Abstract
Background
To identify key microRNAs (miRNAs) and their target mRNAs related to gemcitabine-resistant pancreatic cancer (PC) and investigate the association between gemcitabine-resistant-related miRNAs and mRNAs and immune infiltration.
Methods
Expression profiles of miRNAs and mRNAs were obtained from the Gene Expression Omnibus (GEO) database. The differentially expressed miRNAs and mRNAs (referred to as “DEmiRNAs” and “DEmRNAs”, respectively) were distinguished between gemcitabine-resistant PC cells and its parental cells. The DEmRNAs targeted by the DEmiRNAs were retrieved using miRDB, microT, and Targetscan. Furthermore, GO and KEGG pathway enrichment analysis and GSEA were performed. The Kaplan-Meier plotter was used to analyze the prognosis of key DEmiRNAs and DEmRNAs on PC patients. The relationship between the key DEmRNAs and tumor-infiltrating immune cells in PC was investigated using CIBERSORT method using the LM22 signature as reference. Key infiltrating immune cells were further analyzed for the associations with prognosis of TCGA PAAD patients.
Results
Four DEmiRNAs, including hsa-miR-3178, hsa-miR-485-3p, hsa-miR-574-5p, and hsa-miR-584-5p, were identified to target seven DEmRNAs, including MSI2, TEAD1, GNPDA1, RND3, PRKACB, TRIM68, and YKT6, individually, in gemcitabine-resistant PC cells versus parental cells. Gemcitabine-resistant PC cells were enriched in proteasome-related, immune-related, and memory CD4+ T cell-related pathways, indicating a gemcitabine therapeutic effect on PC cells. All four DEmiRNAs and almost all DEmRNAs had an impact on the prognosis of PC patients. All seven DEmRNAs had remarkable effects on CD4+ memory T cells, which were affected by the gemcitabine therapeutic effect. Effector memory CD4+ T cells rather than central memory CD4+ T cells predicted a good prognosis according to the TCGA PAAD dataset.
Conclusions
Gemcitabine resistance can alter the fraction of memory CD4+ T cells via hsa-miR-3178, hsa-miR-485-3p, hsa-miR-574-5p and hsa-miR-584-5p targeted MSI2, TEAD1, GNPDA1, RND3, PRKACB, TRIM68, and YKT6 network in PC.
Keywords: Gemcitabine resistance, microRNAs, tumor-infiltrating immune cells, CIBERSORT, CD4+ memory T cells
Introduction
Pancreatic cancer (PC) is the fourth most lethal malignancy, with a poor prognosis and a 5-year relative survival rate of only 9% (1). In patients with inoperable or unresectable PC, gemcitabine treatment is the frontline chemotherapy in PDA, though its mechanism of resistance remains unclear.
MicroRNAs (miRNAs), a class of small, endogenous single-stranded RNAs of approximately 22 nucleotides in length initially discovered in 1993 (2), often play a negative regulatory effect in animals and plants via targeting specific mRNAs for degradation or translation suppression (3). However, the emerging role of miRNAs as activators of gene expression by targeting gene regulatory sequences (4) has been fully recognized just recently. Place et al. described this novel miRNA-induced RNA activation (miRNAa) phenomena and discovered that miR-373 bound the E-cadherin promoter sequences and induced gene expression (5). Interestingly, its mechanism has since then been elucidated by several subsequent studies. A series of such studies were followed to elucidate the mechanism. Xiao et al. reported that miR-24-1 could serve as an enhancer trigger by modifying chromatin status favorable for transcriptional gene activation (6). In support to this view, Huang et al. revealed an endogenous function for miRNA in gene activation as miR-744 and miR-1186 could induce CCNB1 expression and reinforce cancer cell growth (7).
Notably, aberrant expression of miRNAs is linked to gemcitabine sensitivity/resistance (8,9). Wang et al. validated the role of miR-30a in PC sensitization to gemcitabine (10). Another study reported that gemcitabine-resistant cells exhibited upregulated miR-301 expression and downregulated gemcitabine-induced apoptosis (11).
The differential expression of miRNAs has also been reported in modulation of immune infiltration. Frank et al. provided evidence for tumor-immune cell interactions shaping the immune cell phenotype and miR-375 acting as a crucial regulator of phagocyte infiltration and the subsequent development of a tumor-promoting microenvironment (12). In addition, Pyfferoen et al. reported that dendritic cell-derived miR-31 promoted lung cancer progression (13).
Unfortunately, the underlying mechanisms of gemcitabine resistance in PC are poorly understood, and the impact of gemcitabine resistance in tumor-associated immune cells is implicit as well.
CIBERSORT is utilized to explore the relationship between immune infiltration and gemcitabine resistance. The bioinformatics tool of CIBERSORT was developed to deconvolve the expression matrix of immune cell subtype based on the principle of linear support vector regression (14). This deconvolution algorithm characterizes cell composition of complex tissues based on their gene expression profiles. In this study, CIBERSORT was used to assess the relative proportions of 22 tumor-infiltrating immune cells in PC and to investigate the relationship between the composition of tumor-infiltrating immune cells and gemcitabine therapeutic effect.
Here, we identified four differentially expressed miRNAs and their targeted DEmRNAs in gemcitabine resistant PC cells from the Gene Expression Omnibus (GEO) database using integrated bioinformatics analysis. Subsequently, through the bioinformatics tool of CIBERSORT, we explored the relationship between immune infiltration and gemcitabine resistance according to the gene expression profiles of PC from the TCGA database and found that gemcitabine therapeutic effects were closely associated to memory CD4+ T cells. Collectively, this study showed a T cell immune-related miRNAa regulatory network. Additionally, our findings provide insights into the role of memory CD4+ T cells in PC chemotherapy and can potentially help in the design of future treatments.
Methods
Cell lines and cell culture
Gemcitabine-resistant PC cells (PANC-1-GEM) and its parental cells (PANC-1) used in this research were purchase from Suyan Co (Guangzhou, China). All cells were maintained in RPMI medium 1640 (Gibco Invitrogen, Grand Island, NY, USA) with 10% fetal bovine serum (FBS) (Gibco Invitrogen, Grand Island, NY, USA) and 1% antibiotics, in a humidified atmosphere containing 5% carbondioxide (CO2) and 95% air at 37 °C. Additionally, the PANC-1-GEM cells was cultured with 80 µmol/L gemcitabine to maintain drug resistance and gemcitabine was removed from RPMI medium 48 hours before any experiment was performed.
