Abstract
Among vertebrates, turtles have many unique characteristics providing biologists with opportunities to study novel evolutionary innovations and processes. We present here a high-quality, partially phased, and chromosome-level Red-Eared Slider (Trachemys scripta elegans, TSE) genome as a reference for future research on turtle and tetrapod evolution. This TSE assembly is 2.269 Gb in length, has one of the highest scaffold N50 and N90 values of any published turtle genome to date (N50 = 129.68 Mb and N90 = 19 Mb), and has a total of 28,415 annotated genes. We introduce synteny analyses using BUSCO single-copy orthologs, which reveal two chromosome fusion events accounting for differences in chromosome counts between emydids and other cryptodire turtles and reveal many fission/fusion events for birds, crocodiles, and snakes relative to TSE. This annotated chromosome-level genome will provide an important reference genome for future studies on turtle, vertebrate, and chromosome evolution.
Keywords: reference genome, Hi-C, linked-reads, IsoSEQ, turtle, synteny, chromosome, assembly
Introduction
The application of whole-genome sequencing to non-model organisms is providing new insights into the genome evolution of tetrapods (Shedlock et al. 2007; Ellegren 2014). Because turtles (Testudines) are one of the three main groups of reptiles, they represent an important lineage for comparison. Within turtles, studies of genomic evolution have contributed to a broader understanding of many turtle questions, including sex determination mechanisms (Bachtrog et al. 2014; Montiel et al. 2016; Platt et al. 2017; Lee et al. 2019). Turtle genomes are also important because turtles are renowned for their ability to hybridize across distantly related lineages (Buskirk et al. 2005). The high-quality, annotated, and chromosome-level genome of the Red-Eared Slider (Trachemys scripta elegans, TSE hereafter) (NCBI BioProject PRJNA552319) presented here is an important source of data for future research into these and other questions relating to the evolution of tetrapod genomes. This genome assembly includes 10× linked-reads, Hi-C data (Lieberman-Aiden et al. 2009), mate-pair data, and PacBio Iso-Seq data. We chose to sequence TSE for this study because it is the most abundant turtle on Earth, has a long history as a comparative subject in turtle studies (Gibbons 1990), and is of conservation and evolutionary interest as a hybridizing introduced species (Parham et al. 2013, 2020).
We apply Benchmarking Universal Single-Copy Ortholog v2.0.1 (BUSCO) synteny analyses to the TSE genome compared with two other turtles (a tortoise and a sea turtle) and three other diapsids (alligator, chicken, and python) and identify patterns of chromosomal fission and fusion events across these diapsids.
Materials and Methods
DNA and RNA Sample Collection
DNA extractions for the 10× genomics linked-reads and RNA extractions for the PacBio Iso-Seq libraries were from freshly collected liver tissue sent to Genewiz (www.genewiz.com, last accessed April 8, 2020) for high-molecular weight DNA extractions. The mate-pair library was from freshly collected liver tissue from a different TSE specimen (CAS 252980) and performed by NGX Bio (ngxbio.com, last accessed April 8, 2020). DNA extraction for the Hi-C library was performed in the Center for Comparative Genomics at the California Academy of Sciences from 200 mg of freshly harvested liver tissue of a TSE from Texas (MVZ 292727) (For more detail on library preparation and sequencing, see supplementary text, Supplementary Material online).
Genome Assembly
Our de novo assembly began with the assembly of the 10× reads using Supernova release 2.0.1 (Weisenfeld et al. 2017) to generate a draft genome assembly. We began with a library of 429 M read pairs of Illumina 2×150 data barcoded by the 10× Genomics Chromium instrument.
Next, we used the 4-kb insert mate-pair reads, adapter trimmed and classified by Illumina’s NxTrim (O’Connell et al. 2015), to scaffold the Supernova generated assembly with BESST version 2.2.8 (Sahlin et al. 2014, 2016); default parameters were used except opts=“–iter 20000000” which sets a maximum of 20 million iterations.
The third step was to use ARKS v1.0.2 (Coombe et al. 2018), which is an alignment-free assembler using a k-mer-based mapping approach for 10× linked-read data. ARKS reuses the original 10× Illumina reads for k-mer mapping against the Supernova/BESST assembly in three steps. The first step uses a k-mer approach to map the linked barcodes to the Supernova/BESST contigs. ARKS then scores contig pairs, and finally, it produces a scaffold graph with estimated distances. The companion LINKS program (Warren et al. 2015) applies this graph to create the longer scaffolded assembly.
