Abstract
Background
Long noncoding RNAs (lncRNAs) are emerging as critical regulators of gene expression in the immune system, but their impact on neuropsychiatric systemic lupus erythematosus (NPSLE) remains unknown.
Methods
RNA sequencing analysis was used to screen the comprehensive expression profile of lncRNAs and messenger RNAs (mRNAs) in peripheral blood mononuclear cells (PBMCs) from NPSLE patients, active SLE patients who had never experienced neuropsychiatric manifestations (Non-NPSLE) and healthy controls. Differentially expressed (DE) lncRNA levels were validated by qRT-PCR in 26 NPSLE patients, 31 Non-NPSLE patients and 30 healthy controls. Further, correlations of DE lncRNAs with clinical manifestations of NPSLE patients were analyzed. Finally, a bioinformatic analysis was performed to investigate the potential functions of DE genes.
Results
Four hundred and fifty-one lncRNAs and 272 mRNAs were DE between the NPSLE patients and Non-NPSLE patients, among which, significantly upregulated expression levels of NONHSAT208182.1, NONHSAT182114.1, NONHSAT106801.2, NONHSAT039491.2, ENST00000356215, NONHSAT087499.2 and NONHSAT207026.1 while downregulated expression levels of NONHSAT001281.2 and NONHSAT024353.2 were further validated in PBMCs from NPSLE patients by qRT-PCR. Bioinformatic analysis suggested several gene ontology (GO) terms and signal pathways may play important roles in NPSLE development. Co-expression networks analysis indicated that 170 lncRNAs and 46 mRNAs were included in the co-expression network. The expression level of NONHSAT039491.2 was associated with the activity of SLE and the presence of anti-dsDNA, anti-RNP antibody, dizziness and headache. NONHSAT087499.2 level correlated with anti-RNA antibody, ENST00000356215 level correlated with olfactory threshold and oral ulcer. NONHSAT208182.1 level correlated with the presence of fever, unstable walking and urinary red blood cells. NONHSAT106801.2 correlated with frequency of B cells and the presence of fever. NONHSAT024353.2 level was associated with serum IgG levels and the presence of anti-SSA and disorder of consciousness.
Conclusions
Our data provided comprehensive evidence regarding the differential expression of lncRNAs in PBMCs from NPSLE patients, indicating that these DE lncRNAs may play roles in NPSLE development. Our finding shed light on the understanding of the molecular mechanisms of lncRNAs in the pathogenesis of NPSLE.
Keywords: Long non-coding RNA, neuropsychiatric systemic lupus erythematosus (NPSLE), gene ontology terms, kyoto encyclopedia of genes and genomes, co-expression networks
Introduction
Systemic lupus erythematosus (SLE) is a prototypic systemic autoimmune disease characterized by autoantibody production and immune complex deposition, resulting in multiple organ damages including kidneys, heart, lungs, and nervous system (1). Neuropsychiatric SLE (NPSLE) is one of the most severe manifestations of SLE and encompasses a variety of neurological and psychiatric disorders, leading to substantial morbidity and mortality (2). Complex interrelated pathogenic mechanisms have been implicated in NPSLE pathogenesis, including genetic factors, vasculopathy, neuroendocrine-immune imbalance, neuronal damage mediated by autoantibodies, inflammatory mediators and blood brain barrier dysfunction (2). Recent studies found that epigenetic alterations, such as altered DNA methylation, histone modifications, and one class of non-coding RNAs (ncRNAs) named microRNAs mediated posttranscriptional gene silencing may affect the initiation and progression of NPSLE (3). However, the potential role of other types of ncRNAs such as long non-coding RNAs (lncRNAs) in etiology and pathogenesis of NPSLE still largely unknown.
LncRNAs are generally defined as non-protein-coding transcripts larger than 200 nucleotides, located within the intergenic stretches or overlapping antisense transcripts of protein coding genes (4). Although without protein coding capability, accumulating evidence suggested that lncRNAs participate in diverse biological processes, including genomic imprinting, chromosome modification, transcriptional activation, protein stability, and miRNAs inhibition. LncRNAs were reported to be involved in pathogenesis of multiple diseases such as neurodegenerative diseases, diabetic mellitus and cancer (5-7). Recently, the roles of lncRNAs in immune responses have aroused increasing attention (8,9). LncRNAs have been found to be differentially expressed (DE) in peripheral blood mononuclear cells (PBMCs), immortalized B cells, and kidney biopsy specimens from SLE patients (10-13), and play crucial roles in autoimmune diseases including SLE (14,15). However, the precise role of lncRNAs in NPSLE pathogenesis remains unclear.
In the present study, we performed high-throughput RNA sequencing assays on PBMCs from NPSLE patients. Outstanding lncRNAs functions were annotated based on co-expression genes and a gene ontology (GO) biological analysis process. The correlations of lncRNAs with disease activity indicators, system and visceral involvement indicators including the nervous system of NPSLE patients were revealed through correlation analyses.
Methods
Study subjects
Totally 57 active SLE patients and 30 age-matched healthy subjects with no history of autoimmune diseases were included. All patients fulfilled the 1997 American College of Rheumatology (ACR) revised criteria for SLE (16). SLE disease activity was recorded by Systemic Lupus Erythematosus Disease Activity Index 2000 (SLEDAI) score at the time of blood draw. Patients with SLEDAI ≥8 were categorized as active SLE patients. Characteristically, active SLE patients can be classified into two groups, NPSLE group are 26 patients with acute symptoms of NPSLE, and Non-NPSLE group are 31 SLE patients who never experienced neuropsychiatric manifestations (17). NPSLE was diagnosed if the patent had at least one of 19 neuropsychiatric syndromes according to ACR classification and no other explanation related with SLE. Demographic and clinical characteristics of these patient samples are shown in the Table S1. All subjects were given informed consents for the collection of peripheral blood. This study was approved by the Ethics Committee of the Affiliated Drum Tower Hospital of Nanjing University Medical School (ID: SC201700201) and was conducted in accordance with the principle set forth under the 1989 Declaration of Helsinki.
Table S1. Demographic and clinical characteristics of SLE patients.
| Characteristics | SLE, n=57 | Non-NPSLE, n=31 | NPSLE, n=26 | HC, n=30 |
|---|---|---|---|---|
| Demographic | ||||
| Sex (female/male) | 50/7 | 25/6 | 25/1 | 24/6 |
| Age, yrs | 37.8±2.5 | 37.7±2.3 | 38.0±2.7 | 34.7±1.5 |
| Current smoking habit (%) | 7.3 | 10.1 | 4.0 | 16.7 |
| Age at onset, yrs | 33.5±2.6 | 33.2±2.5 | 33.8±2.8 | – |
| Disease duration, month | 54.3±10.4 | 53.7±8.8 | 55.1±12.4 | – |
| SLEDAI | 11.8 ±0.9 | 10.7±0.8 | 13.2±0.9 | – |
| Clinical chart review (%) | – | |||
| Arthritis | 52.6 | 54.8 | 50.0 | – |
| Malar rash | 40.3 | 38.7 | 42.3 | – |
| Discoid lesions | 10.5 | 9.6 | 11.5 | – |
| Photosensitivity | 22.8 | 25.8 | 19.2 | – |
| Oral ulcers | 12.3 | 12.9 | 11.5 | – |
| Pericarditis | 31.6 | 32.3 | 30.7 | – |
| Pleuritis | 21.0 | 22.5 | 19.2 | – |
| Proteinuria | 56.2 | 64.5 | 46.2 | – |
| Leuco/erythrocyturia | 28.1 | 32.3 | 23.1 | – |
| Thrombopenia | 35.1 | 38.7 | 30.8 | – |
| Leucopenia | 26.3 | 29.0 | 23.1 | – |
| Fever | 66.6 | 64.5 | 69.2 | – |
| History of consciousness disorder | 14.0 | 0 | 30.8 | – |
| History of dizziness and headache | 21.0 | 6.4 | 38.5 | – |
| History of unstable walking | 5.2 | 0.0 | 11.5 | – |
| History of convulsion | 10.5 | 0.0 | 23.1 | – |
| History of unclear vision | 3.5 | 3.2 | 3.9 | – |
| History of reynold phenomenon | 8.7 | 9.6 | 7.7 | – |
| History of weak limbs | 31.6 | 29.0 | 34.6 | – |
| History of limb numbness | 17.6 | 19.4 | 15.4 | – |
| History of urinary retention | 1.7 | 0 | 3.8 | – |
| Autoantibodies | ||||
| Anti-nuclear antibody | 89.5 | 90.3 | 88.5 | – |
| Anti-SSA antibody | 67.9 | 70.0 | 65.4 | – |
| Anti-SSB antibody | 17.6 | 19.4 | 15.4 | – |
| Anti-ds-DNA | 45.6 | 48.4 | 42.3 | – |
| Anti-Sm antibody | 40.3 | 38.7 | 42.3 | – |
| Anti-cardiolipin antibody | 17.8 | 12.9 | 23.7 | – |
| Anti-ribonucleoprotein antibody | 36.9 | 35.5 | 38.5 | – |
| Anti-histone antibody | 43.9 | 41.9 | 46.2 | – |
| Anti-ribosomal P antibody | 19.3 | 19.4 | 19.2 | – |
| Anti-nucleosome antibody | 36.8 | 38.7 | 34.6 | – |
| Current medication (%) | ||||
| Acetylsalicylic acid | 75.4 | 64.5 | 88.4 | – |
| Antidepressant | 17.6 | 12.9 | 23.1 | – |
| Benzodiazepine | 7.0 | 3.2 | 11.5 | – |
| Hydroxychloroquine | 80.7 | 83.9 | 76.9 | – |
| Immunosuppressant | 70.2 | 64.5 | 76.9 | – |
| Prednisone | 94.7 | 90.3 | 100.0 | – |
SLE, systemic lupus erythematosus; SLEDAI, SLE disease activity index; yrs, years; NPSLE, neuropsychiatric systemic lupus erythematosus; Non-NPSLE, active SLE patients had never experienced neuropsychiatric manifestations; HC, heathy controls.
Cell isolation and RNA extraction
10 ml EDTA-anticoagulated blood were collected from each subject. PBMCs were prepared by Ficoll-Hypaque density gradient centrifugation. Total RNA was extracted from PBMCs by using Trizol Reagent (Invitrogen, Carlsbad, USA) according to the manufacture’s protocol. The purity and concentration of RNA was evaluated with the NanoDrop ND-1000 (Thermo Fisher Scientific, Wilmington, DE).
