Abstract
Background
Colorectal signet-ring cell carcinoma (SRCC) is characterized as a rare subset of colorectal cancer with extremely poor prognosis and it is known to have low or negative 18F fluorodeoxyglucose (18F-FDG) uptake. To date, no in-depth study revealing the metabolic features of colorectal SRCC has been conducted for the lack of reliable study model. The aim of this study was to explore the distinct characteristics of energy utilization for colorectal SRCC based on organoid model.
Methods
Three organoids were derived from colorectal SRCC patients with low or negative FDG uptake and three organoids were derived from colorectal adenocarcinoma (AC) patients. Glucose, fatty acid and glutamine uptake assays were performed to reveal the different metabolic features of SRCC organoids. Immunohistochemistry (IHC), western blotting and real-time PCR were used to test the expression of critical transporters and enzymes of energy metabolism. Glutamine deprivation analyses were used to confirm the dependence of colorectal SRCC on glutamine.
Results
Glucose, fatty acid and glutamine uptake assays showed that only glutamine uptake was significantly increased in colorectal SRCC organoids compared with paired normal organoids. Comparing SRCC organoids with AC organoids indicated that glucose and fatty acid uptake were strikingly higher in AC organoids while glutamine uptake was notably lower. Gene expression analyses confirmed that the glutamine transporter SLC1A5 and glutaminolysis enzyme GPT2 were significantly unregulated in colorectal SRCC. Silencing of SLC1A5 or GPT2 could suppress the proliferation of SRCC organoids but attenuating the sensitivity of SRCC to glutamine deprivation. Administration of SLC1A5 or GPT2 inhibitor could prohibit SRCC growth and significantly enhance the sensitivity of SRCC to the treatment of 5-fu and L-OHP.
Conclusions
This study highlights enhanced glutamine uptake and glutaminolysis as a metabolic feature of colorectal SRCC and a potential therapeutic target.
Keywords: Colorectal signet-ring cell carcinoma, organoid, glutamine metabolism
Introduction
Colorectal signet-ring cell carcinoma (SRCC) is a rare type of adenocarcinoma (AC) and only accounts for approximately 1% of colorectal cancer (1,2). Patients diagnosed with SRCC are always characterized with extremely dismal prognosis. Though it has been established that colorectal SRCC patients have significantly higher rate of relapse and lower rate of response to standard chemoradiotherapy than other common adenocarcinoma, few previous studies have been conducted to explore the underlying mechanisms promoting colorectal SRCC initiation and progression (3).
Lacking appropriate models for research is one of the primary reasons why the study of colorectal SRCC remains scarce. Recently, organoids have been broadly used to model different cancers for they can retain the structure, morphology, genetic mutations and heterogeneity of the parental tumor (4). By using this technology, we standardized the procedure of isolating and culturing SRCC organoids and established the first organoid line allowing the possibility of in-depth study of SRCC biology (5).
Cancer cells always exhibit more enhanced metabolic dependence than normal cellular counterparts, displaying augmented nutrient acquisition and uncontrolled cell proliferation. Metabolic reprogramming during tumorigenesis is an essential process in nearly all cancers (6). The Warburg effect is the first example of metabolic reprogramming that Otto Warburg discovered in 1920s (7). Specifically, cancer cells generate energy for biological activities by preferring glycolysis to mitochondrial oxidative phosphorylation, regardless of the availability of oxygen. Based on the metabolic features, positron emission tomography (PET) of 18F fluorodeoxyglucose (18F-FDG) uptake has been utilized as an important imaging tool for the diagnosis and staging of various cancers. However, SRCC is known to have low fluorodeoxyglucose (FDG) uptake and PET/CT usually cannot successfully detect the primary or metastatic site of SRCC (8). Therefore, we hypothesized that rapid SRCC growth and progression may not be attributed to enhanced glucose utilization and SRCC cells may preferentially leverage other energy sources. Glutamine and fatty acid are two critical substrates for the bioenergetic pathways that support tumor growth (9), which therefore might be involved in the energy metabolism of colorectal SRCC. However, further experiments should be conducted to depict the landscape of energy utilization for colorectal SRCC.
In this study, we compared the expression of critical genes responsible for glucose, fatty acid and glutamine metabolism between colorectal SRCC tissues and adjacent normal tissues, which demonstrated that enhanced glutamine utilization is an essential metabolic feature of colorectal SRCC with therapeutic potential.