We then incorporated the Hi-C data for super-scaffolding using the Proximo assembly pipeline, performed by Phase Genomics. Chromatin conformation capture data were generated using a Phase Genomics (Seattle, WA) Proximo Hi-C Animal Kit, which is a commercially available version of the Hi-C protocol (Lieberman-Aiden et al. 2009). Following the manufacturer’s instructions for the kit, intact cells from two samples were crosslinked using a formaldehyde solution, digested using the Sau3AI (MboI) restriction enzyme, and proximity ligated with biotinylated nucleotides to create chimeric molecules composed of fragments from different regions of the genome that were physically proximal in vivo, but not necessarily genomically proximal. Molecules were pulled down with streptavidin beads and processed into an Illumina-compatible sequencing library. Sequencing was performed on an Illumina HiSeq4000, generating a total of 442,350,436 PE150 read pairs.
Briefly, reads were aligned using BWA-MEM (Li and Durbin 2009) with the -5SP and -t 8 options specified, all others default. SAMBLASTER (Faust and Hall 2014) was used to flag PCR duplicates, which were later excluded from analysis. Alignments were then filtered with SAMtools (Li et al. 2009) using the -F 2304 filtering flag to remove nonprimary and secondary alignments.
The Phase Genomics’ Proximo Hi-C genome scaffolding platform was used to create chromosome-scale scaffolds from the corrected assembly as described in Bickhart et al. (2017). As in the LACHESIS method (Burton et al. 2013), this process computes a contact frequency matrix from the aligned Hi-C read pairs, normalized by the number of Sau3AI restriction sites (GATC) on each contig, and constructs scaffolds in such a way as to optimize expected contact frequency and other statistical patterns in Hi-C data. Approximately 100,000 separate Proximo runs were performed to optimize the number of scaffolds and scaffold construction in order to make the scaffolds as concordant with the observed Hi-C data as possible.
We then ran SOAP GapCloser version 1.12 (Luo et al. 2012) with barcode and adapter trimmed 10× paired-reads together with the mate-pair reads for gap closing; parameters -l 152 -p 31 were used. This was followed by a decontamination and duplicate identification step using NCBI’s tbl2asn script, which generates a “.val” error file listing potential contaminants, mitochondrial sequences, and duplicates. We used this information to manually remove contaminants and duplicates identified by tbl2asn. This step also serves to prepare the sequence data for easier submission to GenBank. Along with the removal of exact duplicates identified by tbl2asn, we also removed near-duplicate contigs that differed by a small number of bases from each other and records with 90% or more Ns; presumably, these are an artifact of the Supernova program’s attempt to phase the genome. This was followed by two more SOAP GapCloser runs.
We made manual Hi-C scaffold adjustments with Juicebox (Durand et al. 2016) (supplementary text and fig. S1, Supplementary Material online) and by using BUSCO v2.01 (Simao et al. 2015) with the 3,950 ortholog Tetrapoda odb9 database for synteny comparisons of TSE with other Archelosauria (supplementary table S1, Supplementary Material online, exclusive of Python). This assembly is named Tse_1.0.fasta (NCBI BioProject PRJNA552319).
To assess the quality and completeness of our assembly, we used BUSCO. We employed the reference gene set of Tetropoda odb9 (a total of 3,950 orthologs) and ran the genome option of the program using the –limit 20 parameter. We also ran BUSCO with the same parameters on a reverse complement of the assembly (For details on genome size estimation see supplementary text, Supplementary Material online).
Genome Annotation and Analyses
Repeat Analysis
For TSE and three other turtles’ assemblies, Chelonia mydas, Chrysemys picta bellii, and Gopherus evgoodei, we created a species specific de novo repeat library file by running RepeatModeler version open-1.0.11 (Smit and Hubley 2008–2015 www.repeatmasker.org, last accessed April 8, 2020). RepeatModeler marshals RECON version 1.08 (Bao and Eddy 2002), RepeatScout version 1.0.5 (Price et al. 2005), and Tandem repeats finder version 4.09 (Benson 1999) to create a de novo repeat library. We ran RepeatMasker on each assembly with options -nolow and -lib referencing a custom library combining its de novo repeat families with the vertebrate RepBase Combined Database (Dfam_3.0 from RepeatMasker and RepBase-20181026 input to rmblastn version 2.9.0+).