High-throughput RNA sequencing and analysis
Briefly, total RNA was extracted using RNeasy mini kit (Qiagen, Germany) according to the manufacturer’s instructions, quantified using NanoDrop ND-2000, and checked for RNA integrity by an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). RNA libraries were generated using TruSeq Stranded RNA Sample Prep Kit (Illumina) following manufacturer’s protocol, quantified with Qubit 2.0 Fluorometer (Life Technologies, USA), validated by Agilent 2100 bioanalyzer (Agilent Technologies, USA) to confirm the insert size and calculate the mole concentration, and subjected to sequencing using Illumina HiSeq 2500 (Illumina, USA). Acquired data were processed to obtain raw reads, which were then extracted for genome mapping with TopHat (version 2.0.9). After genome mapping, Cufflinks v2.1.1 was run with a reference annotation to generate FPKM values for known gene models. DE genes were identified using Cuffdiff. The P value significance threshold in multiple tests was set by the false discovery rate (FDR). The fold changes (FC) were also estimated according to the FPKM in each sample. The DE genes were selected using the following filter criteria: FDR ≤0.01 and FC ≥2. Genes with differential expression levels were identified using edgeR, with GO enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. The library construction and sequencing were performed at Shanghai Biotechnology Corporation.
Screening of DE lncRNAs for validation
We screened DE lncRNAs for further validation using the following approaches to validate the results of sequencing experiments using the same cohort together with another independent cohort and investigate the correlations between gene expression levels and clinical characteristics.
First, lncRNAs were evaluated based on the data revealed by sequencing experiments using the following criteria: (I) the FC of DE lncRNAs (P<0.01) in NPSLE group must be greater than two-fold compared to Non-NPSLE group; (II) lncRNA levels may have no significant difference between Non-NPSLE group and HC group; or if the difference is significant, the change tendencies of abnormally expressed lncRNAs between these two group should be same as that between NPSLE-Non-NPSLE-HC group; (III) the contexts of lncRNA-mRNA network are associated with immune system; and (IV) genes with lncRNA-mRNA repeated sequences and without information in databases were excluded.
Second, the degree of DE genes between NPSLE patients and active SLE patients was compared based on the co-expression network results. The top 10 genes with high different degrees and the qualified requirements for signal values were selected.
Quantitative real-time PCR (qRT-PCR) validation
Extracted total RNA of 87 participants was reverse-transcribed to complementary DNA (cDNA) with random primer using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). The qRT-PCR reaction was performed in duplicate with a total volume of 10 µL, including 5 µL Maxima SYBR Green/ROX qPCR master mix (Thermo Scientific), 0.3 µL PCR Forward Primer (10 µM), 0.3 µL PCR Reverse Primer (10 µM), 2 µL cDNA, and 2.4 µL double-distilled water. ViiA 7 Real-Time PCR System (Applied Biosystems) was used and the samples were subjected to the following thermal cycling: 95 °C for 10 min; 40 cycles of 95 °C for 15 s and annealing/extension for 1 min at 60 °C, followed by a melting curve from 65 °C to 95 °C in 0.5 °C increments. The qRT-PCR primer sequences were displayed in Table S2. Beta-actin was used as internal control and the relative gene expression was calculated using 2−ΔCt comparative threshold cycle method.
Table S2. Primers sequences for qRT-PCR.
| LncRNA ID | Forward primer | Reverse primer |
|---|---|---|
| NONHSAT208182.1 | TGTCCTTTGAATCGGGTCAT | TCCATGCCAAGAAGCCTTAC |
| NONHSAT001281.2 | CTGGGACAGCTAAGGACGAC | AATGAGGGCAATCCAAACAG |
| NONHSAT024353.2 | AGCACAGGGTGGAGACACTT | CTTGGCTTCTCGTCCAATTT |
| NONHSAT182114.1 | CGTAACAACTTCGGCAAGGT | TGAGGGAGAAAGACGACCAG |
| NONHSAT106801.2 | GCCGTGACTTCCAAACAACT | CGAAGCGATGTATGGTTTGA |
| NONHSAT039491.2 | ACGTGCAGCTCAAGGAAAG | CCAGGTGCGTGACCATCTAT |
| ENST00000356215 | GTGAAGCCCTAACCCACCTT | GGTGATCCTTGATGCTCCTT |
| ENST00000414046 | CTGTGGATGACATGGCCTTA | GATGCCAGCTTTGAGTGGA |
| NONHSAT087499.2 | CCCTGGAAAGTACCGTGTGT | ATGACATGGAAGTGCTGCTG |
| NONHSAT207026.1 | CCTGGCGCATTACAACAAG | TTTCTCTGGAAGCGTGAGTG |
| β-actin | GTGGCCGAGGACTTTGATTG | CCTGTAACAACGCATCTCATATT |
| CFH | TTGTCTCCTGACCTCCCAA | TCCACCACTTCACTGTGTC |
| P2RY14 | ATCACTCAGCAGATCATTCC | TCCAGAAGATGGCCACGAAG |
| NAV3 | GCTGCATCACAATTTCAGGTGGGT | TTGAGATGATGCCACGTCTTCGGA |
| VNN1 | GAACCCAGTATGTCTTTCCT | CATACAACCTCCCAAACAGA |
| IDO1 | GGTCATGGAGATGTCCGTAA | ACCAATAGAGAGACCAGGAAGAA |
| CHIT1 | ACCCTGTTAGCCATCGGAGCTGG | TGCACAGCAGCATCCACCTTGAGG |
| BCL2A1 | TCATATTTTGTTGCGGAGTTC | TCCAGCCAGATTTAGGTTCAA |
| KCNJ15 | CCACATCAGAACTCCCTTCAAACA | AGTTCACTTTCAGACGAAGCACCTA |
| KRT27 | CAAGAGGCCTCAAGGGATCT | GGCTCGGAGAGGTTATGACC |
| HIST1H2BC | ACGAGGAGTAGACCTGATGATGT | GTATCACCTATTTCCATTGTCTCAATTGC |
CFH, complement factor H; P2RY14, purinergic receptor P2Y14; NAV3, neuron navigator 3; VNN1, vanin 1; IDO1, Indoleamine 2,3-dioxygenase 1;CHIT1, Chitinase 1; KCNJ15, Potassium voltage-gated channel subfamily J member 15; BCL2A1, BCL2 related protein A1; KRT27, keratin 72; HIST1H2BC, histone cluster 1 H2B family member c.
Bioinformatic analysis
Pathway analysis and GO analysis were applied to explore the potential roles that the DE mRNAs played in biological pathways or GO terms, including the following three categories: biological process, cellular component, and molecular function. Fisher’s exact test was used to find if there was more overlap between the DE list and the GO annotation list than would be expected by chance. The P value denoted the significance of GO terms enrichment in the DE genes. The KEGG (http://www.genome.jp/kegg) database was used to determine the pathways associated with DE gene. The enrichment and statistics calculation of KEGG pathway were similar to the GO analysis. A co-expression analysis was performed by associating the expression profiles of DE-lncRNAs with DE-mRNAs. This association was based on the information of the lncRNA-associated coding regions, which was supplied by the sequencing analysis.
Construction of coding-non-coding gene (CNC) coexpression network
Taking the role of lncRNAs in modulating transcription into account, we analyzed the relationship between DE lncRNAs and mRNAs in hope of uncovering possible mRNA targets for differential lncRNAs. Given the normalized signal intensity (>6), FC (≥2), FDR (<0.01), and the relevance of mRNA in SLE, 36 DE lncRNAs and 151 DE mRNAs [these mRNAs were broadly selected for several reasons: (I) their abnormal expressions in SLE are validated in other studies; (II) their corresponding genes are susceptible genes in SLE; (III) their novel functions in immune response are established by others.] were selected for construction of CNC gene coexpression network. The network construction was based on the correlation analysis between these DE lncRNAs and mRNAs. Pearson correlation coefficients over 0.90 were selected for the construction with Cytoscape software (The Cytoscape Consortium, San Diego, USA) and shown in Supplementary material.
Olfactory function assessment by computerized testing
The odor threshold test measures a person’s ability to detect an odor. Thirteen concentrations of an industry standard odorant are used. The participant smells two samples, one blank and one with an odorant. They are then asked to determine which of the two samples is the stronger. The concentrations are decreased or increased depending on whether the stimulus is identified correctly.
The odor memory test assesses a person’s ability to remember odors that were previously presented. The test begins with the presentation of 10 odors followed by a 10-minute break. Next, 20 odors containing the 10 original and 10 new odors are presented. After each odor presentation, the participant is asked to identify the odor and to determine if the odor is one of the original odors or a new odor. This provides measures of both semantic and episodic odor memory.
All the tests were done according to the manual of OLFACTTM as described in the website: http://www.osmicenterprises.com/index.html.
Statistical analysis
All data represent mean ± SEM. Differences between two groups were determined by unpaired Student’s t-test if the variance was normally distributed. Comparisons among three or more groups were conducted using one-way ANOVA. Randomization and blinding strategy were used whenever possible. Data were analyzed and visualized with GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, USA) and a two-tailed P value <0.05 was considered statistically significant.
