Abstract
Background
Type 2 diabetes mellitus (T2DM) and hepatocellular carcinoma (HCC) are both major health problems throughout the world. It has been reported that T2DM is an independent risk factor for HCC, although the pathophysiology is still unclear.
Methods
In order to identify differentially expressed genes (DEGs) in T2DM and HCC, gene expression datasets for T2DM (GSE15653), HCC (GSE60502) and metformin-treated cells (GSE69850) were obtained from the Gene Expression Omnibus database repository. Protein-protein interaction (PPI) networks for the DEGs were constructed and gene clusters selected for functional enrichment analysis. Ten genes with the highest degree of connectivity were selected as hub genes and prognostic analysis together with analysis of gene expression and protein distribution were performed for these genes. Lastly, we investigated associations between the hub genes and genes associated with metformin treatment in hepatocarcinoma cells.
Results
In total, 256 common DEGs, including 155 up-regulated genes and 101 down-regulated genes, were identified. Enrichment analyses showed that the genes of the major module were largely associated with the cell cycle. All of the 10 hub genes (CCNA2, CCNB1, MAD2L1, BU1B, RACGAP1, CHEK1, BUB1, ASPM, NCAPG and TTK) have a strong association with lower overall survival in liver cancer patients and four genes (CCNA2, CCNB1, CHEK1 and BUB1) have reduced expression in metformin-treated samples.
Conclusions
This study identified a number of genes that may play important roles in the association of T2DM and HCC, including four genes which may be the target of metformin treatment for diabetes and HCC. The specific mechanisms involved remain to be identified.
Keywords: Type 2 diabetes mellitus (T2DM), hepatocellular carcinoma (HCC), differentially expressed gene, Metformin
Introduction
Globally, liver cancers rank seventh for cancer incidence while their mortality ranks fourth among all types of cancer (1). Hepatocellular carcinoma (HCC) is a malignant tumor originating from hepatocytes and it is the most common cancer in the liver. Chronic viral hepatitis and alcoholism are the two main causes of HCC. Non-alcoholic fatty liver disease (NAFLD) is also recognized as an important risk factor for HCC due to the increasing prevalence of obesity and Type 2 diabetes mellitus (T2DM) (2). T2DM is a disease of the endocrine system and is common, with one T2DM patient per every 11 adults (3). Recently, there have been a number of reports researches showing that T2DM has a negative influence on the survival of HCC patients (4,5).
Metformin is frequently used to treat hyperglycemia associated with T2DM and has also been reported to have anti-hepatocellular carcinoma effects (6). However, most of these studies have focused on epidemiology with only a few concentrating on possible genetic connections between HCC and T2DM.
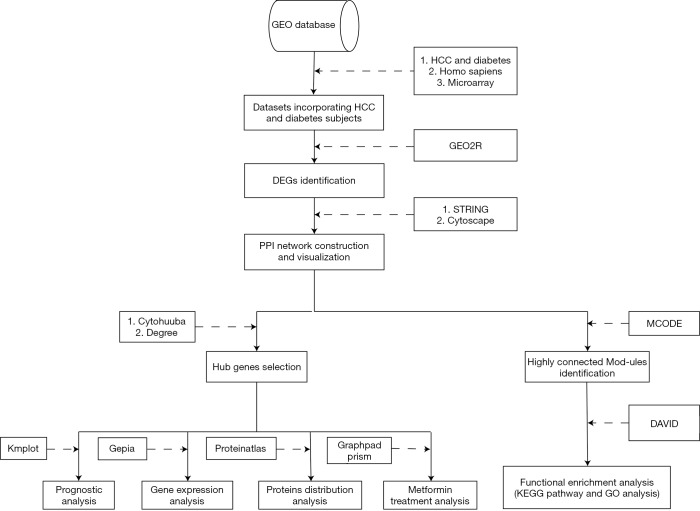
We, therefore, undertook this study to explore the specific mechanism of the association between T2DM and HCC. We analyzed the gene expression profiles of T2DM and HCC tissue to identify differentially expressed genes (DEGs) in both diseases, followed by the construction of protein-protein interaction (PPI) networks for genes observed to be up-regulated or down-regulated in both T2DM and HCC. After construction of the PPI network, a gene cluster would be selected for functional enrichment analyses. Ten genes with the highest degree of connectivity were selected as hub genes. Prognostic analysis, gene expression analysis and protein distribution analysis were performed on the hub genes. We also investigated associations between the hub genes and genes implicated in metformin treatment for HCC. Lastly, we discuss our results in relation to published findings in an attempt to elucidate the relationship between T2DM and HCC.
Methods
Microarray data
In order to avoid errors caused by platform differences, the datasets containing T2DM samples and HCC samples should use the same platform. Two gene expression datasets GSE15653 (GPL96; T2DM) and GSE60502 (GPL96; HCC) were chosen from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database repository. In addition, the dataset GSE69850 (GPL13667), a HepG2 hepatocarcinoma cell dataset which contains metformin-treated samples, was also downloaded from the GEO (Table 1).
Table 1. Description of data used in this study.
| Accession | Platform | Normal/control | Cancer/treat | Type |
|---|---|---|---|---|
| GSE15653 | GPL96 | 5 normal | 4 diabetes | Liver tissues |
| GSE60502 | GPL96 | 18 normal | 18 cancer | HCC tissues |
| GSE69850 | GPL13667 | 39 control | 9 Metformin treated | HepG2 cells |
HCC, hepatocellular carcinoma.
Identification of DEGs
We used GRO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/), a web tool that allows users to identify DEGs between two or more groups of samples in a GEO series, analyze gene expression in GSE15653 and GSE60502. |logFC| ≥1.0 and P<0.05 were the criteria used for gene selection as DEGs and statistical significance, respectively. Genes showing up-regulation or down-regulation in both T2DM and HCC were investigated by Venn diagram, using the Venn diagram web tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
PPI network construction
We used STRING (https://string-db.org), a database of known and predicted protein-protein interactions, to construct the PPI network for genes that were up-regulated or down-regulated in both T2DM and HCC. Interactions with a score >0.7 were considered statistically significant. We visualized the PPI networks by Cytoscape, an open-source software platform for visualizing molecular interaction networks and biological pathways. Ten genes with the highest degree of connectivity were selected as hub genes. The MCODE plugin in Cytoscape was used to find clusters with the degree cut-off, haircut on, k-core, node score cut-off, max depth set as 10, 0.2, 2, 0.2 and 100 in PPI network. Modules with the highest scores were selected for functional enrichment analyses.