We annotated the TSE assembly using Maker version 3.01.02 (Holt and Yandell 2011; Campbell et al. 2014) to predict gene models and predict functional annotations. We ran MAKER in two runs with both homology-based and ab initio gene modelers (for complete details see supplementary text, Supplementary Material online).
BUSCO Synteny Analyses
The term “synteny” has been applied to different types of genetic patterns (Renwick 1972; Passarge et al. 1999). We use the term “synteny” sensu Shields (2001), “conservation of order of orthologous genes between different species.” In order to analyze synteny among lineages, we used the results of BUSCO analyses and custom scripts (github.com/calacademy-research/ccgutils/tree/master/assembly_scripts, last accessed April 8, 2020) to generate single-copy ortholog (SCO) Circos (Krzywinski et al. 2009) synteny “links” files between TSE and two other turtles (a tortoise, G. evgoodei and a sea turtle, C. mydas), the chicken (Gallus), a crocodilian (Alligator), and a snake (Python) (supplementary table S1, Supplementary Material online). Although we have a comprehensive annotation of the TSE genome, many available genomes are not annotated and cannot be included in synteny analyses based on annotations; or, unlike these chromosomal level assemblies, are so fragmented that the synteny matches are not insightful. Because BUSCO analyses can be run relatively easily and quickly (∼24 h) relative to a full annotation (typically weeks), we propose and demonstrate the utility of BUSCO-based synteny analyses. We chose genomes with high-quality chromosome-level assemblies because synteny analyses are sensitive to fragmented assemblies (Liu et al. 2018). For G. evgoodei, we are uncertain of the karyotype; the 24 reported scaffolds likely reflect chromosomes, but further assembly could identify additional smaller chromosomes. Several studies (reviewed by Bickham and Carr 1983) show that most testugurians (the group that includes G. evgoodei and other tortoises) are 2n = 52 and that this condition is likely ancestral for that clade (Bickham and Carr 1983).
Results and Discussion
Genome Assembly
Proximo Hi-C scaffolding resulted in an initial set of additional scaffolds, with which Juicebox and BUSCO synteny analysis with four other archelosaurs were used to correct scaffolding errors as well as introduce eight new breaks into putative misjoined scaffolds from the original assembly. A total of 414 scaffolds were placed and oriented by these methods into 27 new scaffolds; 395 of these comprise the 25 haploid chromosomes of TSE, and 19 of these were used in two unplaced scaffolds.
The resulting Tse_1.0 assembly is 2.269 Gb in length, where the GenomeScope (Vurture et al. 2017) k-mer frequency estimate was just over 2 Gb and Supernova estimated 2.41 Gb (supplementary fig. S2, Supplementary Material online). This assembly size is consistent with other published turtle genome’s sizes (supplementary table S2, Supplementary Material online), the average of which for six assemblies is 2.33 Gb. Tse_1.0 scaffold N50 is 129.68 Mb occurring at chromosome 6 (chr1–chr6 contain 54.84% of the bases) and 19-Mb scaffold N90 at chr21 (chr1–chr21 contain 90.51% of total bases). Contig N50 is 189,165 bp; contig N90 is 32,113 bp. The 25 assembled haploid chromosomes contain 92.92% of the full assembly leaving 7.08% currently unplaced. This is one of the highest scoring turtle genomes published to date (supplementary table S2, Supplementary Material online). We used a modified Assemblathon script (github.com/calacademy-research/ccgutils/tree/master/asmstats, last accessed April 8, 2020) to calculate TSE assembly statistics (table 1).
Table 1.