Results
LncRNAs and mRNAs expression profiles in NPSLE patients
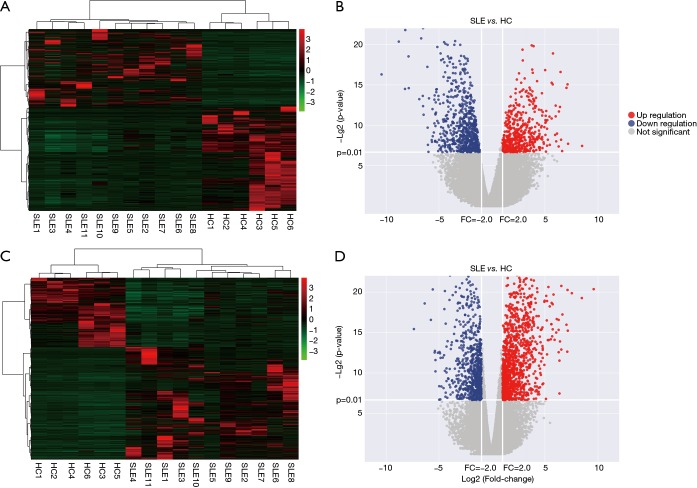
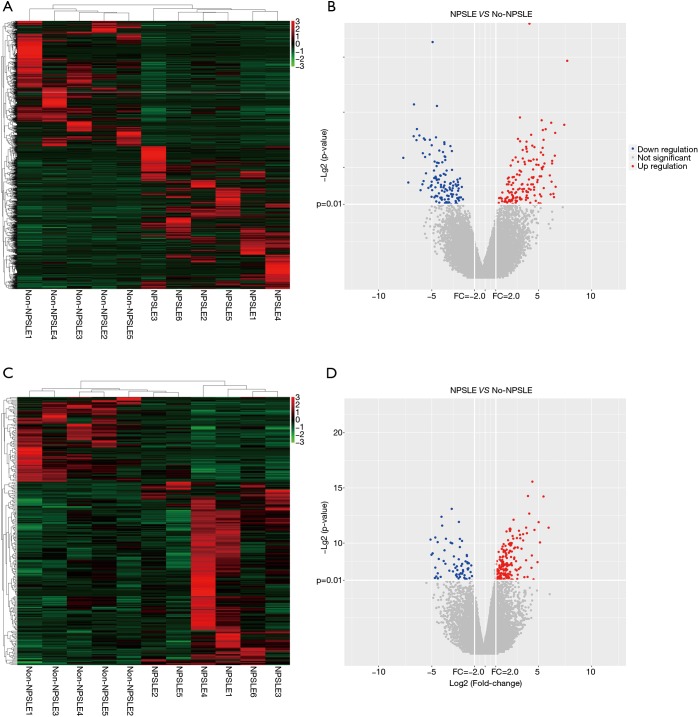
To determine whether the expression levels of lncRNAs were associated with SLE pathogenesis, we firstly performed whole-transcriptome sequencing (RNA-Seq) on the peripheral blood mononuclear cells (PBMCs) of 11 SLE patients and 6 healthy controls. A total of 1,515 lncRNAs and 2,098 mRNAs were DE between these two groups using scatter plot and hierarchical clustering analyses (Figure 1, http://cdn.amegroups.cn/static/other/b191243af9a0d8a7f8fa5ee14a9a4620/atm.2020.03.25-1.xls, http://cdn.amegroups.cn/static/other/2284aef2c7af028a615bd36c215970a9/atm.2020.03.25-2.xlsx; FC ≥2, P<0.01). To reveal DE lncRNA profiles in NPSLE patients, these 11 SLE patients were classified into NPSLE and Non-NPSLE group, and DE lncRNAs in Figure 1 were further analyzed between the two groups. As shown in Figure 2 and http://cdn.amegroups.cn/static/other/ea6fab857009712577755c3a449f940a/atm.2020.03.25-3.xlsx, http://cdn.amegroups.cn/static/other/ae9e70ffaa1ad61a20e131857d1e8749/atm.2020.03.25-4.xlsx, 451 lncRNAs and 272 mRNAs were DE between NPSLE and Non-NPSLE group, among which, 29 lncRNAs and 43 mRNAs were upregulated (http://cdn.amegroups.cn/static/other/a690c7fc04662153fadadc94c29ae7f1/atm.2020.03.25-5.xls, http://cdn.amegroups.cn/static/other/03bf6ea47a10ad0b983eaadb34f49b0b/atm.2020.03.25-6.xls), and 19 lncRNAs and 5 mRNAs were downregulated (http://cdn.amegroups.cn/static/other/a690c7fc04662153fadadc94c29ae7f1/atm.2020.03.25-5.xls, http://cdn.amegroups.cn/static/other/03bf6ea47a10ad0b983eaadb34f49b0b/atm.2020.03.25-6.xls) both in Figure 1 (http://cdn.amegroups.cn/static/other/b191243af9a0d8a7f8fa5ee14a9a4620/atm.2020.03.25-1.xls, http://cdn.amegroups.cn/static/other/2284aef2c7af028a615bd36c215970a9/atm.2020.03.25-2.xlsx) and Figure 2 (http://cdn.amegroups.cn/static/other/ea6fab857009712577755c3a449f940a/atm.2020.03.25-3.xlsx, http://cdn.amegroups.cn/static/other/ae9e70ffaa1ad61a20e131857d1e8749/atm.2020.03.25-4.xlsx), suggesting that these lncRNAs and mRNAs might play roles in NPSLE pathogenesis.
Figure 1.
Expression profiles of lncRNAs and mRNAs in SLE patients and HCs. (A,C) Heat map showing hierarchical clustering of lncRNAs and mRNAs with expression changes greater than two-fold and P value <0.01. SLE patient group: SLE1-11; healthy control group: HC1-6. Red and green colors represent up- and downregulated genes, respectively; (B,D) Volcano plot of differentially expressed lncRNAs and mRNAs. The horizontal white line represents a P value of 0.01, and vertical white lines represent 2.0-fold changes up and down. X axes are the fold change values (log2 scaled), and Y axes are the P values (log2 scaled). Red and blue plots represent up- and downregulated genes, respectively.
Figure 2.
Expression profiles of lncRNAs and mRNAs in NPSLE patients and Non-NPSLE patients. (A,C) Hierarchical clustering of lncRNAs and mRNAs with expression changes greater than two-fold and P value <0.01. NPSLE patient group: NPSLE1-6; Non-NPSLE patient group: Non-NPSLE1-5. Red and green colors represent up- and downregulated genes, respectively; (B,D) Volcano plot of differentially expressed lncRNAs and mRNAs. The horizontal white line represents a P value of 0.01, and vertical white lines represent 2.0-fold changes up and down. X axes are the fold change values (log2 scaled), and Y axes are the P values (log2 scaled). Red and blue plots represent up- and downregulated genes, respectively.
The top 20 DE lncRNAs/mRNAs between NPSLE and Non-NPSLE groups are listed in Table 1 and Table 2, respectively. The RNA-seq data have been submitted to NCBI Gene Expression Omnibus and are accessible with the GEO Series accession number GSE90524 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90524).
Table 1. Top 20 DE lncRNAs between NPSLE and Non-NPSLE patients.
| LncRNA_ID | FPKM (NPSLE) | FPKM (Non-NPSLE) | P value | Up/Down |
|---|---|---|---|---|
| ENST00000414046 | 24.265 | 1.364 | 1.16E-07 | Up |
| ENST00000619625 | 2.566 | 0.002 | 2.31E-08 | Up |
| NONHSAT002346.2 | 15.147 | 0.000 | 1.09E-07 | Up |
| NONHSAT094223.2 | 0.031 | 4.643 | 7.89E-08 | Down |
| NONHSAT169171.1 | 0.268 | 5.482 | 6.46E-08 | Down |
| ENST00000602495 | 0.197 | 5.993 | 3.70E-07 | Down |
| NONHSAT218538.1 | 12.362 | 0.060 | 1.20E-06 | Up |
| NONHSAT076583.2 | 0.481 | 0.000 | 2.32E-06 | Up |
| NONHSAT118801.2 | 0.000 | 0.977 | 2.77E-06 | Down |
| NONHSAT106801.2 | 2.179 | 0.000 | 4.75E-06 | Up |
| NONHSAT147938.2 | 5.265 | 0.000 | 6.07E-06 | Up |
| NONHSAT179347.1 | 0.000 | 13.139 | 6.94E-06 | Down |
| NONHSAT167098.1 | 0.962 | 0.000 | 8.61E-06 | Up |
| NONHSAT001188.2 | 1.186 | 0.000 | 1.08E-05 | Up |
| NONHSAT083781.2 | 0.271 | 27.809 | 1.87E-05 | Down |
| ENST00000499762 | 0.000 | 0.548 | 2.17E-05 | Down |
| NONHSAT068072.2 | 0.239 | 5.485 | 2.06E-05 | Down |
| NONHSAT147817.2 | 0.000 | 0.221 | 2.36E-05 | Down |
| NONHSAT152857.1 | 0.247 | 0.000 | 2.55E-05 | Up |
| NONHSAT044057.2 | 1.112 | 0.000 | 2.88E-05 | Up |
DE, differentially expressed; NPSLE, neuropsychiatric systemic lupus erythematosus.
Table 2. Top 20 DE mRNAs between NPSLE and Non-NPSLE patients.
| Gene id | Gene name | FPKM(NPSLE) | FPKM(Non-NPSLE) | P value | Up/Down |
|---|---|---|---|---|---|
| ENSG00000000971 | CFH | 5.430 | 0.335 | 5.07E-05 | Up |
| ENSG00000174944 | P2RY14 | 6.522 | 0.303 | 2.06E-05 | Up |
| ENSG00000204422 | XXbac-BPG32J3.20 | 0.000 | 2.234 | 6.65E-05 | Down |
| ENSG00000279339 | CTD-2373H9.3 | 0.205 | 0.005 | 5.22E-05 | Up |
| ENSG00000067798 | NAV3 | 0.068 | 0.003 | 4.26E-04 | Up |
| ENSG00000112299 | VNN1 | 29.434 | 5.851 | 4.55E-04 | Up |
| ENSG00000131203 | IDO1 | 1.159 | 0.036 | 2.61E-04 | Up |
| ENSG00000133063 | CHIT1 | 2.497 | 0.511 | 4.00E-04 | Up |
| ENSG00000140379 | BCL2A1 | 188.683 | 37.180 | 3.90E-04 | Up |
| ENSG00000157551 | KCNJ15 | 7.988 | 0.454 | 1.53E-04 | Up |
| ENSG00000170486 | KRT72 | 0.262 | 2.305 | 1.13E-04 | Down |
| ENSG00000180596 | HIST1H2BC | 89.945 | 14.199 | 2.25E-04 | Up |
| ENSG00000186049 | KRT73 | 0.419 | 2.315 | 2.58E-04 | Down |
| ENSG00000204936 | CD177 | 89.236 | 9.899 | 4.48E-04 | Up |
| ENSG00000205611 | LINC01597 | 0.000 | 0.129 | 2.57E-04 | Down |
| ENSG00000211675 | IGLC1 | 10.934 | 0.177 | 3.71E-04 | Up |
| ENSG00000215837 | SDHAP2 | 0.344 | 5.894 | 1.87E-04 | Down |
| ENSG00000229644 | NAMPTP1 | 96.435 | 18.623 | 4.10E-04 | Up |
| ENSG00000242616 | GNG10 | 48.562 | 12.134 | 4.69E-04 | Up |
| ENSG00000244270 | RPL32P29 | 6.749 | 0.470 | 3.68E-04 | Up |
DE, differentially expressed; NPSLE, neuropsychiatric systemic lupus erythematosus.
