Abstract
The fungal-interactive (fungiphilic) strains BS001, BS007, BS110, and BS437 have previously been preliminarily assigned to the species Paraburkholderia terrae. However, in the (novel) genus Paraburkholderia, an as-yet unresolved subgroup exists, that clusters around Paraburkholderia hospita (containing the species P. terrae, P. hospita, and Paraburkholderia caribensis). To shed light on the precise relationships across the respective type strains and the novel fungiphiles, we here compare their genomic and ecophysiological features. To reach this goal, the genomes of the three type strains, with sizes ranging from 9.0 to 11.5 Mb, were de novo sequenced and the high-quality genomes analyzed. Using whole-genome, ribosomal RNA and marker-gene-concatenate analyses, close relationships between P. hospita DSM 17164T and P. terrae DSM 17804T, versus more remote relationships to P. caribensis DSM 13236T, were found. All four fungiphilic strains clustered closely to the two-species cluster. Analyses of average nucleotide identities (ANIm) and tetranucleotide frequencies (TETRA) confirmed the close relationships between P. hospita DSM 17164T and P. terrae DSM 17804T (ANIm = 95.42; TETRA = 0.99784), as compared with the similarities of each one of these strains to P. caribensis DSM 13236T. A species cluster was thus proposed. Furthermore, high similarities of the fungiphilic strains BS001, BS007, BS110, and BS437 with this cluster were found, indicating that these strains also make part of it, being closely linked to P. hospita DSM 17164T (ANIm = 99%; TETRA = 0.99). We propose to coin this cluster the P. hospita species cluster (containing P. hospita DSM 17164T, P. terrae DSM 17804T, and strains BS001, BS007, BS110, and BS437), being clearly divergent from the closely related species P. caribensis (type strain DSM 13236T). Moreover, given their close relatedness to P. hospita DSM 17164T within the cluster, we propose to rename the four fungiphilic strains as members of P. hospita. Analysis of migratory behavior along with fungal growth through soil revealed both P. terrae DSM 17804T and P. hospita DSM 17164T (next to the four fungiphilic strains) to be migration-proficient, whereas P. caribensis DSM 13236T was a relatively poor migrator. Examination of predicted functions across the genomes of the seven investigated strains, next to several selected additional ones, revealed the common presence of features in the P. hospita cluster strains that are potentially important in interactions with soil fungi. Thus, genes encoding specific metabolic functions, biofilm formation (pelABCDEFG, pgaABCD, alginate-related genes), motility/chemotaxis, type-4 pili, and diverse secretion systems were found.
Keywords: average nucleotide identity, comparative genomics, Paraburkholderia hospita, soil bacteria, species cluster
Introduction
A suite of studies has revealed members of the genus Burkholderia to be ubiquitous in soils (Salles et al. 2002) and plants (Stoyanova et al. 2007; Estrada-de los Santos et al. 2013; Sahl et al. 2015). However, key uncertainties have recently been identified with respect to the species boundaries inside and outside of this genus (Sawana et al. 2014; Beukes et al. 2017). Thus, Sawana et al. (2014) proposed a split of the genus Burkholderia into two novel genera, denoted Burkholderia and Paraburkholderia. Whereas the first genus comprises many human-associated and/or -pathogenic species, the second one encompasses mainly environmental (next to poorly characterized) species. A follow-up study by Estrada-de los Santos et al. (2016) revised this split and proposed two “transition” groups, denoted groups 1 and 2, in addition to two established clades (and a Burkholderia andropogonis group). Shortly following this, Beukes et al. (2017) showed evidence for the definition of four distinct genera: 1) Paraburkholderia, 2) Caballeronia (formerly transition group 2 of Estrada-de los Santos et al.), 3) Burkholderia, and 4) Robbsia (andropogonis). Very recently, Estrada-de los Santos et al. (2018) showed that two additional novel genera, that is, Trinickia and Mycetohabitans, also make part of Burkholderia “sensu lato.”
Within the genus Paraburkholderia, as defined by Sawana et al. (2014) and Beukes et al. (2017), a particularly interesting cluster of bacteria relevant for soil settings is formed by the species Paraburkholderia terrae, Paraburkholderia hospita, and Paraburkholderia caribensis. The type strains of these species, that is, P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T, have been used as taxonomical and ecophysiological reference strains (Achouak et al. 1999; Goris et al. 2002; Yang et al. 2006). Paraburkholderia terrae DSM 17804T was originally isolated from a forest soil in Daejeon, South Korea (Yang et al. 2006), whereas P. hospita DSM 17164T came from an agricultural soil in Pittem, Belgium (Goris et al. 2002). The latter strain was isolated by virtue of its prominence as a key in situ recipient of the soil-introduced 2,4-dichlorophenoxyacetic acid (2,4-D) catabolic broad-host-range plasmids pJP4 and pEMT1 (Goris et al. 2002). Lastly, P. caribensis DSM 13236T was first isolated from a vertisol soil in Martinique (French West Indies). Members of this species were found to produce high amounts of exopolysaccharides on carbon-rich media, indicating avid biofilm formation properties (Achouak et al. 1999).
In our initial studies on bacteria that can interact with soil fungi like Laccaria proxima and/or Lyophyllum sp. strain Karsten, particularly dominant bacteria were found to be able to migrate along fungal hyphae, form biofilms on these and grow on compounds present in fungal exudates (Warmink et al. 2011; Nazir et al. 2012). Many of the strains with these characteristics (notably BS001, BS007, BS110, and BS437) were initially assigned to the species P. terrae (Warmink et al. 2011; Nazir et al. 2012). All aforementioned strains were genome-sequenced in our lab, and the genome of one, BS001, was extensively examined (Haq et al. 2014). In this genome, several genes or operons were found to encode traits that enable establishment at, and interaction with, the hyphae of soil fungi. Thus, genes or operons encoding biofilm formation (Warmink et al. 2011), a type-three secretion (T3S) system (Yang et al. 2016), chemotaxis/flagellar movement, type-4 pili (T4P), and adherence traits were detected (Haq et al. 2014, 2016). Moreover, it was shown that comigration of strain BS001 with hyphae of Lyophyllum sp. strain Karsten growing through soil was critically dependent on the presence of flagella, with the T3S and T4P systems having minor effects (Yang et al. 2017). A newly found five-gene cluster that was presumably involved in energy generation from small molecules such as oxalate turned out to be highly upregulated when strain BS001 was placed in contact with Lyophyllum sp. strain Karsten (Haq et al. 2017), highlighting the importance of this gene cluster for fitness in the mycosphere.
Here, we hypothesize that an organismal clade related to the “classical” species P. terrae and P. hospita may have evolved in soil that shares genetic systems enabling its members to interact with soil fungi. At the time of writing of this manuscript, genomic data of the respective type strains of these two species, as well as of the comparator P. caribensis type strain, were unavailable. Thus, to enable genomic comparisons across these organisms, we determined their complete genome sequences using a combination of short- and long-read sequencing. Subsequently, we explored both the evolutionary relationships and ecological versatilities across all sequenced genomes and strains. The questions posed were: How related are the three type strains to one another and to the four aforedescribed fungiphilic Paraburkholderia strains? What are their unique versus common features? How may their evolutionary trajectory have shaped their modes of interaction with soil fungi? Can we find sets of genes or gene clusters that may be ascribed to such bacterial–fungal interactions?
Materials and Methods
Growth Conditions and Genomic DNA Extraction
Paraburkholderia terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T were cultured aerobically in Luria–Bertani (LB) medium, with shaking at 28 °C, 180 rpm (overnight). Genomic DNA was extracted using a modified protocol based on the UltraClean microbial DNA isolation kit (MOBio Laboratories Inc., Carlsbad, CA). The modification consisted of adding glass beads to the cultures in order to spur mechanical cell lysis. The extracted DNA was purified with the Wizard DNA cleanup system (Promega, Madison, WI), after which DNA quality and quantity were determined with a Nanodrop spectrometer (Thermo Scientific, Wilmington, DE). The qualities (degree of shearing) and quantities of the extracted DNAs were assessed using electrophoresis in 1% agarose gels.
Metabolic Capacities of Strains and Interaction with Soil Fungi
Metabolic tests using BIOLOG GN2 (Biolog Inc., Hayward, CA) were performed for the strains according to the manufacturer’s protocol (Nazir et al. 2012). Briefly, early exponential-phase cultures were used as inocula for the Biolog test plates (150 µl per well). Each plate contained 96 microwells with one out of 95 different carbon sources in each and tetrazolium as an indicator of metabolic activity. Plates were incubated at 28 °C for up to 48 h to allow the observation of a purple color as an indicator of metabolic activity.
