Abstract
Background
Glioblastoma multiforme (GBM) is the most common primary brain tumor, and is associated with a poor prognosis. Hypoxia is prevalent in the microenvironment of GBM, and promotes tumorigenesis and resistance to anticancer therapy. However, its mechanism remains incompletely understood.
Methods
We used immunohistochemistry, quantitative real-time PCR, and Western blots to assess c-Myc and Rab7a expression levels in 12 GBM specimens from a single institution. A luciferase reporter assay was conducted to confirmed whether Rab7a is transcriptionally regulated by c-Myc. To clarify the precise role of c-Myc/Rab7a on GBM cell proliferation, we did in vitro and in vivo analyses with lentivirus vectors. Cell viability was assessed using a cell counting kit-8 assay in the context of hypoxia. Autophagy was measured using transmission electron microscopy and Western blot, and apoptosis was measured using flow cytometry and Western blot.
Results
Gene and protein expression of c-Myc and Rab7a were significantly upregulated in GBM specimens. Moreover, c-Myc regulated Rab7a by specifically interacting with the Rab7a promoter. Furthermore, hypoxia activated the c-Myc/Rab7a pathway, which protects GBM cells from damage caused by hypoxia. Importantly, c-Myc/Rab7a inhibited apoptosis and induced autophagy in vitro and in vivo.
Conclusions
Collectively, our results suggest that the c-Myc/Rab7a pathway protects GBM cells from hypoxic injury via regulation of apoptosis and autophagy, contributing to the growth of GBM.
Keywords: Apoptosis, autophagy, c-Myc, hypoxia, Rab7a
Introduction
Glioblastoma multiforme (GBM) is the most common primary brain tumor, characterized by hypoxic necrotic regions and hypervascular areas (1). Survival after a diagnosis of GBM is usually less than 20 months, although the addition of tumor treating fields to temozolomide has demonstrated moderate improvements in survival (2-4). Multiple studies have shown that the hypoxic microenvironment promotes tumorigenesis, metastasis, and resistance to anticancer therapy, including chemotherapy and radiotherapy (5). A previous study showed that hypoxia reduces the response to temozolomide by inhibiting apoptosis in GBM (6).
The c-Myc oncoprotein is a DNA binding transcription factor that regulates the expression of many genes involved in cell growth and proliferation. Transcriptional activation by c-Myc requires hetero-dimerization with MYC-associated factor X (MAX), and binding to E-box sequences (7). Deregulated activity of c-Myc found in cancer cells induces transcription of genes involved in hypoxic adaptation, such as angiogenesis and metabolic reprogramming (8). However, the role of c-Myc in hypoxic microenvironment remains unknown (8). Hypoxia is prevalent in the microenvironment of GBM (9), and thus GBM represents a convenient paradigm to research the transcriptional response of c-Myc to hypoxia.
The Ras-related in brain (RAB) protein family comprises small guanosine triphosphatases (GTPases), which are known to be important for vesicular membrane trafficking (10). Rab7a is a small GTPase of the RAB family, and is primarily localized to the late endosomes with a key role in endosomal membrane traffic. In addition to these functions, Rab7a has many other cellular roles pertaining to autophagy (11), apoptosis (12), and cell migration (13). Computational analyses of the Rab7a promoter identified the presence of eight E-boxes, which are recognized by c-Myc (14). Rab7a expression is strongly increased and regulated by c-Myc and SOX10 in melanomas (14). Rab7a can function as both oncogenic protein and oncosuppressor protein, depending on the cellular context. In our previous work, we observed increased Rab7a expression in GBM under hypoxia condition. However, the correlation between them remains unclear.
We hypothesized that the c-Myc/Rab7a signaling pathway protected GBM cells from hypoxia-induced cell death. Here, we studied the effect of this pathway on the viability of GBM cells under hypoxia. We showed that the hypoxic environment triggered the c-Myc/Rab7a signal pathway, initiated protective autophagy, and inhibited cell apoptosis, thus helping GBM adapt to a hypoxic environment.
Methods
Tumor specimens
GBM specimens were obtained from 12 GBM patients who underwent surgery at the Second Affiliated Hospital of Zhejiang University Medical College in China from 2018 to 2019. Tissue were immediately frozen in liquid nitrogen and stored at −80 °C. Normal brain tissues (NBTs) were obtained from normal tissue adjacent to tumor tissues, and underwent histological confirmation to verify that the tissues were free of any pathological lesions. Among them, seven gliomas showed obvious central necrosis on magnetic resonance imaging (MRI), and five gliomas showed little or no necrosis. The study was approved by the Human Research Ethics Committee of the Second Affiliated Hospital of Zhejiang University Medical College, and the participants gave informed consent.
Cell lines and reagents
Human GBM cell lines (U87 and U251), G422 mouse GBM cell line, and human embryonic kidney (HEK) 293T cell line were all obtained from BeNa Culture Collection (Beijing, China). Cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (Invitrogen, USA) with 10% fetal bovine serum (Invitrogen, USA), 1% streptomycin and penicillin at 37 °C in 5% CO2.
Hypoxia cell culture
GBM cells were seeded on six-well plates at a density of 2×105 cells/well and allowed to adhere overnight under normoxia. Then, cells were incubated in the BINDER hypoxic incubator (Germany) in 1% or 5% O2, with 5% CO2 in a humidified atmosphere at 37 °C. Confirmation of hypoxic conditions was conducted using Western blotting to detect expression of HIF-1α.