Functional enrichment analyses for module genes
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database resource for understanding high-level functions of biological systems from molecular-level information. The Gene Ontology (GO) resource can be used to perform enrichment analysis. We used DAVID (https://david.ncifcrf.gov/) for both KEGG pathway and GO enrichment analysis, including Biological Process, Cellular Component and Molecular Function for the selected module genes.
Analysis of hub genes
To identify the prognostic values of the hub genes, we used Kaplan-Meier Plotter database (https://kmplot.com) to construct Kaplan-Meier survival plots for 10 hub genes. GEPIA (http://gepia.cancer-pku.cn), a web server for cancer and normal gene expression profiling and interactive analysis, was used to analyze the expression of hub genes in HCC and normal tissue and to verify the prognostic values of hub genes in the Cancer Genome Atlas (TCGA) database. Lastly, the Human Protein Atlas (https://www.proteinatlas.org) was used to analyze the protein distribution of hub genes in HCC and normal liver tissue.
Analysis of genes associated with metformin treatment
Unpaired t-tests were used in GraphPad Prism to analyze the expression of 10 hub genes in GSE69850, which contains nine samples of HepG2 hepatocarcinoma cells treated with metformin and 39 control samples treated with dimethyl sulfoxide (DMSO), in order to determine associations between the hub genes and metformin treatment for HCC. Results with P<0.05 were considered statistically significant (Figure 1).
Figure 1.
Graphical abstract.
Results
Identification of DEGs in HCC and T2DM
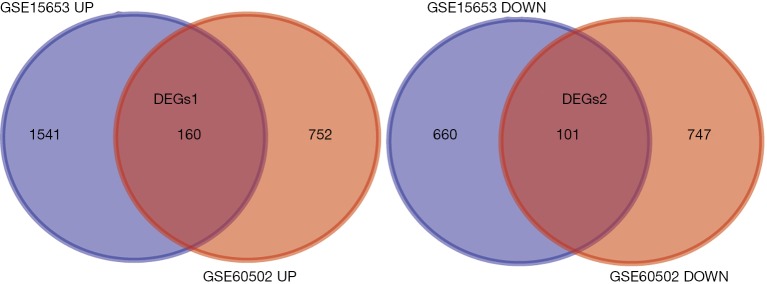
In total, 2,462 DGEs were identified from four T2DM samples and five control samples in dataset GSE15653 and 1,760 DGEs were identified from 18 HCC samples and 18 adjacent non-tumorous liver samples in dataset GSE60502. Then, 256 genes were found to overlap between the T2DM and HCC DGEs, including 155 up-regulated genes and 101 down-regulated genes (Figure 2).
Figure 2.
Differentially expressed genes among diabetes DEGs and HCC DEGs. DEGs1 represents the genes that up-regulated in both diabetes and HCC. DEGs2 represents the genes that down-regulated in both diabetes and HCC. DEGs, differentially expressed genes; HCC, hepatocellular carcinoma.
PPI network construction
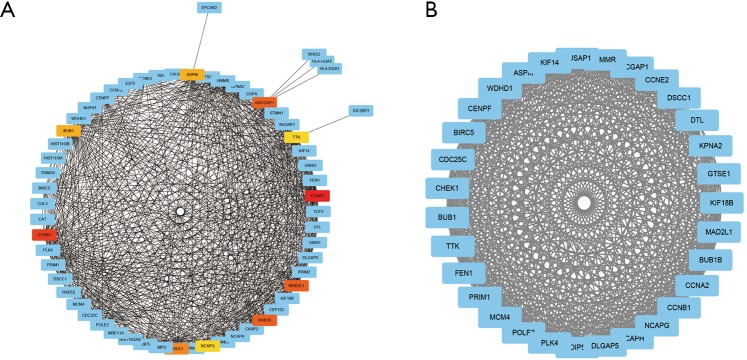
PPI network maps yielded 223 nodes and 531 edges, indicating the interactions between the 256 overlapping genes. The ten hub genes with the highest degree of connectivity were CCNA2, CCNB1, MAD2L1, BUB1B, RACGAP1, CHEK1, BUB1, ASPM, NCAPG, TTK (Figure 3A, Table 2). We use the MCODE plugin in Cytoscape to analyze the PPI networks and choose the module which has a highest score to create a cluster network including 31 nodes and 433 edges (Figure 3B).
Figure 3.
PPI network construction and module analysis on the DEGs1 and DEGs2. (A) PPI network on the DEGs1 and DEGs2, the red, orange and yellow nodes represent hub genes; (B) the chosen module which has a highest score using MCODE. DEGs, differentially expressed genes; PPI, protein-protein interaction.
Table 2. Top 10 hub genes.
| Rank | Name |
|---|---|
| 1 | CCNA2 |
| 2 | CCNB1 |
| 3 | MAD2L1 |
| 3 | BUB1B |
| 3 | RACGAP1 |
| 6 | CHEK1 |
| 7 | BUB1 |
| 7 | ASPM |
| 9 | NCAPG |
| 9 | TTK |
Functional enrichment analyses for module genes
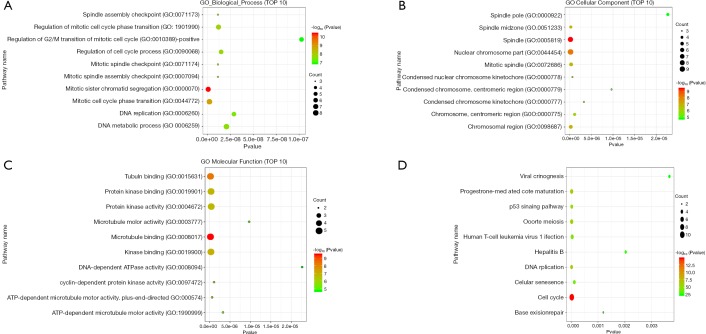
We used DAVID to make functional enrichment analyses for the 31 module genes selected by MCODE. The major biological processes identified for these genes are “mitotic sister chromatid segregation” and “mitotic cell cycle phase transition” (Figure 4A). Cellular component analysis indicated that these genes are mainly enriched in “spindle” and “nuclear chromosome part” (Figure 4B). In terms of molecular function, these genes are mainly associated with “microtubule-binding” and “tubulin-binding” (Figure 4C). Lastly, through KEGG pathway analysis, we observed that these genes are mostly enriched in “cell cycle” (Figure 4D) (Tables 3-6).