Interaction assays with soil fungi, in particular Lyophyllum sp. strain Karsten, were done according to Nazir et al. (2012). Briefly, single-strain migratory assays were done using Petri dishes with three compartments (Greiner Bio one, Frickenhausen, Germany), of which two were filled with presterilized (autoclaved) field soil (at 60% of water holding capacity, bulk density of ∼1.3 g/cm3 and 8 mm depth). The third compartment was filled with oat flake agar (OFA, 30 g/l oat flakes, 15 g/l agar) (Warmink et al. 2009) and served as a nutrient source for the fungus. Fresh (overnight) bacterial cultures were washed by centrifugation and resuspension in water, and then introduced evenly in a 3 mm-wide streak in the soil compartment directly adjacent to the front of the growing fungal hyphae, as well as in a similar system without fungal growth (negative control). The systems were incubated at 23 °C for 12–14 days of incubation. Following incubation, 100-mg soil portions at the hyphal fronts (“migration sites”) were punched out, shaken in liquid in Eppendorf tubes, and the resulting suspensions dilution-plated onto R2A plates. After incubation for up to 96 h at 28 °C, the plates were used for enumeration of the colony forming units (CFU). Strains with CFU numbers exceeding 107 CFU per g soil at the migration site were considered to be good migrators, whereas strains with numbers below 105 CFU per g represented poor migrators. In a second assay, Lyophyllum sp. strain Karsten was grown in propionate-containing minimal medium for two weeks at 28 °C. The culture was then harvested by centrifugation, after which the supernatant was filtered (0.45 µm pore size) and subsequently used as a medium to monitor the growth of selected Paraburkholderia strains (propionate cannot be utilized by the selected strains).
Genome Sequencing and Assembly
Complete genome sequences of all three type strains (P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T) were determined using a combination of two genomic libraries, of which one was prepared for sequencing with the PacBio RSII (Pacific Biosciences, Menlo Park, CA) platform. This SMRTbell template library was prepared and sequenced according to the instructions from Pacific Biosciences following the Procedure and Checklist “Greater than 10 kb Template Preparation and Sequencing.” Briefly, for preparation of 15 kb libraries, 5 µg genomic DNA was end-repaired and ligated overnight to hairpin adapters, applying components from the DNA/Polymerase Binding Kit P6 (Pacific Biosciences). Reactions were carried out according to the instructions of the manufacturer. BluePippin size selection to >4 kb was then performed (cf. Sage Science, Beverly, MA). Conditions for annealing of sequencing primers and binding of polymerase to purified SMRTbell template were assessed with the Calculator in RS Remote (Pacific Biosciences). SMRT sequencing was carried out on the PacBio RSII (Pacific Biosciences) taking one 240-min movie for one SMRT cell using the P6 Chemistry. Totals of around 712, 722, and 689 million bases were produced for P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T, respectively. Paired-end short-read libraries for hybrid error correction were generated and sequenced on the Illumina HiSeq 2500 (Illumina, San Diego, CA) with 200 cycles resulting in ∼3.5 million paired-end reads per genome.
Long-read genome assemblies were generated using the “RS_HGAP_Assembly.3” protocol included in SMRTPortal version 2.3.0, applying default parameters, with the exception of P. hospita DSM 17164T, where the target genome size was set to 20 Mb. For P. terrae DSM 17804T and P. caribensis DSM 13236T, four chromosomal contigs could be assembled, whereas the assembly of P. hospita DSM 17164T led to five chromosomal contigs and the additional plasmid pEMT1. All assembled replicons were trimmed, circularized and adjusted to dnaA or their replication gene as the first gene. Total genome coverages of 52–61× were calculated within the long-read assembly process. Hybrid error correction was performed for each of the genomes by mapping of Illumina short-read data onto the draft circular genomes using BWA (Li and Durbin 2009) followed by automated variant calling using VarScan 2 (Koboldt et al. 2009) and GATK (McKenna et al. 2009) for consensus calling.
The genome sequences of P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T have been deposited at NCBI GenBank under accession numbers CP026111–CP026114, CP026105–CP026110, and CP026101–CP026104, respectively.
Phylogenetic and Comparative Genome Analyses
To quickly check the quality of the sequencing and confirm previous (PCR-based) data, phylogenetic analyses were done based on the 16S rRNA genes of P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T. Over 1,000 bp (including the V2–V6 regions) of the sequences were aligned using MUSCLE (Edgar 2004) and edited in accordance with Gblocks (Talavera et al. 2007). A maximum-likelihood tree was built with RAxML v.8.2.11 with nucleotide substitution model (GTRCAT), default algorithm setting (hill-climbing) and bootstrap value of 1,000 replicates (Stamatakis 2006). A second phylogenomics-based tree was constructed using the type (strain) genome server—TYGS (https://tygs.dsmz.de/; Meier-Kolthoff and Göker 2019). TYGS offers pairwise similarity calculation and a standard phylogenetic approach including multiple sequence alignment and analysis under the maximum-likelihood and maximum parsimony criteria. TYGS also allows the Genome BLAST Distance Phylogeny (GBDP) approach to rapidly infer trees with branch support values, which also enables calculation of dDDH values (Meier-Kolthoff and Göker 2019). Furthermore, trees were built by multilocus sequence analysis (MLSA) on the basis of the selected housekeeping genes aroE, dnaE, groEL, gyrB, mutL, recA, and rpoB. Each gene was aligned independently using MUSCLE (Edgar 2004) and edited in accordance with Gblocks (Talavera et al. 2007). The genes were then concatenated, aligned and edited as previously reported (Hug et al. 2016). The tree—based on maximum-likelihood—was built with RAxML v.8.2.11 using the amino acid substitution model PROTGAMMALG, hill-climbing algorithm, with bootstrap value of 1,000 replicates (Stamatakis 2006). Both phylogenetic trees were visualized using the “interactive tree of life” software (iTOL) v3 (Letunic and Bork 2016).
The MicroScope web platform hosted at Genoscope (MaGe) (Vallenet et al. 2017) was then used for genomic comparisons, and so the locus tags used by us are based on MaGe. The annotated high-quality genomes of P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T are publicly available in MaGe (http://www.genoscope.cns.fr/agc/microscope/home/index.php). Additionally, BlastN analyses were done for genomic comparisons and five-gene cluster searches for selected strains, that is, P. hospita (mHSR1 and LMG 20598), P. caribensis (strains MBA4, TJ182, Bcrs1W and MWAP64), and P. terrae (strain NBRC 100964).
Moreover, the gene sequence information of these type strains was further analyzed using TrEMBL, SwissProt, as well as comparisons to the PubMed and InterPro databases. To search for secreted proteins, SignalP was used. Finally, MicroScope also identified the relevant RNA genes (rRNA and tRNA).
Metabolic pathway analyses were done using two approaches. First, through the MicroScope platform, that is, using the microbial pathway/genome databases (PGDBs). Metabolic profile analysis was based on the computation of a “pathway completion” value, that is, the ratio between the number of reactions for pathway X in a given organism and the total number of reactions of pathway X defined in the database. Second, metabolic pathways were also inferred with the KEGG automatic annotation server (KAAS) (Moriya et al. 2007). Additionally, the secondary metabolite detection program AntiSMASH was used (Vallenet et al. 2017). The web server OrthoVenn (Wang et al. 2015) was used to compare the clusters of orthologous genes (COGs) between the genomes of P. terrae DSM 17804T, P. hospita DSM 17164T, P. caribensis DSM 13236T, next to BS007.
Average nucleotide identity values (ANIs) and tetranucleotide frequency correlation coefficients (TETRA) were obtained using JSpeciesWS (Richter et al. 2016). The measures of ANIs were done by the algorithms BLAST+ (ANIb) and MUMmer-Maximum Unique Matches (ANIm). Additionally, TETRA correlation search (TCS) analyses (shown as Z-values) were also done to provide a hit-list for insight into the relationships of the genomes with those of the reference genome database (Richter et al. 2016). Genes encoding carbohydrate-active enzymes (CAZymes; potentially involved in carbohydrate metabolism) were analyzed using dbCAN (Yin et al. 2012).
Identification of Regions of Genomic Plasticity, Prophages, and CRISPR Spacers
Regions of genomic plasticity (RGPs) were predicted using MicroScope (at Genoscope; Vallenet et al. 2017). The platform employs “RGP finder” together with genomic island (GI) identifiers based on hidden Markov models (Waack et al. 2006) and AlienHunter-IVOM (Vernikos and Parkhill 2006). The GI identifier pipeline identified RGPs based on the criteria: 1) RGPs > 5 kb, 2) CDSs not belonging to conserved synteny groups between the compared organisms, and 3) regions with <50% of gene similarity with reference organisms were removed. RGPs in the three type strains were identified in comparison with the fungiphilic strains BS001, BS007, BS110, and BS437. Moreover, prophage sequences and CRISPR spacers were identified with PHAST (Zhou et al. 2011) and CRISPRFinder (Grissa et al. 2008), respectively. Strict criteria were used to identify complete prophages, as in Pratama et al. (2018).
Results
Rationale of This Study
A range of fungiphilic Paraburkholderia strains has been previously described with respect to their “eco-phenotype” (describing their capacities to interact with soil fungi in simulated soil settings). Many of these strains turned out to be loosely allocated in the species P. terrae, and the genome sequences of four such strains, that is, BS001, BS007, BS110, and BS437, have been described (Haq et al. 2014; Pratama et al. 2017). In order to enable a thorough analysis of the relationships of the fungiphilic strains with the three close relatives P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T, determination of the phenotypic and genomic properties of the latter three type strains was a prerequisite. At the onset of this study, no such deeply sequenced genomes were available. Hence, we first assembled the relevant data sets regarding the genotypes (genome sequences), as well as the eco-phenotypes, of these three type strains. In a second stage, we compared the data of the former four fungiphilic strains to those that typify the type strains and determined their grouping.