RNA extraction and real-time PCR (qRT-PCR)
Trizol regent (Invitrogen, Camarillo, CA, USA) was used for total RNA isolation from PC cells according to the manufacturer’s instruction. After extraction, RNA concentration was detected and the RNA was maintained at −80 °C refrigerator to avoid degradation. miRNAs expressions were determined using Mir-X miR FirstStrand Synthesis Kit (TaKaRa) and SYBR Premix Ex Taq II (TaKaRa) and RNU6-2 was used as an internal control. The 2-ΔΔCT method was used to calculate the relative expressions of miRNAs. Genecopoeia Co. (Guangzhou, China) designed the bulge-loop miRNA qRT-PCR Primer Sets specific for four miRNAs.
miRNAs overexpression and silencing
miRNAs inhibitor, inhibitor negative control (Inhi-NC), miRNAs mimics and mimic negative control (Mim-NC) were purchased from Genecopoeia Co. (Guangzhou, China). Transfection was performed using Lipofectamine 3000 (Invitrogen) according to the protocol.
Cell viability assay
Cell viability ability was determined by using CCK-8 assay. CCK-8 assay was performed under the manufacturer’s instructions (Dojinodo, Shanghai, China). PANC-1-GEM and PANC-1 cells with transfection were seeded in 96-well plates at 5×103 cells per well and incubated with gemcitabine for 48 h. CCK-8 (10 µL per 100 µL) was added to each well and incubated for 2 hours at 37 °C before detection.
Data collection
The mRNA and miRNA expression data used in this study were obtained from the GEO database. miRNA expression data were acquired from GSE74562, GSE74565, and GSE79234. mRNA expression data were obtained derived from GSE80617 and GSE79953.
Identification of DEmRNAs and DEmiRNAs in gemcitabine-resistant PC cells compared with its parental cells
We firstly filtered and deleted the difficultly detected mRNAs or miRNAs with read count value =0 or missing values in more than 33.3% of samples. Then, the k-nearest neighbor algorithm (KNN) was used for filling in the missing values. The differentially expressed mRNAs (“DEmRNAs”) and miRNAs (“DEmiRNAs”) between gemcitabine resistant PC cells and their parental cells were screened out individually using the limma package in the R 3.3.2 software. The threshold for the expression of DEmRNAs and DEmiRNAs was P<0.05 and fold change >1.2. The DEmRNAs and DEmiRNAs whose expression trend was inconsistent in two mRNAs datasets or three miRNAs datasets were excluded. With R package pheatmap, hierarchical clustering analysis of DEmRNAs and DEmiRNAs were conducted.
Prediction of DEmiRNAs target mRNAs and construction of the DEmiRNAs-DEmRNAs interaction pairs
Three bioinformatics algorithms, including miRDB, microT, and Targetscan were used to predict the targeted DEmRNAs. In the predicted results by Targetscan, the “Total context+ score” less than −0.2 was selected. Only mRNAs predicted by all three databases were considered as candidate mRNAs. The candidate mRNAs that intersected with DEmRNAs were screened out, and a miRNA-target gene network was constructed. The Cytoscape (version 3.7.1) software was utilized to draw this network.
Potential functional enrichment analysis of key DEmRNAs related to gemcitabine-resistant PC
Functional enrichment analysis of the key DEmRNAs in this network was performed based on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases to identify the genes’ functions and pathways regarding gemcitabine-resistant PC. Metascape tool was used to display network relationships among DEmRNAs.
Gene set enrichment analysis (GSEA)
GSEA was performed using GSEA 3.0 (JAVA) using GSE79953 dataset. Datasets were divided into two subgroups, gemcitabine-resistant group and parental group. Immune-related gene sets were downloaded in MSigDB, including “go response to interferon gamma”, “memory CD4 T cell vs. B cell up”, “naive vs. memory CD4 T cell up”, “hallmark IL-6 JAK STAT3 signaling”, “immune response”, “IL27 pathway”, “reactome cytokine signaling in immune system”, “go cellular response to interferon gamma”, etc. The number of permutations was set up to 1,000.
Protein-protein interaction (PPI) network construction
GeneMANIA online tool (http://genemania.org/) was used to construct and visualize the PPI network of seven targeted genes, including MSI2, TEAD1, GNPDA1, RND3, PRKACB, TRIM68, and YKT6.
Survival analysis of key DEmiRNAs, targeted DEmRNAs, and memory CD4+ T cells
The Kaplan-Meier plotter is a web server for a meta-analysis based on discovery and validation of survival biomarkers (15). Its mRNA subsystems include 54k genes from 21 cancer types, and miRNA subsystems include 11k samples from 20 different cancer types. Gene expression data and relapse free and overall survival (OS) information are derived from GEO, EGA, and TCGA. The survival analysis tool in Kaplan Meier plotter was utilized to analyze the prognostic significance of DEmiRNAs and targeted DEmRNAs. Survival analysis was performed using the Kaplan-Meier method, and the hazard ratio with 95% confidence intervals and logrank P value were calculated.
Besides, survival analysis of high central memory CD4+ T cell patients and low central memory CD4+ T cell patients and high effector memory CD4+ T cell patients and low effector memory CD4+ T cell patients was performed on The Cancer Immunome Atlas (https://tcia.at/home) using TCGA PAAD dataset.
CIBERSORT deconvolution algorithm
To explore the relationship between the targeted DEmRNAs and immune cell infiltration in PC, the CIBERSORT method with 1,000 permutations was applied in R 3.3.2 using the LM22 signature as reference. Samples whose P value of CIBERSORT results were higher than 0.05 were excluded. The proportion of 22 immune cell types in each sample was obtained, including macrophages M2, T cells CD4+ memory resting, monocytes, dendritic cells resting, neutrophils, mast cells resting, B cells naive, eosinophils, B cells memory, NK cells resting, macrophages M1, T cells CD4+ naive, NK cells activated, T cells follicular helper, T cells regulatory, T cells gamma delta, Macrophages M0, dendritic cells activated, plasma cells, mast cells activated, T cells CD4+ memory activated, and T cells CD8. Pearson correlation among 22 immune cell types was calculated, and the visualization was presented using corrplot R package.