Immunohistochemistry
Paraffin-embedded sections of excised GBM specimens were immunostained for c-Myc or Rab7a protein. The sliced samples were first stained with hematoxylin and eosin for histological examination. Tissue sections were incubated with mouse anti-human c-Myc primary antibody (1:50; Cell Signaling Technology) or anti-human Rab7a primary antibody (1:50; Cell Signaling Technology) at 4 °C overnight, followed by secondary goat anti-mouse IgG (Dako, CA, USA) for 30 min. Negative controls (NCs) were performed throughout the entire procedure. All of the staining results were reviewed in double blinded fashion by two investigators.
Cell counting kit-8 (CCK8)
Cell proliferation of GBM cell lines was evaluated using the CCK8 assay (MedChemExpress, China), according to the manufacturer’s instructions. After treatment, GBM cells were seeded into 96-well plates (2×103 cells per well) containing media. Then, the GBM cells were cultured under hypoxic conditions for 48 h, and incubated with CCK8 reagent at a final concentration of 10 µL/mL for 4 h at 37 °C. Lastly, the absorbance was recorded at 450 nm using a microplate reader (Molecular Devices, Sunnyvale, CA, USA). Each experiment was repeated three times.
Plasmid and transfection
The c-Myc, Rab7a, and NC shRNA were constructed according to effective c-Myc siRNA (5'-CGGUGCAGCCGUAUUUCUAdTdT-3'), Rab7a siRNA (5'-GGAGCTGACTTTCTGACCA-3'), GFP siRNA (5'-CAAGCUGACCCUGAAGUUCTT-3'), and NC sequences (5'-GTTCTCCGAACGTGTCACGT-3'). NC-shRNA, GFP shRNA, c-Myc shRNA, Rab7a shRNA and Rab7a-cDNA lentiviral vector were obtained commercially from ZHBY Biotech Corporation (Jiangxi, China). All lentiviral vectors contained the puromycin resistance gene. The lentiviral vector and package vector were co-transfected into 293T cells, and viral supernatant was collected for infection. According to the manufacturer’s instructions, cells were infected using Lipofectamine 2000 (Invitrogen, USA). After 24 h, confirmation of infection was carried out using quantitative real-time PCR.
Western blot analysis
Total protein was extracted from cultured cells and tissues. The cells were lysed in RIPA buffer with a protease inhibitor. The 10–15% SDS-PAGE gels were used to separate the lysates, and were then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore). The membranes were blocked in 5% nonfat milk with Tween for 1 h and incubated with indicated primary antibodies overnight at 4 °C. Then the membranes were incubated with fluorescence-conjugated secondary antibodies (1:5,000, Invitrogen, USA). ECL chemiluminescence system (Merck Millipore, Germany) was used for signal detection. To perform Western blot analysis, the following antibodies were used: rabbit anti-LC3B (ab48394, Abcam, 1:1,000); rabbit anti-PARP (ab32138, Abcam, 1:1,000); rabbit anti-c-Myc (ab32072, Abcam, 1:1,000); rabbit anti-Rab7 (ab137029, Abcam, 1:1,000); rabbit anti-p62 (sc-48402, Santa Cruz, 1:1,000); rabbit anti-HIF-1α (sc-13515, Santa Cruz, 1:1,000); mouse anti-β-actin (sc-58673, Santa Cruz, 1:2,000).
Real-time RT-PCR analysis
Total RNA was isolated from samples using TRIzol standard protocol (Invitrogen, CA, USA), according to manufacturer’s instruction. First-strand cDNA synthesis was performed using RevertAid RT Reverse Transcription Kit (Thermo Fisher, USA). The expression levels of c-Myc and Rab7a were analyzed using Fast SYBR Green PCR Master Mix (Applied Biosystems, USA). β2-MG was used as the internal control for both c-Myc and Rab7a. The qRT-PCR was performed on thermocycler ABI Prism 7500 fast (Applied Biosystems, CA, USA). All of the data were normalized to the internal control. Forward and reverse primers for each gene are listed below c-Myc: 5'-CCCTCAGTGGTCTTTCCCTAC-3', 5'-CTTGTCGTTTTCCTCCGTGT-3'; Rab7a: 5'-ACCAGTACAAAGCCACAATAGG-3', 5'-GGGGCAGTCACATCAAACAC-3'; β2-MG: 5'-CTATCCAGCGTACTCCAAAGAT-3', 5'-ACACGGCAGGCATACTCATC-3'.
Luciferase assay
The luciferase-encoding Rab7a-promoter and control sequences were purchased from Hanbio Biotechnology (Shanghai, China). 293T cells were seeded into a 96-well plate overnight, transfected with luciferase together using lipofectamine 2000. At 48 h after transfection, the culture medium was changed. Luciferase activities were measured through a commercial kit (SpectraMax M5) using the Dual-Luciferase Reporter Assay System (Promega, USA). As an internal control, Renilla luciferase reporters were also included. Each experiment was repeated three times with duplicate samples.