Figure 4.
Functional enrichment analysis for module genes. (A,B,C,D) Represent the results of biological process, cellular component, molecular function and KEGG pathway analyses for module genes.
Table 3. KEGG pathway analysis.
| Term | P value | Genes |
|---|---|---|
| Cell cycle | 2.30E-15 | CCNA2; CCNB1; CCNE2; CHEK1; BUB1B; MCM4; TTK; CDC25C; BUB1; MAD2L1 |
| DNA replication | 2.69E-07 | FEN1; PRIM1; POLE2; MCM4 |
| Progesterone-mediated oocyte maturation | 4.12E-07 | CCNA2; CCNB1; CDC25C; BUB1; MAD2L1 |
| Oocyte meiosis | 1.31E-06 | CCNB1; CCNE2; CDC25C; BUB1; MAD2L1 |
| p53 signaling pathway | 4.51E-06 | CCNB1; CCNE2; CHEK1; GTSE1 |
| Human T-cell leukemia virus 1 infection | 2.03E-05 | CCNA2; CCNE2; CHEK1; BUB1B; MAD2L1 |
| Cellular senescence | 1.05E-04 | CCNA2; CCNB1; CCNE2; CHEK1 |
| Base excision repair | 0.0011914 | FEN1; POLE2 |
| Hepatitis B | 0.0020202 | CCNA2; CCNE2; BIRC5 |
| Viral carcinogenesis | 0.0036535 | CCNA2; CCNE2; CHEK1 |
Table 4. GO biological process analysis.
| Term | P value | Genes |
|---|---|---|
| Mitotic sister chromatid segregation (GO:0000070) | 3.97E-11 | CCNB1; KIF18B; KIF14; NUSAP1; NCAPG; NCAPH; DLGAP5 |
| Mitotic cell cycle phase transition (GO:0044772) | 1.29E-09 | PLK4; CCNA2; CCNB1; CCNE2; PRIM1; POLE2; MCM4; CDC25C |
| Spindle assembly checkpoint (GO:0071173) | 1.11E-08 | CENPF; BUB1B; TTK; BUB1 |
| Mitotic spindle assembly checkpoint (GO:0007094) | 1.11E-08 | CENPF; BUB1B; TTK; BUB1 |
| Mitotic spindle checkpoint (GO:0071174) | 1.11E-08 | CENPF; BUB1B; TTK; BUB1 |
| regulation of mitotic cell cycle phase transition (GO:1901990) | 1.13E-08 | PLK4; CCNB1; CENPF; KIF14; BUB1B; HMMR; MAD2L1 |
| Positive regulation of cell cycle process (GO:0090068) | 1.42E-08 | FEN1; CCNB1; RACGAP1; KIF14; NUSAP1; MAD2L1 |
| DNA metabolic process (GO:0006259) | 1.99E-08 | WDHD1; FEN1; PRIM1; POLE2; CHEK1; MCM4; KPNA2; CDC25C |
| DNA replication (GO:0006260) | 2.82E-08 | WDHD1; FEN1; POLE2; CHEK1; MCM4; CDC25C |
| Regulation of G2/M transition of mitotic cell cycle (GO:0010389) | 1.02E-07 | PLK4; CCNB1; CENPF; KIF14; HMMR; DTL |
Table 5. GO cellular component analysis.
| Term | P value | Genes |
|---|---|---|
| Spindle (GO:0005819) | 3.30E-10 | CCNB1; CENPF; RACGAP1; KIF14; NUSAP1; BUB1B; TTK; MAD2L1 |
| Nuclear chromosome part (GO:0044454) | 5.48E-09 | WDHD1; FEN1; CCNB1; PRIM1; POLE2; BUB1B; MCM4; BUB1; NCAPH |
| Chromosomal region (GO:0098687) | 1.16E-07 | CENPF; DSCC1; CHEK1; BIRC5; OIP5 |
| Spindle midzone (GO:0051233) | 1.44E-07 | KIF18B; RACGAP1; KIF14; BUB1B |
| Mitotic spindle (GO:0072686) | 1.92E-07 | ASPM; KIF18B; RACGAP1; NUSAP1; MAD2L1 |
| Condensed nuclear chromosome kinetochore (GO:0000778) | 5.52E-07 | CCNB1; BUB1B; BUB1 |
| Chromosome, centromeric region (GO:0000775) | 1.03E-06 | CENPF; DSCC1; BIRC5; OIP5 |
| Condensed chromosome kinetochore (GO:0000777) | 3.21E-06 | BUB1B; BIRC5; BUB1 |
| Condensed chromosome, centromeric region (GO:0000779) | 9.62E-06 | BUB1B; BIRC5; BUB1 |
| Spindle pole (GO:0000922) | 2.26E-05 | ASPM; CCNB1; CENPF; DLGAP5 |
Table 6. GO molecular function analysis.