Methods
Patients selection and SRCC-derived organoids culture
Colorectal SRCC and adjacent normal intestinal tissues were obtained from patients diagnosed and treated at Fudan University Shanghai Cancer Center (FUSCC). Written informed consents were obtained on enrollment. The research protocol was reviewed and approved by the ethics committee of FUSCC. The standard protocol for isolating and culturing organoids derived from human SRCC was as previously described (5). For glutamine deprivation, organoids were cultured in glutamine- and pyruvate-free DMEM with high centration of glucose (Fisher Scientific, Catalogue No. SH30081) and 10% dialysed FBS (dFBS; Invitrogen, Catalogue No. 26400). The SRCC organoids used in this study were derived from three patients with low or negative FDG uptake. Another three AC organoids were derived from AC patients with relatively high SUVmax value. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Expression vectors and gene transfection
Short hairpin RNA (shRNA) constructs against SLC1A5 and GPT2 was generated using pLKO.1-TRC cloning vector (Addgene, #10878). shRNAs for GPT2 (sc-45647 and sc-45648) and SLC1A5 (sc-60210 and sc-60211) were purchased from Santa Cruz Biotechnology. Each constructed plasmid was co-transfected with the packaging plasmids psPAX2 and PMD2.G into HEK293T cells using Lipofectamine 3000 reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. Virus particles were harvested 48 h after transfection. Transfection of the target cells with lentiviral particles were performed using polybrene (2 µg/mL, Sigma-Aldrich) pretreatment, and positive cells were selected with puromycin (2 µg/mL, Sigma-Aldrich).
Western blotting and immunohistochemistry (IHC)
Western blotting was performed using whole-cell protein lysates of CRC cells using primary antibodies against SLC1A5 (ab237704, 1:1,000; Abcam) and GPT2 (16757-1-AP, 1:1,000; Proteintech) and a secondary antibody (anti-rabbit IgG, 1:7,500; Cell Signaling Technology). Equal loading of protein samples was monitored using an anti-β-actin antibody (ab8226, 1:2,500; Abcam). Immunostaining of SRCC patients tissues was conducted as previously described (3,10,11).
RNA isolation and quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to isolate total RNA and PrimeScript RT reagent (TaKaRa, Dalian, China) was used to obtain samples. The expression status of specific genes and β-actin were determined by qRT-PCR using an ABI 7900HT Real-Time PCR System (Applied Biosystems, Frederick, MD, USA). All reactions were run in triplicate.
Cell apoptosis and cell viability measurement
According to the manufacturer’s instructions, FITC Annexin V Apoptosis Detection Kit (BD, La Jolla, CA, USA) was used to detect apoptotic rate of cells. Cell viability was measured by CCK-8 assay.
Glucose, fatty acid and glutamine uptake assay
There are 5×103 organoids were cultured in 96-well plates containing advanced DMEM (Thermo Fisher Scientific) with 10% fetal bovine serum (FBS, Thermo Fisher Scientific) and 2 mM glutamine and then transferred to a CO2 incubator set at 37 °C and 5% CO2 for 24 hours. Spent media were collected to measure remaining glucose, fatty acid and glutamine by using a glucose colorimetric/fluorometric assay kit (Abcam), free fatty acid uptake assay kit (Abcam) and glutamine assay kit (Abcam) respectively following the manufacturer's instruction.
Statistical analysis
All statistical analyses were performed using R software (R version 3.2.5, https://www.r-project.org/). Significant differences between two groups were computed using Wilcoxon rank-sum test for data with skewed distribution or Student’s test for data with normal distribution.
