Abstract
Background
N6-methyladenosine (m6A) modification holds an important position in tumorigenesis and metastasis because it can change gene expression and even function in multiple levels including RNA splicing, stability, translocation and translation. In present study, we aim to conducted comprehensive investigation on m6A RNA methylation regulators and m6A-related genes in pancreatic cancer and their association with survival time.
Methods
Based on Univariate Cox regression analysis, protein-protein interaction analysis, LASSO Cox regression, a risk prognostic model, STRING, Spearman and consensus clustering analysis, data from The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC) database was used to analyze 15 m6A RNA methylation regulators that were widely reported and 1,393 m6A-related genes in m6Avar.
Results
We found that 283 candidate m6A RNA methylation-related genes and 4 m6A RNA methylation regulatory factors, including RNA binding motif protein 15 (RBM15), methyltransferase like 14 (METTL14), fat mass and obesity-associated protein (FTO), and α‐ketoglutarate‐dependent dioxygenase AlkB homolog 5 (ALKBH5), differed significantly among different stages of the American Joint Committee on Cancer (AJCC) staging system. Protein-protein interaction analysis indicated epidermal growth factor receptor (EGFR), plectin-1 (PLEC), BLM RecQ like helicase (BLM), and polo like kinase 1 (PLK1) were closely related to other genes and could be considered as hub genes in the network. The results of LASSO Cox regression and the risk prognostic model indicated that AJCC stage, stage T and N, KRAS mutation status and x8q23.3 CNV fragment mutation differed significantly between the high-risk and the low-risk subgroups. The AUCs of 1 to 5 years after surgery were all more than 0.7 and increased year by year. Finally, we found KRAS mutation status and AJCC stage differed significantly among these groups after TCGA samples divided into subgroups with k=7. Moreover, we identified four m6A RNA methylation related genes expressed significantly differently among these seven subgroups, including collagen type VII alpha 1 chain (COL7A1), branched chain amino acid transaminase 1 (BCAT1), zinc finger protein 596 (ZNF596), and PLK1.
Conclusions
Our study systematically analyzed the m6A RNA methylation related genes, including expression, protein-protein interaction, potential function, and prognostic value and provides important clues to further research on the function of RNA m6A methylation and its related genes in pancreatic cancer.
Keywords: m6A modification, m6A relative genes, m6A RNA methylation regulators, pancreatic cancer, survival
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a highly aggressive, cancerous tumor found in the digestive system. Due to its deep anatomical position and occult onset, the symptoms of early-stage PDAC are often indistinct, making diagnosis earlier on challenging. The majority of patients who are diagnosed with PDAC are at the local advanced or distant metastasis stage, and the median survival time of these patients is four to five months. Although major advancements have been made in technology, such as surgical techniques and comprehensive therapy, the 5-year survival rate for PDAC remains at 8.2% (1). Surgical excision is the first choice of treatment for PDAC, although the recurrence rate remains high (at least 60%) (2). Thus, there is an urgent need to identify factors that might affect the prognosis of PDAC patients in clinical practice. Recently, with the help of high-throughput sequencing, studies to identify the molecular markers of PDAC at the cellular and molecular level have made breakthroughs, which will potentially increase the prognostic accuracy and introduce new therapeutic targets for PDAC.
Based on the updated MODOMICS report in 2017, more than 160 kinds of chemical modifications at the post-transcriptional level have been identified among various RNAs (3). Among these chemical modifications, N6-methyladenosine (m6A), which was first reported in the 1970s (4), is considered to be the most common and abundant posttranscriptional modification in eukaryotic mRNA (5). In mammals, the percentage of all adenosine modified by m6A RNA is only 0.1–0.4%, but it is responsible for about 50% of methylated ribonucleotides (6). Almost every aspect of RNA metabolism, from splicing to decay, involves m6A modification (7,8). There is a growing body of literature that recognizes the crucial difference m6A modification makes in many diseases, such as hypertension, cardiovascular diseases, and acute myeloid leukemia (9,10). Moreover, a growing number of studies have shown m6A modification to occupy a prominent position in tumorigenesis and metastasis (11). Therefore, by identifying m6A-related genes and m6A RNA methylation regulators in deadly cancers, new therapeutic targets may be provided. There have also been a number of studies regarding the role of m6A-related genes and m6A RNA methylation regulators, such as methyltransferase like 3 (METTL3) and α-ketoglutarate-dependent dioxygenase AlkB homolog 5 (ALKBH5), in pancreatic cancer (12,13). However, previous studies have only taken into account one or several m6A-related genes or m6A RNA methylation regulators, and few authors have been able to draw on any systematic research into it. Therefore, in this study, we aimed to use RNA-seq data sourced from The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC) databases to conduct a comprehensive investigation of 15 m6A RNA methylation regulators that were widely reported and 1,393 m6A-related genes in the m6Avar database, which contains data on the functional variants involved in m6A modification.
Methods
The standardized RNA-seq data and sample annotation files from 264 pancreatic tumor samples were obtained from the ICGC database. The RPKM data of pancreatic cancer RNA-seq was downloaded from the TCGA database along with the clinical data of the corresponding samples. Fifteen m6A RNA methylation regulatory factors were collected from the known literature (11,14-16) and 1,393 m6A-related genes identified in the m6Avar database (http://m6avar.renlab.org/) (17). Analysis was carried out to investigate the significance between the lines and the difference in survival among different scoring subgroups.
The data sets corresponding to all m6A-related genes were extracted from ICGC and TCGA, and the clinical data were used to study the differences in the expression of m6A-related genes among the pathological samples. To clarify the prognostic effect of m6A correlation in pancreatic cancer patients, we conducted univariate Cox regression analysis of the m6A RNA methylation regulatory factors and m6A-related genes to determine the gene sets related to prognosis. Then, the prognosis-related gene sets from m6A-related genes were further analyzed by LASSO regression analysis. The genes significantly related to prognosis were obtained, and the risk characteristics of each sample were constructed. According to the characteristics and coefficients, the samples were split into two groups: the high-risk group and the low-risk group. Then, the ROC curve of the risk score model was constructed to analyze the one- and five-year survival rates of patients and to evaluate the impact of different clinical characteristics in pancreatic cancer
The candidate gene sets of m6A-related genes were expressed as feature vectors, and the samples were consistently clustered to establish multiple subgroups. We compared the pathological characteristics (grade, TNM staging, age, gene mutation status, specific CNV fragment variation) and survival time between subgroups. Then, the correlation of the m6A candidate gene set was analyzed by STRING and Spearman correlation test, and the function of these differentially expressed genes (DEGs) was analyzed by screening DEGs, among the subgroups.
Data
Expression data
A total of 264 tumor-like RNA-seq data were downloaded from the ICGC database, and the RPKM data of pancreatic cancer RNA-seq were downloaded from the TCGA database. All RNA-seq data was standardized.
Selection of m6A RNA methylation regulatory factors
Fifteen m6A RNA methylation regulators were collected. According to their different roles in the methylation process, they were divided into three types: methyltransferase (writers); binding protein (readers); and demethylase (erasers). The 15 m6A RNA methylation regulative factors were listed in Table 1.