Verification of lncRNA expression profiles and their correlations with clinical characteristics
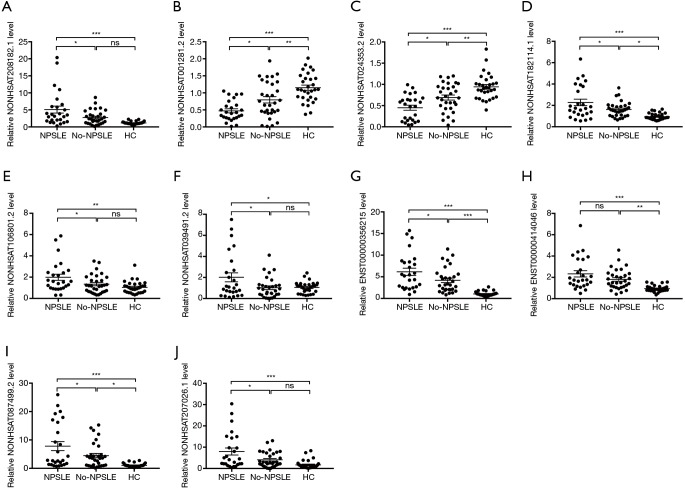
To independently validate our data, DE lncRNAs between NPSLE and Non-NPSLE group were explored using more stringent criteria (as shown in Methods of “Screening of DE lncRNAs for validation”). In this way, 10 aberrantly expressed lncRNAs were selected for qRT-PCR validation in PBMCs from 26 NPSLE patients, 31 Non-NPSLE patients and 30 healthy subjects (Figure 3).
Figure 3.
Relative expression levels of ten selected lncRNAs in PBMCs of NPSLE patients, Non-NPSLE patients and HCs identified using qRT-PCR. (A,D,E,F,G,I,J) the expression levels of seven lncRNAs (NONHSAT208182.1, NONHSAT182114.1, NONHSAT106801.2, NONHSAT039491.2, ENST00000356215, NONHSAT087499.2 and NONHSAT207026.1) were significantly upregulated NPSLE patients compared to Non-NPSLE patients; (B,C) the expression levels of two lncRNAs (NONHSAT001281.2 and NONHSAT024353.2) were significantly downregulated in NPSLE patients compared to Non-NPSLE patients; (H) the difference in lncRNA ENST00000414046 was not significant. n(NPSLE)= 26, n(Non-NPSLE) =31, n (HC) =30. *, P<0.05; **, P<0.01; ***, P<0.001.
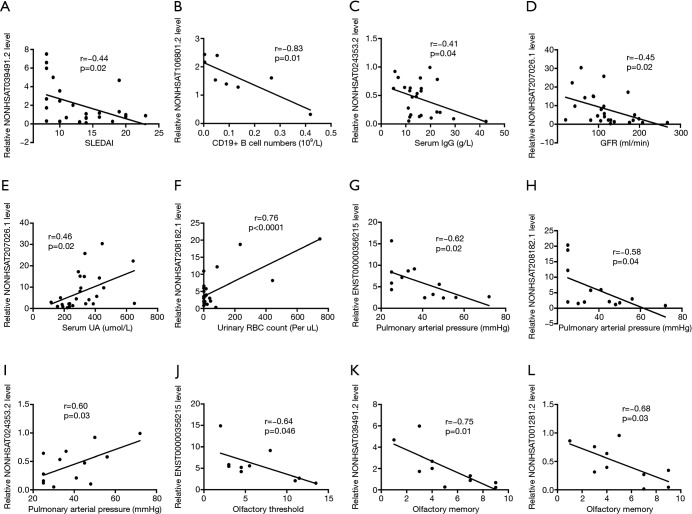
To determine the specific roles of lncRNAs in NPSLE, correlations of these DE lncRNA expression levels with experimental characteristics of NPSLE patients were analyzed. As shown in Figure 4A,B,C,D,E,F, the expression levels of NONHSAT039491.2, NONHSAT106801.2, NONHSAT024353.2 and NONHSAT207026.1 were negatively associated with SLEDAI scores, frequency of CD19+ B cells, serum IgG levels, and glomerular filtration rate (GFR), respectively; whereas NONHSAT207026.1 and NONHSAT208182.1 levels positively correlated with serum uric acid (UA) and urinary red blood cells (RBC) counts, respectively; no significant correlation existed between these DE lncRNAs and characteristics including age, disease duration, serum immunoglobulin (Ig) E, IgA, complement 3, complement 4, C-reactive protein, 24-hour proteinuria, albumin and IgG in cerebrospinal fluid (CSF, the detailed correlation analysis results was shown in Table S3). Further, the role of these DE genes in visceral involvement were analyzed, and found the expression levels of NONHSAT208182.1, NONHSAT024353.2, NONHSAT106801.2 (r=−0.05, P=0.049) and ENST00000356215 were significantly association with pulmonary arterial pressure (PAH) levels of these NPSLE patients (Figure 4G,H,I). Olfactory dysfunction has been confirmed in various central nervous system (CNS) diseases, as well as in NPSLE (18). To explore the roles of lncRNAs in NPSLE, the relationship of lncRNAs with olfactory functions of NPSLE patients was studied, and found negative association of ENST00000356215 with olfactory threshold (Figure 4J), NONHSAT039491.2 and NONHSAT001281.2 with olfactory memory (Figure 4K,L), respectively.
Figure 4.
Correlation between DE lncRNAs and experimental characteristics in NPSLE patients. (A,B,C,D) The expression levels of NONHSAT039491.2, NONHSAT106801.2, NONHSAT024353.2 and NONHSAT207026.1 were negatively associated with systemic lupus erythematosus disease activity index (SLEDAI) scores, frequency of CD19+ B cells, serum IgG levels, and glomerular filtration rate (GFR), respectively. (E,F) NONHSAT207026.1 and NONHSAT208182.1 levels were positively correlated with serum uric acid and urinary red blood cells counts, respectively; (G,H,I) NONHSAT208182.1, NONHSAT024353.2 and ENST00000356215 showed significant association with pulmonary arterial pressure levels of NPSLE patients; (J,K,L) the negative association of ENST00000356215 with olfactory threshold, NONHSAT039491.2 and NONHSAT001281.2 with olfactory memory.
Table S3. Correlation between expression levels of 9 abnormally expressed lncRNAs and clinical characteristics of NPSLE patients.
| Clinical characteristics | r vs. P | NONHSAT208182.1 | NONHSAT001281.2 | NONHSAT024353.2 | NONHSAT182114.1 | NONHSAT106801.2 | NONHSAT039491.2 | ENST00000356215 | NONHSAT087499.2 | NONHSAT207026.1 |
|---|---|---|---|---|---|---|---|---|---|---|
| SLEDAI | r | –0.17 | –0.09 | –0.28 | –0.07 | –0.36 | –0.44 | 0.09 | 0.09 | 0.30 |
| P | 0.40 | 0.65 | 0.16 | 0.72 | 0.07 | 0.02 | 0.66 | 0.67 | 0.14 | |
| Age | r | –0.001 | –0.22 | –0.20 | 0.04 | –0.03 | 0.15 | –0.19 | –0.11 | –0.12 |
| P | 0.99 | 0.27 | 0.32 | 0.86 | 0.89 | 0.47 | 0.35 | 0.58 | 0.56 | |
| Duration of disease | r | 0.24 | 0.26 | 0.01 | 0.02 | 0.07 | 0.35 | 0.08 | –0.07 | –0.23 |
| P | 0.23 | 0.20 | 0.98 | 0.91 | 0.73 | 0.08 | 0.69 | 0.73 | 0.26 | |
| Olfactory threshold | r | –0.25 | –0.35 | 0.23 | –0.33 | –0.21 | 0.43 | –0.64 | –0.16 | 0.09 |
| P | 0.49 | 0.32 | 0.52 | 0.35 | 0.57 | 0.22 | 0.05 | 0.66 | 0.81 | |
| Olfactory memory | r | 0.13 | –0.68 | –0.27 | 0.07 | –0.53 | –0.75 | –0.20 | –0.23 | 0.04 |
| P | 0.73 | 0.03 | 0.44 | 0.86 | 0.11 | 0.01 | 0.58 | 0.52 | 0.92 | |
| WBC (CSF) | r | –0.07 | 0.15 | –0.56 | –0.33 | –0.16 | 0.26 | –0.25 | –0.36 | 0.17 |
| P | 0.82 | 0.63 | 0.06 | 0.29 | 0.62 | 0.42 | 0.43 | 0.26 | 0.60 | |
| Protein (CSF) | r | 0.18 | –0.14 | –0.40 | 0.50 | –0.03 | –0.07 | –0.09 | 0.18 | –0.34 |
| P | 0.58 | 0.65 | 0.20 | 0.10 | 0.92 | 0.82 | 0.78 | 0.58 | 0.28 | |
| Albumin (CSF) | r | 0.36 | 0.08 | –0.36 | 0.50 | 0.29 | 0.01 | 0.04 | 0.28 | –0.39 |
| P | 0.28 | 0.81 | 0.28 | 0.12 | 0.38 | 0.98 | 0.90 | 0.41 | 0.24 | |
| IgG (CSF) | r | 0.01 | –0.22 | –0.36 | 0.39 | –0.26 | –0.21 | –0.10 | 0.03 | –0.39 |
| P | 0.99 | 0.51 | 0.28 | 0.24 | 0.44 | 0.54 | 0.76 | 0.93 | 0.24 | |
| C3 | r | –0.02 | 0.02 | 0.08 | –0.11 | 0.11 | 0.24 | –0.25 | –0.09 | –0.14 |