Results
Glutamine uptake was significantly enhanced in SRCC
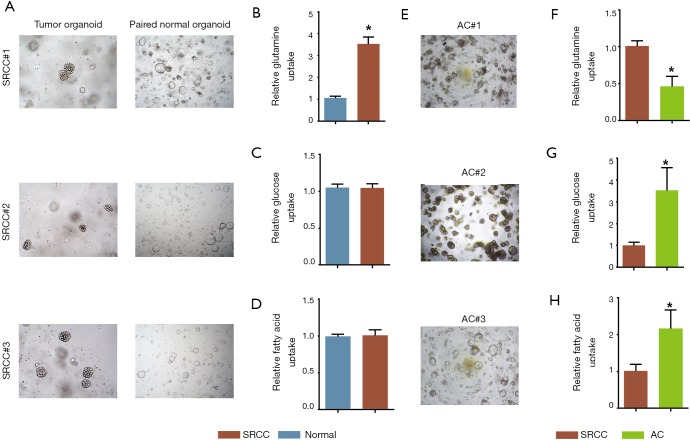
To elucidate the preference of energy source usage in SRCC, we isolated and developed three paired SRCC organoids from cancer and adjacent normal tissues (Figure 1A) and conducted glucose, fatty acid and glutamine uptake analyses, which showed that SRCC significantly increased the glutamine uptake compared with its paired normal organoids (Figure 1B). Instead, glucose (Figure 1C) and fatty acid (Figure 1D) uptake were not notably discrepant between the two groups. Further, we compared SRCC organoids with AC organoids (Figure 1E) and found that glutamine uptake was still elevated in SRCC organoids (Figure 1F) while AC organoids showed strikingly higher rate of glucose (Figure 1G) and fatty acid uptake (Figure 1H).
Figure 1.
Glutamine uptake rate was enhanced in SRCC organoid. (A) Representative images of three SRCC organoids and paired normal colon organoids (light microscope, 200×); comparison of glutamine (B), glucose (C) and fatty acid (D) uptake rate between SRCC organoids and paired normal colon organoids; (E) representative images of three AC organoids (light microscope, 200×); comparison of glutamine (F), glucose (G) and fatty acid (H) uptake rate between SRCC organoids and AC organoids. *, P<0.05. SRCC, signet-ring cell carcinoma; AC, adenocarcinoma.
Metabolic genes responsible for glutamine uptake and glutaminolysis were notably highly expressed in colorectal SRCC
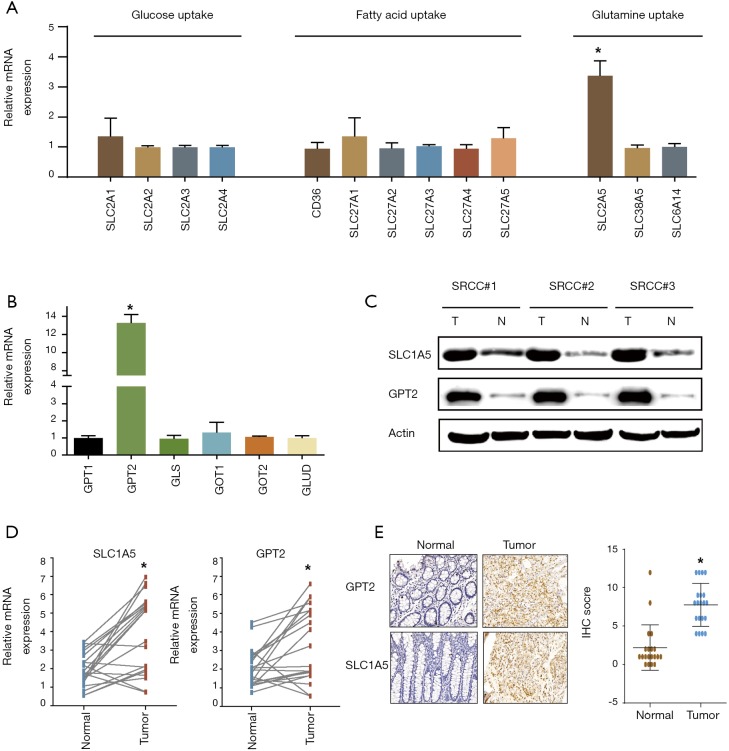
qRT-PCR was next used to detect the mRNA expression of critical genes contributing to glucose, fatty acid and glutamine uptake. The results showed that only SLC1A5 mediating glutamine uptake was strikingly unregulated in SRCC while the other transporters driving glucose and fatty acid uptake were not significantly different between the cancer and normal organoids (Figure 2A). We further tested the pivotal enzymes catalyzing glutaminolysis in SRCC, revealing that GPT2 was dramatically unregulated (Figure 2B). Further western blotting confirmed that the protein level of SLC1A5 and GPT2 were highly expressed in SRCC (Figure 2C). We next validated the results from organoids in another cohort consisting of twenty paired SRCC patient tissues. qRT–PCR and IHC analysis confirmed the markedly higher expression of SLC1A5 and GPT2 in cancer tissues (Figure 2D,E).