Table 1. The list of the 15 m6A RNA methylation regulative factors from publications.
| Regulaors | Full name | Type |
|---|---|---|
| METTL3 | Methyltransferase like 3 | "writers" |
| METTL14 | Methyltransferase like 14 | "writers" |
| METTL16 | Methyltransferase like 16 | "writers" |
| WTAP | WT1 associated protein | "writers" |
| KIAA1429 | Vir like m6A methyltransferase associated | "writers" |
| RBM15 | RNA binding motif protein 15 | "writers" |
| ZC3H13 | Zinc finger CCCH-type containing 13 | "writers" |
| YTHDC1 | YTH domain containing 1 | "readers" |
| YTHDC2 | YTH domain containing 2 | "readers" |
| YTHDF1 | YTH N6-methyladenosine RNA binding protein 1 | "readers" |
| YTHDF2 | YTH N6-methyladenosine RNA binding protein 2 | "readers" |
| YTHDF3 | YTH N6-methyladenosine RNA binding protein 3 | "readers" |
| HNRNPC | Heterogeneous nuclear ribonucleoprotein C | "readers" |
| FTO | Fat mass and obesity-associated protein | "erasers" |
| ALKBH5 | α-ketoglutarate-dependent dioxygenase AlkB homolog 5 | "erasers" |
A total of 1,393 M6A-related genes related to pancreatic adenocarcinoma (PAAD) were identified in the m6Avar database, and two data sets (m6A RNA methylation regulative factor and m6A RNA methylation related genes associated with PAAD), were integrated. By removing the repeated genes and the genes which had no expression value in the sample or less than 80% of the total expression value in the sample, the candidate m6A-related gene set was obtained. This contained 1,302 candidate genes.
Pathological characteristic data
We downloaded all the clinical data of the corresponding samples from the TCGA database, including the typing data from the time of diagnosis. After deleting the samples which were not with specific pathological stages, a total of 181 samples with pathological stage remained. Then, we divided the samples into four groups according to their different stages. The sample sizes of stages I, II, III, and IV were 21, 151, 4, and 5, respectively. The TCGA samples included 82 females and 99 males, of whom 83 were under 65-year-old and 98 were older than 65-year-old.
Analytical methods
One-way ANOVA
One-way analysis of variance (ANOVA) was employed to compare the difference in expression of m6A RNA methylation regulatory factor between different stages of pancreatic cancer based on the TCGA data samples.
Differential expression data
To determine the m6A RNA methylation regulatory factors that were differentially expressed according to different pathological features, including gender and age, we compared the expression values of the ICGC and TCGA data using the one-way ANOVA and unpaired t-test methods (P<0.05). Both methods were implemented using R language, version 3.6.0.
Consensus clustering for m6A RNA methylation regulatory factor
The samples were analyzed by the unsupervised clustering method using ConsensuClusterPlus R-package, and all the tumor samples corresponding to every candidate M6A methylation regulatory factors were consistently clustered. The pancreatic cancer samples were divided into several subgroups.
Functional and pathway enrichment analysis
Enrichr (http://amp.pharm.mssm.edu/Enrichr) was used to identify the functions and pathways of DEGs that may be associated with pancreatic cancer across the different subgroups. At the same time, GSEA was used to study the related functions of different subtypes of pancreatic cancer.
Survival analysis and construction of risk model
The Survival package of R software was used to analyze the survival of pancreatic cancer samples in the different clusters, and the survival curves were drawn. Kaplan-Meier was used to test the significance of the survival curves and analyze the difference in survival between different subgroups. To construct the risk characteristics and calculate the risk score, the formula was as follows:
According to the median risk score, all of the samples were divided into low and high subtypes. Survival analysis was performed on the low and high subtypes using the R software ‘survival’ package. The Kaplan-Meier method was employed to examine the survival curves and compare the difference in survival across different scoring subgroups.
Results
Identification of m6A RNA methylation related genes which correlated with AJCC stage
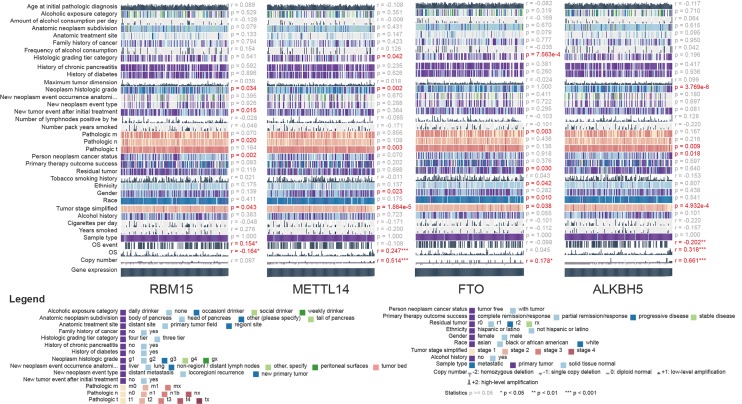
After overlapping 1,393 m6A-related genes related to PAAD and 15 m6A RNA methylation regulative factors, we selected those were with specific information about pathological stage and RNA-seq expression data. We obtained a total of 1,302 candidate m6A RNA methylation-related genes. After dividing the patients into subgroups according to AJCC stage, we identified the m6A RNA methylation-related genes which were expressed differently at different stages of pancreatic cancer. Results showed there to be 283 candidate m6A RNA methylation-related genes that differed significantly among these subgroups (Figure 1). Notably, four of the m6A RNA methylation regulatory factors, RNA binding motif protein 15 (RBM15), METTL14, fat mass and obesity-associated protein (FTO), and ALKBH5, differed significantly between the subgroups. In addition, the expressions of RBM15, METTL14, and ALKBH5 were significantly associated with overall survival in the PAAD cohort (all P<0.05), whereas the expressions of METTL14, FTO, and ALKBH5 had significant correlation with copy number variation (all P<0.05) (Figure S1). Additionally, the expression of RNA binding motif protein 17 (RBM17) was significantly associated with pathologic N status (P=0.02) and new tumor event (NTE) after initial treatment (P=0.015). There was also a significant association between METTL14 (P=0.003) and ALKBH5 (P=0.009) expression and pathologic T status (Figure S1).
Figure 1.
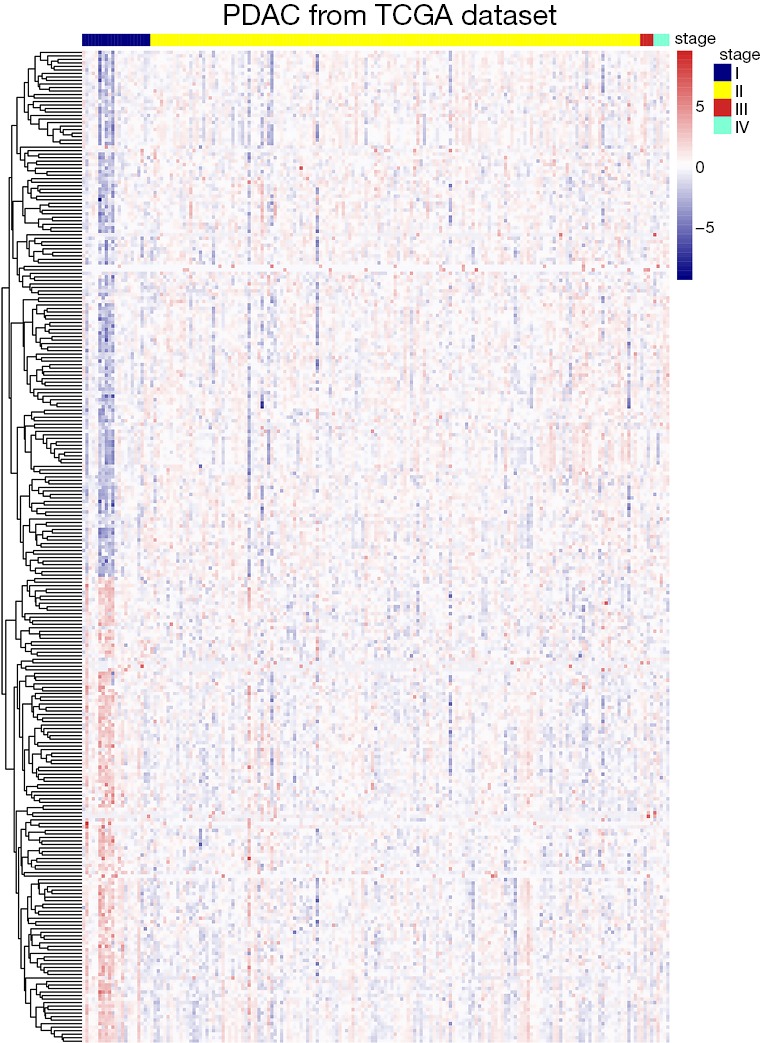
Expression of 283 m6A RNA methylation related-genes at different stages (I–IV) of pancreatic carcinoma. m6A, N6-methyladenosine.