Summary of Phenotypic Traits of Type Strains P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T
Microscopic studies of P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T cells confirmed that all three type strains had Gram-negative, rod-shaped and motile cells, as described (Achouak et al. 1999; Goris et al. 2002; Yang et al. 2006). On R2A plates, all three strains grew between 15 and 37 °C, and optimally at 28 °C. Based on BIOLOG GN2 assays, the three type strains, next to the fungiphilic strains BS001, BS007, BS110, and BS437, consistently utilized the following 53 of the 95 GN2 carbon sources tested: Tween-40, Tween-80, N-acetyl-d-glucosamine, adonitol, l-arabinose, d-arabitol, d-fructose, l-fucose, d-galactose, α-d-glucose, m-inositol, lactulose, d-mannitol, d-mannose, l-rhamnose, d-sorbitol, cis-aconitic acid, citric acid, formic acid, d-galactonic acid, lactose, d-galacturonic acid, d-gluconic acid, d-glucosaminic acid, d-glucuronic acid, α-hydroxy butyric acid, β-hydroxy butyric acid, p-hydroxy phenyl acetic acid, α-keto butyric acid, dl-lactic acid, quinic acid, d-saccharic acid, sebacic acid, succinic acid, bromo succinic acid, l-alaninamide, d-alanine, l-alanine, l-alanyl-glycine, l-asparagine, l-aspartic acid, l-glutamic acid, glycyl-l-glutamic acid, l-histidine, hydroxy-l-proline, l-ornithine, l-phenylalanine, l-proline, l-pyroglutamic acid, l-threonine, dl-carnithine, γ-amino butyric acid, urocanic acid, 2-aminoethanol, dl-α-glycerol phosphate, and glucose-6-phosphate (supplementary table 1A, Supplementary Material online).
Two additional compounds, that is, N-acetyl-d-galactosamine and manolic acid, were utilized by P. terrae DSM 17804T, P. hospita DSM 17164T, and the four fungiphiles, but not by P. caribensis DSM 13236T. In contrast, the compounds inosine, maltose, d-trehalose and methyl pyruvate could only be utilized by P. caribensis DSM 13236T.
These data indicate the metabolic versatility of these strains, in that most strains utilized a majority of the carbon sources of the BIOLOG system. A hierarchical cluster analysis based on the carbon compound utilization patterns showed a division in two clusters, one with P. terrae DSM 17804T, P. hospita DSM 17164T, and the four fungiphilic strains, and a distant one containing P. caribensis DSM 13236T (supplementary fig. 1, Supplementary Material online).
Furthermore, P. terrae DSM 17804T and P. hospita DSM 17164T showed responses to compounds released by the fungus Lyophyllum sp. strain Karsten into propionate-supplemented mineral medium, which include glycerol, oxalate, citric acid, acetate and formate (>5-fold increased population sizes), whereas P. caribensis DSM 13236T did not show such growth responses (table 1). This finding was consistent with data from experiments that showed P. terrae DSM 17804T and P. hospita DSM 17164T to be able to actively interact with Lyophyllum sp. strain Karsten in soil, in terms of showing single-strain migratory capabilities along with the soil-exploring fungal hyphae (table 1); this was similar to the behavior of fungiphilic strains BS001, BS007, and BS110 (here used as controls, previously observed as in Nazir et al. 2012). In contrast, P. caribensis DSM 13236T was a poor migrator, with the connotation that its abundance at the inoculation site in soil increased in the presence of Lyophyllum sp. strain Karsten hyphae (see table 1).
Table 1.
Fungal-Interactive Traits (Partially Modified From Nazir et al. [2012])
| Strains | Survival at Inoculation Sitea | Migration to Distal Sitea | Response to Fungal Exudate (Propionate)b |
|---|---|---|---|
| Paraburkholderia terrae DSM 17804T | + | ++ | 13 |
| Paraburkholderia hospita DSM 17164T | + | +++ | 15 |
| Paraburkholderia caribensis DSM 13236T | + | – | 0 |
| Strain BS001 | + | +++ | 17 |
| Strain BS007 | + | ++ | 16 |
| Strain BS110 | + | +++ | 23 |
| Strain BS437 | + | + | 11 |
Population sizes are given. +, log CFU/g 6.0–6.5; ++, log CFU/g 6.5–7.5; +++, log CFU/g 7.5–8.5.
Approximate fold increase compared with P. caribensis.
On the basis of these collective data, we conclude that P. terrae DSM 17804T and P. hospita DSM 17164T had a very similar eco-phenotype, in particular with respect to their metabolic and fungal-responsive capacities, being akin to the four fungiphiles. This eco-phenotype was clearly divergent from that of P. caribensis DSM 13236T.
Overall Analysis of the Genomes of P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T
Deep sequencing and high-quality assembly of the three genomes revealed total genome sizes of 9.0–11.5 Mb. In detail, the size of the P. terrae DSM 17804T genome was 10,062,489 bp (G + C content 61.79%), that of P. hospita DSM 17164T 11,527,706 bp (G + C content 61.79%) and that of P. caribensis DSM 13236T 9,032,490 bp (G + C content 62.58%). The genome of P. terrae DSM 17804T was assembled into four, that of P. hospita DSM 17164T into six (including the introduced plasmid pEMT1) and that of P. caribensis DSM 13236T into four circular contigs, highlighting the number of separately replicating entities (supplementary table 2, Supplementary Material online). All assembled contigs were deposited as circular bacterial chromosomes, with the exception of the circular ∼100 kb contig in P. hospita strain DSM 17164T, which represents the full 2,4-D degradative plasmid pEMT1 that had been introduced earlier (Goris et al. 2002) (GenBank accession no. CP026110).
Totals of 8,752, 10,009, and 7,761 coding sequences (CDSs) were found on the genomes of P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T, respectively. Moreover, the genome of P. terrae DSM 17804T contained 18 ribosomal RNA (rRNA) and 60 tRNA encoding genes, that of P. hospita DSM 17164T 21 and 67 and that of P. caribensis DSM 13236T 18 and 61, respectively. A summary of these genome features is shown in supplementary table 1A, Supplementary Material online, the project information is in supplementary table 1B, Supplementary Material online and the genome statistics in supplementary table 1C, Supplementary Material online.
Phylogenetic Analyses of the Set of Strains
To verify and confirm the phylogenetic placement of the investigated strains on the basis of alignments of the 16S rRNA genes, a suite of relevant sequences across a selection of related Paraburkholderia species, including the close relative Paraburkholderia phymatum, was used. The analyses showed that P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T indeed clustered as a tight group within the genus Paraburkholderia (fig. 1). The former two species were most tightly knit (>98.67% reciprocal similarity), with the latter one being more distant (98.19% similarity with P. terrae DSM 17804T and 98.47% with P. hospita DSM 17164T, see supplementary table 2, Supplementary Material online). The analysis confirmed P. phymatum to be a close relative to the whole species cluster. Moreover, P. terrae DSM 17804T and P. hospita DSM 17164T showed close relatedness to other (similarly named) Paraburkholderia strains, that is, P. terrae NBRC 100964 (AB201285.1) and P. hospita LMG 20598 (NR025656.1), respectively. Hence, this preliminary phylogenetic analysis produced a first glimpse of a possibly “tight” two-species cluster.
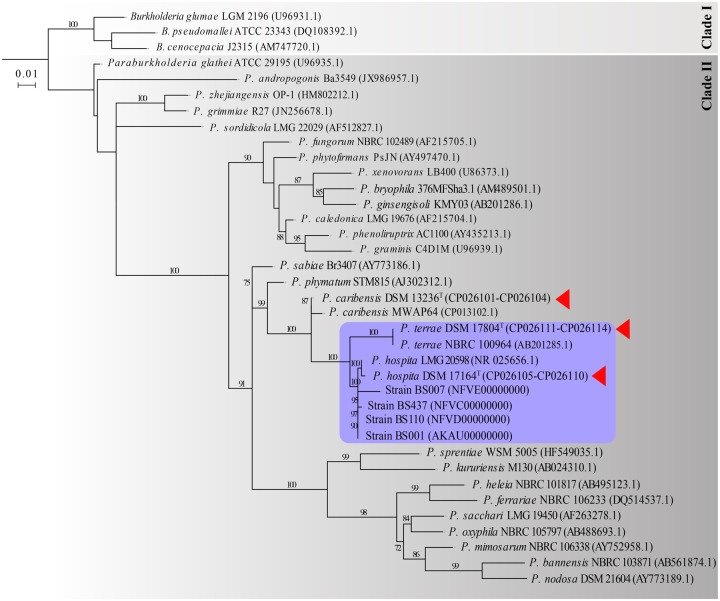
Fig. 1.
—Maximum-likelihood tree based on the V2–V6 (>1,000 bp) regions of the 16S rRNA gene. Accession numbers for the sequences used for each organism are provided in brackets after the organism’s name. The tree was constructed using RAxML (nucleotide substitution model GTRCAT), default algorithm settings (hill-climbing), with bootstrap value of 1,000 replicates. Bootstrap confidence values ≥ 70% are indicated. Purple box: proposed “species cluster.” Red triangles: type strains.