Figure 2.
Metabolic genes responsible for glutamine uptake and glutaminolysis were notably highly expressed in colorectal SRCC. (A) fold change of transcripts of genes driving glucose, fatty acid and glutamine uptake were detected by qRT-PCR in SRCC organoids and paired normal organoids; (B) qRT-PCR showing change of genes mediating glutaminolysis in SRCC organoids. (C) western blot showing SLC1A5 and GPT2 expression in SRCC organoids and paired normal organoids; (D) SLC1A5 and GPT2 expression between paired normal tissues and SRCC tumor tissues in twenty patients from FUSCC database; (E) IHC staining of SRCC tumor tissues and paired normal tissues microarrays using SLC1A5 and GPT2 antibody for patients from FUSCC (light microscope, 200×). *, P<0.05. SRCC, signet-ring cell carcinoma.
Upregulation of SLC1A5 and GPT2 rendered SRCC organoid more sensitive to glutamine deprivation
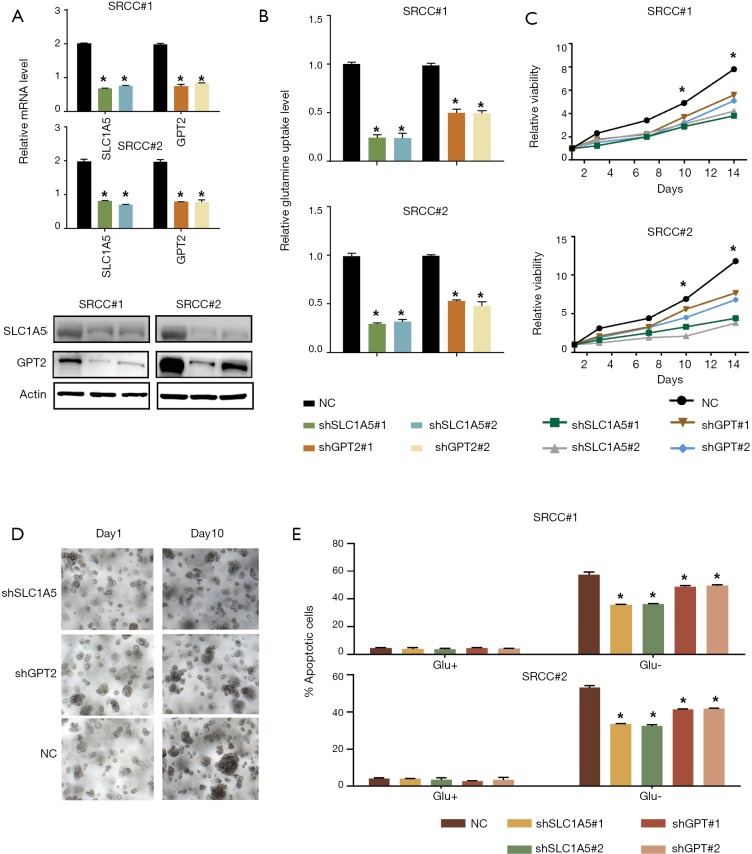
To determine whether the upregulation of SLC1A5 and GPT2 can contribute to the dependence of SRCC organoids growth on glutamine, we knocked down SLC1A5 or GPT2 in two organoids using two independent shRNAs (Figure 3A). Compared with the control organoids, SLC1A5 or GPT2 knockdown organoids showed significantly decreased glutamine uptake (Figure 3B), grew more slowly under normal culture conditions (Figure 3C,D), but they were less sensitive to glutamine deprivation as tested by apoptosis assay (Figure 3E), suggesting that SRCC was dependent on glutamine for rapid growth.
Figure 3.
High expression of SLC1A5 and GPT2 rendered SRCC organoid more sensitive to glutamine deprivation. (A) Western blot showing short hairpin RNA-mediated deletion of SLC1A5 and GPT2 in two organoids; (B) glutamine uptake was examined in Control and GPT2 or SLC1A5 knockdown organoids; (C,D) silence of SLC1A5 and GPT2 significantly inhibited growth of SRCC organoids (light microscope, 200×); (E) control and GPT2 or SLC1A5 knockdown organoids were grown with or without glutamine for 72 h. Cell apoptosis was quantified. *, P<0.05. SRCC, signet-ring cell carcinoma.