| Term | P value | Genes |
|---|---|---|
| Microtubule binding (GO:0008017) | 1.19E-05 | KIF18B; RACGAP1; KIF14; NUSAP1; BIRC5 |
| Tubulin binding (GO:0015631) | 4.28E-05 | KIF18B; RACGAP1; KIF14; NUSAP1; BIRC5 |
| Kinase binding (GO:0019900) | 4.27E-04 | CCNA2; CCNB1; RACGAP1; KIF14; CDC25C |
| Protein kinase binding (GO:0019901) | 9.17E-04 | CCNA2; CCNB1; RACGAP1; KIF14; CDC25C |
| Protein kinase activity (GO:0004672) | 0.0010748 | PLK4; CCNA2; CHEK1; BUB1B; BUB1 |
| ATP-dependent microtubule motor activity, plus-end-directed (GO:0008574) | 0.0011202 | KIF18B; KIF14 |
| cyclin-dependent protein kinase activity (GO:0097472) | 0.0013400 | CCNA2; CCNB1 |
| ATP-dependent microtubule motor activity (GO:1990939) | 0.0026130 | KIF18B; KIF14 |
| Microtubule motor activity (GO:0003777) | 0.0040194 | KIF18B; KIF14 |
| DNA-dependent ATPase activity (GO:0008094) | 0.0057059 | DSCC1; MCM4 |
Hub genes analysis
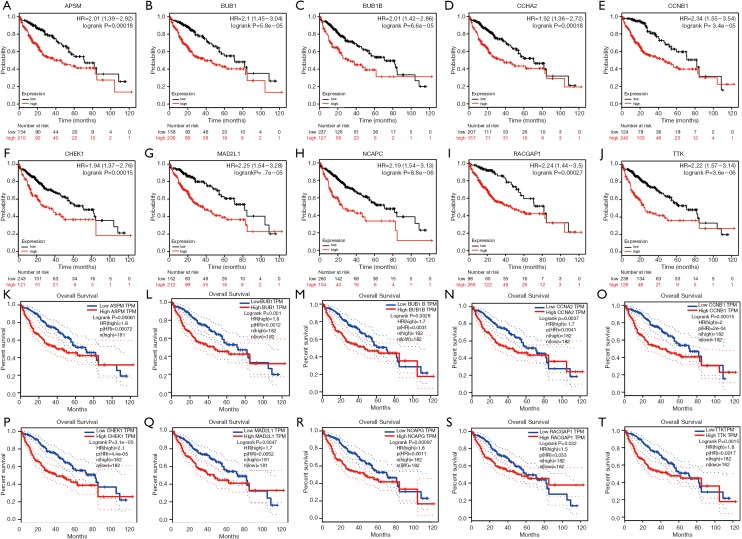
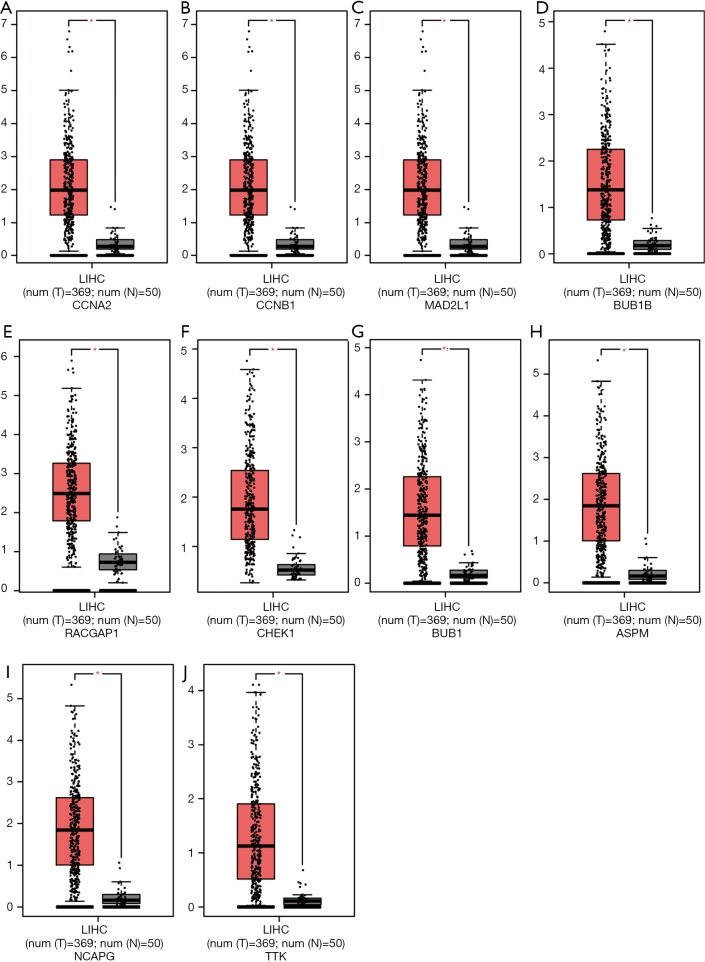
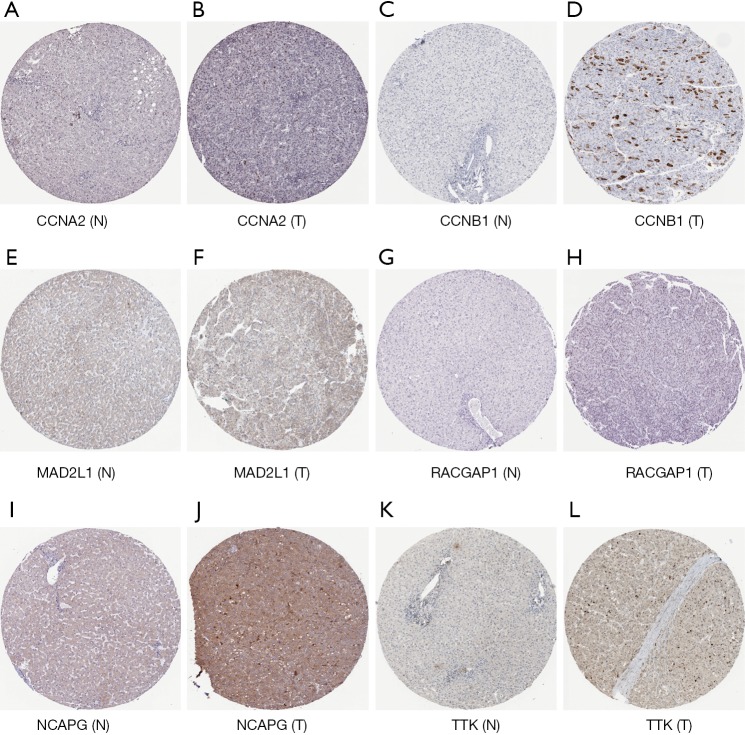
The results of the prognostic analysis showed that all of the 10 hub genes (CCNA2, CCNB1, MAD2L1, BU1B, RACGAP1, CHEK1, BUB1, ASPM, NCAPG and TTK) have strong associations with a lower overall survival rate in liver cancer patients (P<0.05) (Figure 5). Likewise, GEPIA analysis showed that the 10 hub genes are all overexpressed in HCC (Figure 6). Lastly, using The Human Protein Atlas, we investigated the protein distribution data of six genes: CCNA2, CCNB1, MAD2L1, RACGAP1, NCAPG, TTK. All were found to be overexpressed in HCC (Figure 7).