We then examined the precise placement of the four fungiphilic strains BS001 (NZ_AKAU00000000), BS007 (NFVE00000000), BS110 (NFVD00000000), and BS437 (NFVC00000000) versus this two-species cluster. Clearly, all four fungiphiles fell inside the cluster. Thus, using the strict criterion for species delineations, that is, up to 1–1.5% divergence of the 16S rRNA gene sequence, a tightly knit cluster of sequences, possibly indicating a “species cluster,” became discernible (fig. 1). This cluster showed a rather distant relatedness (96–98% similarity) to P. caribensis strains DSM13236T and MWAP64 (CP013102.1), P. phymatum AJ302312.1 and P. sabiae AY773186.1. Interestingly, this is in agreement with data from Estrada-de los Santos et al. (2018).
We then examined the whole-genome sequences across these strains to assess the validity of the initial analyses (fig. 2). This approach, as well as MLST analysis based on seven concatenated housekeeping genes (fig. 3A) yielded results that were fully consistent with the aforeshown clustering. Thus, the close relatedness of P. terrae DSM 17804T and P. hospita DSM 17164T, as well as the divergence of P. caribensis DSM 13236T from this two-species cluster, were confirmed. The two approaches (figs. 2 and 3A) also confirmed the tight relatedness between P. hospita DSM 17164T and the four fungiphilic strains.
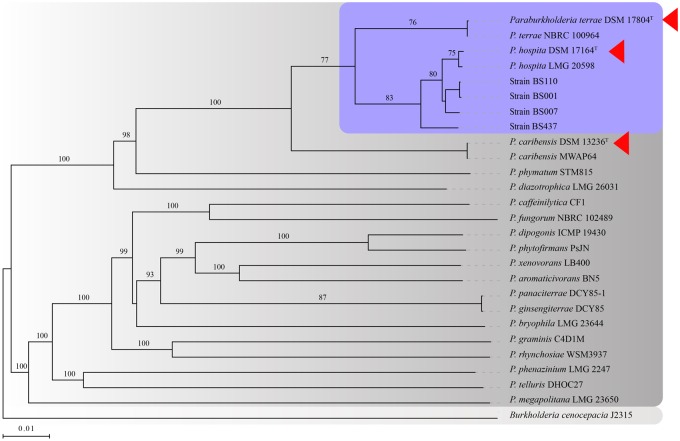
Fig. 2.
—Phylogenomics-based tree constructed using the type (strain) genome server—TYGS (https://tygs.dsmz.de/). Genome BLAST Distance Phylogeny (GBDP) distances were calculated from genome sequences. Branch lengths are scaled in terms of GBDP distance formula d5. Numbers above branches: GBDP pseudobootstrap support values from 100 replications. Bootstrap confidence values ≥ 70% are indicated. Purple box: proposed “species-cluster.” Red triangles: type strains.
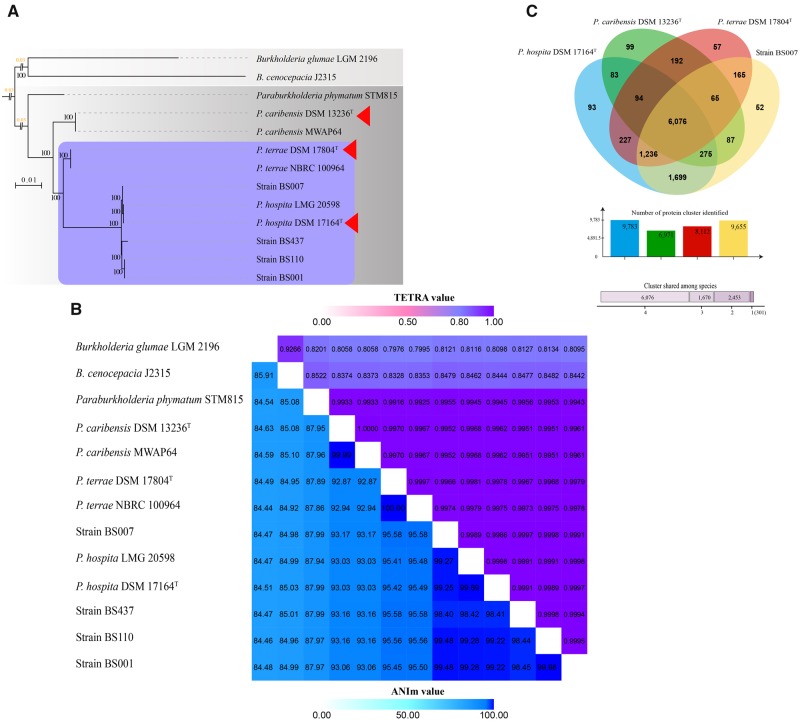
Fig. 3.
—(A) Maximum-likelihood phylogenetic tree based multilocus sequence analysis (MLSA) using seven concatenated core genes (aroE, dnaE, groeL, gyrB, mutL, recA, and rpoB). The tree was built with RAxML using amino acid substitution model PROTGAMMA, default matrix setting (Dayhoff) and algorithm (hill-climbing), with bootstrap value of 1,000 replicates. Bootstrap confidence values ≥ 70% are indicated. Purple box represents the proposed “species-cluster,” and red triangles indicate the type strains. (B) Heat maps of average nucleotide identity (ANIm) and tetranucleotide frequency (TETRA) analyses. The ANI (threshold 95–96%) and TETRA (>0.99) values were used for species circumscriptions (Richter and Rossello-Mora 2009). The ≥70% ANI coverage values were indicated. (C) Venn diagram of the orthologous clusters of proteins of P. terrae DSM 17804T, strain BS007, P. hospita DSM 17164T, and P. caribensis DSM 13236T. The number of protein clusters identified in each strain and shared protein clusters are indicated.
ANI and TETRA Analyses
To explore the findings of similarity from the trees built on the basis of the 16S rRNA gene, the whole-genome sequences and the seven-gene concatenates, we assessed the evolutionary relationships between the three novel genomes under study (fig. 3B). As outgroups, we used genome sequences of P. phymatum, Burkholderia cenocepacia, and Burkholderia glumae (see fig. 3B and supplementary table 2, Supplementary Material online). The data indicated that P. hospita DSM 17164T, P. terrae DSM 17804T, and P. caribensis DSM 13236T are only remotely related to either P. phymatum (ANIm: 87.89–87.99%, alignment: 60.36–61.91%) or the two Burkholderia strains (ANIm: 84–85%, alignment: 16–21%). Furthermore, the genomes of P. hospita DSM 17164T and P. terrae DSM 17804T were found to be highly similar across each other, with an ANIm of 95.42% (71.94% alignment). In contrast, the ANIm values between, on the one hand, P. hospita DSM 17164T and P. terrae DSM 17804T and, on the other hand, P. caribensis DSM 13236T were only 93.03% (60.97% alignment) and 92.97% (69.61% alignment), respectively. These latter values are below those used for species delineations (threshold 95–96%) (Chun et al. 2018), and so the collective data confirm that P. hospita DSM 17164T and P. terrae DSM 17804T are 1) tightly related, at, or even within, species borders, and 2) divergent from P. caribensis DSM 13236T.
In addition, all four fungiphilic strains, that is, BS001, BS007, BS110, and BS437, fell inside this two-species cluster, being closer to P. hospita DSM 17164T than to P. terrae DSM 17804T (ANIm > 99%, 82–85% alignment, TETRA > 0.999). In detail, the genome of strain BS001 showed Z-score values of 0.99974 and 0.99789 against the genomes of P. hospita DSM 17164T and P. terrae DSM 17804T, versus 0.99613 against that of P. caribensis DSM 13236T (supplementary tables 3 and 4, Supplementary Material online).
Clustering of Genes for Orthologous Proteins
Analyses of the COGs of the three type strains, next to the closely related strain BS007 (here selected for comparison) showed that the P. terrae and P. hospita type strains had high relatedness, with 227 COGs shared between them. Remarkably, 1,699 COGs were shared between P. hospita DSM 17164T and strain BS007 (fig. 3C). In contrast, P. hospita DSM 17164T and P. caribensis DSM 13236T had only 83 COGs in common.
Furthermore, COG functional category analyses revealed the number of secondary metabolite biosynthesis functions (category Q) to be lower in P. caribensis DSM 13236T (260 genes, 2.82%) than in both P. hospita DSM 17164T (376, 3.12%) and P. terrae DSM 17804T (344, 3.34%) (supplementary fig. 2, Supplementary Material online and supplementary table 5, Supplementary Material online).