Chemical inhibition of SLC1A5 or GPT2 synergistically enhanced the treatment effect of chemotherapy in SRCC
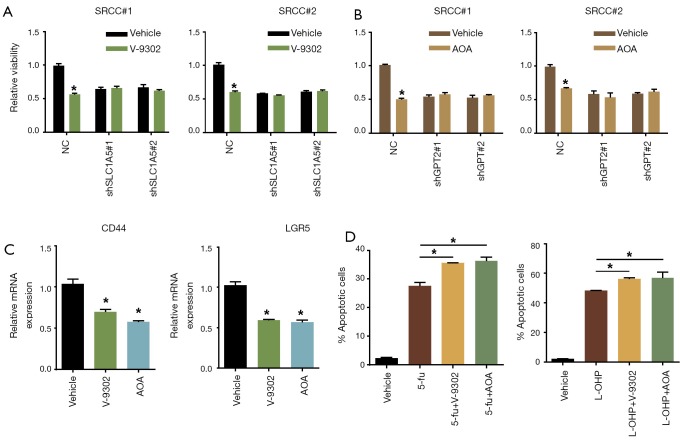
As inhibitors targeting SLC1A5 and GPT2 have been developed, we explored whether the corresponding inhibitor could be effective to treat SRCC. The results suggested that V-9302 (SLC1A5 inhibitor) and AOA (GPT2 inhibitor) could significantly suppress SRCC organoids proliferation (Figure 4A,B). In contrast, V-9302 and AOA had no effect on organoids with silenced SLC1A5 and GPT2 expression, respectively (Figure 4A,B). We next used inhibitors to treat the all three organoids and examined the expression of Wnt target genes that were significantly associated with 'stem-like'. It showed that LGR5 and CD44 expression were dramatically downregulated in SRCC organoids upon being treated with V-9302 or AOA (Figure 4C). Further, we combined separately inhibitors with 5-fu and L-OHP, two commonly used chemotherapeutic drugs for colorectal cancer to treat SRCC organoids and found that combined administration of any inhibitors would significantly increase the sensitivity of SRCC organoids to 5-fu and L-OHP (Figure 4D).
Figure 4.
Chemical inhibition of SLC1A5 or GPT2 synergistically enhanced the treatment effect of chemotherapy in SRCC. (A) V-9302 (20 µM) administration significantly inhibited SRCC organoids growth but did not impact the organoids with SLC1A5 depletion; (B) AOA (10 mM) administration significantly inhibited SRCC organoids growth but did not impact the organoids with GPT2 depletion; (C) qRT-PCR showing V-9302 or AOA administration strikingly reduced expression of CD44 and LGR5; (D) apoptosis analysis of SRCC organoids treated with single 5-fu (1 mM) or L-OHP (20 µM) and inhibitors (V-9302 and AOA). *, P<0.05. SRCC, signet-ring cell carcinoma.
Discussion
SRCC is a rare subtype of colorectal adenocarcinoma with distinct biological behaviors and extremely poor prognosis. Stomach SRCC accounts for 90% of all SRCCs, while only 1% of all colorectal cancers were SRCC. Previous basic studies mainly focused on gastric SRCC based on gastric cancer cell lines, but no mechanism studies have been conducted in colorectal SRCC for the lack of established colorectal SRCC cell lines. Recently, our team successfully established and verified the first colon signet ring cell organoid line and standardized its isolating and culturing procedures. By using this technology, we performed the first basic research aiming to reveal the metabolic features of colorectal SRCC based on organoid model.
Organoids grew from organ-specific stem cells and showed three-dimensional (3D) structures (12). They have currently been used to model different cancers, including those of the colon, stomach, lung, pancreas, liver, breast, prostate and fallopian tube for entirely retaining the structure, morphology, genetic mutations and heterogeneity of the parental tumor (4). By using a 3D culture system, high-throughput drug screening coupled with genomic and biological behavior analyses might be able to individualize therapeutic regimens, regardless of the prevalence of the tumor.