Results
DEmiRNAs in gemcitabine-resistant PC cells compared to their parental cells
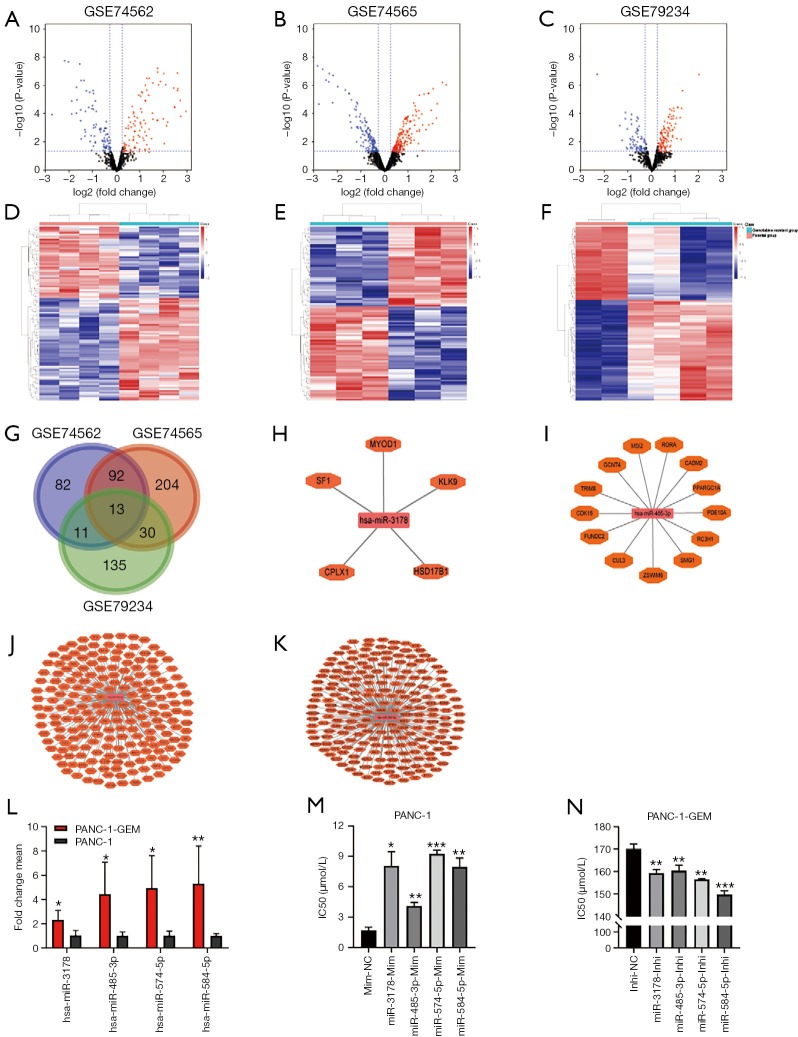
Compared with the parental cells, a total of 198 (108 up- and 90 down-regulated miRNAs), 339 (194 up- and 145 down-regulated miRNAs), and 189 (115 up- and 74 down-regulated miRNAs) DEmiRNAs were identified from GSE74562, GSE74565, and GSE79234 in gemcitabine-resistant PC cells (Figure 1A,B,C,D,E,F), respectively. Unsupervised hierarchical clustering analysis was performed for the top 100 up-regulated and top 100 down-regulated miRNAs. The heatmaps showed that the gemcitabine-resistant PC cell samples could be clearly distinguished from the parental cells with respect to the expression of these differentially expressed miRNAs (Figure 1D,E,F).
Figure 1.
DEmiRNAs between gemcitabine-resistant pancreatic cancer cells and parental cells. (A-C) Volcano plots showed DEmiRNAs in GEO datasets GSE79234, GSE74565, and GSE74562; (D-F) the cluster heatmap showed the top 100 up-regulated and top 100 down-regulated miRNAs in GEO datasets GSE79234, GSE74565, and GSE74562. Red color indicates high expression level, and green color indicates low expression level; (G) Venn diagram showed that 13 DEmiRNAs overlapped in GSE79234, GSE74565, and GSE74562; (H-K) miRNA-target gene network of hsa-miR-3178, hsa-miR-485-3p, hsa-miR-574-5p, and hsa-miR-584-5p, which had a consistent expression trend in three GEO datasets. The targeted genes were mapped into miRDB, microT, and Targetscan databases; (L) qRT-PCR analyses showed that the four miRNAs were upregulated in gemcitabine-resistant PC cells (PANC-1-GEM) compared with its parental cells (PANC-1); (M) miRNAs transfection in PANC-1 cells led to increased IC50 to gemcitabine compared to control cells; (N) knockdown of miRNAs in PANC-1-GEM cells led to decreased IC50 to gemcitabine compared to control cells. Each bar represents the mean ± SD. *, P<0.05; **, P<0.01; ***, P<0.001. PANC-1-GEM, gemcitabine-resistant PANC-1 cells; IC50, half-inhibitory concentration; Inhi-NC, inhibitor negative control; Mim-NC, mimic negative control.
Further intersection analysis showed that 13 miRNAs overlapped in three datasets (Figure 1G). We excluded the DEmiRNAs whose expression trend was inconsistent, and four miRNAs (hsa-miR-3178, hsa-miR-485-3p, hsa-miR-574-5p, and hsa-miR-584-5p) were defined as key miRNAs related to PC gemcitabine resistance (Figure 1H,I,J,K).
Consistently, qRT-PCR analyses confirmed that the four miRNAs were upregulated in gemcitabine-resistant PC cells (PANC-1-GEM) compared with its parental cell (PANC-1) (Figure 1L). We next studied the effect of these four miRNAs on the sensitivity of gemcitabine in PC gemcitabine resistance and found that all of these four miRNAs conferred gemcitabine resistance in PANC-1 cell (Figure 1M) and knockdown of four miRNAs re-sensitized PANC-1-GEM cells (Figure 1N).
The four DEmiRNAs recognized above were mapped into miRDB, microT, and Targetscan databases to search for their target mRNAs according to the inclusion criteria. Their respective miRNA-target gene network was generated using Cytoscape software (Figure 1H,I,J,K).
DEmRNAs in gemcitabine-resistant PC cells compared with their parental cells
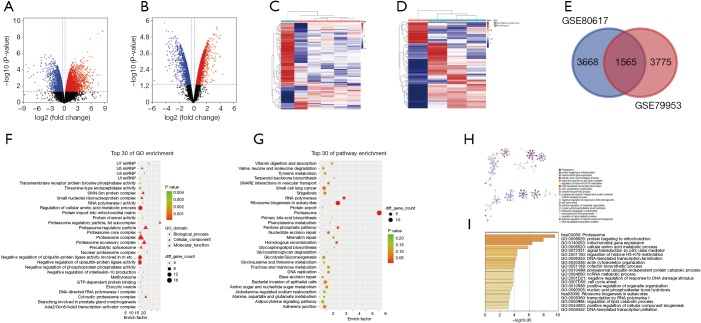
Compared with the parental cells, a total of 5,340 (3,084 up- and 2,256 down-regulated mRNAs) and 5,774 (3,518 up- and 1,715 down-regulated mRNAs) DEmRNAs were identified from GSE79953 and GSE80617 in gemcitabine-resistant PC cells (Figure 2A,B,C,D), respectively. Further intersection analysis showed that 1,565 mRNAs overlapped in two datasets (Figure 2E). Similarly, DEmRNAs with inconsistent expression trend were excluded. Furthermore, the remaining 952 DEmRNAs were performed for next analysis.