Transmission electron microscopy (TEM)
After treatment, GBM cells were fixed with 2.5% ice-cold glutaraldehyde in phosphate buffered saline (PBS), and preserved at 4 °C for further processing. Next, cells were post-fixed with 1% osmium tetroxide, followed by an increasing gradient dehydration step using ethanol and acetone (30%, 50%, 70%, 80%, 90%, 100%). The cells were then embedded in araldite, and ultrathin sections were obtained (50–60 nm) with a microtome, placed on uncoated copper grids, and stained with 3% lead citrate-uranyl acetate. The structure and morphology of sections were examined by a TEM [JEM-1230 (80 kV), JEOL].
Cell apoptosis assay by flow cytometry
The annexin V-fluorescein isothiocyanate (FITC) assay was used to measure apoptosis ratio according to the manufacturer’s instruction (Invitrogen, USA). Briefly, cells (2×105 per well) with stable transduction were incubated in six-well plates to 70–80% confluence. After cultured under hypoxic condition for 48 h, the cells were collected by trypsin, washed with ice-cold phosphate-buffered saline (PBS) twice, and resuspended in annexin V-labeling solution with FITC-conjugated anti-annexin V. After 15 min incubation at room temperature, the cells were centrifuged and resuspended with propidium iodide (PI). Apoptotic cells were quantified by flow cytometry (BD FACS, USA). Data were expressed as the percentage of apoptotic cells at early stage (PI negative and annexin V positive) and late stage (PI positive and annexin V positive).
Tumorigenesis assay in nude mice
G422 GBM cell suspension (5×106) with stable transduction was injected subcutaneously into the left flank of 5-week-old female BALB/c nude mice (n=6/group; 5 groups; Hunan SLAC Laboratory Animal Company). The tumor volume was monitored once every three days using the following formula: volume = (length × width2)/2. The resected xenografted tumors were weighed using an electronic balance. Total protein and RNA were extracted from resected tumor on the eighteenth day. The qRT-PCR analysis was performed to detect c-Myc and Rab7a expression. Western blot analysis was performed to detect the expressions of hallmark of apoptosis and autophagy. The Experimental Animal Ethics Committee of the Second Affiliated Hospital of Zhejiang University Medical College approved all animal procedures.
Statistical analysis
The data from triplicate experiments were presented as the mean ± SD. Statistical analysis between groups was performed using two-tailed Student’s t-test. For multiple groups, significant differences were determined using one-way ANOVA (GraphPad Prism 5.0). P<0.05 was considered statistically significant (indicated by single asterisk in the Figures), P<0.01 was strongly significant (indicated by double asterisks).
Results
High expression of c-Myc and Rab7a in GBM specimens
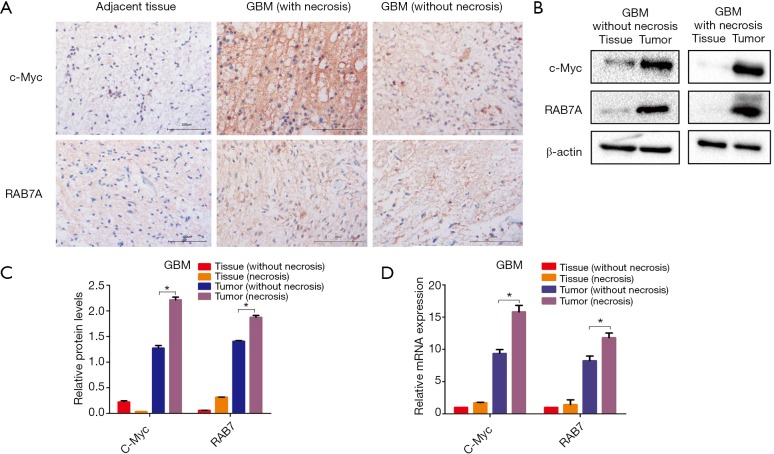
To investigate the function of c-Myc and Rab7a in GBMs, we first detected the expression levels of c-Myc and Rab7a in GBM specimens (n=12) via immunohistochemical analysis. We found that c-Myc and Rab7a expression levels were significantly upregulated in GBM tissues compared to the NBTs. It is noteworthy that the GBM with necrotic/hypoxic area (n=7) had higher c-Myc and Rab7a expression levels compared to GBMs without necrotic area (n=5; Figure 1A). The increased protein expression of both c-Myc and Rab7a was also verified via Western blot (Figure 1B,C). Moreover, qRT-PCR results showed that the mRNA expression level of both c-Myc and Rab7a were higher in GBM specimens, when compared with NBTs. Remarkably, the mRNA expression levels of both c-Myc and Rab7a were higher in GBM that had obvious necrotic area than GBM without necrotic area (Figure 1D). Together, these data demonstrated that both c-Myc and Rab7a were highly expressed in GBM specimens, especially in tumors with necrosis.
Figure 1.
High expression of c-Myc and Rab7a in GBM specimens. (A) Photomicrographs show Immunohistochemical staining of c-Myc and Rab7a protein of GBM samples with or without necrosis, and NBTs. Original of magnification ×200. Scale bars represent 100 µm; (B) Western blot analysis of c-Myc and Rab7a expression in GBM samples with or without necrosis, and their relative NBTs. β-actin is shown as the loading control; (C) graphical representation of the Western blot analysis; (D) c-Myc and Rab7a mRNA expression in GBM samples with or without necrosis, and their relative NBTs. Data are presented as mean ± SEM for three separate experiments performed in duplicate. *, P<0.05. GBM, glioblastoma multiforme; NBTs, normal brain tissues; SEM, standard error of the mean.