Figure S1.
Association between 4 m6A RNA methylation-related genes, including RBM15, METTL14, FTO, and ALKBH5, and various clinical features in the PAAD cohort from TCGA. Statistics P≥0.05; *, P<0.05; **, P<0.01; ***, P<0.001; r representing coefficient index when analyzing continuous parameters. m6A, N6-methyladenosine; PAAD, pancreatic adenocarcinoma; TCGA, The Cancer Genome Atlas.
Functional annotation of the survival-related m6A RNA methylation-related gene set
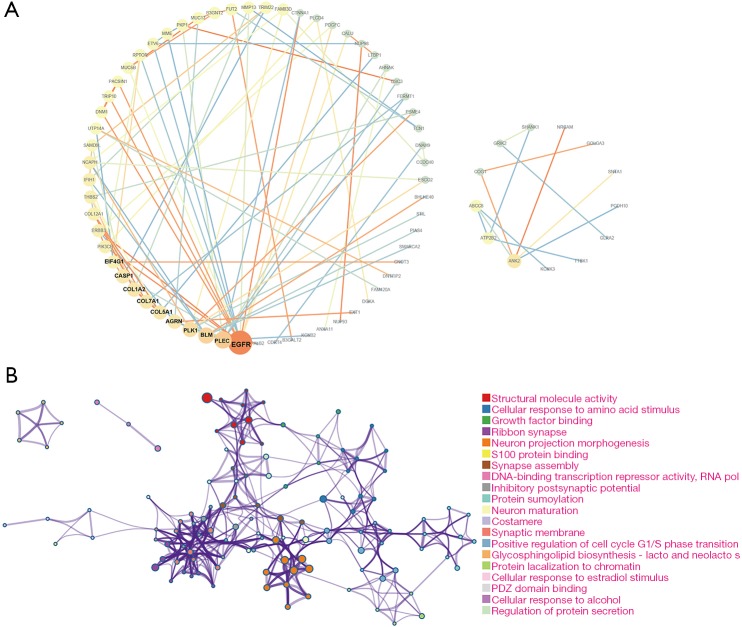
We performed the univariate analysis of the PAAD cohort from TCGA to explore the prognostic potential of the candidate m6A RNA methylation-related gene set. The results showed that 148 out of 283 candidate genes, including m6A RNA methylation regulatory factor ALKBH5, were significantly associated with overall survival of pancreatic cancer. ALKBH5 was indicated to predict favorable overall survival (HR =0.45; 95% CI, 0.27 to 0.74). We then used the bioinformatic tool STRING to analyze functional protein association networks between these 148 candidate genes. The results indicated that epidermal growth factor receptor (EGFR), plectin-1 (PLEC), BLM RecQ like helicase (BLM), and polo like kinase 1 (PLK1) were the hub genes (Figure 2). All statistically enriched terms (Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms) by Metascape, and accumulative hypergeometric p-values and enrichment factors were calculated and used for filtering. The remaining significant terms were then hierarchically clustered into a tree, based on Kappa statistical similarities among their gene memberships. The results indicated that structural molecule activity, cellular response to amino acid stimulus, and growth factor binding pathways were the most significantly enriched (all P<0.001).
Figure 2.
Protein-protein interaction and enrichment analysis of survival-associated m6A RNA methylation-related genes. (A) An interactive network of survival-associated m6A RNA methylation-related genes was created by Cytoscape. Genes are represented as nodes in the diagram, and their interactions are represented by lines. The size and color of the node denotes the degree values, respectively. Genes with lighter colors and larger circles show higher degree value in the network, while darker colors and smaller circles show lower degree values in the network. (B) Enriched Ontology Clusters. Each term is represented by a circle node, where its size is proportional to the number of survival-related genes, and its color represents its cluster identity. The thickness of the edge represents the similarity score. m6A, N6-methyladenosine.
The prognostic value of m6A RNA methylation regulators, and a risk signature built using sixteen selected m6A RNA methylation genes
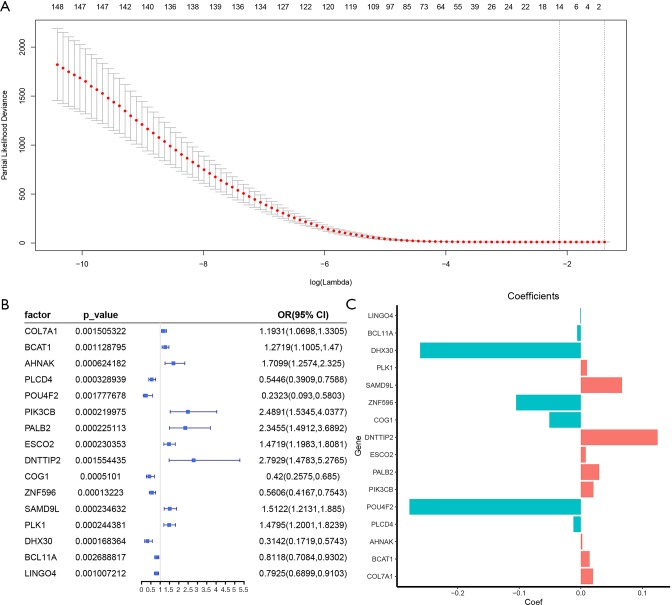
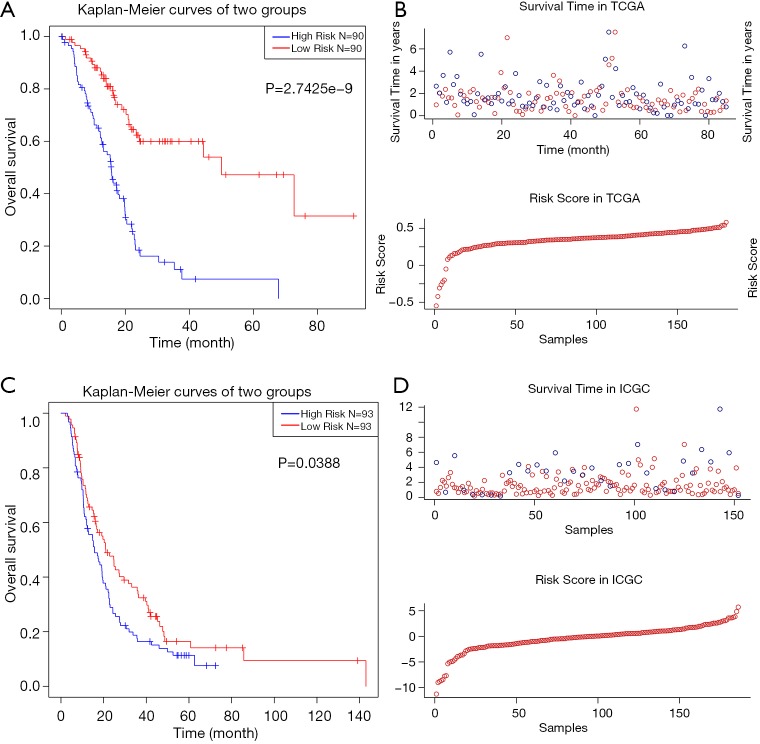
We used LASSO Cox regression to build a prognostic model, which chose 16 m6A RNA methylation-related genes from the PAAD cohort from TCGA (Figure S2). Of these 16 candidate genes for the prognostic model, those found to be significantly associated with poor overall survival were: collagen type VII alpha 1 chain (COL7A1); branched chain amino acid transaminase 1 (BCAT1); AHNAK nucleoprotein (AHNAK); phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta (PIK3CB); partner and localizer of BRCA2 (PALB2); establishment of sister chromatid cohesion N-acetyltransferase 2 (ESCO2); deoxynucleotidyl transferase terminal interacting protein 2 (DNTTIP2); sterile alpha motif domain containing 9 like (SAMD9L); and PLK1. The remaining genes predicted better overall survival of PAAD (Figure S2B,C). The PAAD cohort was then split into the high- and low-risk groups according to the median value of prognostic risk score. Kaplan-Meier survival analysis suggested that overall survival was significantly better in patients with low risk scores than those with high risk scores in the training cohort (P=2.7425e-9) (Figure 3A,B). To verify the use of these candidate prognostic genes as independent biomarkers, and to ensure their prognostic value was not affected by clinical factors, such as age, stage, TNM grade, or gender, we carried out multivariate Cox regression analysis. The results indicated that these clinical features had no significant effect on the prognosis of PAAD (Table S1), which further emphasized the significant value of the prognostic model. The prognostic model was verified with data from the ICGC serving as the validation set. The results showed that this risk model could effectively distinguish the survival of the high- and low-risk groups in the validation set (P=0.0388), as shown in Figure 3C,D.