Figure 2.
DEmRNAs between gemcitabine-resistant pancreatic cancer cells and parental cells and significantly enriched GO terms and KEGG pathways of DEmRNAs. (A,B) Volcano plots showed DEmRNAs in GEO datasets GSE79953 and GSE80617; (C,D) the cluster heatmap showed the top 100 up-regulated and top 100 down-regulated mRNAs in GEO datasets GSE79953 and GSE80617, respectively. Red color indicates high expression level, and green color indicates low expression level; (E) Venn diagram demonstrated 1,565 DEmRNAs overlapped in GSE79953 and GSE80617; (F-I) considerably enriched GO terms and KEGG pathways based on the 952 DEmRNAs with a consistent expression trend in GSE79953 and GSE80617 datasets.
Functional enrichment analysis
Functional enrichment analysis based on 952 DEmRNAs showed that the DEmRNAs related to PC gemcitabine resistance were involved in biological processes (BPs) such as regulation of cellular amino acid metabolic process and negative regulation of ubiquitin-protein ligase activity and molecular functions (MFs) such as protein channel activity in GO database (Figure 2F). In addition, they had an impact on cell components (CCs) such as proteasome complex, proteasome accessory complex, proteasome regulatory particle, proteasome regulatory particle, and lid subcomplex (Figure 2F). As for the KEGG database, proteasome and ribosome biogenesis in eukaryotes and RNA polymerase-related pathways were significantly enriched (Figure 2G). Consistent with GO and KEGG results, hsa03050:proteasome was enriched best using the tool of Metascape (Figure 2H,I)
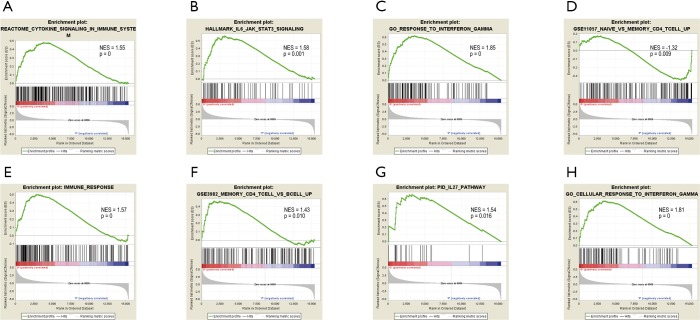
Interestingly, GSEA revealed that chemotherapy resistance to gemcitabine in PC was positively correlated with gene signatures of immune related and memory CD4+ T cell-related pathways (Figure 3). “Go response to interferon gamma” (Normalized Enrichment Score,NES =1.85, P=0), “memory CD4 T cell vs. B cell up” (NES =1.43, P=0.010), “hallmark IL-6 JAK STAT3 signaling” (NES =1.58, P=0.001), “immune response” (NES =1.57, P=0), “IL-27 pathway” (NES =1.54, P=0.016), “reactome cytokine signaling in immune system” (NES =1.55, P=0), and “go cellular response to interferon gamma” (NES =1.81, P=0) were significantly enriched in the gemcitabine-resistant group, whereas “naive vs. memory CD4 T cell up” (NES =−1.32, P=0.009) was well enriched in the parental group.
Figure 3.
Gene set enrichment analysis using GSE79953 dataset revealed that gemcitabine resistance to PC strongly correlated positively with gene signatures associated with immune related and memory CD4+ T cell-related pathways. NES, normalized enrichment score.
The DEmiRNAs-DEmRNAs interaction pairs
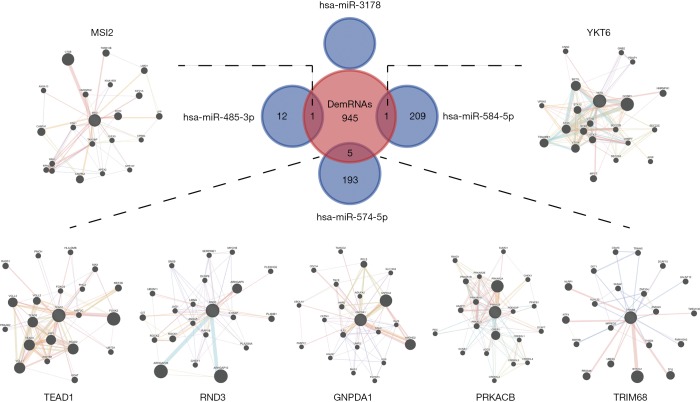
The candidate mRNAs targeted by the four DEmiRNAs were intersected with the 952 DEmRNAs, and zero, one, five, and one target mRNAs were identified by hsa-miR-3178, hsa-miR-485-3p, hsa-miR-574-5p, and hsa-miR-584-5p, respectively. Particularly, the expression tendency of all four DEmiRNAs and their target mRNAs has increased in gemcitabine-resistant PC cells compared with their parental cells, which was consistent with miRNAa phenomena. The PPI network of seven targeted genes is displayed in Figure 4.
Figure 4.
The targeted genes of four DEmiRNAs and the PPI network of these targeted genes. These targeted genes predicted by miRDB, microT, and Targetscan databases of four DEmiRNAs were intersected with the 952 DEmRNAs, and zero, one (MSI2), five (TEAD1, RND3, GNPDA1, PRKACB, and TRIM68), one (YKT6) target mRNAs were identified by hsa-miR-3178, hsa-miR-485-3p, hsa-miR-574-5p, and hsa-miR-584-5p, respectively.