Assemblathon+ Statistics for TSE Genome Assembly (generated with custom asmstats.pla)
| Number of scaffolds >1K nt | 26,710 | 66.60% | |
| Number of scaffolds >10K nt | 2,988 | 7.50% | |
| Number of scaffolds >100K nt | 32 | 0.10% | |
| Number of scaffolds >1M nt | 28 | 0.10% | |
| Number of scaffolds >10M nt | 24 | 0.10% | |
| Mean scaffold size | 56,571 | ||
| Median scaffold size | 1,555 | ||
| N50 scaffold length | 129,675,691 | L50 scaffold count | 6 |
| N60 scaffold length | 126,808,733 | L60 scaffold count | 7 |
| N70 scaffold length | 85,829,911 | L70 scaffold count | 10 |
| N80 scaffold length | 43,716,676 | L80 scaffold count | 13 |
| N90 scaffold length | 19,049,219 | L90 scaffold count | 21 |
| Scaffold %A | 27 | Number of A | 609,556,304 |
| Scaffold %C | 21 | Number of C | 482,905,406 |
| Scaffold %G | 21 | Number of G | 483,034,848 |
| Scaffold %T | 27 | Number of T | 609,529,607 |
| Scaffold %N | 4 | Number of N | 83,700,144 |
| Scaffold %non-ACGTN | 0 | ||
| Number of scaffold non-ACGTN nt | 0 | ||
| Percentage of assembly in scaffolded contigs | 94.50 | ||
| Percentage of assembly in unscaffolded contigs | 5.50 | ||
| Average number of contigs per scaffold | 1.5 | ||
| Average length of break (≥10 N) between contigs in scaffold | 4,165 | ||
| Number of contigs | 60,193 | ||
| Number of contigs in scaffolds | 22,314 | ||
| Number of contigs not in scaffolds | 37,879 | ||
| Total size of contigs | 2,185,039,207 | ||
| Longest contig | 1,642,093 | ||
| Shortest contig | 48 | ||
| Number of contigs >1K nt | 45,044 | 74.80% | |
| Number of contigs >10K nt | 19,706 | 32.70% | |
| Number of contigs >100K nt | 6,735 | 11.20% | |
| Number of contigs >1M nt | 21 | 0.00% | |
| Number of contigs >10M nt | 0 | 0.00% | |
| Mean contig size | 36,301 | ||
| Median contig size | 3,258 | ||
| N50 contig length | 189,165 | L50 contig count | 3,255 |
| N60 contig length | 146,678 | L60 contig count | 4,570 |
| N70 contig length | 108,417 | L70 contig count | 6,295 |
| N80 contig length | 71,323 | L80 contig count | 8,775 |
| N90 contig length | 32,113 | L90 contig count | 13,220 |
| Contig %A | 28 | Number of A | 609,556,304 |
| Contig %C | 22 | Number of C | 482,905,406 |
| Contig %G | 22 | Number of G | 483,034,848 |
| Contig %T | 27.9 | Number of T | 609,529,607 |
| Contig %N | 0 | Number of N | 13,042 |
| Contig %non-ACGTN | 0 | ||
| Number of contig non-ACGTN nt | 0 | ||
asmstats.pl is a modification of github.com/ucdavis-bioinformatics/assemblathon2-analysis/blob/master/assemblathon_stats.pl (last accessed April 8, 2020) available at github.com/calacademy-research/ccgutils/tree/master/asmstats (last accessed April 8, 2020).
Genome Annotation and Analyses
Genome Annotation
After the MAKER runs and InterProScan results, we determined the TSE set of 28,415 predicted gene models in the assembly and added functional annotations to them based on homology results.
We assigned homology-based annotation to 27,439 (96.33%) of the proteins for which the genes code, with 57.26% of those assigned to a Chrysemys picta bellii homolog and a total of 86% (23,883) assigned to homologs of one of five turtle species. About 28,903 Pfam domains were found with 6,039 of them unique; 43,835 Gene Ontology (GO) terms, 3,703 unique; 77,913 InterPro matches, 14,749 unique. Pathways from three pathway databases show 3,287 KEGG pathways, 904 unique; 3,179 MetaCyc, 622 unique; and 27,381, 1,606 unique, from the Reactome database.
Repeat Analysis
The results from the RepeatMasker analyses for TSE, C. mydas, Chrysemys picta belli, and G. evgoodei are listed in supplementary table S3, Supplementary Material online.
Quality Assessment
The BUSCO analyses were run twice, in the forward direction and on the reverse complement. The reverse complement run produced an additional 29 complete and 1 fragmented BUSCO, thus reducing missing ones by 30 from the forward run. About 95.8% complete BUSCOs were found, 3,783 with 25 duplicates, 106 fragmented, and 61 missing from the reference gene set 3,950 in Tetropoda odb9 (supplementary table S4, Supplementary Material online).