PET/CT has been widely adopted to detect primary and metastatic cancer. Its application is based upon the strikingly enhanced glucose uptake in malignant tumors. Nevertheless, previous clinical research suggested that SRCC usually showed low FDG uptake, making it’s infeasible to be detected by PET/CT. Therefore, we speculated that glucose utilization may not be significantly up-regulated in colorectal SRCC. In this study, we discovered that compared with adjacent normal organoids and AC organoids, SRCC organoids showed dramatically increased glutamine uptake rate, suggesting that glutamine utilization may be an important metabolic feature for SRCC. Considering that energy source uptake was regulated by specific transporters, we conducted further experiments which showed that SLC1A5 contributing to glutamine uptake was indeed unregulated in SRCC. For the higher rate of glutamine uptake, we next examined the expression of critical enzymes catalyzing the glutaminolysis and found that GPT2 showed notably higher expression in SRCC.
Circulating glutamine is the most abundant amino acid, making up over 20% of the free amino acid pool in the blood (13,14). The maintenance of high serum levels of glutamine provides a readily accessible source of carbon and nitrogen for biosynthesis, energetics and cellular homeostasis that cancer cells may exploit for growth (15). SLC1A5 mediates the exchange of amino acid substrates and is critical for glutamine uptake in rapidly growing cancer cells. The expression of SLC1A5 has been found to be elevated in many types of cancer including colorectal cancer and its higher expression was confirmed to be related to poor prognosis (16-18). GPT2 is an aminotransaminase that converts glutamate to a-KG, which is a TCA cycle intermediate. Its expression was also found to be upregulated in many cancers and was confirmed as an important factor contributing to rapid cell growth (19-23). Consistent with previous studies, we found that knock down of or pharmacological inhibition of SLC1A5 or GPT2 could significantly suppress SRCC proliferation, suggesting that the glutamine metabolism mediated by SLC1A5 and GPT2 might be critically important for SRCC. In addition, we also uncovered that glutamine deprivation or metabolism inhibition exert no effects on SRCC organoids with attenuated SLC1A5 or GPT2 expression. Taken together, pharmacological use of inhibitors was dependent on the expression of SLC1A5 or GPT2.
Considering the optimal treatment effects of glutamine metabolism inhibitors, we combined inhibitors separately with L-OHP and found that addition of inhibitors significantly increased the antitumor effects of L-OHP by augmenting apoptosis inducing effects. However, further research is still warranted to reveal the underlying mechanisms.
In this study, we, for the first time, explored partial metabolic features of energy source utilization for SRCC organoid. However, we should admit that a large number of problems pertinent to SRCC energy metabolism remain to be elucidated. For example, we did not examine the glycolysis or fatty acid oxidization rate in SRCC though no significant differences upon glucose and fatty acid uptake were found between SRCC and normal organoids. Moreover, intracellular glycolysis or fatty acid oxidization have not been clarified. On the other hand, the SRCC organoids used in this study were derived from three patients with low or negative FDG uptake, thus the metabolic features of those SRCC patients with significantly high FDG uptake are still unknown.
In summary, we highlight that enhanced glutamine uptake and glutaminolysis is a metabolic feature of colorectal SRCC and SLC1A5 or GPT2 might be novel therapeutic targets for colorectal SRCC.
Acknowledgments
Funding: This study was supported by the Grant of Science and Technology Commission of Shanghai Municipality (No. 16401970502 and No.17411951100 and No. 19140902100), the Grant of National Natural Science Foundation of China (No. 81572351 and No. 81871958)
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The research protocol was reviewed and approved by the ethics committee of FUSCC (050432-4-1212B). Written informed consents were obtained on enrollment.
Footnotes
Conflicts of Interest: QL serves as an unpaid editorial board member of Annals of Translational Medicine from Oct 2019 to Sep 2020. The other authors have no conflicts of interest to declare.