Survival analysis of four DEmiRNAs and seven targeted DEmRNAs
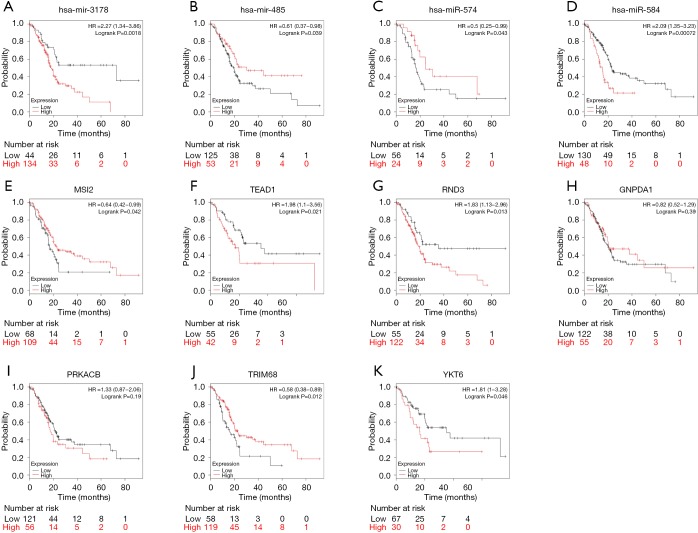
The prognostic significance of four DEmiRNAs and seven targeted DEmRNAs was analyzed using Kaplan-Meier plotter. As shown in Figure 5, data of 177 PAAD patients were incorporated in the survival analysis. Three DEmiRNAs, hsa-miR-3178 (logrank P=0.0018, HR =2.27), hsa-miR-485-3p (logrank P=0.039, HR =0.61), and hsa-miR-584-5p (logrank P=0.00072, HR =2.09) were significantly associated with the OS of PAAD patients. Another DEmiRNA, hsa-miR-574-5p (logrank P=0.043, HR =0.5), was significantly linked to OS of female PDAC patients. Among seven targeted DEmRNAs, three DEmRNAs, MSI2 (logrank P=0.042, HR =0.64), RND3 (logrank P=0.013, HR =1.83), and TRIM68 (logrank P=0.012, HR =0.58) were significantly associated with the OS of PDAC patients. Two DEmRNAs, TEAD1 (logrank P=0.021, HR =1.98) and YKT6 (logrank P=0.046, HR =1.81), were significantly linked to OS of male PAAD patients. In addition, PRKACB and GNPDA1 were not qualified predictors for PAAD patient survival.
Figure 5.
Kaplan-Meier survival curves of PAAD patients from Kaplan-Meier plotter for the four DEmiRNAs (A-D) and seven targeted DEmRNAs (F-K). P value <0.05 was considered statistically significant. Three DEmiRNAs, hsa-miR-3178 (A), hsa-miR-485-3p (B), and hsa-miR-584-5p (D), were significantly associated with the OS of PAAD patients. hsa-miR-574-5p (C) was significantly linked to OS of female PDAC patients. Three DEmRNAs, MSI2 (E), RND3 (G), and TRIM68 (J), were significantly associated with the OS of PDAC patients. Two DEmRNAs, TEAD1 (F) and YKT6 (K), were significantly linked to OS of male PAAD patients. In addition, GNPDA1 (H) and PRKACB (I) were not qualified predictors for PAAD patient survival.
Immune infiltration analysis of TCGA PAAD patients
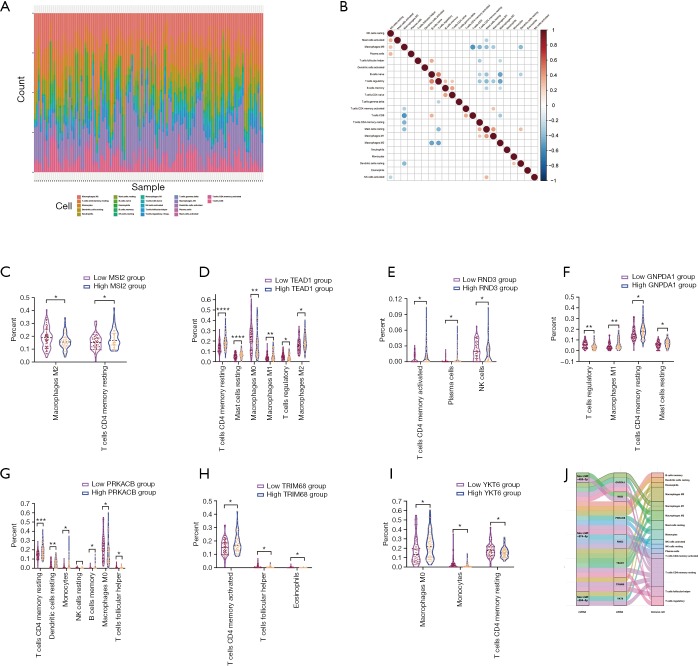
Given that gemcitabine might affect immune-related pathways of PC, we investigated whether immune cell fractions were altered by DEmRNAs. TCGA PAAD patients’ immune infiltration status was analyzed using CIBERSORT using RNA-seq data (Figure 6A). Most patients exhibited heterogeneity of immune infiltration. However, some of immune cell types showed high correlation with the others (Figure 6B). Activated memory CD4+ T cells showed positive correlation with CD8 T cells, whereas resting memory CD4+ T cells negatively correlated with macrophages M0, T cells follicular helper, B cells naïve, and T cells regulatory, indicating totally different roles of activated and resting memory CD4+ T cells. Regrouping PAAD dataset by level of seven DEmRNAs expression, we found that the fraction of CD4+ memory T cells significantly changed (Figure 6C,D,E,F,G,H,I). Resting memory CD4+ T cells were significantly altered by MSI2, TEAD1, GNPDA1, PRKACB, TRIM68, and YKT6, whereas activated memory CD4+ T cells were significantly altered by RND3.
Figure 6.
TCGA PAAD patients’ immune cell fractions were altered by seven targeted DEmRNAs. (A) TCGA PAAD patients’ immune infiltration status was analyzed by CIBERSORT using RNA-seq data; (B) some immune cell types showed high correlation with others. T cells CD4+ memory activated showed positive correlation with T cells CD8, whereas T cells CD4+ memory resting negatively correlated with macrophages M0, T cells follicular helper, B cells naïve, and T cells regulatory, indicating totally different roles of resting and activating CD4+ memory T cells; (C-I) regrouping PAAD dataset by level of seven DEmRNAs expression, the fraction of CD4+ memory T cells significantly changed. T cells CD4+ memory resting was significantly altered by MSI2, TEAD1, GNPDA1, PRKACB, TRIM68, and YKT6, whereas T cells CD4+ memory activated were significantly altered by RND3; (J) alteration of infiltrating immune cells through seven DEmRNAs targeted by hsa-miR-485-3p, hsa-miR-574-5p, and hsa-miR-584-5p.