c-Myc directly regulated Rab7a expression in GBM
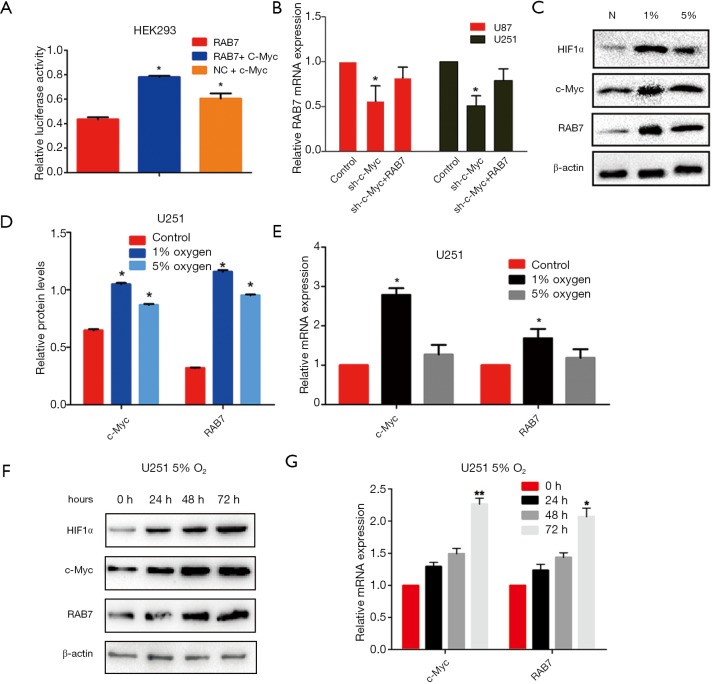
To assess the relationship between the oncogenic transcription factors, c-Myc and Rab7a, luciferase reporters containing human Rab7a promoter or NC were transfected into 293T cells together using an overexpressing c-Myc vector. As shown in Figure 2A, overexpression of c-Myc upregulated the activity of Rab7a promoter reporters, suggesting that Rab7a was transcriptionally regulated by c-Myc. The U87MG and U251MG cells were stable transfected with lentiviruses encoding c-Myc shRNA or co-infected with c-Myc shRNA and Rab7a overexpression vector. RT-PCR showed that Rab7a mRNA expression was downregulated after transfection of c-Myc shRNA, and rescued by co-infection with overexpressed Rab7a, confirming the regulatory function of c-Myc and its transcriptional effect (Figure 2B). Together, these data suggested that c-Myc regulated Rab7a by specifically interacting with the Rab7a promoter.
Figure 2.
Hypoxia activates c-Myc/Rab7a pathway in GBM. (A) Dural luciferase assay on 293T cells cotransfected with firefly luciferase constructs containing the Rab7a promoter or NC and c-Myc-pcDNA3.1 vector. Renilla luciferase served as the transfection control; (B) real-time RT-PCR analysis to determine mRNA expression of Rab7a in U87 and U251 cells infected with NC, c-Myc shRNA or virus expressing Rab7a. Data are shown at 72 h post-transfection; (C) immunoblot analysis of HIF1-α, c-Myc, and Rab7a in U251 cells treated with 1% or 5% hypoxia. β-actin is shown as the loading control; (D) graphical representation of the Western blot analysis; (E) real-time RT-PCR analysis to determine mRNA expression of c-Myc and Rab7a in U251 cells treated with 1% or 5% hypoxia. Control group was treated with 20% O2 for 48 h; (F) immunoblot analysis of HIF1-α, c-Myc and Rab7a in U251 cells treated with 0 h, 24, 48, or 72 h of moderate hypoxia. β-actin is shown as the loading control; (G) real-time RT-PCR analysis to determine mRNA expression of c-Myc and Rab7a in U251 cells treated with 0, 24, 48, or 72 h of moderate hypoxia. Data are presented as mean ± SEM for three separate experiments performed in duplicate. *, P<0.05; **, P<0.01. GBM, glioblastoma multiforme; NC, negative control.
Hypoxia initiated c-Myc/Rab7a pathway
Since both c-Myc and Rab7a were highly expressed in GBM with necrotic/hypoxic area, we hypothesized that hypoxia activates the c-Myc/Rab7a pathway. U251MG cells were exposed to normoxia, severe hypoxia (1% O2), or moderate hypoxia (5% O2) for 48 h. As expected, hypoxia increased the protein expression of both c-Myc and Rab7a (Figure 2C,D). We also performed an RT-PCR analysis to measure the mRNA expression of c-Myc and Rab7a in hypoxia-treated U251MG cells. The mRNA levels of these genes were significantly increased in hypoxia-treated cells compared to the normoxia-treated cells (Figure 2E). Notably, protein and mRNA expression levels of both c-Myc and Rab7a were higher in severe hypoxia compared to moderate hypoxia. In addition, U251 MG cells were treated with different durations of moderate hypoxia ranging from 0 to 72 h. We observed that moderate hypoxia increased the expression of c-Myc and Rab7a in a time-dependent manner (Figure 2F,G). Similar results were found in U87MG cells. Together, these data show that hypoxia initiates c-Myc/Rab7a signaling pathway in GBM cells.