Figure S2.
Sixteen survival related m6A RNA methylation related-genes were screened. (A) Lasso regression analysis was used to screen 148 m6A RNA methylation related-genes. (B,C) The OR, 95% CI calculated by univariate Cox regression and the coefficients calculated by multivariate Cox regression using funnel plot as shown. m6A, N6-methyladenosine; OR, odds ratio; CI, confidence intervals.
Figure 3.
Multivariate Cox regression analysis was used to construct and analyze risk scores based on 16 survival-related m6A RNA methylation-related genes. According to the median risk score, PAAD patients in the TCGA and ICGC datasets were divided into low- and high-risk groups. The Kaplan-Meier curve described the significant survival difference between the high- and low-risk groups in the prognostic model, and the top of each assembly diagram represents the survival state and survival time of PAAD patients, which was distributed according to the risk score. At the bottom is the risk score curve for PAAD patients. Risk scores were constructed using the TCGA (A,B) and ICGC (C,D) databases. m6A, N6-methyladenosine; PAAD, pancreatic adenocarcinoma; TCGA, The Cancer Genome Atlas; ICGC, the International Cancer Genome Consortium.
Table S1. Multivariate Cox regression analysis for the association between clinical features and the prognosis of PAAD.
| Factor | coef | P value |
|---|---|---|
| Age | 0.018171783 | 0.225533978 |
| Gender_level | −0.185130283 | 0.557321381 |
| Stage_level | −0.142796625 | 0.623627393 |
| M | −0.240936156 | 0.741356109 |
| N | 0.540217548 | 0.152289831 |
| T | −0.032060004 | 0.942718563 |
| Grade | 0.364250232 | 0.088945274 |
PAAD, pancreatic adenocarcinoma.
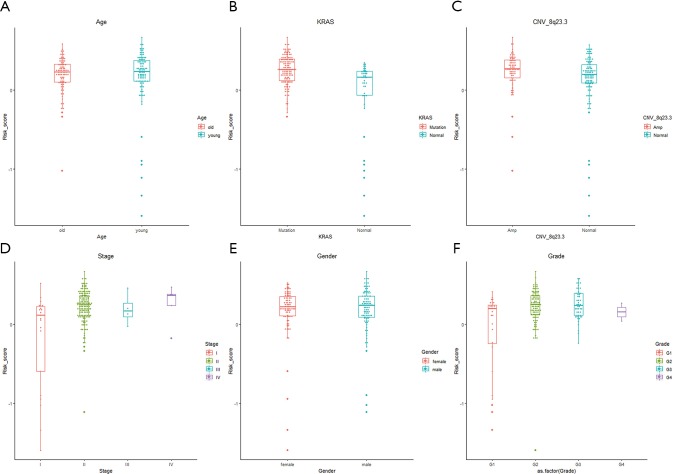
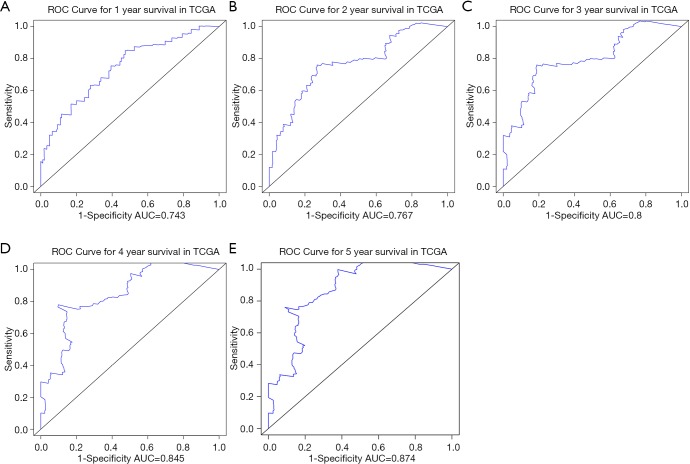
We further explored the correlations between the prognostic model and various clinical features. Gender, age, grade and M stage did not significantly differ between the subgroups with high and low risk, although AJCC stage, stages T and N, KRAS mutation status, and x8q23.3 CNV fragment mutation were found to be significantly different in these two subgroups (all P<0.05) (Figure 4 and Figure S3). ROC curves were used to demonstrate the sensitivity and specificity in predicting the overall survival of patients, and we found that the present risk model depicted a good accuracy of survival prediction at 1 year (AUC =0.743) and 5 years (AUC =0.874) after surgery, suggesting it could be used to predict the prognostic survival of PAAD patients with high accuracy (Figure 5).
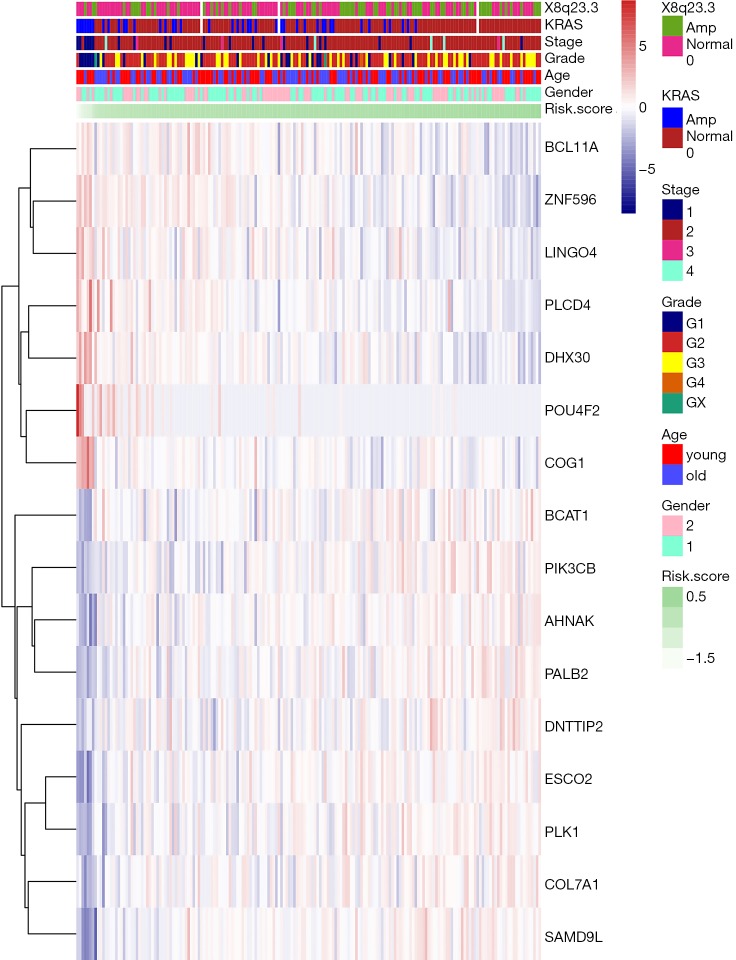
Figure 4.
