could spare these patients the need to undergo a potentially painful tumor biopsy procedure or to be treated with androgen receptor pathway inhibitors, as the trial results indicate a better radiographic progression-free survival rate with olaparib. Likewise, we observed PTEN or PIK3CA changes in 25 patients, who may be suitable candidates for inhibitors targeting the PI3K/AKT pathway, such as ipatasertib, which is currently being studied in phase III clinical trials in patients with mCRPC [40].
Additionally, ctDNA is advantageous because serial blood samples are readily obtainable, allowing for the identification of patients with early treatment failure due to the rapid emergence of adaptive resistance, such as specific AR mutations in biochemically progressive mCRPC state. Notably, in the mHSPC state, early genomic changes also arose in TP53 and ATM even when patients experienced biochemical responses with declining serum PSA levels. These early genomic changes emphasize the genomic plasticity of prostate cancer under testosterone suppression. These changes may also play a key biological role in tumor suppressor and DDR genes in advanced prostate cancer and may be used to monitor minimal residual disease. However, realizing the clinical value of ctDNA-based profiling for this indication will require greater future effort.
4.1. Limitations
We were unable to evaluate the effect of treatments using serial sampling in most of our patients to determine therapy selection pressure on clonal evolutionary perturbations. This will require a systematic study in future. We also did not evaluate intronic structural variations, which can be twice as common in genes such as RB1 and PTEN. Our attempt was to go beyond cataloging aberrations in single genes and pathways, and thus, we used a custom panel of 120 cancer-related genes. At the same time, this cancer panel did not include all possible cancer genomic aberrations, such as ERG gene rearrangements. There is at least a risk of underestimation of tumor fraction due to missing tumor DNA that does not harbor alterations in any of the genes included on the custom panel. A more comprehensive genome-wide determination will need to be conducted to more accurately capture somatic variants and tumor fractions. Additionally, in our study, we observed when the ctDNA fraction was low, as noted for 15% of our study cohort, that ctDNA profiling may not have been sensitive enough to detect key alterations. Nevertheless, the use of ctDNA profiling to follow the emergence of cumulative genomic alterations in the blood is a potentially powerful tool for monitoring minimal residual disease, therapeutic targeting, and accurate prognostication of survival in metastatic-stage prostate cancer; however, further validation is needed.
5. Conclusions
In our study, ctDNA fraction before, during, and after ADT failure may appear to refine known clinical prognostic factors across the spectrum of metastatic prostate cancer, and this will need future validation. We also detected specific ctDNA alterations in mCRPC that correlate with poorer survival, as do mutations in multiple DNA repair genes observed to be predictive of ADT efficacy and survival in the hormone-sensitive state. The molecular characterization of progressive metastatic prostate cancer also supports the rational matching of a growing armamentarium of drug choices for metastatic prostate cancer states. Finally, molecular profiling, with or without nucleic acid yields, may be useful for monitoring minimal residual disease in metastatic prostate cancer, for which further validation is needed.
Declaration of Competing Interest
Manish Kohli received travel/accommodation from Celgene; Tiantian Zheng, Amy Wang, Carlos Montesinos, Calven Wong, Pan Du, Shidong Jia, and Jianjun Yu are stockholders in Predicine, Inc.; Lisa Horvath received research funding from Astellas Pharma, travel/accommodation from Astellas Pharma and Pfizer, and is on the Scientific Advisory Board for Imagion; Kate Mahon received travel/accommodation from Astellas Pharma; Edmond M Kwan received honoraria from Janssen, research funding from Astellas Pharma and AstraZeneca, and travel /accommodations from Astellas Pharma, Pfizer, and Ipsen; Arun A Azad is a consultant for Astellas Pharma, Janssen, and Novartis, is on the speakers bureau for Astellas Pharma, Janssen, Novartis and Amgen received honoraria from Astellas Pharma, Janssen, Novartis, Tolmar, Amgen, Pfizer, and Telix, is on the Scientific Advisory Board for Astellas Pharma, Novartis, Sanofi, AstraZeneca, Tolmar, Pfizer, and Telix, and received research funding from Astellas Pharma, and Merck Serono.
Acknowledgments
Acknowledgments
We thank Paul Fletcher and Daley Drucker (Moffitt Cancer Center) for editorial assistance. They were not compensated beyond their regular salaries.
Funding
This study was based on a non-commercial collaboration between academic institutions. No funding was received from Predicine, Inc. during the conduct of the study.
This study was supported by: NHMRC Postgraduate Scholarship (to Edmond M Kwan); Movember/Prostate Cancer Foundation of Australia Clinical Scientist Fellow (CSA 0515; to Kate Mahon); the Astellas Investigator-Initiated Grant, Cancer Institute NSW Translational Program Grant (2018/TPG001; to Lisa G Horvath); the NHMRC Project Grant, Victorian Cancer Agency Clinical Research Fellowship-CRF14009, Astellas Investigator-Initiated Grant, NHMRC Project Grant - GNT1098647 (to Arun A Azad); and the National Institute of Health- RO1-CA212097 (to Manish Kohli).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102728.