Figure 5.
Prognostic analysis for hub genes. Using the Kaplan-Meier plotter database to make overall survival time analyses of the hub genes. (A) ASPM; (B) BUB1; (C) BUB1B; (D) CCNA2; (E) CCNB1; (F) CHEK1; (G) MAD2L1; (H) NCAPG; (I) RACGAP1; (J) TTK; (A-J) is in kmplot database. (K) APSM; (L) BUB1; (M) BUB1B; (N) CCNA2; (O) CCNB1; (P) CHEK1; (Q) MAD2L1; (R) NCAPG; (S) RACGAP1; (T) TTK; (K-T) is in TCGA database.
Figure 6.
mRNA expression of hub genes. mRNA expression of hub genes in HCC tissue (red; n=369) and normal tissues (black; n=50). (A) CCNA2; (B) CCNB1; (C) MAD2L1; (D) BUB1B; (E) RACGAP1; (F) CHEK1; (G) BUB1; (H) ASPM; (I) NCAPG; (J) TTK; * represents P value <0.05.
Figure 7.
Protein expression of CCNA2, CCNB1, MAD2L1, RACGAP1, NCAPG, TTK. The tissue microarray using paired tumor tissue and adjacent normal tissue from the margin of the tumor. Immunohistochemistry of tissue microarray was performed by using the antibody (CAB000114, CAB003804, HPA003348, HPA043912, HPA039613, CAB013229, Abcam). (A) Protein expression of CCNA2 in normal tissue (not detected); (B) protein expression of CCNA2 in tumor tissue (staining: medium; intensity: strong; quantity: <25%); (C) protein expression of CCNB1 in normal tissue (not detected); (D) protein expression of CCNB1 in tumor tissue (staining: medium; intensity: strong; quantity: <25%); (E) protein expression of MAD2L1 in normal tissue (not detected); (F) protein expression of MAD2L1 in tumor tissue (staining: low; intensity: weak; quantity: 75–25%); (G) protein expression of RACGAP1 in normal tissue (not detected); (H) protein expression of RACGAP1 in tumor tissue (staining: medium; intensity: moderate; quantity: 75–25%); (I) protein expression of NCAPG in normal tissue (not detected); (J) protein expression of NCAPG in tumor tissue (staining: medium; intensity: moderate; quantity: >75%); (K) protein expression of TTK in normal tissue(not detected); (L) protein expression of TTK in tumor tissue (staining: low; intensity: moderate; quantity: <25%).
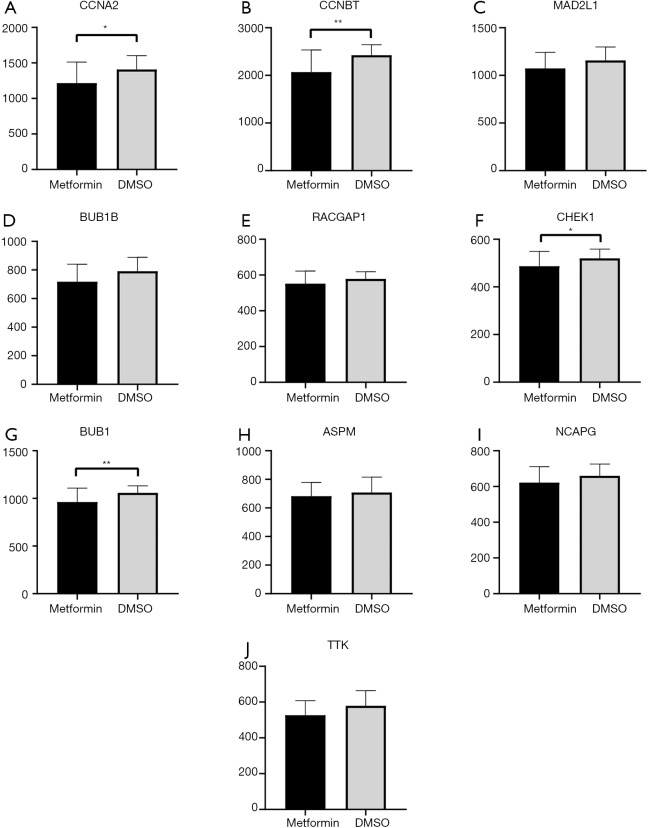
Association between hub genes and metformin treatment
The GSE69850 dataset contains nine HepG2 samples treated by metformin and 39 controlled samples treated by DMSO. After analyzing the expression of the 10 hub genes in metformin-treated and control samples, we found that four genes, CCNA2, CCNB1, CHEK1 and BUB1, have a significantly lower expression in metformin-treated samples (P<0.05) (Figure 8), suggesting that these genes may be involved in the mechanism of metformin treatment for T2DM and HCC.
Figure 8.
Changes of gene expression of HepG2 cell lines after Metformin treatment. (A-J) Represent the gene expression in 9 Metformin-treated HepG2 cell lines (black) and 39 controlled HepG2 cell lines (grey). (A) CCNA2; (B) CCNB1; (C) MAD2L1; (D) BUB1B; (E) RACGAP1; (F) CHEK1; (G) BUB1; (H) ASPM; (I) NCAPG; (J) TTK. * represents P value <0.05; ** represents P value <0.01.