Comparative Genomic Analyses
BUSCO Synteny
We mapped shared orthologs from the Tetrapoda odb9 database of BUSCOs to each of the genomes used in this study (TSE, G. evgoodei, C. mydas, Alligator mississippiensis, Gallus gallus, and Python bivittatus) and created Circos links files to generate Circos synteny diagrams (supplementary figs. S4–S6, Supplementary Material online). We demonstrate here the utility of BUSCO synteny analyses by identifying clear chromosomal fission/fusion events and patterns (creation and loss of chromosomes) across the diapsids (squamates, birds, crocodiles, and turtles).
Chromosome Fission/Fusion
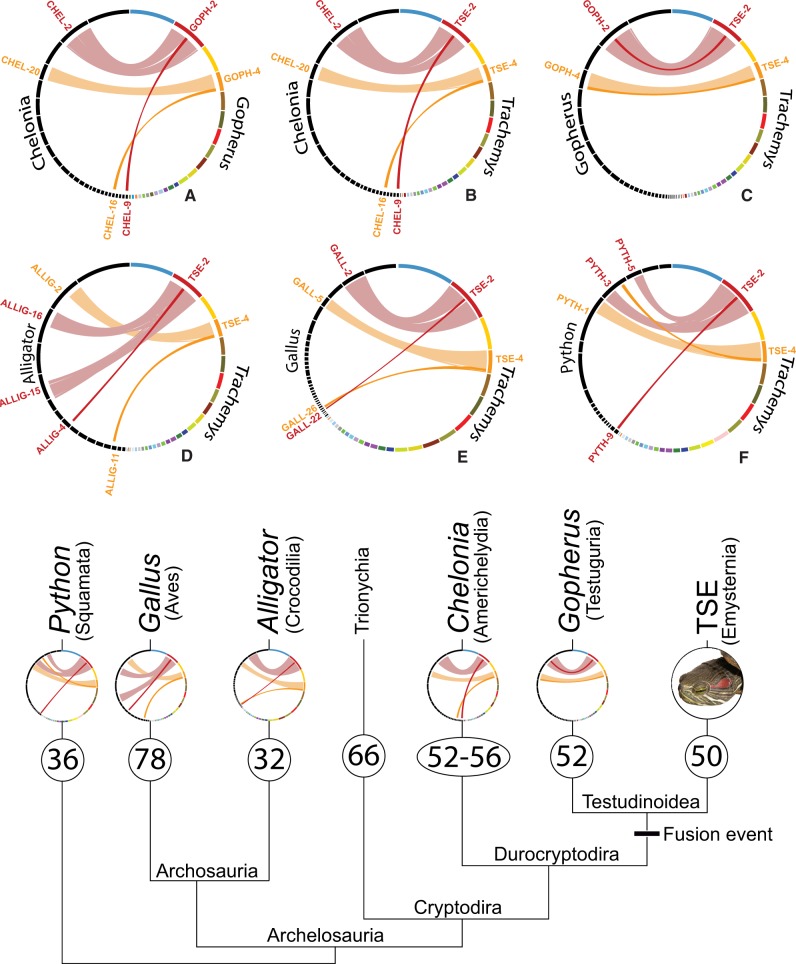
Using BUSCO synteny analyses, we were able to identify various chromosomal fission/fusion events and the lineages where they occurred. We identify two clusters of BUSCOs shared among all diapsids examined in this study. For TSE and Gopherus, these BUSCO clusters are part of two pair of homologous chromosomes TSE-2 to GOPH-2, and TSE-4 to GOPH-4 (fig. 1C), for all other taxa, these clusters are found on different and nonhomologous chromosomes (fig. 1A, B, and D–F). Of the archelosaurian genomes studied here, only Alligator has fewer chromosomes (32) than TSE and Gopherus (50 and 52 respectively, supplementary table S1, Supplementary Material online), which suggests that two chromosomal fusions occurred sometime since the most recent common ancestor of Testudinoidea (fig. 1 tree).
Fig. 1.