c-Myc/Rab7a pathway promoted GBM cells growth under hypoxia
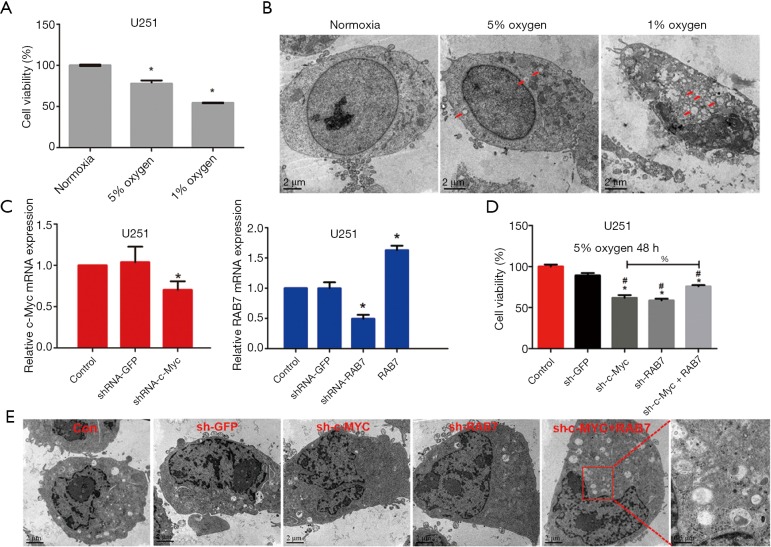
To investigate the role of c-Myc/Rab7a under hypoxia, U251MG cells were cultured under hypoxic conditions with 5% or 1% concentrations, respectively. After 48 h, the CCK8 assay was performed and cell viability was estimated. As shown in Figure 3A, hypoxia inhibited cell viability in a concentration-dependent manner. TEM analysis was performed to observe the cell morphology changes of U251. We found that apoptosis-like changes appeared in cell morphology under hypoxia, such as nuclear chromosome agglutination and marginalization, as well as apoptotic bodies. Meanwhile, an increase of autophagic vacuoles was observed under hypoxia, suggesting that autophagy may be activated (Figure 3B). We generated lentiviruses expressing c-Myc shRNA, Rab7a shRNA, GFP shRNA, and Rab7a overexpressing vector, and confirmed their transduction effect by PCR in U251MG cells (Figure 3C). After being cultured under hypoxic conditions for 48 h, knockdown of either c-Myc or Rab7a decreased cell viability significantly compared to the NC or GFP shRNA group. However, cell viability of U251 cells co-transduced with c-Myc shRNA and overexpressed Rab7a was rescued compared to the c-Myc shRNA group, suggesting that c-Myc/Rab7a may promote GBM cell growth under hypoxic conditions (Figure 3D). We also observed the cell morphology of each group using TEM, and found that the autophagic vacuoles increased significantly in the co-transduction group compared to the c-Myc shRNA group (Figure 3E). Together, these data suggest that the c-Myc/Rab7a pathway promotes the growth of GBM cells under hypoxic conditions, which may involve apoptosis and autophagy.
Figure 3.
Hypoxia promoted GBM cell growth in vitro depending on the c-Myc/Rab7a pathway. (A) CCK8 assay to determine the cell viability of U251 cells treated with 1% or 5% hypoxia for 48 h; (B) TEM photomicrographs showing the morphology of U251 cells treated with 1% or 5% hypoxia for 48 h. Red arrows represent autophagic vacuole. Scale bars represent 2 µm; (C) real-time RT-PCR analysis to determine mRNA expression of c-Myc and Rab7a in U251 cells infected with GFP shRNA, c-Myc shRNA, Rab7a shRNA, or Rab7a overexpression vector; (D) CCK8 assay to determine the cell viability of U251 cells infected with GFP shRNA, c-Myc shRNA, and Rab7a shRNA or co-infected with c-Myc shRNA and Rab7a overexpression vector; (E) TEM photomicrographs showing the morphology of U251 cells infected with GFP shRNA, c-Myc shRNA, Rab7a shRNA or co-infected with c-Myc shRNA and Rab7a overexpression vector under 5% hypoxia for 48 h. Data are presented as mean ± SEM for three separate experiments performed in duplicate. *, compared to negative control, P<0.05; #, compared to GFP shRNA, P<0.05. GBM, glioblastoma multiforme; TEM, transmission electron microscopy.