Appendix. Supplementary materials
References
- 1.Gandara D.R., Paul S.M., Kowanetz M., Schleifman E., Zou W., Li Y. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24(9):1441–1448. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 2.Gandara D.R., Paul S.M., Kowanetz M., Schleifman E., Zou W., Li Y. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018;24(9):1441. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 3.Basch E., Loblaw D.A., Rumble R.B. Systemic therapy in men with metastatic castration-resistant prostate cancer: American Society of Clinical Oncology and Cancer Care Ontario Clinical Practice guideline summary. J Oncol Pract. 2014;10(6):e418–ee20. doi: 10.1200/JOP.2014.001701. [DOI] [PubMed] [Google Scholar]
- 4.Mateo J., Carreira S., Sandhu S., Miranda S., Mossop H., Perez-Lopez R. DNA-Repair defects and Olaparib in metastatic prostate cancer. N Engl J Med. 2015;373(18):1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis I.D., Martin A.J., Stockler M.R., Begbie S., Chi K.N., Chowdhury S. Enzalutamide with standard first-line therapy in metastatic prostate cancer. New Engl J Med. 2019;381(2):121–131. doi: 10.1056/NEJMoa1903835. [DOI] [PubMed] [Google Scholar]
- 6.Sweeney C.J., Chen Y.H., Carducci M., Liu G., Jarrard D.F., Eisenberger M. Chemohormonal therapy in metastatic hormone-sensitive prostate cancer. N Engl J Med. 2015;373(8):737–746. doi: 10.1056/NEJMoa1503747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fizazi K., Tran N., Fein L., Matsubara N., Rodriguez-Antolin A., Alekseev B.Y. Abiraterone acetate plus prednisone in patients with newly diagnosed high-risk metastatic castration-sensitive prostate cancer (LATITUDE): final overall survival analysis of a randomised, double-blind, phase 3 trial. Lancet Oncol. 2019;20(5):686–700. doi: 10.1016/S1470-2045(19)30082-8. [DOI] [PubMed] [Google Scholar]
- 8.James N.D., de Bono J.S., Spears M.R., Clarke N.W., Mason M.D., Dearnaley D.P. Abiraterone for prostate cancer not previously treated with hormone therapy. N Engl J Med. 2017;377(4):377338–377351. doi: 10.1056/NEJMoa1702900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith M.R., Saad F., Chowdhury S., Oudard S., Hadaschik B.A., Graff J.N. Apalutamide treatment and metastasis-free survival in prostate cancer. N Engl J Med. 2018;378(15):1408–1418. doi: 10.1056/NEJMoa1715546. [DOI] [PubMed] [Google Scholar]
- 10.James N.D., Sydes M.R., Clarke N.W., Mason M.D., Dearnaley D.P., Spears M.R. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet. 2016;387(10024):1163–1177. doi: 10.1016/S0140-6736(15)01037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L., Dehm S.M., Hillman D.W., Sicotte H., Tan W., Gormley M. A prospective genome-wide study of prostate cancer metastases reveals association of WNT pathway activation and increased cell cycle proliferation with primary resistance to abiraterone acetate-prednisone. Ann Oncol. 2017;29(2):352–360. doi: 10.1093/annonc/mdx689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson D., Van Allen E.M., Wu Y.M., Schultz N., Lonigro R.J., Mosquera J.M. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grasso C.S., Wu Y.M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jimenez R.E., Atwell T.D., Sicotte H., Eckloff B., Wang L., Barman P. A prospective correlation of tissue histopathology with nucleic acid yield in metastatic castration-resistant prostate cancer biopsy specimens. Mayo Clin Proc Innov Qual Outcomes. 2019;3(1):14–22. doi: 10.1016/j.mayocpiqo.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia S., Kohli M., Du M., Dittmar R.L., Lee A., Nandy D. Plasma genetic and genomic abnormalities predict treatment response and clinical outcome in advanced prostate cancer. Oncotarget. 2015;6(18):16411–16421. doi: 10.18632/oncotarget.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binder M., Zhang B.Y., Hillman D.W., Kohli R., Kohli T., Lee A. Common genetic variation in CYP17A1 and response to abiraterone acetate in patients with metastatic castration-resistant prostate cancer. Int J Mol Sci. 2016;17(7):1097. doi: 10.3390/ijms17071097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang B.Y., Riska S.M., Mahoney D.W., Costello B.A., Kohli R., Quevedo J.F. Germline genetic variation in JAK2 as a prognostic marker in castration-resistant prostate cancer. BJU Int. 2016 doi: 10.1111/bju.13584. [DOI] [PubMed] [Google Scholar]
- 18.Huang X., Yuan T., Liang M., Du M., Xia S., Dittmar R. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 2015;67(1):33–41. doi: 10.1016/j.eururo.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman A.M., Lovejoy A.F., Klass D.M., Kurtz D.M., Chabon J.J., Scherer F. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34(5):547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandekerkhove G., Todenhofer T., Annala M., Struss W.J., Wong A., Beja K. Circulating tumor DNA reveals clinically actionable somatic genome of metastatic bladder cancer. Clin Cancer Res. 2017;23(21):6487–6497. doi: 10.1158/1078-0432.CCR-17-1140. [DOI] [PubMed] [Google Scholar]