| P | 0.93 | 0.92 | 0.70 | 0.62 | 0.61 | 0.26 | 0.25 | 0.66 | 0.52 | |
| C4 | r | –0.18 | –0.04 | 0.20 | 0.02 | –0.11 | 0.10 | –0.25 | –0.10 | –0.16 |
| P | 0.41 | 0.84 | 0.34 | 0.93 | 0.60 | 0.64 | 0.24 | 0.64 | 0.46 | |
| Globulin | r | –0.08 | –0.21 | –0.39 | 0.02 | –0.10 | –0.14 | –0.23 | 0.03 | –0.04 |
| P | 0.70 | 0.29 | 0.05 | 0.92 | 0.64 | 0.49 | 0.25 | 0.87 | 0.86 | |
| Albumin | r | 0.09 | 0.26 | 0.22 | –0.20 | 0.29 | 0.38 | 0.06 | –0.16 | –0.02 |
| P | 0.67 | 0.20 | 0.27 | 0.32 | 0.15 | 0.05 | 0.77 | 0.45 | 0.91 | |
| IgG | r | 0.01 | –0.33 | –0.41 | 0.11 | –0.18 | –0.10 | –0.17 | –0.14 | –0.12 |
| P | 0.96 | 0.11 | 0.04 | 0.60 | 0.38 | 0.64 | 0.40 | 0.50 | 0.57 | |
| IgA | r | –0.13 | 0.08 | –0.14 | 0.0008 | –0.03 | –0.08 | –0.20 | 0.26 | 0.07 |
| P | 0.52 | 0.71 | 0.51 | 1.00 | 0.90 | 0.69 | 0.34 | 0.20 | 0.75 | |
| IgM | r | –0.18 | 0.05 | 0.01 | –0.29 | –0.07 | 0.10 | –0.15 | 0.17 | 0.18 |
| P | 0.40 | 0.82 | 0.98 | 0.17 | 0.74 | 0.63 | 0.47 | 0.42 | 0.39 | |
| IgE | r | –0.18 | –0.40 | –0.26 | 0.24 | –0.03 | –0.02 | –0.04 | 0.04 | 0.03 |
| P | 0.40 | 0.05 | 0.21 | 0.24 | 0.87 | 0.91 | 0.86 | 0.85 | 0.88 | |
| 24 h proteinuria | r | 0.22 | –0.19 | –0.37 | –0.02 | –0.28 | –0.30 | 0.10 | 0.11 | 0.40 |
| P | 0.31 | 0.37 | 0.08 | 0.93 | 0.19 | 0.17 | 0.65 | 0.61 | 0.06 | |
| Cr | r | 0.03 | 0.17 | 0.19 | –0.05 | –0.15 | –0.26 | 0.12 | 0.09 | 0.23 |
| P | 0.90 | 0.41 | 0.36 | 0.79 | 0.47 | 0.20 | 0.55 | 0.68 | 0.26 | |
| BUN | r | 0.09 | –0.30 | –0.08 | –0.04 | –0.18 | –0.11 | –0.07 | –0.11 | –0.09 |
| P | 0.68 | 0.13 | 0.71 | 0.85 | 0.37 | 0.58 | 0.72 | 0.58 | 0.66 | |
| UA | r | 0.06 | 0.09 | 0.15 | –0.11 | –0.20 | –0.21 | 0.12 | 0.12 | 0.46 |
| P | 0.79 | 0.65 | 0.47 | 0.60 | 0.33 | 0.29 | 0.54 | 0.57 | 0.02 | |
| GFR | r | –0.03 | –0.18 | –0.02 | –0.08 | 0.19 | 0.15 | –0.30 | –0.14 | –0.45 |
| P | 0.90 | 0.37 | 0.94 | 0.71 | 0.35 | 0.46 | 0.14 | 0.48 | 0.02 | |
| CRP | r | –0.32 | –0.20 | –0.12 | –0.06 | –0.35 | –0.004 | –0.16 | –0.13 | –0.18 |
| P | 0.11 | 0.32 | 0.55 | 0.78 | 0.08 | 0.99 | 0.43 | 0.52 | 0.38 | |
| ESR | r | –0.24 | –0.32 | –0.20 | –0.17 | –0.30 | –0.27 | –0.28 | 0.10 | 0.09 |
| P | 0.26 | 0.12 | 0.35 | 0.41 | 0.15 | 0.19 | 0.18 | 0.63 | 0.66 | |
| WBC | r | –0.09 | 0.29 | –0.05 | –0.14 | –0.11 | 0.07 | 0.11 | –0.21 | –0.09 |
| P | 0.68 | 0.15 | 0.83 | 0.49 | 0.61 | 0.74 | 0.60 | 0.31 | 0.67 | |
| HB | r | 0.03 | 0.21 | 0.35 | –0.15 | 0.22 | 0.38 | –0.19 | –0.04 | –0.02 |
| P | 0.89 | 0.31 | 0.08 | 0.46 | 0.28 | 0.05 | 0.35 | 0.85 | 0.91 | |
| PLT | r | –0.10 | 0.25 | –0.16 | –0.02 | –0.08 | 0.04 | –0.05 | 0.07 | 0.31 |
| P | 0.61 | 0.23 | 0.43 | 0.92 | 0.69 | 0.86 | 0.80 | 0.73 | 0.12 | |
| Urinary RBC | r | 0.72 | 0.26 | –0.32 | 0.24 | 0.30 | –0.04 | 0.17 | 0.35 | –0.10 |
| P | <0.0001 | 0.19 | 0.11 | 0.23 | 0.14 | 0.85 | 0.41 | 0.08 | 0.63 | |
| D-Dmier | r | 0.17 | 0.08 | 0.0008 | –0.16 | –0.09 | 0.05 | –0.23 | –0.19 | –0.14 |
| P | 0.45 | 0.73 | 1.00 | 0.47 | 0.69 | 0.83 | 0.30 | 0.39 | 0.53 | |
| CD4+ T cells | r | –0.60 | 0.18 | 0.28 | –0.24 | –0.14 | 0.08 | –0.51 | 0.05 | –0.03 |
| P | 0.12 | 0.67 | 0.50 | 0.56 | 0.74 | 0.84 | 0.20 | 0.90 | 0.94 | |
| CD8+ T cells | r | –0.55 | 0.08 | 0.27 | 0.29 | 0.06 | 0.36 | 0.03 | –0.03 | –0.16 |
| P | 0.15 | 0.84 | 0.52 | 0.48 | 0.89 | 0.38 | 0.95 | 0.94 | 0.71 | |
| NK cells | r | 0.11 | 0.36 | 0.35 | 0.05 | 0.41 | 0.58 | –0.05 | –0.26 | 0.002 |
| P | 0.79 | 0.38 | 0.40 | 0.90 | 0.31 | 0.13 | 0.91 | 0.53 | 1.00 | |
| B cells | r | –0.44 | –0.31 | –0.58 | 0.04 | –0.83 | –0.00008 | –0.45 | –0.04 | –0.01 |
| P | 0.79 | 0.38 | 0.40 | 0.90 | 0.31 | 0.13 | 0.91 | 0.53 | 1.00 | |
| PAH | r | –0.58 | –0.41 | 0.60 | –0.41 | –0.55 | –0.08 | –0.62 | –0.52 | –0.46 |
| P | 0.04 | 0.16 | 0.03 | 0.17 | 0.05 | 0.80 | 0.02 | 0.07 | 0.12 | |
| CFH | r | –0.10 | 0.08 | 0.31 | –0.32 | –0.03 | –0.50 | 0.43 | 0.21 | 0.03 |
| P | 0.24 | 0.45 | 0.14 | 0.21 | 0.13 | 0.12 | 0.08 | 0.13 | 0.90 | |
| P2RY14 | r | –0.34 | 0.23 | 0.32 | 0.34 | –0.34 | –0.38 | 0.42 | –0.06 | –0.32 |
| P | 0.24 | 0.34 | 0.35 | 0.12 | 0.09 | 0.34 | 0.23 | 0.92 | 0.13 | |
| VNN1 | r | 0.34 | 0.24 | 0.03 | 0.08 | 0.23 | 0.53 | –0.23 | –0.024 | 0.03 |
| P | 0.22 | 0.25 | 0.48 | 0.83 | 0.32 | 0.39 | 0.07 | 0.98 | 0.83 | |
| IDO1 | r | –0.61 | –0.09 | –0.28 | 0.08 | –0.43 | 0.09 | 0.32 | –0.05 | –0.54 |
| P | 0.02 | 0.19 | 0.32 | 0.92 | 0.09 | 0.84 | 0.34 | 0.67 | 0.09 | |
| BCL2A1 | r | 0.45 | 0.38 | 0.42 | 0.37 | –0.33 | 0.93 | –0.92 | –0.43 | –0.42 |
| P | 0.03 | 0.05 | 0.32 | 0.22 | 0.06 | 0.10 | 0.35 | 0.93 | 0.23 | |
| KCNJ15 | r | –0.34 | 0.54 | 0.23 | –0.84 | –0.03 | 0.23 | –0.33 | 0.45 | –0.32 |
| P | 0.50 | 0.14 | 0.06 | 0.32 | 0.94 | 0.24 | 0.09 | 0.34 | 0.32 |
The r and P values of nonparametric analysis are listed. SLEDAI, SLE disease activity index; yrs, years; NPSLE, neuropsychiatric systemic lupus erythematosus; WBC (CSF), white blood cells in cerebrospinal fluid; Protein (CSF), protein in cerebrospinal fluid; Albumin (CSF), albumin in cerebrospinal fluid; IgG (CSF), immunoglobulin G in cerebrospinal fluid; C3, complement 3; C4, complement 4; IgG, immunoglobulin G; IgA, immunoglobulin A; IgM, immunoglobulin M; IgE, immunoglobulin E; 24 h proteinuria, 24 hour proteinuria; Cr, creatinine; BUN, urea nitrogen; UA, uric acid; GFR, glomerular filtration rate; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; WBC, white blood cells; HB, hemoglobin; PLT, platelet; Urinary RBC, urinary red blood cells; NK cells, natural killer cell; PAH, pulmonary hypertension; CFH, complement factor H; P2RY14, purinergic receptor P2Y14; VNN1, vanin 1; IDO1, Indoleamine 2,3-dioxygenase 1; BCL2A1, BCL2 related protein A1; KCNJ15, Potassium voltage-gated channel subfamily J member 15.