Insights into Potential Functions in P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T as Compared with Selected Fungiphilic Strains
Metabolic and Ecological Competence Traits
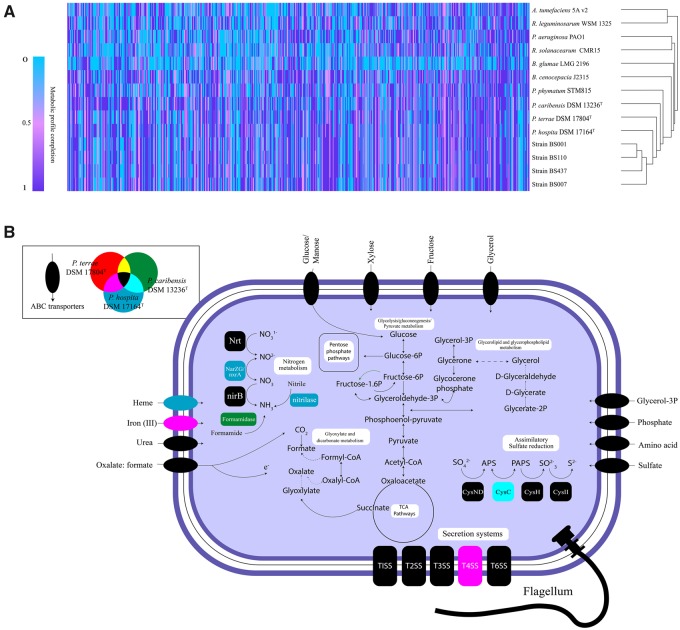
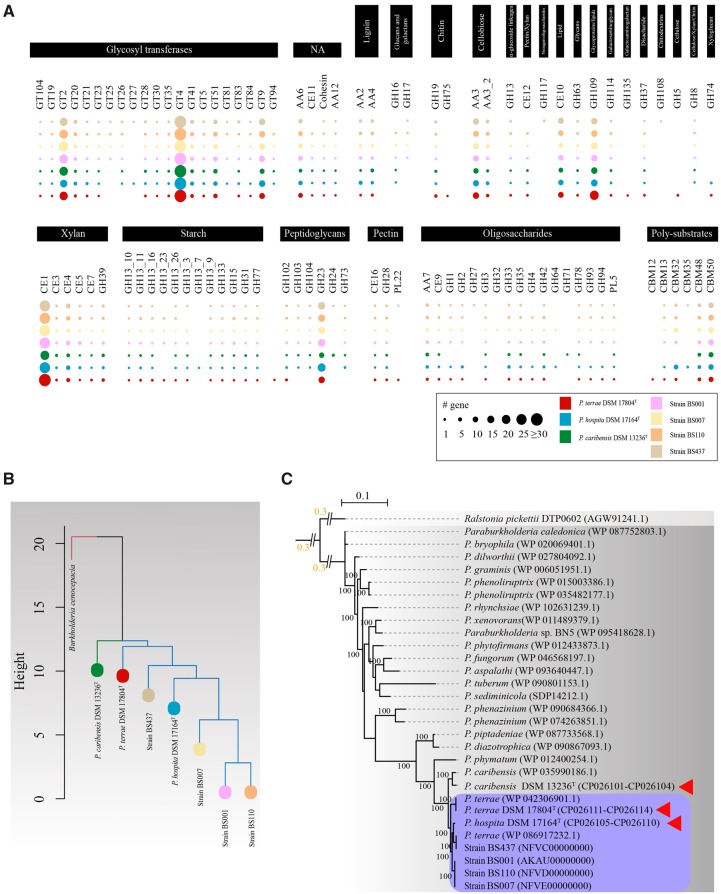
Expectedly, the genomes of the three type strains contained genes or operons for numerous diverse (primary and secondary) metabolic capacities (supplementary tables 6 and 7, Supplementary Material online), however without a clear distinction as to metabolic ranges. Across the genomes, sets of varying carbohydrate metabolism genes were found, indicating the presence of capacities to utilize simple (e.g., glucose, fructose) to complex (e.g., cellulose and hemicellulose) carbohydrates. Profile analyses showed that the genomes of the three type strains, in addition to those of all fungiphilic strains, possessed similar numbers and types of metabolic pathways, including the TCA cycle, glycolysis, the Entner–Doudoroff pathway, and gluconeogenesis (fig. 4A and B). Furthermore, they were predicted to be able to synthesize the essential amino acids histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan and valine, the “conditional” amino acids arginine, cysteine, glutamine, glycine, proline and tyrosine, and the nonessential amino acids alanine, asparagine, glutamic acid, serine and selenocysteine. Evidence for the presence of several fermentation pathways was also found across the three genomes (fig. 4B and supplementary table 6, Supplementary Material online). Biofilm synthetic systems, that is, 1) pellicle formation (Pel—glucose-rich biofilm matrix exopolysaccharide), 2) poly-beta-1,6-N-acetyl-d-glucosamine (PGA—biofilm adhesin polysaccharide), and 3) alginate-related biofilm formation genes were found consistently (see fig. 5A). Likewise, sets of flagellar genes (fig. 5B) were found across the three type strains as well as the four fungiphilic strains. Diverse siderophore biosynthesis systems were also found (supplementary table 6, Supplementary Material online).
Fig. 4.
—(A) Hierarchical cluster analysis of metabolic profile completeness based on the Microscope platform. Computation of “pathway completeness” value: see Material and Methods. (B) Metabolic reconstruction of Paraburkholderia terrae DSM 17804T, Paraburkholderia hospita DSM 17164T and Paraburkholderia caribensis DSM 13236T. The text in the white bubbles depicts names of pathways and metabolic processes. Pathways and corresponding enzymes were colored based on in which genome they were found. Moreover, incomplete pathways are indicated with dotted lines.
Fig. 5.
—(A) Gene number profile of selected genes in Paraburkholderia terrae DSM 17804T, Paraburkholderia hospita DSM 17164T, Paraburkholderia caribensis DSM 13236T and four fungiphilic strains, i.e. BS001, BS007, BS110 and BS437. Color code based on the strain is indicated (P. terrae DSM 17804T: red; P. hospita DSM 17164T: blue; P. caribensis DSM 13236T: green; fungiphilic strains BS001: pink; BS007: yellow; BS110: orange; and BS437: brown). Number of genes is indicated by the dot size. (B) Synteny reconstruction of flagellar genes of P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T. Color code based on the strain is indicated.
The genomes of P. hospita DSM 17164T and P. terrae DSM 17804T, but not that of P. caribensis DSM 13236T, further revealed the presence of genes encoding the capacity to degrade 2-nitrobenzoate, anthranilate, alanine, 4-aminobutyrate and to synthesize trehalose. For more details, see supplementary table 6, Supplementary Material online. The capacity to degrade anthranilate has been linked to the early stage of biofilm formation in Pseudomonas aeruginosa (Costaglioli et al. 2012). This trait can also affect the structure of mushrooms at a later stage of biofilm formation (Kim et al. 2015).
Distinctive metabolic traits that were found to be uniquely encoded in the P. caribensis DSM 13236T genome were: putrescine biosynthesis, oxidation of methanol to formaldehyde, fructose degradation, intra-aerobic nitrite reduction, methane sulfonate degradation, dissimilatory nitrate reduction, oxidation of GTP and dGTP, and hydrogen production.
There was a conspicuous absence of any gene encoding 6-phosphofructo-1-kinase (PFK-1; glycolysis pathway) from the genomes of P. hospita DSM 17164T and P. terrae DSM 17804T as well as from those of the fungiphilic strains BS001, BS007, BS110, and BS437. PFK-1 phosphorylates fructose 6-phosphate to fructose 1,6-bisphosphate. Conversely, this gene was found to be present in P. caribensis DSM 13236T (fig. 4B). In all former strains, we assume this function to be taken over by the predicted kinase encoded by the pfkB gene.
Genes encoding CAZymes
The genomes of P. terrae DSM 17804T and P. hospita DSM 17164T contained, respectively, 302 and 308 genes encoding CAZymes, whereas those of fungiphilic strains BS001, BS007, BS110, and BS437 had 300, 310, 298, and 298 such genes (fig. 6A). In contrast, P. caribensis DSM 13236T revealed a substantially lower number (253) of such genes. However, there was no major difference regarding the CAZyme or carbohydrate-binding module (CBM) profiles among all strains, including the type strains. Thus, all genomes revealed the presence of 31–39 genes for “auxiliary activity” (AA) family proteins, 14–26 for CBMs, 45–59 for carbohydrate esterases (CEs), 69–89 for glycosyl hydrolases (GHs), 92–109 for glycosyl transferases (GTs), and 1–2 for polysaccharide lyases (PLs). Overall, the most abundant genes predicted to encode CAZy family proteins were associated with: GT2, GT4, GT9 (family GTs), CE1 (carbohydrate esterases; involved in hydrolysis of xylan; acetyl xylan esterase [EC 3.1.1.72]), GH109 (glycosyl hydrolases, in particular glycoproteins; α-N-acetyl galactosaminidases involved in degradation of glycoproteins, EC 3.2.1.49), and AA3 (cellobiose dehydrogenase, EC 1.1.99.18) (fig. 6A).
Fig. 6.