- 23.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Ser B (Methodol) 1995;57(1):289–300. [Google Scholar]
- 24.Quintanal-Villalonga A., Molina-Pinelo S., Cirauqui C., Ojeda-Marquez L., Marrugal A., Suarez R. FGFR1 cooperates with EGFR in lung cancer oncogenesis, and their combined inhibition shows improved efficacy. J Thorac Oncol. 2019;14(4):641–655. doi: 10.1016/j.jtho.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 25.Halabi S., Lin C.Y., Small E.J., Armstrong A.J., Kaplan E.B., Petrylak D. Prognostic model predicting metastatic castration-resistant prostate cancer survival in men treated with second-line chemotherapy. J Natl Cancer Inst. 2013;105(22):1729–1737. doi: 10.1093/jnci/djt280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sumiyoshi T., Mizuno K., Yamasaki T., Miyazaki Y., Makino Y., Okasho K. Clinical utility of androgen receptor gene aberrations in circulating cell-free DNA as a biomarker for treatment of castration-resistant prostate cancer. Sci Rep. 2019;9(1):4030. doi: 10.1038/s41598-019-40719-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beltran H., Eng K., Mosquera J.M., Sigaras A., Romanel A., Rennert H. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol. 2015;1(4):466–474. doi: 10.1001/jamaoncol.2015.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quigley D.A., Dang H.X., Zhao S.G., Lloyd P., Aggarwal R., Alumkal J.J. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell. 2018;174(3):758–769. doi: 10.1016/j.cell.2018.06.039. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armenia J., Wankowicz S.A.M., Liu D., Gao J., Kundra R., Reznik E. The long tail of oncogenic drivers in prostate cancer. Nat Genet. 2018;50(5):645–651. doi: 10.1038/s41588-018-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abida W., Cyrta J., Heller G., Prandi D., Armenia J., Coleman I. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A. 2019;116(23):11428–11436. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chi K.N., Agarwal N., Bjartell A., Chung B.H., Pereira de Santana Gomes A.J., Given R. Apalutamide for metastatic, castration-sensitive prostate cancer. N Engl J Med. 2019;381(1):13–24. doi: 10.1056/NEJMoa1903307. [DOI] [PubMed] [Google Scholar]
- 32.Chung J.H., Dewal N., Sokol E., Mathew P., Whitehead R., Millis S.Z. Prospective comprehensive genomic profiling of primary and metastatic prostate tumors. JCO Precis Oncol. 2019;(3):1–23. doi: 10.1200/PO.18.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamid A.A., Gray K.P., Shaw G., MacConaill L.E., Evan C., Bernard B. Compound genomic alterations of TP53, PTEN, and RB1 tumor suppressors in localized and metastatic prostate cancer. Eur Urol. 2019;76(1):89–97. doi: 10.1016/j.eururo.2018.11.045. [DOI] [PubMed] [Google Scholar]
- 35.De Laere B., Oeyen S., Mayrhofer M., Whitington T., van Dam P.J., Van Oyen P. TP53 outperforms other androgen receptor biomarkers to predict abiraterone or enzalutamide outcome in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2019;25(6):1766–1773. doi: 10.1158/1078-0432.CCR-18-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nombela P., Lozano R., Aytes A., Mateo J., Olmos D., Castro E. BRCA2 and other DDR genes in prostate cancer. Cancers (Basel) 2019;11(3):352. doi: 10.3390/cancers11030352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schweizer M.T., Antonarakis E.S. Prognostic and therapeutic implications of DNA repair gene mutations in advanced prostate cancer. Clin Adv Hematol Oncol. 2017;15(10):785–795. [PubMed] [Google Scholar]
- 38.Mayrhofer M., De Laere B., Whitington T., Van Oyen P., Ghysel C., Ampe J. Cell-free DNA profiling of metastatic prostate cancer reveals microsatellite instability, structural rearrangements and clonal hematopoiesis. Genome Med. 2018;10(1):85. doi: 10.1186/s13073-018-0595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hussain M., Mateo J., Fizazi K., Saad F., Shore N.D., Sandhu S. LBA12_PRPROfound: phase III study of olaparib versus enzalutamide or abiraterone for metastatic castration-resistant prostate cancer (mCRPC) with homologous recombination repair (HRR) gene alterations. Ann Oncol. 2019;30(Suppl 5) mdz394.039. [Google Scholar]
- 40.Bono J.S.D., Giorgi U.D., Massard C., Bracarda S., Kocak I., Font A. Randomized phase II study of AKT blockade with ipatasertib (GDC-0068) and abiraterone (Abi) vs. Abi alone in patients with metastatic castration-resistant prostate cancer (mCRPC) after docetaxel chemotherapy (A. Martin study) J Clin Oncol. 2016;34(15_suppl):5017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.