The heat map showing the expression levels of 16 m6A RNA methylation-related genes in the low- and high-risk groups. The distribution of clinicopathological features was compared between the low- and high-risk groups. m6A, N6-methyladenosine.
Figure S3.
Risk score distribution corresponding to different clinicopathological features of TCGA dataset stratified by age (A), KRAS status (B), CNV_8q23.3 status (C), WHO stage (D), gender (E), WHO grade (F). TCGA, The Cancer Genome Atlas.
Figure 5.
ROC curve showed the predictive efficiency of the 1- to 5-year survival rates (A,B,C,D,E) in the prognostic model of TCGA dataset, respectively. TCGA, The Cancer Genome Atlas.
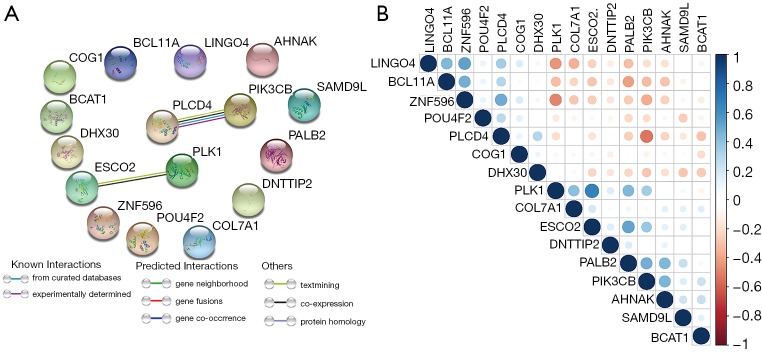
To better understand the interactions among the 16 m6A RNA methylation-related genes, analysis was also carried out on the interactions and correlations between these genes (Figure 6A,B). According to the STRING database, interaction existed between PLK1 and ESCO2, and phospholipase C delta 4 (PLCD4) and PIK3CB. However, 23 pairs with significant correlating factors were identified after the Spearman’s correlation test was carried out on PAAD cohort from TCGA, as shown in Table S2. Notably, we found that zinc finger protein 596 (ZNF596) was positively associated with PLCD4, leucine rich repeat and Ig domain containing 4 (LINGO4) and BCL11A, but negatively associated with PK1 and PIK3CB (all P<0.05). Expression of PLK1 was positively associated with ESCO2, PALB2, COL7A1, and PIK3CB, but negatively associated with LINGO4, BAF chromatin remodeling complex subunit BCL11A (BCL11A), and ZNF596 (all P<0.05). Therefore, little is known about the interaction among these 16 m6A RNA methylation-related genes, and further well-designed studies are warranted.
Figure 6.
Interactions and correlations among 16 survival-related m6A RNA methylation-related genes. (A) m6A modification-related interactions between 16 m6A RNA methylation-related genes. (B) Spearman correlation analysis of 16 m6A RNA methylation-related genes. m6A, N6-methyladenosine.
Table S2. Twenty-three pairs of significant correlation factors identified by the Spearman correlation test from PAAD cohort of TCGA.
| Gene1 | Gene2 | PP | Estimate | abs(estimate) | P value |
|---|---|---|---|---|---|
| COL7A1 | PLK1 | COL7A1(pp)PLK1 | 0.417993 | 0.417993 | 5.27E-09 |
| COL7A1 | LINGO4 | COL7A1(pp)LINGO4 | −0.35413 | 0.354129 | 1.08E-06 |
| AHNAK | PIK3CB | AHNAK(pp)PIK3CB | 0.458289 | 0.458289 | 9.86E-11 |
| AHNAK | PALB2 | AHNAK(pp)PALB2 | 0.44091 | 0.44091 | 5.85E-10 |
| PLCD4 | PIK3CB | PLCD4(pp)PIK3CB | −0.53479 | 0.534792 | 1.05E-14 |
| PLCD4 | ZNF596 | PLCD4(pp)ZNF596 | 0.487728 | 0.487728 | 3.81E-12 |
| PLCD4 | LINGO4 | PLCD4(pp)LINGO4 | 0.342979 | 0.342979 | 2.44E-06 |
| PIK3CB | PALB2 | PIK3CB(pp)PALB2 | 0.472086 | 0.472086 | 2.23E-11 |
| PIK3CB | ESCO2 | PIK3CB(pp)ESCO2 | 0.373616 | 0.373616 | 2.39E-07 |
| PIK3CB | ZNF596 | PIK3CB(pp)ZNF596 | −0.37598 | 0.375981 | 1.98E-07 |
| PIK3CB | PLK1 | PIK3CB(pp)PLK1 | 0.341393 | 0.341393 | 2.73E-06 |
| PIK3CB | BCL11A | PIK3CB(pp)BCL11A | −0.31308 | 0.31308 | 1.88E-05 |
| PALB2 | ESCO2 | PALB2(pp)ESCO2 | 0.511654 | 0.511654 | 2.13E-13 |
| PALB2 | PLK1 | PALB2(pp)PLK1 | 0.446083 | 0.446083 | 3.48E-10 |
| PALB2 | BCL11A | PALB2(pp)BCL11A | −0.41452 | 0.414518 | 7.25E-09 |
| PALB2 | LINGO4 | PALB2(pp)LINGO4 | −0.30305 | 0.303054 | 3.55E-05 |
| ESCO2 | PLK1 | ESCO2(pp)PLK1 | 0.660751 | 0.660751 | 5.97E-24 |
| ZNF596 | PLK1 | ZNF596(pp)PLK1 | −0.48507 | 0.485069 | 5.17E-12 |
| ZNF596 | BCL11A | ZNF596(pp)BCL11A | 0.472231 | 0.472231 | 2.19E-11 |
| ZNF596 | LINGO4 | ZNF596(pp)LINGO4 | 0.520534 | 0.520534 | 6.89E-14 |
| PLK1 | BCL11A | PLK1(pp)BCL11A | −0.30743 | 0.307431 | 2.69E-05 |
| PLK1 | LINGO4 | PLK1(pp)LINGO4 | −0.44449 | 0.444494 | 4.08E-10 |
| BCL11A | LINGO4 | BCL11A(pp)LINGO4 | 0.448756 | 0.448756 | 2.65E-10 |
PAAD, pancreatic adenocarcinoma; TCGA, The Cancer Genome Atlas.