c-Myc/Rab7a pathway induced autophagy, but inhibited apoptosis in vitro and in vivo
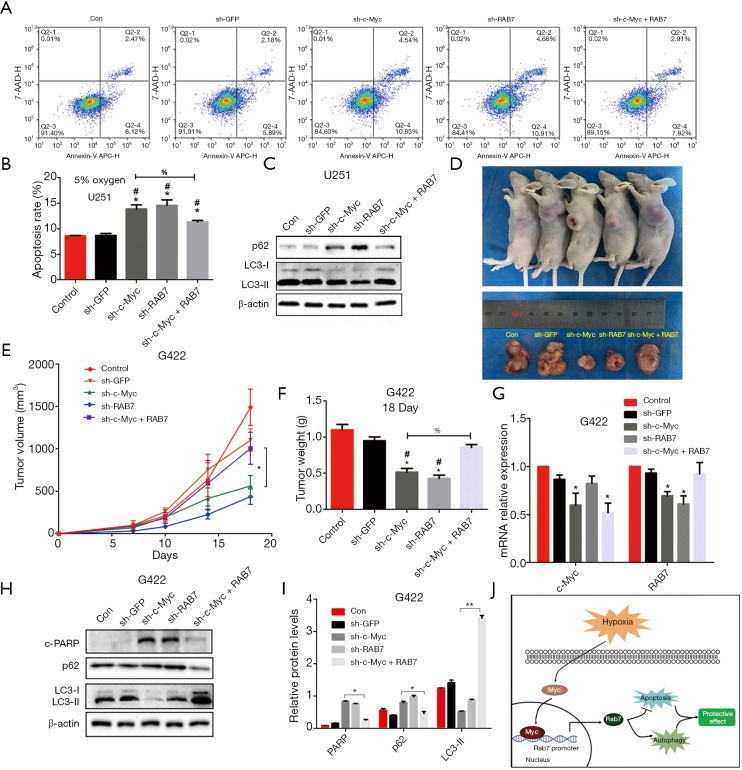
Next, we sought to dissect the mechanism by which c-Myc/Rab7a could promote GBM cell growth. First, we detected apoptosis of U251MG cells transduced with vectors as described above during moderate hypoxia. Flow cytometry analysis showed that the rate of apoptosis increased after knockdown of c-Myc or Rab7a, but that it also significantly decreased in cells that had been co-transduced with c-Myc shRNA and overexpressed Rab7a compared to c-Myc shRNA. Then, Western blot analyses with LC3 and p62 antibodies were conducted to detect autophagy. Low levels of autophagy were found in c-Myc shRNA and Rab7a shRNA (Figure 4A,B). However, the level of autophagy increased in the co-transduction group compared to the c-Myc shRNA group (Figure 4C). These data support the notion that the c-Myc/Rab7a pathway inhibits apoptosis, but induces autophagy in GBM cells.
Figure 4.
The c-Myc/Rab7a pathway induced autophagy, but inhibited apoptosis in vitro and in vivo. (A) Flow cytometry was performed to analyze apoptosis of U251 cell infected with GFP shRNA, c-Myc shRNA, Rab7a shRNA or co-infected with c-Myc shRNA and Rab7a overexpression vector under 5% hypoxia for 48 h; (B) statistical analysis of apoptosis rate, which was detected in (A); (C) immunoblot analysis of p62, LC3, and β-actin in U251 cells treated as described in (A). β-actin is shown as the loading control; (D) photograph showing excised tumors from representative mice in each group implanted with infected G422 cells (n=6 mice per treatment group) on day 18 after tumor implantation; (E) graph showing tumor volume curves of mice with xenografts; (F) graph illustrating the excised tumor weight on day 18 after tumor implantation; (G) real-time RT-PCR analysis to determine mRNA expression of c-Myc and Rab7a in xenograft tumor specimens. Data are presented as mean ± SEM for three separate experiments performed in duplicate; (H) immunoblot analysis of c-PARP, p62, LC3, and β-actin in xenograft tumor specimens. β-actin is shown as the loading control; (I) graphical representation of the Western blot analysis. *, P<0.05; **, P<0.01; (J) a schematic model summarizing key aspects of function of c-Myc/Rab7a described in this study. In (B,F), *, compared to negative control, P<0.05; #, compared to GFP shRNA, P<0.05; %, comparison between sh-MYC and sh-MYC + Rab7a group, P<0.05. SEM, standard error of the mean.
To investigate the role of c-Myc/Rab7a in tumor growth in vivo, the G422 cells transduced with the above lentiviruses were subcutaneously implanted in the flank of nude mice (Figure 4D). The tumor volume and weight were substantially lower in the c-Myc shRNA and Rab7a shRNA groups than that in GFP shRNA group and NC group. However, compared with the c-Myc knockdown group, the tumor volume and weight in Rab7a replenished group were significantly higher, and reached a similar level with the control group (Figure 4E,F). The qRT-PCR analysis was also performed to detect c-Myc and Rab7a expression in tumor xenografts (Figure 4G). Western blot analysis was performed to detect the expressions of hallmark of apoptosis and autophagy. The changes of autophagic proteins, p62 and LC3, were similar to the Western blot analyses in vitro. The level of apoptotic protein, c-PARP, increased in c-Myc shRNA group or Rab7a shRNA group. However, compared with the c-Myc knockdown group, the c-PARP level decreased significantly in the co-transduction group (Figure 4H,I). The results showed that knockdown of c-Myc or Rab7a inhibited autophagy and increased apoptosis. Taken together, our results validate the protective role of the c-Myc/Rab7a pathway in GBM in vitro and in vivo under hypoxia.
Discussion
Hypoxia is a recognised characteristic of the specialised microenvironment that is beneficial for tumorigenesis, and is responsible for resistance to anticancer treatment (15). Here, we confirmed that c-Myc and Rab7a are upregulated in clinical GBM specimens, particularly in GBMs with necrotic area. We further presented evidence that the c-Myc/Rab7a signaling pathway is activated in GBM cells under hypoxic conditions. Specifically, we have shown that c-Myc/Rab7a promotes proliferation of GBM cells by inducing protective autophagy and inhibiting apoptosis both in vitro and in vivo. Thus, our findings support the protective role of c-Myc/Rab7a in GBM under hypoxia.