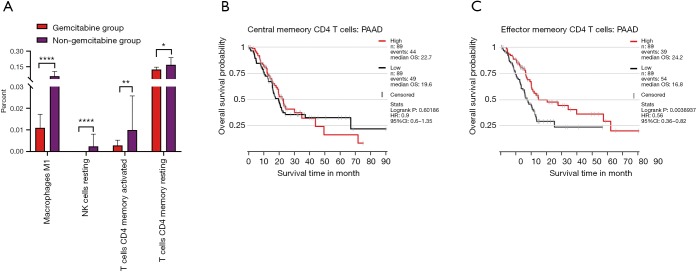
Besides, fractions of macrophages M1, NK cells resting, activated memory CD4+ T cells, and resting memory CD4+ T cells were altered in the presence of gemcitabine according to TCGA PAAD patients’ RNA-seq data who took gemcitabine or not (Figure 7A). Finally, we tested whether memory CD4+ T cell faction could influence prognosis of PC patients and found that effector memory CD4+ T cells had a great effect on prognosis, whereas central memory CD4+ T cells had no effect (Figure 7B).
Figure 7.
Gemcitabine therapeutic effect influenced fractions of immune cell fractions. (A) Fractions of macrophages M1, NK cells resting, T cells CD4+ memory activated, and T cells CD4+ memory resting were altered in the presence of gemcitabine according to TCGA PAAD patients’ RNA-seq data who took gemcitabine or not; (B) central memory CD4+ T cells had no influence on prognosis of PC patients; (C) effector memory CD4+ T cells had a positive effect on prognosis of PC patients.
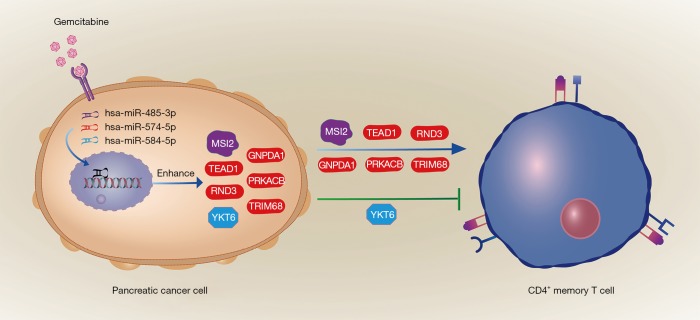
As Figure 8 shows (16), gemcitabine resistance could alter memory CD4+ T cell faction of PC patients via regulating MSI2, TEAD1, GNPDA1, RND3, PRKACB, TRIM68, and YKT6 by miRNAs. Gemcitabine therapeutic effect could lead to a better prognosis in high effector memory CD4+ T cell fraction patients.
Figure 8.
Schematic illustration of a CD4+ memory T cell immune-related miRNAa regulatory network associated with PC gemcitabine resistance. In gemcitabine-resistant pancreatic cancer cells, hsa-miR-485-3p, hsa-miR-574-5p, and hsa-miR-584-5p were up-regulated, served as activators of gene expression according to miRNAa hypothesis, promoted the expression of seven targeted mRNAs, including MSI2, TEAD1, GNPDA1, RND3, PRKACB, TRIM68, and YKT6. Complicatedly, MSI2, TEAD1, GNPDA1, RND3, PRKACB, and TRIM68 increased the fractions of CD4+ memory T cells, while YKT6 suppressed them.
Discussion
Three notable findings stand out in the present study. First, our study identified four DEmiRNAs (hsa-miR-3178, hsa-miR-485-3p, hsa-miR-574-5p, and hsa-miR-584-5p) and seven targeted DEmRNAs (MSI2, TEAD1, RND3, GNPDA1, PRKACB, TRIM68, and YKT6), all upregulated, which was consistent with novel hypothesis of miRNAa (4,6,17), in gemcitabine resistant PC cells compared with their parental cells. Second, we discovered that these seven targeted DEmRNAs in chemotherapy-resistant cells were associated with immune infiltration status, which was analyzed using CIBERSORT algorithm according to gene expression profiles of PAAD from TCGA database. Finally, we found that the gemcitabine therapeutic effect was closely associated with effector memory CD4+ T cells in PC patients.
It is generally accepted that miRNAs inhibit translation by mRNA degradation or destabilization and translational repression (18). Moreover, researches over the past decade have shown that miRNAs can not only suppress but also activate the expression of target genes (4,6,17), namely, miRNAa phenomena. In the present study, we identified four DEmiRNAs (hsa-miR-3178, hsa-miR-485-3p, hsa-miR-574-5p, and hsa-miR-584-5p) and their targeted DEmRNAs (MSI2, TEAD1, RND3, GNPDA1, PRKACB, TRIM68, and YKT6) from GEO database and found, unexpectedly, their expression tendency increased in the gemcitabine treatment group, which was consistent with miRNAa phenomena.
Several studies have testified that above gemcitabine-resistant-related DEmiRNAs and DEmRNAs are associated with chemotherapy resistance in different cancers. hsa-miR-3178 is significantly up-regulated after pre-neoadjuvant chemotherapy, and its overexpression is linked to drug resistance in breast cancer (19,20). Similarly, Lucotti et al. showed that hsa-miR-485-3p was highly expressed in prostate cancer cells and enabled the cancer cells to survive the treatment with fludarabine (21). MSI2 promotes the resistance to gemcitabine and cisplatin and the development of PC (22). TEAD1 is associated with chemotherapy resistance in patients with advanced bladder cancer (23). Dysregulation of RND3 is related to chemotherapy resistance in gastric cancer, nasopharyngeal carcinoma, and lung cancer cell human gastric carcinoma (24). PRKACB plays key roles in the development of multidrug resistance of Hodgkin’s lymphoma (25). YKT6 is significantly up-regulated in p53-mutated breast tumors and associated with a low response rate to docetaxel (26).
Gemcitabine therapeutic effect was proven to decrease memory T cells and promote naive T cell activation (27), consistent with our results. However, the intrinsic mechanism is totally unclear. In our study, we propose a completely fresh approach that gemcitabine therapeutic effect may alter fraction of CD4+ memory T cell through miRNA-induced RNA activation-targeted mRNA network in PC. None of the four DEmiRNAs (hsa-miR-3178, hsa-miR-485-3p, hsa-miR-574-5p, and hsa-miR-584-5p) had been reported to have an impact on CD4+ memory T cell. However, hsa-miR-3178 played a role in cancer immune surveillance by regulating DUSP1, PD-L1, and MUC1 (28). miR-485 restricts influenza virus infection by priming antiviral immune response (29,30) and is associated with the immune system in Alzheimer’s disease (31). miR-584 is significantly up-regulated in inflamed pulps and plays an intricate and specific role in inflammation and immunity (32). All seven DEmRNAs (MSI2, TEAD1, RND3, GNPDA1, PRKACB, TRIM68, and YKT6) did not show any direct effects on CD4+ memory T cell. Nevertheless, TEAD1/p65 complex could regulate innate immune response (33). PRKACB-encoding Cβ2 regulates innate as well as adaptive immune sensitivity in vivo (34). TRIM68 could regulate IFN production by targeting TFG (35). In addition, DEmRNAs can have an indirect effect on CD4+ memory T cell through PPI network. Most of DEmiRNAs and DEmRNAs influence immune system through a variety of ways.