—(A) Gene number profile of predicted CAZYmes family proteins in Paraburkholderia terrae DSM 17804T, Paraburkholderia hospita DSM 17164T, and Paraburkholderia caribensis DSM 13236T, next to fungiphilic strains BS001, BS007, BS110, and BS437. Color code based on the strain is indicated. Number of genes is indicated by the dot size. (B) Hierarchical cluster analysis based on CAZyme gene number for P. terrae DSM 17804T, P. hospita DSM 17164T, P. caribensis DSM 13236T, strains BS001, BS007, BS110, and BS437. B. cenocapacia J2315 was used as an outgroup. (C) Phylogenetic analysis of GH3 type proteins across all strains, and the top hits of PSI-BLASTP. The tree was built with RAxML using amino acid substitution model PROTGAMMA, default matrix setting (Dayhoff) and hill-climbing algorithm, with bootstrap value of 1,000 replicates. Bootstrap confidence values ≥ 70% are indicated. Purple box represents the proposed “species-cluster,” and red triangles indicate the type strains.
Another suite of genes encoding CAZymes or CBMs was only found in P. terrae DSM 17804T and P. hospita DSM 17164T and not in P. caribensis DSM 13236T. The predicted proteins belonged to glycosyl hydrolase classes GH1, GH2, GH27, GH4, GH93, GH94, and GH74 and to CBM (carbohydrate binding moiety) classes CBM12, CBM13, and CBM32. In detail, moderate numbers (19–26) of genes encoding CBM family proteins were observed in both P. hospita DSM 17164T and strain BS007, in particular those for CBM32 proteins (8). Interestingly, genes encoding CBM32 proteins were completely absent from the P. caribensis DSM 13236T genome (fig. 6A).
A hierarchical cluster analysis based on the number of each of such genes per genome showed a close grouping of P. terrae DSM 17804Tand P. hospita DSM 17164T together with strains BS001, BS007, BS110, and BS437. In contrast, P. caribensis DSM 13236T showed distant relatedness to this cluster (fig. 6B).
To address potential functional divergences, we then examined one example of a key GH3 family enzyme, that is, a glycoside hydrolase that removes single glycosyl residues and hydrolyzes bonds in cellulose, hemicellulose and starch (amylase) (Kusaoke et al. 2017). Genes for GH3 family proteins were found across all genomes examined here. The sequences of the identified genes were closely related, both between each other and with those of a suite of comparator Paraburkholderia strains. Based on the tree resulting from this analysis (see fig. 6C), the predicted GH3 protein in P. caribensis DSM 13236T was strongly divergent from those of both P. terrae DSM 17804T and P. hospita DSM 17164T.
Genes Encoding Membrane Transporters
The genomes of P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T contained genes predicted to encode a plethora of different membrane transporters. In particular, energy-dependent membrane transporter proteins, that is, ABC transporters, phosphotransferase systems (PTS), secondary membrane transporters (i.e., major facilitator superfamily; MFS), and solute carrier family (SLC) proteins were found (supplementary table 7, Supplementary Material online). We also found similar genes for aquaporins and small neutral solute transporters (supplementary table 8, Supplementary Material online). Finally, we found similar glycerol transporter and oxalate: formate antiporter (OFA) family transporters across all three type strains (see fig. 4B and supplementary table 8, Supplementary Material online).
Remarkably, a particular suite of genes encoding membrane transporters was only found in the P. terrae DSM 17804T and P. hospita DSM 17164T genomes, but not in that of P. caribensis DSM 13236T. This gene set included an iron (III) transporter system (afuA/fbpA, afuB/fbpB, afuC/fbpC) and the transmembrane electron carrier torZ (trimethylamine-N-oxide reductase; cytochrome c). Moreover, some genes for a few specific membrane transporters were unique per genome, that is, a predicted erythritol transporter in P. terrae DSM 17804T, arginine/ornithine and heme transporters in P. hospita DSM 17164T and an organophosphate: Pi antiporter (OPA) family transporter in P. caribensis DSM 13236T (supplementary table 8, Supplementary Material online).
Genes Encoding Motility Complexes
As expected, rather similar flagella-, T4P-, and chemotaxis-encoding gene complexes were found in the genomes of the three type strains, as well as in those of the four fungiphiles (fig. 5 and supplementary table 9, Supplementary Material online). The flagellar biosynthetic cluster was found to stretch over ∼45.3 kb, with high similarities across the whole region in P. terrae DSM 17804T, P. hospita DSM 17164T, all fungiphilic comparator strains, and P. caribensis DSM 13236T (>90% similarity) (supplementary fig. 3, Supplementary Material online). Moreover, sets of chemotaxis genes, that is, cheA, cheW, cheD, cheR, cheB, cheBR, cheY, cheZ, and cheV, were found across these genomes. Genes encoding the T4P assembly proteins pilABCDQW were also found across the type strain genomes. However, a serine sensor receptor, that is, methyl-accepting chemotaxis protein I (tsr), could only be found in P. hospita DSM 17164T (see fig. 5A and supplementary table 9, Supplementary Material online).
Traits Predicted to Confer Associative Behavior with Soil Fungi
In this section, we analyze the genetic systems that are potentially related to mycosphere competence across the three type strains as compared with the fungal-interactive strain BS001. The mycosphere competence traits of strains BS007, BS110, and BS437 have been discussed earlier (Pratama et al. 2017).
Genes Encoding Secretion Systems
Genes/operons for type-1, -2, -3, and -6 secretion systems (T1SS, T2SS, T3SS, T6SS) were found across the genomes of all three type strains (see supplementary fig. 3, Supplementary Material online). In contrast, T4SS complexes, consisting of VirB1 through VirB11, in addition to VirD4 (gene encoding the coupling protein key to conjugational DNA transfer) were only present in P. terrae DSM17804T and P. hospita DSM17164T, but absent from the P. caribensis DSM 13236T genome.
In detail, a complete T1SS system was found in P. terrae DSM 17804T (i.e., OMP: tolC; MFP: raxA; and ABC: raxB, cvaB). In contrast, P. hospita DSM 17164T and P. caribensis DSM 13236T revealed genes for the ABC (raxB, cvaB), next to hemolysin D, proteins (fig. 5A and supplementary fig. 3, Supplementary Material online). With respect to the T2SSs, all three type strains contained genes for the nine canonical gsp genes (gspD, gspE, gspF, gspG, gspH, gspJ, gspK, gspL, and gspN). These T2SSs also contained tight adherence (Tad) export apparatuses, that is, cpaA/tadV, cpaB/rcpC, cpaC/rcpA, cpaE/tadZ, cpaF/tadA, tadB, and tadC (fig. 5A). With respect to the T3SSs, clusters containing the 19 canonical T3SS genes were identified across all three type strains. Ten of the 19 genes (sctC, sctD, sctJ, sctL, sctN, sctQ, sctR, sctT, sctU, and sctV) were highly syntenous, at ∼17–60% similarity (supplementary fig. 3, Supplementary Material online).
Finally, copies of the T6SS (consisting of the core component of the imp/vas secretion system) were found across the three type strains (supplementary fig. 3, Supplementary Material online). With respect to the imp system, one copy was found in P. terrae DSM 17804T, three in P. hospita DSM 17164T and two in P. caribensis DSM 13236T. High synteny was found among the copies (denoted as cluster) in P. terrae DSM 17804T, P. hospita DSM 17164T (cluster 3) and fungiphilic strain BS001 (cluster 1), indicating high relatedness. In addition, the T6SS cluster 1 of P. hospita DSM 17164T was highly syntenous to that of P. caribensis DSM 13236T, as well as strain BS001 cluster 3 (Haq et al. 2014). Also, high synteny was observed between the T6SS clusters 2 in P. hospita DSM 17164T and in strain BS001 (supplementary fig. 3, Supplementary Material online).
Genes Encoding Glycerol and Oxalate Metabolism, and Five-Gene Cluster
The analyses of the genomes of the three type strains did not detect the glycerol uptake gene gup that was previously discovered in the strain BS001 genome (Haq et al. 2014). However, glycerol uptake/transporter genes glpV, glpP, glpO, and glpS and putative sn-glycerol 3-phosphate transporter genes (ugpB, ugpA, ugpE, and ugpC) were found across the genomes of all three type strains (see supplementary fig. 4, Supplementary Material online).
We further found sets of genes predicted to encode proteins involved in oxalate (and formate) oxidation in all three type strains (supplementary fig. 4B, Supplementary Material online). A high similarity of one gene, that is, oxalyl-CoA decarboxylase, was found in the genomes of P. terrae DSM 17804T (99%) and P. hospita DSM 17164T (100%), next to that of strain BS001, suggesting these organisms have similar responsive behavior to oxalate. The relatedness of this gene sequence to the copy in P. caribensis DSM 13236T was lower (98%).