—Circos synteny diagrams displaying chromosomal fissions/fusions within diapsids. The dark red and dark orange lines represent homologous clusters of SCOs found on chromosomes not found on testudinoid genomes (TSE and Gopherus) and likely fused with TSE and Gopherus chromosomes 2 and 4. Synteny diagram (A) reveals the fusion of two Chelonia chromosomes 16 and 9 with Gopherus chromosomes 2 and 4 respectively. Synteny diagram (B) reveals the fusion of two Chelonia chromosomes 16 and 9 with TSE chromosomes 2 and 4 respectively. Synteny diagram (C) reveals that for both Gopherus and TSE the dark red and dark orange cluster of SCOs have fused with TSE and Gopherus chromosomes 2 and 4. Synteny diagram (D) and (E) reveal the archosaurian Alligator chromosomes 4 and 11 and Gallus chromosomes 26 and 22 have fused with testudinoid (Gopherus and TSE) chromosomes. Synteny diagram (F) reveals that Python chromosome 9 has fused with Gopherus and TSE chromosome 2. The dark orange cluster of SCOs from TSE chromosome 4 are part of Python chromosome 5. The colored bars in rings represent TSE chromosomes except in (A), where colored bar represent Gopherus. Black bars represent other chromosomes. Note that all single relocations have been removed for clarity. For the phylogenetic tree of reptile and avian genomes used in this study, the number in ovals represents the hypothesized ancestral 2n number of chromosome for each lineage based on Bickham and Carr (1983) and the circular diagrams represent the synteny diagrams highlighting the chromosomes involved in the indicated fusion event. TSE and Gopherus share the same fusion of these two clusters (diagram C).
The genomic evolution of birds has produced genomes with many small chromosomes (Burt 2002). The syntenic comparison between TSE and Gallus reveals at least six fusion events relative to TSE (supplementary fig. S5A, Supplementary Material online), which accounts for six additional chromosomes for Gallus relative to TSE. In contrast to birds, crocodiles have a reduced number of chromosomes (Gallus [2n = 78], Alligator [2n = 32]). We see seven clear fission events relative to TSE and account for a greater number of chromosomes in TSE (supplementary fig. S5B, Supplementary Material online). All six of the TSE chromosomes involved in fission/fusion with Gallus are shared by the Alligator versus TSE analysis. The only exception is the addition of TSE chromosome 1 (TSE-1) in the Alligator versus TSE analysis (supplementary fig. S5B, Supplementary Material online). A BUSCO synteny analysis between the squamate Python (2n = 36) and TSE reveals more than a dozen clear fission/fusion events relative to TSE. Python chromosomes 1, 2, and 3 each appear to be comprised at least four large syntenic blocks from separate TSE chromosomes (supplementary fig. S6, Supplementary Material online). TSE chromosomes 2, 4, 8, 9, and 10 each have large syntenic blocks that map to two different Python chromosomes (supplementary fig. S6, Supplementary Material online).
Supplementary Material
Acknowledgments
We acknowledge and thank the Museum of Vertebrate Zoology at University of California Berkeley and the California Academy of Science's Herpetology Department and their staff, in particular Carol Spencer (MVZ) and Lauren Scheinberg (CAS); the staff of the CAS Center for Comparative Genomics for their help with lab work; the CAS Center for Comparative Genomics’ computational resources; Shawn Sullivan and Phase Genomics for their help with Proximo and Juicebox; the Vertebrate Genome Project for the release of the Gopherus evgoodei genome and for making so many new genomes publicly available; and the DNA Zoo for releasing the Python, Alligator, and Chelonia genomes and for creating so many publicly available chromosome-level (Hi-C scaffolding) genomes. This research was supported by the California Academy of Science's annual Resarch Allotment (2018, 2019).
Author Contributions
W.B.S., J.B.H., and J.F.P. conceived the study. T.J.P. collected the specimen and extracted tissues. A.W.L. performed all lab work in the Center for Comparative Genomics under supervision of W.B.S. J.B.H. and W.B.S. performed computational analyses. W.B.S. wrote the article with contributions from J.F.P. and J.B.H. All authors read, revised, and approved the article.
Literature Cited
- Bachtrog D, et al. 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12(7):e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Eddy SR.. 2002. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12(8):1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickham JW, Carr JL.. 1983. Taxonomy and phylogeny of the higher categories of Cryptodiran turtles based on a cladistic analysis of chromosomal data. Copeia 1983(4):918–932.