DEmRNAs can influence fraction of CD4+ memory T cells through several pathways, including “IL-6-JAK-STAT3 signaling pathway” (NES =1.58, P=0.001), “IL-27 pathway” (NES =1.54, P=0.016), and “cytokine signaling in immune system” (NES =1.55, P=0). STAT3 transcription factor signaling is critical in human memory T cell formation (36). IL-27R signaling plays a role in altering fraction of CD4+ memory T cells and regulating the magnitude and quality of secondary immune responses during rechallenge infections (37). These data indicate that transformation of CD4 memory T cell number can be relevant with these signal pathways. Besides, gemcitabine-resistant PC cell group showed a CD4+ memory T cell signature while “memory CD4 T cell vs. B cell up” (NES =1.43, P=0.010) were significantly enriched in the gemcitabine-resistant group, and “naive vs. memory CD4 T cell up” (NES =−1.32, P=0.009) was well enriched in the parental group. These findings confirm alteration of CD4+ memory T cells and potential mechanism under gemcitabine resistance.
Finally, we found that effector memory CD4+ T cells and not central memory CD4+ T cells had a positive effect on the prognosis of PC patients, indicating a substantial role of effector memory CD4+ T cells. IFN-γ priming is regarded as a mechanism affecting effector memory CD4+ T cells (37). Our results also showed that “go response to interferon gamma” (NES =1.85, P=0) and “go cellular response to interferon gamma” (NES =1.81, P=0) were significantly enriched in the gemcitabine-resistant group. Together, these findings suggested that gemcitabine therapeutic effect altered effector memory CD4+ T cells by IFN-γ priming and responding to related pathways.
Memory CD4+ T cells has been reported to be an important part in tumor microenvironment. Memory CD4+ T cells has been identified in more infiltrated in colorectal cancer than normal tissue, particularly in T1-2 tumor stage (38). Central memory CD4+ T cells enrichment score seem higher in invasive tumors that were not ductal/lobular carcinoma in triple-negative breast cancer (39). Central memory CD4+ T cells seem relative hypometabolism and favorable prognosis in lung adenocarcinoma (40). These reports suggest the clinical relevance of memory CD4+ T cells in various tumors and important role in tumor microenvironment. In patients with advanced PC treated with gemcitabine, a slight increase in the absolute numbers of effector memory CD4+ T cells is noted during treatment, suggesting the potential key role of memory CD4+ T cells in treatment with gemcitabine (41). Gemcitabine treatment is regard to promote immunosuppressive microenvironment in PC (42-44). However, no researches about memory CD4+ T cells alteration in gemcitabine resistant PC patients and their clinical relevance are reported, which has been discussed in our study.
Conclusions
Our study provides a splendid insight on gemcitabine resistance and association between chemotherapy resistance and tumor immune infiltration status in PC. However, further experiments are required, as we did not completely prove our conclusion.
Acknowledgments
Funding: This work was supported by the National Key R&D Program of China (No. 2017YFC1308600) and the National Natural Science Foundation of China (No. 81672382) for Huaizhi Wang and the Science and Technology Plan Project of Guangdong Province (No. 2015A020214012, 2016B090917001, and 2015B020214005) for Yingfang Fan.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 4.Vaschetto LM. miRNA activation is an endogenous gene expression pathway. RNA Biol 2018;15:826-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Place RF, Li LC, Pookot D, et al. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A 2008;105:1608-13. 10.1073/pnas.0707594105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao M, Li J, Li W, et al. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol 2017;14:1326-34. 10.1080/15476286.2015.1112487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang V, Place RF, Portnoy V, et al. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res 2012;40:1695-707. 10.1093/nar/gkr934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madurantakam Royam M, Ramesh R, Shanker R, et al. miRNA Predictors of Pancreatic Cancer Chemotherapeutic Response: A Systematic Review and Meta-Analysis. Cancers (Basel) 2019. doi: . 10.3390/cancers11070900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binenbaum Y, Na'ara S, Gil Z. Gemcitabine resistance in pancreatic ductal adenocarcinoma. Drug Resist Updat 2015;23:55-68. 10.1016/j.drup.2015.10.002 [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Chen G, Ma X, et al. MiR-30a regulates cancer cell response to chemotherapy through SNAI1/IRS1/AKT pathway. Cell Death Dis 2019;10:153. 10.1038/s41419-019-1326-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funamizu N, Lacy CR, Kamada M, et al. MicroRNA-200b and -301 are associated with gemcitabine response as biomarkers in pancreatic carcinoma cells. Int J Oncol 2019;54:991-1000. [DOI] [PubMed] [Google Scholar]
- 12.Frank AC, Ebersberger S, Fink AF, et al. Apoptotic tumor cell-derived microRNA-375 uses CD36 to alter the tumor-associated macrophage phenotype. Nat Commun 2019;10:1135. 10.1038/s41467-019-08989-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pyfferoen L, Brabants E, Everaert C, et al. The transcriptome of lung tumor-infiltrating dendritic cells reveals a tumor-supporting phenotype and a microRNA signature with negative impact on clinical outcome. Oncoimmunology 2016;6:e1253655. 10.1080/2162402X.2016.1253655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy A, Lanczky A, Menyhart O, et al. Author Correction: Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:11515. 10.1038/s41598-018-29514-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sidiropoulos K, Viteri G, Sevilla C, et al. Reactome enhanced pathway visualization. Bioinformatics 2017;33:3461-7. 10.1093/bioinformatics/btx441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu M, Roth A, Yu M, et al. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev 2013;27:2543-8. 10.1101/gad.224170.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catalanotto C, Cogoni C, Zardo G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int J Mol Sci 2016;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong S, Chen X, Wang D, et al. MicroRNA expression profiles of drug-resistance breast cancer cells and their exosomes. Oncotarget 2016;7:19601-9. 10.18632/oncotarget.7481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Lu P, Wang DD, et al. The role of miRNAs in drug resistance and prognosis of breast cancer formalin-fixed paraffin-embedded tissues. Gene 2016;595:221-6. 10.1016/j.gene.2016.10.015 [DOI] [PubMed] [Google Scholar]