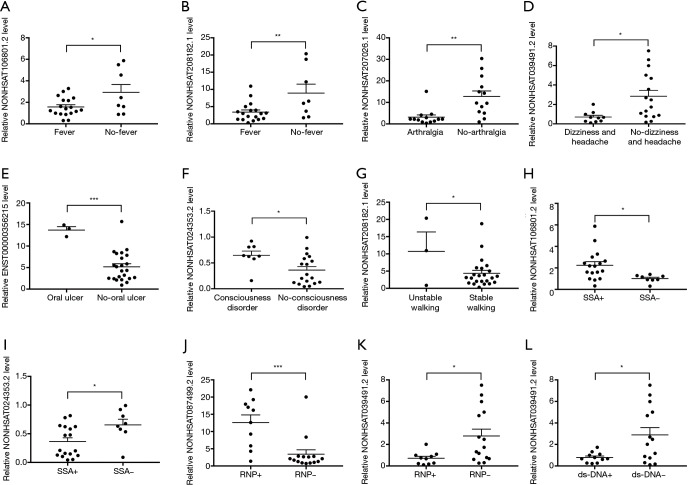
Given the multitude of clinical manifestations related to NPSLE, the correlations of expression levels of DE lncRNAs with clinical manifestations were analyzed. Compared with patients without such symptoms, expression levels of NONHSAT106801.2 and NONHSAT208182.1, NONHSAT207026.1, and NONHSAT039491.2 were significantly downregulated in those with fever (Figure 5A,B), arthralgia (Figure 5C), dizziness and headache (Figure 5D), respectively; while expression levels of ENST00000356215, NONHSAT024353.2 and NONHSAT208182.1 were upregulated in patients with oral ulcer (Figure 5E), consciousness disorder (Figure 5F) and unstable walking (Figure 5G), respectively.
Figure 5.
Correlation between DE lncRNAs and clinical characteristics in NPSLE patients. (A,B) NONHSAT106801.2 and NONHSAT208182.1 expression levels in NPSLE patients with and without fever; (C) NONHSAT207026.1 expression level in NPSLE patients with and without arthralgia; (D) NONHSAT039491.2 expression level in NPSLE patients with and without dizziness and headache symptom; (E) ENST00000356215 expression level in NPSLE patients with and without oral ulcer; (F) NONHSAT024353.2 expression level in NPSLE patients with and without consciousness disorder; (G) NONHSAT208182.1 expression level in NPSLE patients with and without unstable walking symptom; (H,I) NONHSAT106801.2 and NONHSAT024353.2 expression levels in anti-SSA+ and anti-SSA- NPSLE patients; (J,K) NONHSAT087499.2 and NONHSAT039491.2 expression levels in anti-RNP+ and anti-RNP- NPSLE patients; (L) NONHSAT039491.2 expression level in anti-dsDNA+ and anti-dsDNA- NPSLE patients. *, P<0.05; **, P<0.01; ***, P<0.001.
Available studies on the significance of different autoantibodies in NPSLE have shown controversial results (19). In our study, NONHSAT106801.2 was significantly upregulated while NONHSAT024353.2 was downregulated in anti-SSA+ patients compared to anti-SSA- patients (Figure 5H,I); NONHSAT087499.2 was significantly upregulated in anti-ribonucleoprotein antibody (anti-RNP)+ patients compared to anti-RNP- patients while NONHSAT039491.2 was down-regulated both in anti-RNP+ and anti-dsDNA+ patients compared to anti-RNP- and anti-dsDNA- patients, respectively (Figure 5J,K,L); no difference of lncRNA levels was found between groups positive and negative for anti-cardiolipin antibody, anti-nuclear antibody, anti-smith antibody, anti-histone antibody, anti-ribosomal P antibody and anti-nucleosome antibody, respectively.
Considering that clinical treatment including corticosteroids and immunosuppressants may affect the expression of lncRNAs, we analyzed the relationship between the expression level of lncRNA and the therapeutic dosage of glucocorticoid (GLU) and immunosuppressive agents including hydroxychloroquine (HCQ), cyclophosphamide (CYC), and mycophenolate (MMF). It was found that only NONHSAT024353.2 was highly expressed in CTX-treated patients compared with patients without CTX treatment (0.232±0.07019, n=9 vs. 0.5652±0.06684, n=17, P=0.0042), but no relationship was found in the expression of other lncRNAs between immunosuppressive and non-immunosuppressive group, as well as different doses of corticosteroids treatment group (Table S4), these above findings indicating that these abnormally expressed lncRNAs between NPSLE and Non-NPSLE group might not be influenced by clinical therapy.
Table S4. Correlation between expression levels of 9 abnormally expressed lncRNAs and clinical treatments of NPSLE patients.
| Clinical treatment | r vs. P | NONHSAT208182.1 | NONHSAT001281.2 | NONHSAT024353.2 | NONHSAT182114.1 | NONHSAT106801.2 | NONHSAT039491.2 | ENST00000356215 | NONHSAT087499.2 | NONHSAT207026.1 |
|---|---|---|---|---|---|---|---|---|---|---|
| Pred | r | 0.27 | 0.27 | -0.15 | -0.09 | -0.02 | -0.01 | 0.03 | -0.17 | -0.22 |
| P | 0.18 | 0.19 | 0.45 | 0.66 | 0.93 | 0.98 | 0.90 | 0.41 | 0.29 | |
| HCQ | r | -0.09 | 0.10 | 0.13 | -0.05 | -0.05 | -0.24 | 0.14 | 0.34 | 0.35 |
| P | 0.68 | 0.63 | 0.52 | 0.80 | 0.81 | 0.25 | 0.49 | 0.09 | 0.08 | |
| CTX | r | 0.35 | 0.17 | -0.54 | 0.05 | -0.19 | -0.21 | 0.27 | 0.12 | 0.21 |
| P | 0.08 | 0.40 | 0.004 | 0.81 | 0.36 | 0.31 | 0.19 | 0.57 | 0.29 | |
| MMF | r | -0.004 | -0.13 | 0.16 | -0.30 | -0.01 | -0.09 | -0.07 | -0.21 | -0.20 |
| P | 0.99 | 0.52 | 0.44 | 0.13 | 0.98 | 0.67 | 0.73 | 0.30 | 0.32 |
The r and P values of nonparametric analysis are listed. GLU, glucocorticoid; HCQ, hydroxychloroquine; CYC, cyclophosphamide; MMF, mycophenolate.
GO and KEGG pathway analyses of DE mRNAs
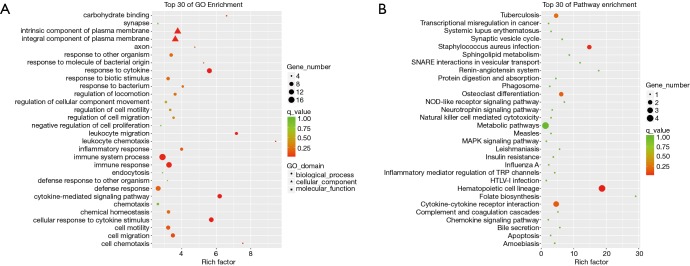
Next, to further examine the potential mechanism of NPSLE, GO enrichment analysis was performed to investigate the over-representation of biological processes, cellular components, and specific molecular function associating protein-coding mRNAs. As shown in Figure 6, the top 30 GO from the DE mRNAs were listed. Our data demonstrated that the most significantly enriched cellular components of DE mRNAs in PBMCs of NPSLE were the external side of the axon, intrinsic and integral component of plasma membrane (Figure 6A, http://cdn.amegroups.cn/static/other/5984c3b0471eb93f988c76e7878328c7/atm.2020.03.25-7.xls). The most significantly enriched molecular functions of DE mRNAs were carbon hydrate binding, protein homodimerization activity and substrate-specific transmembrane transporter activity (Figure 6A, http://cdn.amegroups.cn/static/other/5984c3b0471eb93f988c76e7878328c7/atm.2020.03.25-7.xls); The most significantly enriched biological processes of DE mRNAs were leukocyte chemotaxis and migration, and cell chemotaxis (Figure 6A, http://cdn.amegroups.cn/static/other/5984c3b0471eb93f988c76e7878328c7/atm.2020.03.25-7.xls); these together reveal the roles of significantly dysregulated mRNAs in NPSLE.
Figure 6.
Enrichment analysis of GO terms and pathways for DE mRNAs between NPSLE and Non-NPSLE group. (A) The top 30 GO analyses which consisted of significant molecular function, cellular component and biological process of DE mRNAs; (B) significantly enriched pathways of DE mRNAs. P <0.01, FC >2.0 or <−2.0.
KEGG pathway enrichment analysis for significantly dysregulated mRNAs is useful to reveal related pathways and molecular interactions among genes. The results revealed that the DE mRNAs in PBMCs of NPSLE were significantly involved in hematopoietic cell lineage, staphylococcus aureus infection, and osteoclast differentiation (Figure 6B, http://cdn.amegroups.cn/static/other/a916f2ba1dfe9c79658b5d5b12c5539b/atm.2020.03.25-8.xls). Our data showed that the DE mRNAs were each individually associated with 11 pathways, among them, the metabolic pathway, neurotrophin signaling pathway, and chemokine signaling pathway were three of the most enriched pathways within the set of DE mRNAs (Figure 6B, http://cdn.amegroups.cn/static/other/a916f2ba1dfe9c79658b5d5b12c5539b/atm.2020.03.25-8.xls), suggesting that these pathways may be involved in the development of NPSLE.