Discussion
Both T2DM and HCC are globally epidemic diseases that place a huge burden on the patients and health services throughout the world. Many reports have indicated that T2DM is an independent risk factor for HCC. In this study, we have identified 256 genes that overlapped between T2DM DGEs and HCC DGEs, including 155 up-regulated genes and 101 down-regulated genes. Enrichment analyses show that the genes of the major module are largely associated with the cell cycle. Considering the ten identified hub genes, Cyclin A2, encoded by the cyclin A2 (CCNA2) gene, is a regulator of cell division and which activates Cdk1 and Cdk2, playing an important role in G1/S transition (7). Several studies have found that high levels of Cyclin A2 may be associated with carcinogenesis (8,9). It has also been reported that CCNA2 affects β-Cell proliferation (10) in the pancreas. Cyclin B1 (CCNB1) is a mitotic regulatory protein which complexes with CDK1 (CDC2) to form the maturation phase-promoting factor (MPF) and induces the G2/M phase transition in G2 phase cells (11). A meta-analysis has suggested that elevated expression of cyclin B1 may be associated with poor outcomes in lung cancer and esophageal carcinoma (12). It has also been found that increased cyclin B1 may promote the proliferation of HCC cells (13) and that it may be a biomarker and therapeutic target for recurrent HBV-related HCC (14). Mitotic arrest deficient 2 protein (MAD2L1) is an essential part of the mitotic spindle assembly checkpoint that ensures the proper alignment of chromosomes, strongly suggesting that MAD2L1 has a function in tumor suppression (15). Many studies have shown that the overexpression of MAD2L1 promotes the proliferation and metastasis of HCC (16). In addition, interactions between MAD2L1 and the insulin receptor have been reported, suggesting that MAD2L1 may influence the progression of diabetes (17,18). Genes budding uninhibited by benzimidazoles homolog 1 (BUB1) and budding uninhibited by benzimidazoles 1 homolog beta (BUB1B) encode serine/threonine protein kinases that play important roles in mitosis and are also essential components of the spindle checkpoint (19), with most studies suggesting both BUB1 and BUB1B may contribute to the proliferation and metastasis of some types of cancer including HCC (20-22). Gene Rac GTPase activating protein 1 (RACGAP1) encodes a GTPase-activating protein (GAP) which is an essential component of the central spindle complex and plays an important role in cytokinesis its GAP activity (23). RACGAP1 can maintain cell survival (24) and has been reported to be associated with several cancers including HCC, particularly with the migration and invasion process (25-28). Gene non-SMC condensin I complex subunit G (NCAPG) encodes a subunit of the condensin complex, which plays a central role in mitotic chromosome assembly and segregation (29). Recently, some studies have suggested that NCAPG is overexpressed in HCC and suppression of NCAPG inhibits proliferation and induces apoptosis in HCC cells (30-32), while normal hepatocytes show low expression of NCAPG (31). CHEK1 encodes a serine/threonine kinase, which is required for checkpoint-mediated cell cycle arrest in response to DNA damage or the presence of unreplicated DNA. It is reported that CHEK1 plays an essential role in cell cycle progression and survival whether in the presence or absence of DNA damage (33). CHEK1 is reported to be significantly overexpressed in HCC (34), which may be the result of reduced miR-497 in HCC (35). Interestingly, another study has suggested that CHEK1 may induce the overexpression of CCNB1 to promote tumor growth in human colorectal cancer (36), while a CHEK1 inhibitor has been observed to have a significant effect in treating tumors (37). Abnormal spindle microtubule assembly (ASPM), a key gene for microcephaly (38), plays an essential role in regulating spindle poles (39). Several studies have suggested that ASPM is closely related to HCC and can be used as a marker for HCC metastasis and early recurrence (40,41). Gene TTK encodes a dual-specificity protein kinase with the ability to phosphorylate tyrosine, serine and threonine. It is an essential component of the spindle assembly checkpoint and is commonly overexpressed in many human tumors (42). TTK has been shown to have significant effects on HCC proliferation and sorafenib resistance, and it can serve as a biomarker and potential target for HCC treatment (43,44). We found that all ten hub genes are overexpressed in cancers, including HCC, and most of them play an important role in cell division. However, there are few reports of an association between these genes and T2DM.
Metformin is a commonly used hyperglycemic drug. It has been reported to have inhibiting effect on many types of tumors including colorectal cancer, prostate cancer (45), lung cancer (46), pancreatic cancer (47) and HCC (6). It appears that its anti-tumorigenic effect may be related to the activation of AMP-activated protein kinase (AMPK) (48). In this study, we found that four of the hub genes, CCNA2, CCNB1, CHEK1 and BUB1, may participate in the metformin treatment mechanism for T2DM and HCC and that these genes are all enriched in cell cycle control.
It is possible that T2DM could promote the genesis of HCC via insulin resistance (49), over-activation of mTOR and chronic inflammation caused by adipokines (50). Besides, T2DM can also lead to NAFLD and non-alcoholic steatohepatitis (NASH), which are the main causes of HCC (51). Very few studies discuss the specific mechanism of the correlation between T2DM and HCC. Li et al. has suggested that high glucose promotes HCC cell proliferation and migration through the MEG3-MiR-483-3p-ERp29 pathway (52). The current study is the first exploration of the role of these 10 genes implicated in the combination of HCC and T2DM together with genes associated with metformin treatment for T2DM and HCC using a bioinformatics analysis.
This study has several limitations. Firstly, the sample sizes of the datasets we used are small and, secondly, our findings still need to be verified in clinical practice.
In conclusion, we have identified 256 DEGs and selected 10 genes (CCNA2, CCNB1, MAD2L1, BUB1B, RACGAP1, CHEK1, BUB1, ASPM, NCAPG and TTK) which may function biomarkers in both HCC and T2DM. We found four genes (CCNA2, CCNB1, CHEK1 and BUB) which may be the target of metformin treatment for T2DM and HCC. The specific mechanisms of action of these genes, however, require further investigation.
Acknowledgments
Funding: This study was supported by Key Laboratory of Tumor Molecular Diagnosis and Individualized Medicine of Zhejiang Province, Zhejiang Provincial People’s Hospital (People’s Hospital of Hangzhou Medical College). This study was supported by grants from the National Science and Technology Major Project for New Drug (No. 2017ZX09302003); the National Natural Science Foundation of China (81874049, 81602179); the Co-construction of Provincial and Department Project (WKJ-ZJ-1919); the Zhejiang Provincial Natural Science Foundation of China (LY19H160036); the Public Projects of Zhejiang Province (2018C37033). General scientific research project of education department of Zhejiang province (Y201534516); Zhejiang Administration of Traditional Chinese Medicine (2016A029).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: QX serves as an unpaid section editor of Annals of Translational Medicine from Oct 2019 to Sep 2020. The other authors have no conflicts of interest to declare.