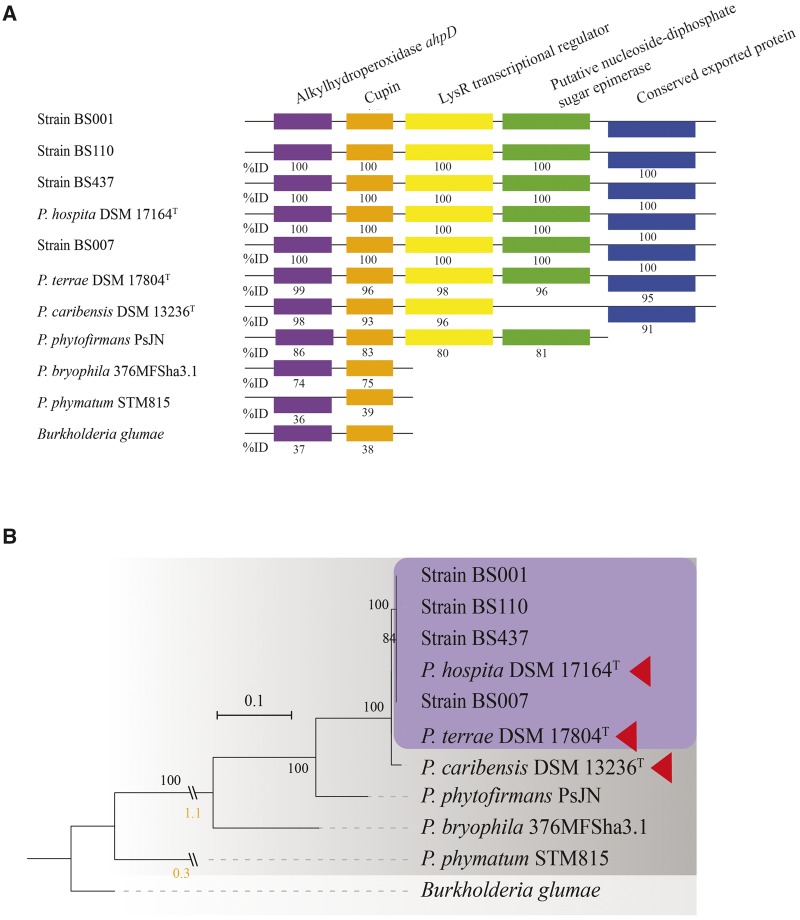
Remarkably, full copies of the five-gene cluster first found in strain BS001 and hypothesized by Haq et al. (2017) to be involved in the generation of energy from small carbonaceous molecules released by soil fungi (e.g., oxalate), as well as removal of concomitant oxidative toxicity, were found in both P. terrae DSM 17804T and P. hospita DSM 17164T. Moreover, the genomes of strains BS007, BS110, and BS437 also contained the full gene cluster (fig. 7A and B). Although the cluster was also found in the P. caribensis DSM 13236T genome, one gene (IV, encoding a putative nucleoside-diphosphate sugar epimerase) was lacking (fig. 7A). The relatedness of the five-gene cluster was close between P. terrae DSM 17804T and P. hospita DSM 17164T and more distant when compared with P. caribensis DSM 13236T, as illustrated in the phylogenetic analysis of the gene for alkyl hydroperoxide ahpD across the Paraburkholderia species (fig. 7B).
Fig. 7.
—(A) Comparison of the five-gene cluster among the strains. Comparison percentage using Paraburkholderia terrae strain BS001 as a reference and based on the Microscope genoscope platform. (B) Phylogenetic analysis of alkyl hydroperoxidase AhpD across Paraburkholderia species, including type strains and mycosphere-derived strains. Burkholderia glumae was used as an outgroup. The tree was built with RAxML using the amino acid substitution model PROTGAMMA, default matrix setting (Dayhoff) and hill-climbing algorithm. Bootstrap value 1,000 replicates. Bootstrap confidence values ≥ 70% are indicated. Purple box represents the proposed “species-cluster,” and red triangles indicate the type strains.
RGPs, Prophage-Related Sequences and CRISPR-Cas Arrays
The genomes of all three type strains were found to contain multiple sets of RGPs. In detail, P. terrae DSM 17804T and P. hospita DSM 17164T had 97 and 99 RGPs, whereas P. caribensis DSM 13236T had only 76 (supplementary table 9, Supplementary Material online). The total sizes of the RGPs were 3,009,744 (29.9% of the genome), 4,401,854 (38.2%) and 2,133,117 bp (23.6%), respectively.
The largest RGP (RGP72; 308 CDS) in P. terrae DSM 17804T was 283,846 bp in size, that in P. hospita DSM 17164T 1,365,074 bp (spanning one of the six contigs, and so resembling a megaplasmid—RGP98; 1,480 CDS) and that in P. caribensis DSM 13236T 267,272 bp (RGP75; 302 CDS). RGPs containing complete T4SSs, thus indicating integrated plasmids (like RGP97 in BS001; Haq et al. 2014) were found in P. hospita DSM 17164T (RGP99: 419,946 bp) as well as P. terrae DSM 17804T (RGP97: 157,478 bp), but not in P. caribensis DSM 13236T.
Furthermore, genes encoding both predicted transposases and integrases were amply found in the P. terrae DSM 17804T (42 and 11, respectively), P. hospita DSM 17164T (154/44), and P. caribensis DSM 13236T genomes (36/8). In all three type strains, these were located inside RGPs (supplementary table 9, Supplementary Material online).
Analyses of putative prophage (PP) regions across the type strains using PHAST (Zhou et al. 2011) identified one (25 kb) in P. terrae DSM 17804T, two in P. hospita DSM 17164T (total size 40.7 kb), and two (total size 89.9 kb) in P. caribensis DSM 13236T (supplementary table 10, Supplementary Material online). The genes in the two PP regions in P. hospita DSM 17164T were assigned as mobile genetic elements (MGEs) related genes (no specific hits in the database), with mainly genes encoding hypothetical proteins and integrases, without phage structural genes (e.g., capsid, tail, terminase) being identified. Furthermore, the single 25 kb PP region in P. terrae DSM 17804T probably represents an intact prophage (denoted as ϕPt17804), similarly to the two sequences in P. caribensis DSM 13236T (yielding ϕPcari1DS and ϕPcari2DS).
Finally, we analyzed the three type strain genomes for the presence of CRISPR-Cas spacer sequences. Using CRISPR-Finder, we found 13, 14 and 9 CRISPR spacer sequences in P. terrae DSM 17804T, P. hospita DSM 17164T, and P. caribensis DSM 13236T, respectively. These spacer sequences matched sequences in a large variety of phage families, mostly identified as Myoviridae. Detailed analyses of the evolutionary history of prophages in the genomes of Paraburkholderia species are discussed in Pratama et al. (2018).
Genome Comparison with Other Strains in the Species P. hospita, P. caribensis, and P. terrae
In the course of this work, several new genome sequences of P. hospita, P. terrae, and P. caribensis became available, next to sequences of some newly identified Paraburkholderia spp. Hence, we extended our comparative whole-genome analyses to these strains, in a separate analysis. The additional analyses included P. caribensis strains MBA4, TJ182, Bcrs1W, and MWAP64, P. terrae NBRC 100964 and P. hospita strains mHSR1 and LMG 20598 (supplementary fig. 5, Supplementary Material online). The data provide strong evidence for the contention that P. hospita and P. terrae are indeed tightly linked within one species cluster, which includes the four fungiphilic strains BS001, BS110, BS007, and BS437. Moreover, all P. caribensis strains clustered clearly as a sister group, separate from the former species cluster. All other strains used in the comparison clustered remote from these two sister groups. Moreover, genome size comparisons revealed the two-species cluster strains (especially P. hospita) to have large genomes (up to over 11 Mb), exceeding those of the P. caribensis strains (average ∼8 Mb), next to the other comparator genomes (i.e., of P. phymatum, P. azotifigens, P. piptademiae, and P. diazotrophica; average ∼9 Mb).
A search, across the additional genomes, for traits that might be involved in associative behavior with soil fungi revealed the presence of many such traits, that is, secretion systems, flagella, chemotaxis, glycerol-oxalate related and biofilm formation genes across these. The exception was the aforementioned five-gene cluster, as outlined below. Thus, all genomes did contain genes encoding secretion systems of the T2SS, T3SS, T4SS, and T6SS classes, with just P. hospita LMG 20598 and P. caribensis MWAP64 having a T1SS. Furthermore, across the genomes, we also found biofilm synthesis systems of the Pel and PGA classes. Complete sets of flagellar and chemotaxis genes (i.e., cheD, cheR, cheB, cheA, cheW, cheV, cheZ, and cheY) and genes for glycerol transporters and OFAs were also found across these strains. Specifically, genes glpV, glpP, glpQ, glpS and glpT, and ugpB, ugpA, ugpE and ugpC were found. Very interestingly, the complete five-gene cluster was found only in the additional P. terrae and P. hospita species, with incomplete versions of this cluster being detected in P. caribensis strains Bcrs1W and TJ182. This gene cluster was completely absent from P. caribensis strains MBA4 and MWAP64 (supplementary fig. 5, Supplementary Material online).
Discussion
Phenotypic Traits—Interactivity with Soil Fungi
In this study, we examined the genomic and metabolic/fungal-interactivity traits across the type strains of P. terrae, P. hospita, and P. caribensis, next to selected other (fungiphilic) strains of this genus, in order to delineate species boundaries and analyze fungal interactivity as a potentially common complex trait. All strains had a soil origin, and, to date, it has remained unexplored to what extent they would cluster together, or are divergent, with respect to their genomic and ecophysiological features. Moreover, we here report and analyze the deeply sequenced, assembled and annotated high-quality genomes of the three key type strains.