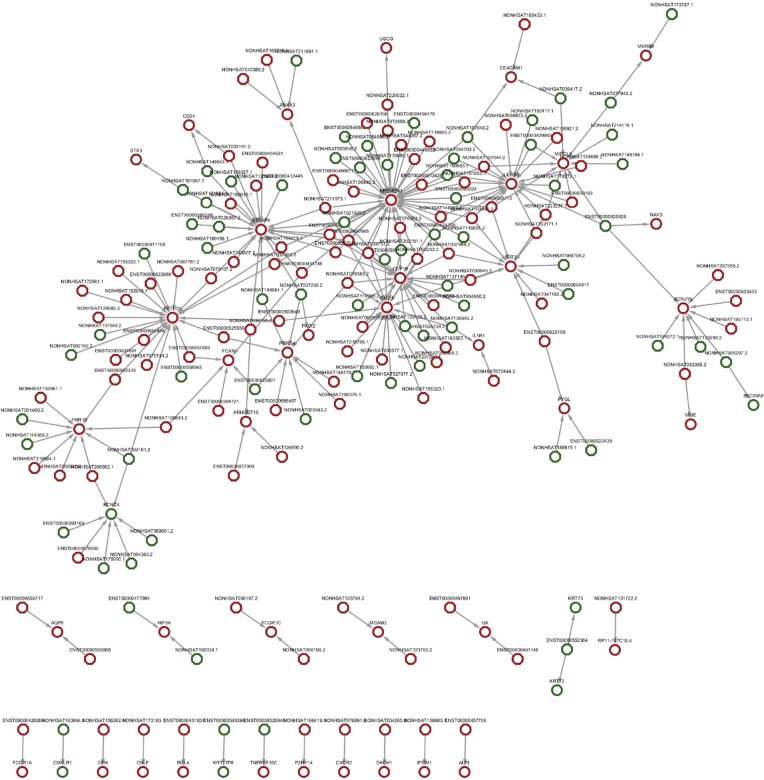
lncRNA-mRNA co-expression network
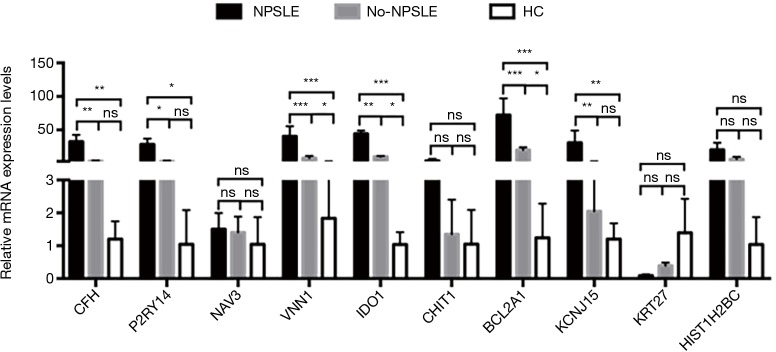
Co-expression network analysis was performed between the 193 DE lncRNAs and the 49 DE mRNAs based on the Max-relevance Min-redundancy (mRMR) method. In total, 170 lncRNAs and 46 mRNAs were included in the co-expression network (Figure 7, http://cdn.amegroups.cn/static/other/a7791bdae56436daf1f955a9b6288064/atm.2020.03.25-9.xls). In addition, our data showed that the co-expression network was composed of 216 network nodes and 251 connections. To further address DE mRNAs as possible predicted target of these DE lncRNAs, the expression levels of top 10 DE mRNAs that have characterized proteins were performed using RT-PCR. Compared with that of Non-NPSLE patients, the relative expression levels of CFH, P2RY14, VNN1, IDO1, BCL2A1and KCNJ15 were significantly increased in PBMCs from NPSLE patients (Figure S1). Considering of the co-expression network of DE lncRNAs with DE mRNAs (http://cdn.amegroups.cn/static/other/a7791bdae56436daf1f955a9b6288064/atm.2020.03.25-9.xls), and the negative correlated role of IDO1 and BCL2A1 with NONHSAT001281.2 (Table S3), our data support that differential lncRNAs might operate via modulating expressions of their correlated, relevant mRNAs in NPSLE.
Figure 7.
Network graph of DE lncRNA-related mRNAs between NPSLE and Non-NPSLE group. Octagon and circles represent DE lncRNAs and their related DE genes, respectively. Red and green represent upregulated and downregulated lncRNAs or genes, respectively. P<0.01, FC >2.0 or <−2.0.
Figure S1.
The expression levels of DE mRNAs in PBMCs from NPSLE, Non-NPSLE, and healthy subjects. All data are means ± SEM. n (NPSLE) =26, n (Non-NPSLE) =31, n (HC) =30. *, P<0.05; **, P<0.01; ***, P<0.001.
Discussion
Recent discoveries have revealed that lncRNAs are aberrantly expressed in a broad spectrum of autoimmune diseases (AD) including SLE (14,15,20), and thus, play key roles in promoting and maintaining AD initiation and progression, demonstrating their clinical potential as biomarkers and therapeutic targets. Studies have shown that serum lnc-gas5, lnc0597, lnc-DC could be used as diagnostic markers for SLE (12). LncRNA NEAT1 acted as an inflammatory regulator to activate the MAPK pathway involved in SLE pathogenesis (21). LncRNA MALAT1 could affect the expression level of IL-21 in monocytes by regulating SIRT1 signaling and participate in SLE development (13). lncRNA-Gas5 binds to the DNA binding region of the glucocorticoid receptor, preventing the receptor from interacting with the glucocorticoid response element of genomic DNA, thereby affecting the patient's response to glucocorticoids (22). These above findings supported the pathogenic role of lncRNAs in SLE pathogenesis (23), but the roles of lncRNAs in NPSLE remain largely unknown.
Growing evidence showed that blood–brain barrier (BBB) dysfunction may be essential to the development of NPSLE (24,25), allowing the passive diffusion of auto-reactive antibodies and cytokines in CSF of SLE patients. Considering of the specific link between PBMCs, serum and CSF (26), CSF are rare, difficult to obtain and traumatic, while peripheral blood is easy to obtain, with more clinical utility. In our study, compared with that of Non-NPSLE patients, hundreds of DE lncRNAs were found by RNA-seq and 10 of them were validated by qRT-PCR in PBMCs from NPSLE patients (Figure 3, Figure S1), and the correlation of expression levels of DE lncRNAs with experimental indicators in CSF from NPSLE patients were explored. Unfortunately, there was no significant correlation of expression levels of lncRNA with protein or IgG levels in CSF of NPSLE patients (Table S3). Since protein or IgG levels in CSF positively associated with SLEDAI of NPSLE patients, while in our study, none of these lncRNAs displayed significantly association with SLEDAI except NONHSAT039491.2, thus the link between these lncRNAs with NPSLE activity still needs further investigation.
Multitude of clinical manifestations were related to NPSLE, each syndrome may present various different patterns, which increases the complexity of clinical outcomes (27). In our study, the expression levels of NONHSAT024353.2, NONHSAT039491.2, NONHSAT208182.1 were associated with clinical index of consciousness, dizziness and headache, as well as unstable walking, respectively (Figure 5D,F,G). To predict their possible target gene, except for lncRNA-mRNA co-expression network analysis (Figure 7), the correlations of DE lncRNAs with DE mRNAs confirmed by RT-PCR were analyzed, and found negative correlation of expression levels IDO1 and BCL2A1 with that of NONHSAT208182.1 in PBMCs from NPSLE patients (Table S3), together with the positive association of NONHSAT208182.1 levels with unstable walking (Figure 5G), it is likely that NONHSAT208182.1 may be involved in the development of unstable walking related-NPSLE through targeting IDO1 or BCL2A1, further RNA-pulldown and CHIP experiments will be performed to support the possible target of NONHSAT001281.2 in our next experiments.
Olfactory disturbance was one of the manifestations of NPSLE (28). Recently, the correlation between decreased olfactory abilities and disease activity, especially CNS manifestations of NPSLE patients is supported by increasing studies, indicating that smell decrement might be a useful and easy tool for the physician in early diagnosis of CNS involvement in autoimmune diseases (18). Interestingly, negative association of ENST00000356215 with olfactory threshold, NONHSAT039491.2 and NONHSAT001281.2 with olfactory memory were found in our NPSLE cohort. Since accumulating studies found that decreased sense of smell was associated with history of neuropsychiatric manifestations and higher disease activity, ENST00000356215, NONHSAT039491.2 and NONHSAT001281.2 may be used as predictors of diffuse disorders in NPSLE patients but still need further investigation.
As one of the major causes of death in SLE, SLE related PAH could also be postulated as vasculitis led by immune-mediated vasculopathy (29), a potential link between PAH and NPSLE manifestations, however, have never been studied. In our study, NONHSAT208182.1, NONHSAT024353.2, NONHSAT106801.2 and ENST00000356215 all showed significant association with PAH levels of these NPSLE patients. Furthermore, NONHSAT106801.2 and NONHSAT024353.2 levels were also associated with the presence of anti-SSA, which had been reported to be an independent variable with PAH in SLE (30), supporting that some specific lncRNAs together with autoantibodies could be recommended in early diagnosis and precise management of NPSLE-associated PAH patients.
Auto-antibody mediated vasculopathy is one of the mechanisms for NPSLE (19). To date, more than 116 auto-antibodies have been reported in SLE, and at least 20 of them, including 11 brain-specific and 9 systemic auto-antibodies, have been controversially associated with NPSLE. Among these auto-antibodies, ACA and anti-RNP were significantly associated with specific manifestations of neuropsychiatric disease attributed to SLE (19). In our study, our data support important correlations of NONHSAT106801.2, NONHSAT024353.2, NONHSAT039491.2 and NONHSAT087499.2 in NPSLE patients with some specific autoantibodies including anti-SSA, anti-RNP, and anti-dsDNA, respectively (Figure 5H,I,J,K,L). Although the detection of auto-antibodies in serum or cerebrospinal fluid has not been included as a diagnostic marker within the ACR classification (19,27), our data indicate that lncRNAs together with their specific autoantibodies might be used for precise management of NPSLE. Various inflammatory cytokines and chemokines, including IL-6, IL-1, IL-8, IL-10, tumor necrosis factor-α (TNF-α), interferon-α (IFN-α), IP-10/CXCL10, MCP-1/CCL2, normal T-cell expressed and secreted (RANTES)/CCL5 as well as CX3CR1, have been reported to increase the permeability of the BBB, providing access for circulating autoantibodies and leukocytes into the CNS, and thus, involved in the pathogenesis of NPSLE (31-33). It is worth noting that, several GO terms were enriched in gene lists through a bioinformatics analysis. Since lncRNAs have been reported to play important roles in immune cell homeostasis (34), and chemokines are crucial for NPSLE development, our results suggest important effects of these lncRNAs together with chemokines in the diagnosis of NPSLE.