References
- 1.Fitzmaurice C, Akinyemiju TF, Al Lami FH, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 2018;4:1553-68. 10.1001/jamaoncol.2018.2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertot LC, Adams LA. Trends in hepatocellular carcinoma due to non-alcoholic fatty liver disease. Expert Rev Gastroenterol Hepatol 2019;13:179-87. 10.1080/17474124.2019.1549989 [DOI] [PubMed] [Google Scholar]
- 3.Zheng Y, Ley SH, Hu FB. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol 2018;14:88-98. 10.1038/nrendo.2017.151 [DOI] [PubMed] [Google Scholar]
- 4.Huang TS, Lin CL, Lu MJ, et al. Diabetes, hepatocellular carcinoma, and mortality in hepatitis C-infected patients: A population-based cohort study. J Gastroenterol Hepatol 2017;32:1355-62. 10.1111/jgh.13670 [DOI] [PubMed] [Google Scholar]
- 5.Shyu YC, Huang TS, Chien CH, et al. Diabetes poses a higher risk of hepatocellular carcinoma and mortality in patients with chronic hepatitis B: A population-based cohort study. J Viral Hepat 2019;26:718-26. 10.1111/jvh.13077 [DOI] [PubMed] [Google Scholar]
- 6.Tseng CH. Metformin and risk of hepatocellular carcinoma in patients with type 2 diabetes. Liver Int 2018;38:2018-27. 10.1111/liv.13872 [DOI] [PubMed] [Google Scholar]
- 7.Blanchard JM. Cyclin A2 transcriptional regulation: modulation of cell cycle control at the G1/S transition by peripheral cues. Biochem Pharmacol 2000;60:1179-84. 10.1016/S0006-2952(00)00384-1 [DOI] [PubMed] [Google Scholar]
- 8.Hung YH, Huang HL, Chen WC, et al. Argininosuccinate lyase interacts with cyclin A2 in cytoplasm and modulates growth of liver tumor cells. Oncol Rep 2017;37:969-78. 10.3892/or.2016.5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayard Q, Meunier L, Peneau C, et al. Cyclin A2/E1 activation defines a hepatocellular carcinoma subclass with a rearrangement signature of replication stress. Nat Commun 2018;9:5235. 10.1038/s41467-018-07552-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song WJ, Schreiber WE, Zhong E, et al. Exendin-4 stimulation of cyclin A2 in beta-cell proliferation. Diabetes 2008;57:2371-81. 10.2337/db07-1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kishimoto T. Entry into mitosis: a solution to the decades-long enigma of MPF. Chromosoma 2015;124:417-28. 10.1007/s00412-015-0508-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye C, Wang J, Wu P, et al. Prognostic role of cyclin B1 in solid tumors: a meta-analysis. Oncotarget 2017;8:2224-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chai N, Xie HH, Yin JP, et al. FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochem Biophys Res Commun 2018;500:924-9. 10.1016/j.bbrc.2018.04.201 [DOI] [PubMed] [Google Scholar]
- 14.Weng L, Du J, Zhou Q, et al. Identification of cyclin B1 and Sec62 as biomarkers for recurrence in patients with HBV-related hepatocellular carcinoma after surgical resection. Mol Cancer 2012;11:39. 10.1186/1476-4598-11-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science 1996;274:246-8. 10.1126/science.274.5285.246 [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Bai W, Zhang J. MiR-200c-5p suppresses proliferation and metastasis of human hepatocellular carcinoma (HCC) via suppressing MAD2L1. Biomed Pharmacother 2017;92:1038-44. 10.1016/j.biopha.2017.05.092 [DOI] [PubMed] [Google Scholar]
- 17.O'Neill TJ, Zhu Y, Gustafson TA. Interaction of MAD2 with the carboxyl terminus of the insulin receptor but not with the IGFIR. Evidence for release from the insulin receptor after activation. J Biol Chem 1997;272:10035-40. 10.1074/jbc.272.15.10035 [DOI] [PubMed] [Google Scholar]
- 18.Choi E, Zhang X, Xing C, et al. Mitotic Checkpoint Regulators Control Insulin Signaling and Metabolic Homeostasis. Cell 2016;166:567-81. 10.1016/j.cell.2016.05.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolanos-Garcia VM, Blundell TL. BUB1 and BUBR1: multifaceted kinases of the cell cycle. Trends Biochem Sci 2011;36:141-50. 10.1016/j.tibs.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Q, Liu Y, Shang L, et al. The FOXM1/BUB1B signaling pathway is essential for the tumorigenicity and radioresistance of glioblastoma. Oncol Rep 2017;38:3367-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Lee J, Kljavin NM, et al. Requirement for BUB1B/BUBR1 in tumor progression of lung adenocarcinoma. Genes Cancer 2015;6:106-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu X, Chen G, Cai ZD, et al. Overexpression of BUB1B contributes to progression of prostate cancer and predicts poor outcome in patients with prostate cancer. Onco Targets Ther 2016;9:2211-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lekomtsev S, Su KC, Pye VE, et al. Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature 2012;492:276-9. 10.1038/nature11773 [DOI] [PubMed] [Google Scholar]
- 24.Yamada T, Kurosaki T, Hikida M. Essential roles of mgcRacGAP in multilineage differentiation and survival of murine hematopoietic cells. Biochem Biophys Res Commun 2008;372:941-6. 10.1016/j.bbrc.2008.05.170 [DOI] [PubMed] [Google Scholar]
- 25.Yeh CM, Sung WW, Lai HW, et al. Opposing prognostic roles of nuclear and cytoplasmic RACGAP1 expression in colorectal cancer patients. Hum Pathol 2016;47:45-51. 10.1016/j.humpath.2015.09.002 [DOI] [PubMed] [Google Scholar]
- 26.Imaoka H, Toiyama Y, Saigusa S, et al. RacGAP1 expression, increasing tumor malignant potential, as a predictive biomarker for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis 2015;36:346-54. 10.1093/carcin/bgu327 [DOI] [PubMed] [Google Scholar]
- 27.Wang SM, Ooi LL, Hui KM. Upregulation of Rac GTPase-activating protein 1 is significantly associated with the early recurrence of human hepatocellular carcinoma. Clin Cancer Res 2011;17:6040-51. 10.1158/1078-0432.CCR-11-0557 [DOI] [PubMed] [Google Scholar]