The prognostic value of the candidate m6A-related genes for pancreatic cancer with distinct clinical outcomes and clinicopathological features
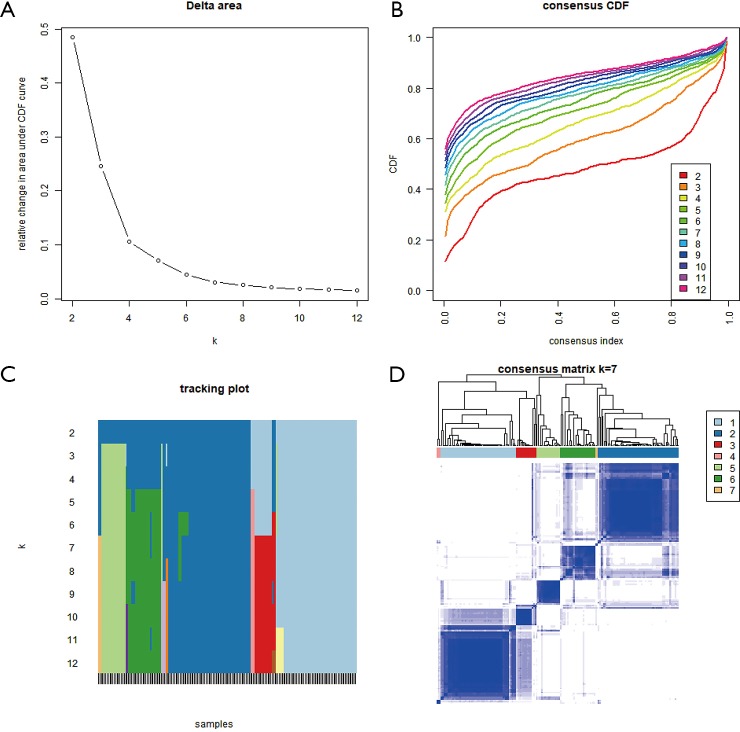
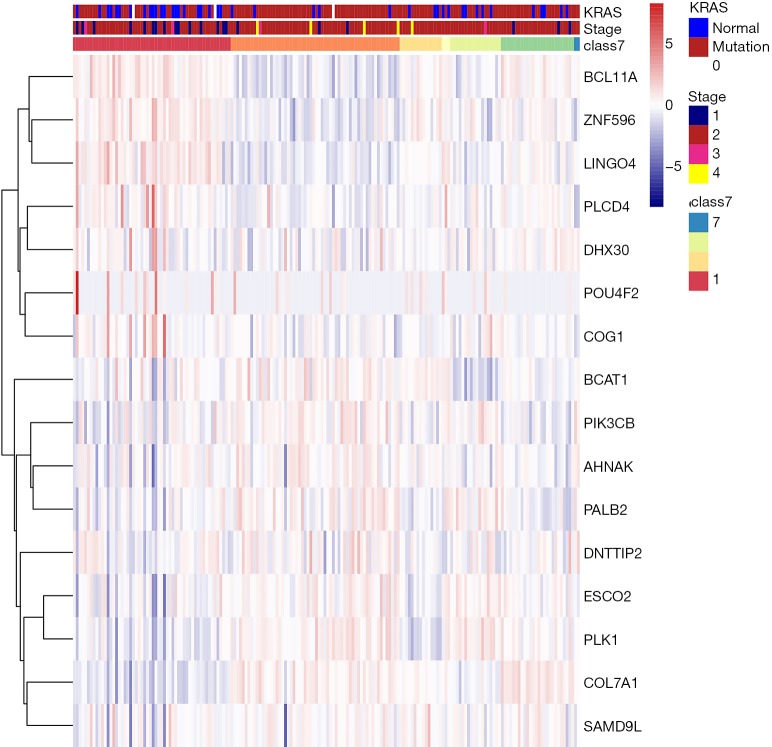
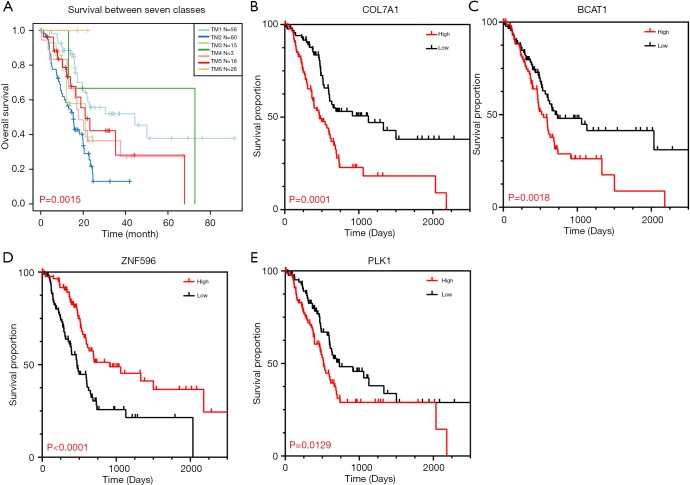
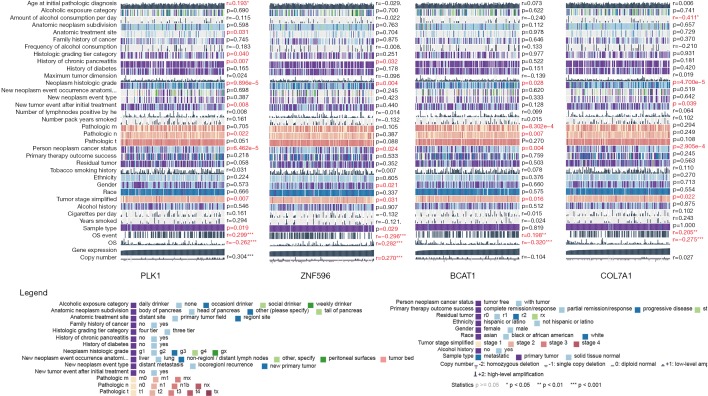
To specifically investigate the function of the candidate m6A-related genes in pancreatic cancer, we divided the TCGA pancreatic cancer samples into different subgroups according to the expression similarity of 16 m6A-related genes using the R package software ConsensusClusterPlus. Based on the expression similarity of m6A RNA methylation regulators, k=7 appeared to be an adequate choice, with clustering stability rising from k=2 to 12 for the TCGA datasets. All subgroups were named TM1 to TM7. Further analysis revealed that cancer stage and KRAS mutation status differed significantly among these seven subgroups (Figure S4). We explored the differences in clinical features among these subgroups, discovering that KRAS mutation status and AJCC stage differed significantly among these groups (both P<0.05) (Figure 7). Moreover, survival analysis indicated that a significant difference in survival between these 7 subgroups (P=0.0015). We analyzed the genes which were expressed differently among these 7 subgroups, and found that the expressions of 6 of the 16 m6A RNA methylation-related genes, including COL7A1, BCAT1, ZNF596, and PLK1, were significantly different. In addition, COL7A1 (P=0.0001), BCAT1 (P=0.0118), and PLK1 (P=0.0129) were associated with poor overall survival, whereas ZNF596 (P<0.0001) was significantly associated with increased overall survival (Figure 8). We further investigated the associations between the clinical features and expression of these six genes. Remarkably, there was a positive association between PLK1 (P<0.001), BCAT1 (P=0.004), and COL7A1 (P<0.001) expression and positive cancer status, which indicated relapse after the initial surgery at a particular time point. Consensus clustering analysis of m6A RNA methylation regulators singled out two cluster points in time. However, expression of ZNF596 (P=0.024) was negatively associated with tumor-free status, which suggested that COL7A1, BCAT1, ZNF596, and PLK1 may be involved in tumor recurrence, and subsequently influence the survival of PAAD (Figure 9).
Figure S4.
Using 16 m6A RNA methylation related-genes, the TCGA dataset was divided into 7 subtypes (TM1–TM7). (A) The relative variation of the area under the Consensus clustering CDF curve for k=2 to 12. (B) CDF curve for k=2 to 12. (C) Tracking plot for k=2 to12. (D) The consensus clustering matrix at k=7. m6A, N6-methyladenosine; TCGA, The Cancer Genome Atlas; CDF, cumulative distribution function.
Figure 7.
Consensus clustering analysis of the 7 subtypes of heat maps and significant clinicopathological features.
Figure 8.
Kaplan-Meier analysis. (A) Kaplan-Meier survival curve of PAAD patients in TCGA database. (B,C,D,E) Survival curve analysis of the m6A RNA methylation-related genes differentially expressed in 7 subgroups, which was significantly related to the prognosis of PAAD. PAAD, pancreatic adenocarcinoma; TCGA, The Cancer Genome Atlas.
Figure 9.