References
- 1.Chew MH, Yeo SA, Ng ZP, et al. Critical analysis of mucin and signet ring cell as prognostic factors in an Asian population of 2,764 sporadic colorectal cancers. Int J Colorectal Dis 2010;25:1221-9. 10.1007/s00384-010-1033-3 [DOI] [PubMed] [Google Scholar]
- 2.Nitsche U, Zimmermann A, Spath C, et al. Mucinous and signet-ring cell colorectal cancers differ from classical adenocarcinomas in tumor biology and prognosis. Ann Surg 2013;258:775-82; discussion 782-3. 10.1097/SLA.0b013e3182a69f7e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang R, Ma X, Li Y, et al. The Characteristics and Prognostic Effect of E-Cadherin Expression in Colorectal Signet Ring Cell Carcinoma. PLoS One 2016;11:e0160527. 10.1371/journal.pone.0160527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clevers H. Modeling Development and Disease with Organoids. Cell 2016;165:1586-97. 10.1016/j.cell.2016.05.082 [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Wang R, Huang D, et al. A novel human colon signet-ring cell carcinoma organoid line: establishment, characterization and application. Carcinogenesis 2019. [Epub ahead of print]. 10.1093/carcin/bgz178 [DOI] [PubMed] [Google Scholar]
- 6.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 2012;21:297-308. 10.1016/j.ccr.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029-33. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi BH, Song HS, An YS, et al. Relation between fluorodeoxyglucose uptake and glucose transporter-1 expression in gastric signet ring cell carcinoma. Nucl Med Mol Imaging 2011;45:30-5. 10.1007/s13139-010-0058-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, et al. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol 2017;14:11-31. 10.1038/nrclinonc.2016.60 [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Liang L, Dai W, et al. Prognostic impact of programed cell death-1 (PD-1) and PD-ligand 1 (PD-L1) expression in cancer cells and tumor infiltrating lymphocytes in colorectal cancer. Mol Cancer 2016;15:55. 10.1186/s12943-016-0539-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q, Liang X, Wang Y, et al. miR-139-5p Inhibits the Epithelial-Mesenchymal Transition and Enhances the Chemotherapeutic Sensitivity of Colorectal Cancer Cells by Downregulating BCL2. Sci Rep 2016;6:27157. 10.1038/srep27157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 2014;345:1247125. 10.1126/science.1247125 [DOI] [PubMed] [Google Scholar]
- 13.Mayers JR, Vander Heiden MG. Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem Sci 2015;40:130-40. 10.1016/j.tibs.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergström J, Furst P, Noree LO, et al. Intracellular free amino acids in muscle tissue of patients with chronic uraemia: effect of peritoneal dialysis and infusion of essential amino acids. Clin Sci Mol Med 1978;54:51-60. 10.1042/cs0540051 [DOI] [PubMed] [Google Scholar]
- 15.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer 2016;16:619-34. 10.1038/nrc.2016.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kekuda R, Prasad PD, Fei YJ, et al. Cloning of the sodium-dependent, broad-scope, neutral amino acid transporter Bo from a human placental choriocarcinoma cell line. J Biol Chem 1996;271:18657-61. 10.1074/jbc.271.31.18657 [DOI] [PubMed] [Google Scholar]
- 17.Fuchs BC, Finger RE, Onan MC, et al. ASCT2 silencing regulates mammalian target-of-rapamycin growth and survival signaling in human hepatoma cells. Am J Physiol Cell Physiol 2007;293:C55-63. 10.1152/ajpcell.00330.2006 [DOI] [PubMed] [Google Scholar]
- 18.Kim S, Kim DH, Jung WH, et al. Expression of glutamine metabolism-related proteins according to molecular subtype of breast cancer. Endocr Relat Cancer 2013;20:339-48. 10.1530/ERC-12-0398 [DOI] [PubMed] [Google Scholar]
- 19.Coloff JL, Murphy JP, Braun CR, et al. Differential Glutamate Metabolism in Proliferating and Quiescent Mammary Epithelial Cells. Cell Metab 2016;23:867-80. 10.1016/j.cmet.2016.03.016 [DOI] [PubMed] [Google Scholar]
- 20.Itkonen HM, Gorad SS, Duveau DY, et al. Inhibition of O-GlcNAc transferase activity reprograms prostate cancer cell metabolism. Oncotarget 2016;7:12464-76. 10.18632/oncotarget.7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 2006;9:121-32. 10.1016/j.ccr.2006.01.013 [DOI] [PubMed] [Google Scholar]
- 22.Cao Y, Lin SH, Wang Y, et al. Glutamic Pyruvate Transaminase GPT2 Promotes Tumorigenesis of Breast Cancer Cells by Activating Sonic Hedgehog Signaling. Theranostics 2017;7:3021-33. 10.7150/thno.18992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao Y, Samuels Y, Li Q, et al. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat Commun 2016;7:11971. 10.1038/ncomms11971 [DOI] [PMC free article] [PubMed] [Google Scholar]