- 28.Zhang P, Bai H, Fu C, et al. RacGAP1-driven focal adhesion formation promotes melanoma transendothelial migration through mediating adherens junction disassembly. Biochem Biophys Res Commun 2015;459:1-9. 10.1016/j.bbrc.2014.11.088 [DOI] [PubMed] [Google Scholar]
- 29.Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev 2012;26:1659-78. 10.1101/gad.194746.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, Liang B, Liu H, et al. Overexpression of non-SMC condensin I complex subunit G serves as a promising prognostic marker and therapeutic target for hepatocellular carcinoma. Int J Mol Med 2017;40:731-8. 10.3892/ijmm.2017.3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q, Su R, Shan C, et al. Non-SMC Condensin I Complex, Subunit G (NCAPG) is a Novel Mitotic Gene Required for Hepatocellular Cancer Cell Proliferation and Migration. Oncol Res 2018;26:269-76. 10.3727/096504017X15075967560980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu K, Li Y, Yu B, et al. Silencing non-SMC chromosome-associated polypeptide G inhibits proliferation and induces apoptosis in hepatocellular carcinoma cells. Can J Physiol Pharmacol 2018;96:1246-54. 10.1139/cjpp-2018-0195 [DOI] [PubMed] [Google Scholar]
- 33.McNeely S, Beckmann R, Bence Lin AK. CHEK again: revisiting the development of CHK1 inhibitors for cancer therapy. Pharmacol Ther 2014;142:1-10. 10.1016/j.pharmthera.2013.10.005 [DOI] [PubMed] [Google Scholar]
- 34.Hong J, Hu K, Yuan Y, et al. CHK1 targets spleen tyrosine kinase (L) for proteolysis in hepatocellular carcinoma. J Clin Invest 2012;122:2165-75. 10.1172/JCI61380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie Y, Wei RR, Huang GL, et al. Checkpoint kinase 1 is negatively regulated by miR-497 in hepatocellular carcinoma. Med Oncol 2014;31:844. 10.1007/s12032-014-0844-4 [DOI] [PubMed] [Google Scholar]
- 36.Fang Y, Yu H, Liang X, et al. Chk1-induced CCNB1 overexpression promotes cell proliferation and tumor growth in human colorectal cancer. Cancer Biol Ther 2014;15:1268-79. 10.4161/cbt.29691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu Z, Oleinick NL, Zhang J. ATR/CHK1 inhibitors and cancer therapy. Radiother Oncol 2018;126:450-64. 10.1016/j.radonc.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Létard P, Drunat S, Vial Y, et al. Autosomal recessive primary microcephaly due to ASPM mutations: An update. Hum Mutat 2018;39:319-32. 10.1002/humu.23381 [DOI] [PubMed] [Google Scholar]
- 39.Jiang K, Rezabkova L, Hua S, et al. Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex. Nat Cell Biol 2017;19:480-92. 10.1038/ncb3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin SY, Pan HW, Liu SH, et al. ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res 2008;14:4814-20. 10.1158/1078-0432.CCR-07-5262 [DOI] [PubMed] [Google Scholar]
- 41.Wang F, Chang Y, Li J, et al. Strong correlation between ASPM gene expression and HCV cirrhosis progression identified by co-expression analysis. Dig Liver Dis 2017;49:70-6. 10.1016/j.dld.2016.10.017 [DOI] [PubMed] [Google Scholar]
- 42.Xie Y, Wang A, Lin J, et al. Mps1/TTK: a novel target and biomarker for cancer. J Drug Target 2017;25:112-8. 10.1080/1061186X.2016.1258568 [DOI] [PubMed] [Google Scholar]
- 43.Liang XD, Dai YC, Li ZY, et al. Expression and function analysis of mitotic checkpoint genes identifies TTK as a potential therapeutic target for human hepatocellular carcinoma. PLoS One 2014;9:e97739. 10.1371/journal.pone.0097739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Liao W, Yuan Q, et al. TTK activates Akt and promotes proliferation and migration of hepatocellular carcinoma cells. Oncotarget 2015;6:34309-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coyle C, Cafferty FH, Vale C, et al. Metformin as an adjuvant treatment for cancer: a systematic review and meta-analysis. Ann Oncol 2016;27:2184-95. 10.1093/annonc/mdw410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin J, Gill A, Zahm SH, et al. Metformin use and survival after non-small cell lung cancer: A cohort study in the US Military health system. Int J Cancer 2017;141:254-63. 10.1002/ijc.30724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Lai ST, Xie L, et al. Metformin is associated with reduced risk of pancreatic cancer in patients with type 2 diabetes mellitus: a systematic review and meta-analysis. Diabetes Res Clin Pract 2014;106:19-26. 10.1016/j.diabres.2014.04.007 [DOI] [PubMed] [Google Scholar]
- 48.Heckman-Stoddard BM, DeCensi A, Sahasrabuddhe VV, et al. Repurposing metformin for the prevention of cancer and cancer recurrence. Diabetologia 2017;60:1639-47. 10.1007/s00125-017-4372-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chettouh H, Lequoy M, Fartoux L, et al. Hyperinsulinaemia and insulin signalling in the pathogenesis and the clinical course of hepatocellular carcinoma. Liver Int 2015;35:2203-17. 10.1111/liv.12903 [DOI] [PubMed] [Google Scholar]
- 50.Fujita K, Iwama H, Miyoshi H, et al. Diabetes mellitus and metformin in hepatocellular carcinoma. World J Gastroenterol 2016;22:6100-13. 10.3748/wjg.v22.i27.6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bedi O, Aggarwal S, Trehanpati N, et al. Molecular and Pathological Events Involved in the Pathogenesis of Diabetes-Associated Nonalcoholic Fatty Liver Disease. J Clin Exp Hepatol 2019;9:607-18. 10.1016/j.jceh.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, Cheng T, He Y, et al. High glucose regulates ERp29 in hepatocellular carcinoma by LncRNA MEG3-miRNA 483-3p pathway. Life Sci 2019;232:116602. 10.1016/j.lfs.2019.116602 [DOI] [PubMed] [Google Scholar]