Association between 4 m6A RNA methylation related-genes, including PLK1, ZXF596, BCAT1 and COL7A1, and various clinical features in PAAD cohort from TCGA. Statistics P≥0.05; *, P<0.05; **, P<0.01; ***, P<0.001; r representing coefficient index when analyzing continuous parameters. m6A, N6-methyladenosine; PAAD, pancreatic adenocarcinoma; TCGA, The Cancer Genome Atlas.
Discussion
PDAC is a highly aggressive and deadly disease. Despite the remarkable technological advancements seen in recent decades, the 5-year survival rate (8.2%) for PDAC remains bleak. Moreover, for PDAC patients who are able to undergo surgical resection, the recurrence rate is at least 60%. Therefore, there is an urgent need to identify novel therapeutic strategies for PDAC as well as factors that might affect its prognosis in clinical practice. Over 160 kinds of chemical modifications have been identified among various RNAs at the post-transcriptional level. Among these, there is mounting evidence to suggest that m6A modification makes a crucial difference not only in hypertension and cardiovascular diseases but also in tumorigenesis and metastasis (18). Thus, identifying m6A-related genes and m6A RNA methylation regulators in deadly pancreatic cancers may provide us with valuable therapeutic targets.
Previous studies that have pointed out the importance of METTL3 and ALKBH5 in the development and progression of pancreatic cancer; however, little is known about the function of m6A-related genes. Therefore, we carried out the present study to identify the key genes from 1,302 candidate m6A RNA methylation-related genes and further explored their significance in relation to the clinical features of pancreatic cancer patients.
Our results showed that 283 candidate m6A RNA methylation-related genes differed significantly across different AJCC stages, which implied that these genes may engage in the development of pancreatic cancer. It is worth noting that our results also identified significant differences among AJCC-stage subgroups with four of the m6A RNA methylation regulatory factors, including RBM15, METTL14, FTO, and ALKBH5.
As a critical demethylase which regulates cellular mRNA stability by removing N-methyladenosine (mA) residues in mRNA, FTO has been demonstrated to be linked to body mass index, risk of obesity, and even cancer (19). Recent evidence has shown the gene polymorphism (rs9939609 T/A) of FTO to be significantly associated with increased risk of pancreatic cancer (20). Other studies have suggested that FTO could enhance its stability by interaction with MYC proto-oncogene, bHLH transcription factor, and thus promote cancer cell growth (21). Cho et al. found that the ALKBH5 gene is a novel biomarker that is a prognostic predictor for pancreatic cancer patients, which supports the reliability and accuracy of our present results (22). In contrast, the functions of RBM15 and METTL14 in pancreatic cancer are yet to be revealed. Remarkably, RBM15 and METTL14 were found to have a significant association with overall survival in our study. In addition, RBM17 expression was significantly associated with pathologic N status and new tumor event (NTE) after initial treatment. Based on their significant association with clinical features, it is reasonable to suspect that RBM15 and METTL14 may engage in the initiation and development of pancreatic cancer, and further well-designed studies are required.
Univariate analysis revealed 148 out of 283 candidate genes to be closely associated with overall survival in pancreatic cancer. Protein-protein interaction analysis between these 148 survival-associated genes was further conducted. The results indicated that EGFR, PLEC, BLM, and PLK1 are closely related to other genes and could be considered as hub genes in the network.
Numerous studies have shown EGFR, which correlated with most of the m6A-related genes among these 148 survival-associated genes, to act as an oncogene and some EGFR inhibitors hold great promise in clinical use (23). EGFR expression has been detected in a high percentage of PDAC (up to 90%) (24), and it occupies a crucial position in PDAC recurrence and liver metastasis. According to clinical data, there is a supposed association between overexpression of EGFR and more aggressive tumor behavior and higher postoperative recurrence rates (25).
As a downstream gene activated by the PI3K/Akt and NF-κB signaling pathways, PLK1 is closely related to poor prognosis in a variety of cancers. Previous studies have demonstrated that by combining PI3K inhibitor LY294002 together with gemcitabine to target the depletion of Plk1, sensitivity to chemotherapy could be enhanced (26). Using the amphiphilic nanocarrier, combined regulation of PLK1 and miR-34a could improve therapeutic response (27), potentially presenting a new therapeutic choice for treating gemcitabine-resistant pancreatic cancer (28).
Furthermore, PLEC is known to be aberrantly expressed in the surface of PDAC cells but negative in benign tissues, which puts forward another ideal target for cancer diagnosis and therapy (29,30).
BLM may act to suppress inappropriate recombination in normal cells and prevent instability of structure-forming DNA sequences, but aberrant BLM expression has been detected in colorectal cancer and leukemia (31,32). However, there is no existing evidence which illustrates the function of BLM in pancreatic cancer. As BLM is important to maintain genome stability, elucidating its function in pancreatic cancer holds immense significance for new therapeutic benefits (33).
Taken together, the expression of m6A RNA methylation-related genes were closely associated with malignant clinicopathological features in pancreatic cancer.
To further explore the potential m6A RNA methylation regulators might hold for PAAD prognosis, LASSO Cox regression was carried out and a risk prognostic model was built by 16 m6A RNA methylation-related genes. The prognostic model was verified with data from ICGC database serving as the validation set. Interestingly, Kaplan-Meier analysis indicated that the survival of high-risk and low- risk groups could be distinguished in both the training group and the validation set. Further analysis revealed that AJCC stage, stages T and N, KRAS mutation status, and x8q23.3 CNV fragment mutation differed significantly in the high-risk and the low-risk subgroups. The AUCs of 1 year to 5 years after surgery were all more than 0.7 and increased year by year, which indicates that the present risk model is an ideal model for predicting survival of PAAD with high accuracy. We also tried to elaborate on the interactions among the 16 m6A RNA methylation-related genes and found interactions between PLK1 and ESCO2, and PLCD4 and PIK3CB according to the STRING database. Correlation analysis was conducted, and we found several significant associations to exist among these genes. However, more needs to be understood about the interaction among these 16 m6A RNA methylation-related genes.
Consensus clustering analysis of m6A RNA methylation regulators was also performed to identify two clusters of pancreatic cancer with defined clinical outcomes and clinicopathological features. After the TCGA pancreatic cancer samples were divided into subgroups with k=7, we found KRAS mutation status and AJCC stage differed significantly between the groups. Moreover, we identified four m6A RNA methylation-related genes that were expressed significantly differently among these seven subgroups, including COL7A1, BCAT1, ZNF596, and PLK1. The association between the expressions of these four m6A RNA methylation-related genes and clinical features was further investigated. Notably, PLK1, BCAT1, and COL7A1 were found to be positively associated with positive cancer status, while ZNF596 were negatively associated with tumor-free status. Previous reports have suggested that inhibition of COL7A1, a TGF-beta-regulated gene, by a TGF-betaRI kinase inhibitor could offer a potential therapeutic benefit for pancreatic cancer (34). BCAT1 is the enzymes responsible for branched-chain amino acids utilization, but loss of it was recently shown to not be required for PDAC tumor formation, which is inconsistent with the present findings (35). The functions of ZNF596, a member of the zinc finger protein family, are still unknown. Recent evidence showed that ZNF596 regulated the EZH2/STAT3 signaling pathway, and thus promoted glioma stem-like cell tumorigenicity (36). A study concerning the function of ZNF596 in pancreatic cancer is yet to be carried out. In our study, ZNF596 was found to be significantly associated with better overall survival, which indicates its anti-tumor function in pancreatic cancer and is inconsistent with previous study (36). Therefore, more studies are warranted to fully elucidate the function of ZNF596 in pancreatic cancer.