Plenty of studies have shown that lncRNAs are expressed in a highly lineage-specific manner. Cao et al. elucidated for the first time that DCs-specific lncRNA lnc-DC can inhibit SHP1-mediated de-phosphorylation by directly binding to the structural region near the STAT3 phosphorylation site Tyr705, thereby enhancing STAT3 signaling and participating in DCs differentiation and functional maintenance (35). Chen et al. found that PU.1 regulated lnc-MC can regulate mononuclear macrophage differentiation by "sponge" adsorption of miRNA-199a-5p (36). In our study, compared with that of Non-NPSLE patients, NONHSAT208182.1 and NONHSAT087499.2 were specifically highly expressed while NONHSAT001281.2 were reduced in CD19+ B cells from NPSLE patients, NONHSAT182.114.1 and ENST00000356215 were specifically highly expressed in CD4+ T cells, while NONHSAT024353.2 were significantly reduced in CD19+ B cells and NK cells (data not shown), supporting that DE lncRNAs may play a role in the development of NPSLE by affecting different subsets of PBMCs from NPSLE patients.
In summary, to the best of our knowledge, this is the first study that describes the expression profiles of human lncRNAs in NPSLE using RNA-seq. Altered lncRNAs may play potential roles in the pathogenesis and/or development of “the most clinical challenging ‘visceral’ involvement of SLE”. To accurately and comprehensively elucidate the role of lncRNAs in NPSLE, more comprehensive laboratory and clinical researches will be needed to confirm whether these lncRNAs play essential roles in the pathogenesis, treatment, and prognosis of NPSLE.
Conclusions
We have demonstrated a comprehensive lncRNA expression profile in PBMCs of NPSLE patients, whose deregulation could be related to the pathophysiology of the disease. More specifically, this study has highlighted some pathways that could be crucial in the onset or the maintenance of the disease. Our finding shed light on the understanding of the molecular mechanisms of lncRNAs in the pathogenesis of NPSLE.
Acknowledgments
The authors thank the patients who were involved in this study and the individuals from the rheumatology departments who took part in the care of these patients.
Funding: The work was supported by the Major International (Regional) Joint Research Project (No. 81720108020), National Natural Science Foundation of China (No. 81871283, 81571586, 81501347 and 81601418), Research Project of Jiangsu Province Health Committee (H2019060), National Natural Science Foundation of Jiangsu (BK20150098), Jiangsu Provincial Special Program of Medical Science (BE2015602), Medical Science and Technology Development Key Project of Nanjing (ZKX15018), and Jiangsu Province Six Talent Peaks Project (No. 2016-WSN-148).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of the Affiliated Drum Tower Hospital of Nanjing University Medical School (SC201700201) and was conducted in accordance with the principle set forth under the 1989 Declaration of Helsinki.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Tsokos GC, Lo MS, Costa Reis P, et al. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat Rev Rheumatol 2016;12:716-30. 10.1038/nrrheum.2016.186 [DOI] [PubMed] [Google Scholar]
- 2.Govoni M, Bortoluzzi A, Padovan M, et al. The diagnosis and clinical management of the neuropsychiatric manifestations of lupus. J Autoimmun 2016;74:41-72. 10.1016/j.jaut.2016.06.013 [DOI] [PubMed] [Google Scholar]
- 3.Sharaf-Eldin WE, Kishk NA, Gad YZ, et al. Extracellular miR-145, miR-223 and miR-326 expression signature allow for differential diagnosis of immune-mediated neuroinflammatory diseases. J Neurol Sci 2017;383:188-98. 10.1016/j.jns.2017.11.014 [DOI] [PubMed] [Google Scholar]
- 4.Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 2018;19:143-57. 10.1038/nrm.2017.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arun G, Diermeier SD, Spector DL. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol Med 2018;24:257-77. 10.1016/j.molmed.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang DQ, Fu P, Yao C, et al. Long Non-coding RNAs, Novel Culprits, or Bodyguards in Neurodegenerative Diseases. Mol Ther Nucleic Acids 2018;10:269-76. 10.1016/j.omtn.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Rosa S, Arcidiacono B, Chiefari E, et al. Type 2 Diabetes Mellitus and Cardiovascular Disease: Genetic and Epigenetic Links. Front Endocrinol (Lausanne) 2018;9:2. 10.3389/fendo.2018.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol 2017;18:962-72. 10.1038/ni.3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ranzani V, Rossetti G, Panzeri I, et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol 2015;16:318-25. 10.1038/ni.3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li LJ, Zhao W, Tao SS, et al. Comprehensive long non-coding RNA expression profiling reveals their potential roles in systemic lupus erythematosus. Cell Immunol 2017;319:17-27. 10.1016/j.cellimm.2017.06.004 [DOI] [PubMed] [Google Scholar]
- 11.Luo Q, Li X, Xu C, et al. Integrative analysis of long non-coding RNAs and messenger RNA expression profiles in systemic lupus erythematosus. Mol Med Rep 2018;17:3489-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu GC, Li J, Leng RX, et al. Identification of long non-coding RNAs GAS5, linc0597 and lnc-DC in plasma as novel biomarkers for systemic lupus erythematosus. Oncotarget 2017;8:23650-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Liang N, Wang M, et al. Long noncoding RNA MALAT-1 is a novel inflammatory regulator in human systemic lupus erythematosus. Oncotarget 2017;8:77400-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Y, Zhou T, Yu X, et al. The role of long non-coding RNAs in rheumatic diseases. Nat Rev Rheumatol 2017;13:657-69. 10.1038/nrrheum.2017.162 [DOI] [PubMed] [Google Scholar]
- 15.Wu GC, Pan HF, Leng RX, et al. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev 2015;14:798-805. 10.1016/j.autrev.2015.05.004 [DOI] [PubMed] [Google Scholar]
- 16.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997;40:1725. 10.1002/art.1780400928 [DOI] [PubMed] [Google Scholar]
- 17.The American College of Rheumatology nomenclature and case definitions for neuropsychiatric lupus syndromes. Arthritis Rheum 1999;42:599-608. [DOI] [PubMed] [Google Scholar]
- 18.Shoenfeld N, Agmon-Levin N, Flitman-Katzevman I, et al. The sense of smell in systemic lupus erythematosus. Arthritis Rheum 2009;60:1484-7. 10.1002/art.24491 [DOI] [PubMed] [Google Scholar]
- 19.Sciascia S, Bertolaccini ML, Roccatello D, et al. Autoantibodies involved in neuropsychiatric manifestations associated with systemic lupus erythematosus: a systematic review. J Neurol 2014;261:1706-14. 10.1007/s00415-014-7406-8 [DOI] [PubMed] [Google Scholar]
- 20.Li J, Wu GC, Zhang TP, et al. Association of long noncoding RNAs expression levels and their gene polymorphisms with systemic lupus erythematosus. Sci Rep 2017;7:15119. 10.1038/s41598-017-15156-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F, Wu L, Qian J, et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun 2016;75:96-104. 10.1016/j.jaut.2016.07.012 [DOI] [PubMed] [Google Scholar]
- 22.Mayama T, Marr AK, Kino T. Differential Expression of Glucocorticoid Receptor Noncoding RNA Repressor Gas5 in Autoimmune and Inflammatory Diseases. Horm Metab Res 2016;48:550-7. 10.1055/s-0042-106898 [DOI] [PubMed] [Google Scholar]
- 23.Long H, Yin H, Wang L, et al. The critical role of epigenetics in systemic lupus erythematosus and autoimmunity. J Autoimmun 2016;74:118-38. 10.1016/j.jaut.2016.06.020 [DOI] [PubMed] [Google Scholar]
- 24.Gelb S, Stock AD, Anzi S, et al. Mechanisms of neuropsychiatric lupus: The relative roles of the blood-cerebrospinal fluid barrier versus blood-brain barrier. J Autoimmun 2018;91:34-44. 10.1016/j.jaut.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirohata S, Sakuma Y, Matsueda Y, et al. Role of serum autoantibodies in blood brain barrier damages in neuropsychiatric systemic lupus erythematosus. Clin Exp Rheumatol 2018;36:1003-7. [PubMed] [Google Scholar]
- 26.Yoshio T, Okamoto H, Kurasawa K, et al. IL-6, IL-8, IP-10, MCP-1 and G-CSF are significantly increased in cerebrospinal fluid but not in sera of patients with central neuropsychiatric lupus erythematosus. Lupus 2016;25:997-1003. 10.1177/0961203316629556 [DOI] [PubMed] [Google Scholar]
- 27.Faria R, Goncalves J, Dias R. Neuropsychiatric Systemic Lupus Erythematosus Involvement: Towards a Tailored Approach to Our Patients? Rambam Maimonides Med J 2017. doi: . 10.5041/RMMJ.10276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bombini MF, Peres FA, Lapa AT, et al. Olfactory function in systemic lupus erythematosus and systemic sclerosis. A longitudinal study and review of the literature. Autoimmun Rev 2018;17:405-12. 10.1016/j.autrev.2018.02.002 [DOI] [PubMed] [Google Scholar]
- 29.Tselios K, Gladman DD, Urowitz MB. Systemic lupus erythematosus and pulmonary arterial hypertension: links, risks, and management strategies. Open Access Rheumatol 2016;9:1-9. 10.2147/OARRR.S123549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang C, Li M, Liu Y, et al. Baseline Characteristics and Risk Factors of Pulmonary Arterial Hypertension in Systemic Lupus Erythematosus Patients. Medicine (Baltimore) 2016;95:e2761. 10.1097/MD.0000000000002761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fragoso-Loyo H, Richaud-Patin Y, Orozco-Narvaez A, et al. Interleukin-6 and chemokines in the neuropsychiatric manifestations of systemic lupus erythematosus. Arthritis Rheum 2007;56:1242-50. 10.1002/art.22451 [DOI] [PubMed] [Google Scholar]
- 32.Guo L, Lu X, Wang Y, et al. Elevated levels of soluble fractalkine and increased expression of CX3CR1 in neuropsychiatric systemic lupus erythematosus. Exp Ther Med 2017;14:3153-8. 10.3892/etm.2017.4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pranzatelli MR. Advances in Biomarker-Guided Therapy for Pediatric- and Adult-Onset Neuroinflammatory Disorders: Targeting Chemokines/Cytokines. Front Immunol 2018;9:557. 10.3389/fimmu.2018.00557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mowel WK, Kotzin JJ, McCright SJ, et al. Control of Immune Cell Homeostasis and Function by lncRNAs. Trends Immunol 2018;39:55-69. 10.1016/j.it.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P, Xue Y, Han Y, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014;344:310-3. 10.1126/science.1251456 [DOI] [PubMed] [Google Scholar]