C-Myc plays an important role in regulating tumor cell growth, proliferation, apoptosis, and metabolism (16). Hypoxia is a common characteristic of tumor microenvironment (17). Inhibition of c-Myc was proven to increase HIF-1α degradation and induce cell death (apoptosis) by increasing mitochondrial reactive oxygen species (ROS) production and ROS-mediated iron transition in colorectal cancer under hypoxia (18). Similarly, under conditions of growth factor withdrawal, c-Myc dysregulation can also promote apoptosis (19). Hypoxia can also induce tumor angiogenesis and growth by promoting c-Myc expression in lung cancer and colorectal carcinoma (20,21). Meanwhile, hypoxia-induced ectopic expression of MYC-nick (a cytoplasmic, transcriptionally inactive cleavage product of c-Myc) promotes anchorage-independent growth and cell survival by promoting autophagy in colon cancer (22). These findings all appear to demonstrate the protective role of c-Myc under hypoxic conditions in tumor cells, which are consistent with our conclusions.
Rab7a is one of the most studied RAB proteins in tumor growth, which is involved with both apoptosis and autophagy in various conditions (13). The Rab7a was downregulated in the hypoxic ovarian cancer cells, and inhibition of the Rab7a upstream activator, STAT3, significantly increased apoptosis, decreased colony formation, and cell proliferation (23). Furthermore, it was found that the hypoxia-induced miR-138-5p inhibited autophagy and tumor growth via inhibition of the SIRT1/FoxO1/Rab7 axis in vivo (24). Meanwhile, LRP6 upregulated Rab7, and further activated autophagy in the trophoblast cells (25).
The data on the tumor inhibition and/or development via autophagy seem contradictory in the past (26,27). During the early stages of tumorigenesis, autophagy appears to limit cell proliferation and DNA damage, therefore tumor growth. When the hypoxia emerges after tumor growth, which can restrict the blood supply, autophagy may promote the survival of tumor cells (28,29). Additionally, excessive autophagy induces cell damage, leading to mechanisms of cell death, such as apoptosis. Apoptosis may begin with autophagy, but autophagy frequently leads to apoptosis (30). When considering our results and those of previous studies, this appears to explain the tumor-promoting effect of c-Myc-induced Rab7a activation via autophagy promotion and apoptosis inhibition in hypoxic conditions. Furthermore, through administration of c-Myc, Rab7a knockdown, and Rab7a overexpression on glioma cells, it was found that autophagy level and cell proliferation ability remained positively correlated, suggesting that autophagy likely contributes to the protective role in this process.
The present study has several limitations. Our method of establishing the animal model is subcutaneous implantation, rather than intracranial in situ implantation. Therefore, it is difficult to simulate the actual GBM microenvironment. Moreover, we have only discovered that c-Myc/Rab7a can initiate autophagy and inhibit apoptosis under hypoxic conditions. However, the specific mechanism by which Rab7a regulates autophagy and apoptosis during this process remains unclear. Besides, the mechanism by which the c-Myc/Rab7a pathway is activated under hypoxic conditions also requires further inquiry. In addition, considering the close complex crosstalk between HIF-1α and c-Myc, it is also worthy of further study to determine whether HIF-1α participates in this process.
Conclusions
In conclusion, our data indicated that the c-Myc/Rab7a pathway protects GBM cells from hypoxic damage by inducing autophagy and inhibiting apoptosis. Hence, our results provide an improved understanding of the microenvironment and tumorigenesis.
Acknowledgments
Funding: This study was funded by National Natural Science Foundation of China (No. 81702462 to C Li, No. 81801162 to S Zhang).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Human Research Ethics Committee (No. 2017-130) and the Experimental Animal Ethics Committee (No. 2017-240) of the Second Affiliated Hospital of Zhejiang University Medical College, and the participants gave informed consent.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Chen X, Yan Y, Zhou J, et al. Clinical prognostic value of isocitrate dehydrogenase mutation, O-6-methylguanine-DNA methyltransferase promoter methylation, and 1p19q co-deletion in glioma patients. Ann Transl Med 2019;7:541. 10.21037/atm.2019.09.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrlinger U, Tzaridis T, Mack F, et al. Lomustine-temozolomide combination therapy versus standard temozolomide therapy in patients with newly diagnosed glioblastoma with methylated MGMT promoter (CeTeG/NOA-09): a randomised, open-label, phase 3 trial. Lancet 2019;393:678-88. 10.1016/S0140-6736(18)31791-4 [DOI] [PubMed] [Google Scholar]
- 3.Desjardins A, Gromeier M, Herndon JE, 2nd, et al. Recurrent Glioblastoma Treated with Recombinant Poliovirus. N Engl J Med 2018;379:150-61. 10.1056/NEJMoa1716435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan Y, Xu Z, Chen X, et al. Novel function of lncrna adamts9-as2 in promoting temozolomide resistance in glioblastoma via upregulating the fus/mdm2 ubiquitination axis. Front Cell Dev Biol 2019;7:217. 10.3389/fcell.2019.00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lebelo MT, Joubert AM, Visagie MH. Warburg effect and its role in tumourigenesis. Arch Pharm Res 2019;42:833-47. 10.1007/s12272-019-01185-2 [DOI] [PubMed] [Google Scholar]