Conclusions
In summary, our study systematically analyzed the m6A RNA methylation-related genes, including their expression, protein-protein interaction, potential function, and prognostic value. We found a close association between the expression of m6A RNA methylation-related genes and malignant clinicopathological features in pancreatic cancer. Several genes were identified, which are useful for identifying novel therapeutic targets, and chemicals modulating the m6A methylation process may offer a new treatment strategy for pancreatic cancer. In summary, our study provides important clues for further investigation into the function of RNA m6A methylation and its related genes in pancreatic cancer.
Acknowledgments
Funding: This study was supported by grants from the National Natural Science Foundation of China (Grant No. 81701560), Guangdong Natural Science Foundation (Grant No. 2017A030313530 and 2019A1515011676), Guangzhou Science and Technology Plan of Scientific Research Projects, People’s Republic of China (Grant No. 201904010021) and High-level Hospital Construction Project (Grant No. DFJH201915). These funds made significant contributions to the study design, data interpretation, and writing of this paper.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Ciofoaia V, Haddad NG, Smith JP. How many pancreatic cysts are out there and how to best manage them? Transl Cancer Res 2018;7:S500-5. 10.21037/tcr.2018.03.30 [DOI] [Google Scholar]
- 3.Boccaletto P, Machnicka MA, Purta E, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 2018;46:D303-7. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A 1974;71:3971-5. 10.1073/pnas.71.10.3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev 2015;29:1343-55. 10.1101/gad.262766.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell 1975;4:379-86. 10.1016/0092-8674(75)90158-0 [DOI] [PubMed] [Google Scholar]
- 7.Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017;169:1187-200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 2017;18:31-42. 10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ianniello Z, Paiardini A, Fatica A. N6-Methyladenosine (m6A): A Promising New Molecular Target in Acute Myeloid Leukemia. Front Oncol 2019;9:251. 10.3389/fonc.2019.00251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paramasivam A, Vijayashree Priyadharsini J, Raghunandhakumar S. N6-adenosine methylation (m6A): a promising new molecular target in hypertension and cardiovascular diseases. Hypertens Res 2020;43:153-4. 10.1038/s41440-019-0338-z [DOI] [PubMed] [Google Scholar]
- 11.Chen XY, Zhang J, Zhu JS. The role of m6A RNA methylation in human cancer. Mol Cancer 2019;18:103. 10.1186/s12943-019-1033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia T, Wu X, Cao M, et al. The RNA m6A methyltransferase METTL3 promotes pancreatic cancer cell proliferation and invasion. Pathol Res Pract 2019;215:152666. 10.1016/j.prp.2019.152666 [DOI] [PubMed] [Google Scholar]
- 13.He Y, Hu H, Wang Y, et al. ALKBH5 Inhibits Pancreatic Cancer Motility by Decreasing Long Non-Coding RNA KCNK15-AS1 Methylation. Cell Physiol Biochem 2018;48:838-46. 10.1159/000491915 [DOI] [PubMed] [Google Scholar]
- 14.Dai D, Wang H, Zhu L, et al. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis 2018;9:124. 10.1038/s41419-017-0129-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet 2014;15:293-306. 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- 16.Lan Q, Liu PY, Haase J, et al. The Critical Role of RNA m6A Methylation in Cancer. Cancer Res 2019;79:1285-92. 10.1158/0008-5472.CAN-18-2965 [DOI] [PubMed] [Google Scholar]
- 17.Zheng Y, Nie P, Peng D, et al. m6AVar: a database of functional variants involved in m6A modification. Nucleic Acids Res 2018;46:D139-45. 10.1093/nar/gkx895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother 2019;112:108613. 10.1016/j.biopha.2019.108613 [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, Ueda J, Yagyu K, et al. Association between variations in the fat mass and obesity-associated gene and pancreatic cancer risk: a case-control study in Japan. Bmc Cancer 2013;13:337. 10.1186/1471-2407-13-337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X, Zhao J, Yang M, et al. Association between FTO gene polymorphism (rs9939609 T/A) and cancer risk: a meta-analysis. Eur J Cancer Care (Engl) 2017. doi: . 10.1111/ecc.12464 [DOI] [PubMed] [Google Scholar]
- 21.Tang X, Liu S, Chen D, et al. The role of the fat mass and obesity-associated protein in the proliferation of pancreatic cancer cells. Oncol Lett 2019;17:2473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho SH, Ha M, Cho YH, et al. ALKBH5 gene is a novel biomarker that predicts the prognosis of pancreatic cancer: A retrospective multicohort study. Ann Hepatobiliary Pancreat Surg 2018;22:305-9. 10.14701/ahbps.2018.22.4.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verma HK, Kampalli PK, Lakkakula S, et al. A Retrospective Look at Anti-EGFR Agents in Pancreatic Cancer Therapy. Curr Drug Metab 2019;20:958-66. 10.2174/1389200220666191122104955 [DOI] [PubMed] [Google Scholar]
- 24.Mollinedo F, Gajate C. Novel therapeutic approaches for pancreatic cancer by combined targeting of RAF→MEK→ERK signaling and autophagy survival response. Ann Transl Med 2019;7:S153. 10.21037/atm.2019.06.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polireddy K, Chen Q. Cancer of the Pancreas: Molecular Pathways and Current Advancement in Treatment. J Cancer 2016;7:1497-514. 10.7150/jca.14922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao Y, Xi L, Li Q, et al. Combination of PI3K/Akt Pathway Inhibition and Plk1 Depletion Can Enhance Chemosensitivity to Gemcitabine in Pancreatic Carcinoma. Transl Oncol 2018;11:852-63. 10.1016/j.tranon.2018.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibori H, Eliyahu S, Krivitsky A, et al. Amphiphilic nanocarrier-induced modulation of PLK1 and miR-34a leads to improved therapeutic response in pancreatic cancer. Nat Commun 2018;9:16. 10.1038/s41467-017-02283-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Wang R, Schweickert PG, et al. Plk1 inhibition enhances the efficacy of gemcitabine in human pancreatic cancer. Cell Cycle 2016;15:711-9. 10.1080/15384101.2016.1148838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Zhou H, Li X, et al. Plectin-1 Targeted Dual-modality Nanoparticles for Pancreatic Cancer Imaging. Ebiomedicine 2018;30:129-37. 10.1016/j.ebiom.2018.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pal K, Al-Suraih F, Gonzalez-Rodriguez R, et al. Multifaceted peptide assisted one-pot synthesis of gold nanoparticles for plectin-1 targeted gemcitabine delivery in pancreatic cancer. Nanoscale 2017;9:15622-34. 10.1039/C7NR03172F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Min J, Wright WE, Shay JW. Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev 2019;33:814-27. 10.1101/gad.324905.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Votino C, Laudanna C, Parcesepe P, et al. Aberrant BLM cytoplasmic expression associates with DNA damage stress and hypersensitivity to DNA-damaging agents in colorectal cancer. J Gastroenterol 2017;52:327-40. 10.1007/s00535-016-1222-0 [DOI] [PubMed] [Google Scholar]
- 33.Tripathi V, Agarwal H, Priya S, et al. MRN complex-dependent recruitment of ubiquitylated BLM helicase to DSBs negatively regulates DNA repair pathways. Nat Commun 2018;9:1016. 10.1038/s41467-018-03393-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medicherla S, Li L, Ma JY, et al. Antitumor activity of TGF-beta inhibitor is dependent on the microenvironment. Anticancer Res 2007;27:4149-57. [PubMed] [Google Scholar]
- 35.Mayers JR, Torrence ME, Danai LV, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 2016;353:1161-5. 10.1126/science.aaf5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang J, Yu B, Li Y, et al. TGF-β-activated lncRNA LINC00115 is a critical regulator of glioma stem-like cell tumorigenicity. EMBO Rep 2019;20:e48170. 10.15252/embr.201948170 [DOI] [PMC free article] [PubMed] [Google Scholar]