- Bickhart DM, et al. 2017. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet. 49(4):643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt DW. 2002. Origin and evolution of avian microchromosomes. Cytogenet Genome Res. 96(1–4):97–112. [DOI] [PubMed] [Google Scholar]
- Burton JN, et al. 2013. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 31(12):1119–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskirk JR, Parham JF, Feldman CR.. 2005. On the hybridisation between two distantly related Asian turtles (Testudines: Sacalia × Mauremys.). Salamandra 41:21–26. [Google Scholar]
- Campbell MS, Holt C, Moore B, Yandell M.. 2014. Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinformatics. 2014:4.11.1–4.11.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombe L, et al. 2018. ARKS: chromosome-scale scaffolding of human genome drafts with linked read kmers. BMC Bioinformatics 19(1):234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, et al. 2016. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3(1):99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. 2014. Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol. 29(1):51–63. [DOI] [PubMed] [Google Scholar]
- Faust GG, Hall IM.. 2014. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 30(17):2503–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons JW. 1990. Life history and ecology of the slider turtle. Washington, DC: Smithsonian Institution Press. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2 : an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12(1):491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9):1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L, Montiel EE, Navarro-Domínguez BM, Valenzuela N.. 2019. Chromosomal rearrangements during turtle evolution altered the synteny of genes involved in vertebrate sex determination. Cytogenet Genome Res. 157(1–2):77–88. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326(5950):289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Hunt M, Tsai IJ.. 2018. Inferring synteny between genome assemblies: a systematic evaluation. BMC Bioinformatics 19(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel EE, et al. 2016. Cytogenetic insights into the evolution of chromosomes and sex determination reveal striking homology of turtle sex chromosomes to amphibian autosomes. Cytogenet Genome Res. 148(4):292–304. [DOI] [PubMed] [Google Scholar]
- O’Connell J, et al. 2015. NxTrim: optimized trimming of Illumina mate pair reads. Bioinformatics 31(12):2035–2037. [DOI] [PubMed] [Google Scholar]
- Parham JF, et al. 2013. Genetic introgression and hybridization in Antillean freshwater turtles (Trachemys) revealed by coalescent analyses of mitochondrial and cloned nuclear markers. Mol Phylogenet Evol. 67(1):176–187. [DOI] [PubMed] [Google Scholar]
- Parham JF, Papenfuss TJ, Sellas AB, Stuart BL, Brian Simison W.. 2020. Genetic variation and admixture of red-eared sliders (Trachemys scripta elegans) in the USA. Mol Phylogenet Evol. 145:106722. [DOI] [PubMed] [Google Scholar]
- Passarge E, Horsthemke B, Farber RA.. 1999. Incorrect use of the term synteny [1]. Nat Genet. 23(4):387–387. [DOI] [PubMed] [Google Scholar]
- Platt RN, et al. 2017. Conflicting evolutionary histories of the mitochondrial and nuclear genomes in New World Myotis bats. Syst Biol. 67(2):236–249. [DOI] [PMC free article] [PubMed]
- Price AL, Jones NC, Pevzner PA.. 2005. De novo identification of repeat families in large genomes. Bioinformatics 21(Suppl 1):i351–i358. [DOI] [PubMed] [Google Scholar]
- Renwick JH. 1972. Human genetics In: Grouchy J, Ebling FJG, Henderson IW, editors. Proceedings of the Fourth International Congress of Human Genetics. Amsterdam: Excerpta Medica. p. 443–444. [Google Scholar]
- Sahlin K, Chikhi R, Arvestad L.. 2016. Assembly scaffolding with PE-contaminated mate-pair libraries. Bioinformatics 32(13):1925–1932. [DOI] [PubMed] [Google Scholar]
- Sahlin K, Vezzi F, Nystedt B, Lundeberg J, Arvestad L.. 2014. BESST – efficient scaffolding of large fragmented assemblies. BMC Bioinformatics 15(1):281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlock AM, et al. 2007. Phylogenomics of nonavian reptiles and the structure of the ancestral amniote genome. Proc Natl Acad Sci U S A. 104(8):2767–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Vurture GW, et al. 2017. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33(14):2202–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RL, et al. 2015. LINKS: scalable, alignment-free scaffolding of draft genomes with long reads. GigaScience 4(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB.. 2017. Direct determination of diploid genome sequences. Genome Res. 27(5):757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.