- 6.Lan J, Xue Y, Chen H, et al. Hypoxia-induced miR-497 decreases glioma cell sensitivity to TMZ by inhibiting apoptosis. FEBS Lett 2014;588:3333-9. 10.1016/j.febslet.2014.07.021 [DOI] [PubMed] [Google Scholar]
- 7.Cascón A, Robledo M. MAX and MYC: a heritable breakup. Cancer Res 2012;72:3119-24. 10.1158/0008-5472.CAN-11-3891 [DOI] [PubMed] [Google Scholar]
- 8.Mongiardi MP, Savino M, Falchetti ML, et al. c-MYC inhibition impairs hypoxia response in glioblastoma multiforme. Oncotarget 2016;7:33257-71. 10.18632/oncotarget.8921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu B, Zhao S, Qi C, et al. Inhibition of metabotropic glutamate receptor 5 facilitates hypoxia-induced glioma cell death. Brain Res 2019;1704:241-8. [DOI] [PubMed]
- 10.Guerra F, Bucci C. Role of the RAB7 Protein in Tumor Progression and Cisplatin Chemoresistance. Cancers (Basel) 2019. doi: . 10.3390/cancers11081096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuchitsu Y, Fukuda M. Revisiting Rab7 Functions in Mammalian Autophagy: Rab7 Knockout Studies. Cells 2018. doi: . 10.3390/cells7110215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snider MD. A role for rab7 GTPase in growth factor-regulated cell nutrition and apoptosis. Mol Cell 2003;12:796-7. 10.1016/S1097-2765(03)00401-5 [DOI] [PubMed] [Google Scholar]
- 13.Guerra F, Bucci C. Multiple Roles of the Small GTPase Rab7. Cells 2016. doi: . 10.3390/cells5030034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alonso-Curbelo D, Riveiro-Falkenbach E, Perez-Guijarro E, et al. RAB7 controls melanoma progression by exploiting a lineage-specific wiring of the endolysosomal pathway. Cancer Cell 2014;26:61-76. 10.1016/j.ccr.2014.04.030 [DOI] [PubMed] [Google Scholar]
- 15.Colegio OR, Chu NQ, Szabo AL, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014;513:559-63. 10.1038/nature13490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gnanaprakasam JNR, Sherman JW, Wang R. MYC and HIF in shaping immune response and immune metabolism. Cytokine Growth Factor Rev 2017; 35: 63-70. 10.1016/j.cytogfr.2017.03.004 [DOI] [PubMed] [Google Scholar]
- 17.Oh ET, Kim JW, Kim JM, et al. NQO1 inhibits proteasome-mediated degradation of HIF-1alpha. Nat Commun 2016;7:13593. 10.1038/ncomms13593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh ET, Kim CW, Kim HG, et al. Brusatol-Mediated Inhibition of c-Myc Increases HIF-1alpha Degradation and Causes Cell Death in Colorectal Cancer under Hypoxia. Theranostics 2017;7:3415-31. 10.7150/thno.20861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson AW, Cheng T, Johnston RN. Apoptosis induced by c-myc overexpression is dependent on growth conditions. Exp Cell Res 1995;218:351-8. 10.1006/excr.1995.1166 [DOI] [PubMed] [Google Scholar]
- 20.Xue G, Yan HL, Zhang Y, et al. c-Myc-mediated repression of miR-15-16 in hypoxia is induced by increased HIF-2alpha and promotes tumor angiogenesis and metastasis by upregulating FGF2. Oncogene 2015;34:1393-406. 10.1038/onc.2014.82 [DOI] [PubMed] [Google Scholar]
- 21.Lee JG, Wu R. Erlotinib-cisplatin combination inhibits growth and angiogenesis through c-MYC and HIF-1alpha in EGFR-mutated lung cancer in vitro and in vivo. Neoplasia 2015;17:190-200. 10.1016/j.neo.2014.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conacci-Sorrell M, Ngouenet C, Anderson S, et al. Stress-induced cleavage of Myc promotes cancer cell survival. Genes Dev 2014;28:689-707. 10.1101/gad.231894.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorayappan KDP, Wanner R, Wallbillich JJ, et al. Hypoxia-induced exosomes contribute to a more aggressive and chemoresistant ovarian cancer phenotype: a novel mechanism linking STAT3/Rab proteins. Oncogene 2018;37:3806-21. 10.1038/s41388-018-0189-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian S, Guo X, Yu C, et al. miR-138-5p suppresses autophagy in pancreatic cancer by targeting SIRT1. Oncotarget 2017;8:11071-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Peng W, Zhou Q, et al. LRP6 regulates Rab7-mediated autophagy through the Wnt/β-catenin pathway to modulate trophoblast cell migration and invasion. J Cell Biochem 2020;121:1599-609. 10.1002/jcb.29394 [DOI] [PubMed] [Google Scholar]
- 26.Xu Z, Yan Y, Qian L, et al. Long non-coding RNAs act as regulators of cell autophagy in diseases (Review). Oncol Rep 2017;37:1359-66. 10.3892/or.2017.5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res 2009;15:5308-16. 10.1158/1078-0432.CCR-07-5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jawhari S, Ratinaud MH, Verdier M. Glioblastoma, hypoxia and autophagy: a survival-prone 'menage-a-trois'. Cell Death Dis 2016;7:e2434. 10.1038/cddis.2016.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan Y, Chen X, Wang X, et al. The effects and the mechanisms of autophagy on the cancer-associated fibroblasts in cancer. J Exp Clin Cancer Res 2019;38:171. 10.1186/s13046-019-1172-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Booth LA, Tavallai S, Hamed HA, et al. The role of cell signalling in the crosstalk between autophagy and apoptosis. Cell Signal 2014;26:549-55. 10.1016/j.cellsig.2013.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]