- 21.Lucotti S, Rainaldi G, Evangelista M, et al. Fludarabine treatment favors the retention of miR-485-3p by prostate cancer cells: implications for survival. Mol Cancer 2013;12:52. 10.1186/1476-4598-12-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheng W, Dong M, Chen C, et al. Cooperation of Musashi-2, Numb, MDM2, and P53 in drug resistance and malignant biology of pancreatic cancer. FASEB J 2017;31:2429-38. 10.1096/fj.201601240R [DOI] [PubMed] [Google Scholar]
- 23.Wang KJ, Wang C, Dai LH, et al. Targeting an Autocrine Regulatory Loop in Cancer Stem-like Cells Impairs the Progression and Chemotherapy Resistance of Bladder Cancer. Clin Cancer Res 2019;25:1070-86. 10.1158/1078-0432.CCR-18-0586 [DOI] [PubMed] [Google Scholar]
- 24.Jie W, Andrade KC, Lin X, et al. Pathophysiological Functions of Rnd3/RhoE. Compr Physiol 2015;6:169-86. 10.1002/cphy.c150018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, Huang Y, Zhang L, et al. Identification of crucial genes and prediction of small molecules for multidrug resistance of Hodgkin's lymphomas. Cancer Biomark 2018;23:495-503. 10.3233/CBM-181496 [DOI] [PubMed] [Google Scholar]
- 26.Ooe A, Kato K, Noguchi S. Possible involvement of CCT5, RGS3, and YKT6 genes up-regulated in p53-mutated tumors in resistance to docetaxel in human breast cancers. Breast Cancer Res Treat 2007;101:305-15. 10.1007/s10549-006-9293-x [DOI] [PubMed] [Google Scholar]
- 27.Plate JM, Plate AE, Shott S, et al. Effect of gemcitabine on immune cells in subjects with adenocarcinoma of the pancreas. Cancer Immunol Immunother 2005;54:915-25. 10.1007/s00262-004-0638-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pascut D, Krmac H, Gilardi F, et al. A comparative characterization of the circulating miRNome in whole blood and serum of HCC patients. Sci Rep 2019;9:8265. 10.1038/s41598-019-44580-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingle H, Kumar S, Raut AA, et al. The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci Signal 2015;8:ra126. 10.1126/scisignal.aab3183 [DOI] [PubMed] [Google Scholar]
- 30.Zheng Y, Fu X, Wang L, et al. Comparative analysis of MicroRNA expression in dog lungs infected with the H3N2 and H5N1 canine influenza viruses. Microb Pathog 2018;121:252-61. 10.1016/j.micpath.2018.05.015 [DOI] [PubMed] [Google Scholar]
- 31.Swarbrick S, Wragg N, Ghosh S, et al. Systematic Review of miRNA as Biomarkers in Alzheimer's Disease. Mol Neurobiol 2019;56:6156-67. 10.1007/s12035-019-1500-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong S, Zhang S, Bair E, et al. Differential expression of microRNAs in normal and inflamed human pulps. J Endod 2012;38:746-52. 10.1016/j.joen.2012.02.020 [DOI] [PubMed] [Google Scholar]
- 33.Chokas AL, Bickford JS, Barilovits SJ, et al. A TEAD1/p65 complex regulates the eutherian-conserved MnSOD intronic enhancer, eRNA transcription and the innate immune response. Biochim Biophys Acta 2014;1839:1205-16. [DOI] [PMC free article] [PubMed]
- 34.Moen LV, Sener Z, Volchenkov R, et al. Ablation of the Cbeta2 subunit of PKA in immune cells leads to increased susceptibility to systemic inflammation in mice. Eur J Immunol 2017;47:1880-9. 10.1002/eji.201646809 [DOI] [PubMed] [Google Scholar]
- 35.Wynne C, Lazzari E, Smith S, et al. TRIM68 negatively regulates IFN-beta production by degrading TRK fused gene, a novel driver of IFN-beta downstream of anti-viral detection systems. PLoS One 2014;9:e101503. 10.1371/journal.pone.0101503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siegel AM, Heimall J, Freeman AF, et al. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity 2011;35:806-18. 10.1016/j.immuni.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borges da Silva H, Fonseca R, Alvarez JM, et al. IFN-gamma Priming Effects on the Maintenance of Effector Memory CD4(+) T Cells and on Phagocyte Function: Evidences from Infectious Diseases. J Immunol Res 2015;2015:202816. [DOI] [PMC free article] [PubMed]
- 38.Ge P, Wang W, Li L, et al. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of colorectal cancer. Biomed Pharmacother 2019;118:109228. 10.1016/j.biopha.2019.109228 [DOI] [PubMed] [Google Scholar]
- 39.Deng L, Lu D, Bai Y, et al. Immune Profiles of Tumor Microenvironment and Clinical Prognosis among Women with Triple-Negative Breast Cancer. Cancer Epidemiol Biomarkers Prev 2019;28:1977-85. 10.1158/1055-9965.EPI-19-0469 [DOI] [PubMed] [Google Scholar]
- 40.Choi H, Na KJ. Integrative analysis of imaging and transcriptomic data of the immune landscape associated with tumor metabolism in lung adenocarcinoma: Clinical and prognostic implications. Theranostics 2018;8:1956-65. 10.7150/thno.23767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ullenhag GJ, Mozaffari F, Broberg M, et al. Clinical and Immune Effects of Lenalidomide in Combination with Gemcitabine in Patients with Advanced Pancreatic Cancer. PLoS One 2017;12:e0169736. 10.1371/journal.pone.0169736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deshmukh SK, Tyagi N, Khan MA, et al. Gemcitabine treatment promotes immunosuppressive microenvironment in pancreatic tumors by supporting the infiltration, growth, and polarization of macrophages. Sci Rep 2018;8:12000. 10.1038/s41598-018-30437-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C, Tan X, Hu X, et al. Tumor Microenvironment following Gemcitabine Treatment Favors Differentiation of Immunosuppressive Ly6C(high) Myeloid Cells. J Immunol 2020;204:212-23. 10.4049/jimmunol.1900930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamath SD, Kalyan A, Kircher S, et al. Ipilimumab and Gemcitabine for Advanced Pancreatic Cancer: A Phase Ib Study. Oncologist 2019. [Epub ahead of print]. 10.1634/theoncologist.2019-0473 [DOI] [PMC free article] [PubMed] [Google Scholar]