First, the clear evidence found for the tenet that P. hospita DSM 17164T and P. terrae DSM 17804T (but not P. caribensis DSM 13236T) can migrate through soil along with the hyphae of Lyophyllum sp. strain Karsten, placed these two strains in the category of “single-strain migratory” fungal-interactive strains, much like the comparator strains BS001, BS007, BS110, and (although weaker) BS437, that had previously been loosely assigned to the species P. terrae (table 1). Additionally, the metabolic complement of the three type strains, although fairly comparable, allowed a grouping in two clusters, one including P. caribensis and the other one all other organisms. It should be noted that P. hospita DSM 17164T and P. terrae DSM 17804T were able to utilize the compounds glycerol, oxalate, citric acid, formic acid and acetic acid, which are commonly found in exudates produced, for instance, by the soil saprotroph Lyophyllum sp. strain Karsten into mineral media (propionate as the carbon source). All or some of these compounds presumably constitute chemoattractants for the fungiphiles studied (Nazir et al. 2012; Zhang et al. 2014; Haq et al. 2018). In contrast, P. caribensis DSM 13236T was much less able to migrate along fungal hyphae, or utilize fungal-released compounds in spent propionate medium. We thus assume that the aforementioned compounds, in their form in the propionate medium, are less palatable for this organism, but, alternatively, compounds hampering the growth of this organism might have been present. Overall, the results suggest that P. hospita DSM 17164T and P. terrae DSM 17804T have behavior—upon confrontation with soil fungi—that allows them to be interactive with these, and is thus similar to that of the aforementioned fungiphilic strains. This was clearly different for P. caribensis DSM 13236T.
Genomic Analyses Identify a Two-Species Cluster in the Genus Paraburkholderia
We surmised that, within the diverse genus Paraburkholderia, evolution may have given rise to clusters of species that are both phylogenetically and ecophysiologically similar. Along time, our understanding of bacterial species evolution and relatedness has been based on 1) DNA–DNA hybridization (DDH), 2) phenotype, 3) the sequence of the 16S rRNA gene, 4) MLSA analysis using concatenated housekeeping genes (Estrada-de los Santos et al. 2013), 5) whole-genome sequence analysis (Meier-Kolthoff and Göker 2019), and 6) ANI/TETRA analyses. With respect to the latter, it has been suggested that it may eventually substitute traditional DDH (Richter and Rossello-Mora 2009; Chun et al. 2018; Ciufo et al. 2018). ANI uses pairwise comparisons of shared orthologous protein-encoding genes. It does not require slicing of the genomes into pieces, thus enabling rapid alignment of large sequences (Richter and Rossello-Mora 2009). By substituting the laborious and error-prone DDH, ANI may even accelerate bacterial taxonomy (Goris et al. 2002). Recently, Ciufo et al. (2018) analyzed the levels of agreement (giving “concordant” data) versus disagreement (discordant) among publicly (NCBI) available type strain genomes using a 96% ANI to define species boundaries. Thus, strains identified as B. cepacia had concordant ANI at 97%, versus discordant ANI (with other species) at 87.63%. We here used comparable ANIm threshold values for our species circumscriptions.
Thus, the ANIm (and TETRA) values found by us confirmed P. hospita DSM 17164T, P. terrae DSM 17804T, and P. caribensis DSM 13236T to be indeed closely related, with a much tighter linkage between P. hospita DSM 17164T and P. terrae DSM 17804T on the one hand than between each of these two and P. caribensis DSM 13236T on the other hand. Paraburkholderia terrae DSM 17804T and P. hospita DSM 17164T (ANIm/TETRA of 95.42/0.99784) had values at the border of those that delineate species. Here, we posit that the two species constitute one larger species “cluster.” Interestingly, all of the additional four strains derived from soil fungi, that is, BS001, BS110, BS007, and BS437, fell inside this species cluster, and so we surmised fungal interactivity may be one key ecoevolutionary driver of the respective genomes (fig. 3B). We propose to coin this species cluster the P. hospita species cluster.
Additional arguments for the existence of a species cluster that encompasses the P. terrae and P. hospita type strains next to strains BS007, BS110, BS437, and BS001, could be found in the great similarities across all of these strains, as evidenced by the phylogenetic analyses and the shared (concatenated) core genes, with a consistent divergence of the cluster from the close relative P. caribensis DSM 13236T as well as P. phymatum (figs. 1 and 2A). Moreover, the species cluster is supported by previous studies that, by other means, also found taxonomic closeness of the two type strains and divergence from both P. caribensis DSM 13236T (Nazir et al. 2012; Estrada-de los Santos et al. 2013; Sawana et al. 2014; Beukes et al. 2017) and P. phymatum (see figs. 1–3A).
There were some other, subtler, differences across the examined strains, for example, the high overlap of COGs between P. terrae DSM 17804T and P. hospita DSM 17164T (227), in contrast to, for instance, between P. terrae DSM 17804T and P. caribensis DSM 13236T (83) (fig. 3C).
Arguments for the Renaming of Fungiphilic Strains BS001, BS110, BS007, and BS437 as Members of Paraburkholderia hospita
In the foregoing, we provided arguments for the creation of a species cluster named the P. hospita species cluster, encompassing P. hospita DSM 17164T, P. terrae DSM 17804T, and the four fungiphilic strains examined here, that is, BS001, BS110, BS007, and BS437. Additional evidence suggests this cluster also encompasses the other two P. hospita and P. terrae strains analyzed. There may be arguments (based on analyses of genome relatedness) that are in favor of mending the two species into just one species. However, we stopped short of redefining the whole cluster as one new species, as more work—based on larger strain/genome numbers—would be required to provide a more solid basis for such a contention. On the basis of the current analyses, strains BS001, BS007, BS110, and BS437 all appeared to be closest related to P. hospita DSM 17164T and hence there are strong arguments for the tenet they belong to this species. Here, we propose to rename the fungiphilic strains BS001, BS110, BS007, and BS437 as members of the species P. hospita.
Ecologically Relevant Traits Were Differentially Found among All Strains
Lifestyle in Soil
As revealed by the in-depth genomic analyses, P. hospita DSM 17164T, P. terrae DSM 17804T, and P. caribensis DSM 13236T, plus the fungal-derived strains, were all predicted to be ecologically very versatile (fig. 4 and supplementary table 6, Supplementary Material online). The finding in the type strains of genes for all major pathways, in particular the TCA, glycolysis, Entner–Doudoroff, and gluconeogenesis pathways, indicated their metabolic versatility in soil settings. Also, there was consistency in the finding of incomplete/partial pathways across the three type strains. With respect to this finding, it is possible that complementary genes that are potentially key for these pathways were not recognized on the basis of the current data and database entries. Moreover, the finding of diverse siderophore biosynthesis systems was interesting (supplementary table 6, Supplementary Material online), as siderophores—as iron capturing systems—are important in most soils, particularly in situations in which iron is limiting. They have been observed in other soil-derived Paraburkholderia strains, for example, Paraburkholderia xenovorans (Vargas-Straube et al. 2016), strain BS001 and even in the horizontal gene pool in mycospheres (Zhang et al. 2016).
The analyses further showed the genomes of P. terrae DSM 17804T and P. hospita DSM 17164T to contain sets of genes potentially involved in fungal-interactive next to saprotrophic behaviors. This indicated that these organisms, notwithstanding their fungal interactivity potential, constitute ecological “generalists” rather than specialists (Haq et al. 2014). Fungal-interactive capacities presumably involve several metabolic response genes (e.g., the five-gene cluster), next to genes for motility/chemotaxis, T3SS, T4P, and biofilm formation, much like found earlier in the fungal-interactive strain BS001 (Warmink and van Elsas 2009; Haq et al. 2014; Yang et al. 2016, 2017). Furthermore, the genomic analyses revealed an enormous capacity in all strains for both primary and secondary metabolisms (supplementary tables 6 and 7, Supplementary Material online). On the basis of these capabilities, the lifestyles of these strains in soil may be depicted as multi-faceted, including saprotrophic next to host-interactive phases. With respect to saprotrophy, the presence of a variety of carbohydrate metabolism genes indicated a clear capacity of the P. hospita species cluster strains to be involved in “spurs” of degradation of the respective polymer substrates (see fig. 6). In detail, the presence of genes related to chitin degradation (CH19 and CH75) was suggestive of fungal-interactive behavior of members of this species cluster, as fungal cell walls contain chitins, next to chitosan and glucans (Zhao et al. 2013; Shinya et al. 2016; Kusaoke et al. 2017). The finding of a gene encoding a GH5 enzyme uniquely in P. terrae DSM 17804T was surprising; this gene endows its host with endo-β-1,4-mannanase (EC: 3.2.1.78) or plant/fungal cell-wall-degrading enzymes, and so backbones of mannan polysaccharides may be transformed into oligosaccharides (Latgé 2007; Engel et al. 2012; Busch et al. 2017).
Secretion Systems and Putative Roles
The finding of copies of the T1SS, T2SS, T3SS, and T6SS across the three type strains as well as the further strains of the same species (next to the four fungiphiles) points to the importance of such systems in the contact of soil-dwelling cells with their surroundings, including living cells (fig. 4A and supplementary fig. 3, Supplementary Material online). Of all of these systems, the T3SS has been indicated to confer an enhanced capacity to the host to adhere to fungal surfaces, endowing cells with an adherence/injection device (Warmink and van Elsas 2008; Haq et al. 2014; Yang et al. 2016, 2017). The finding of ample T3SSs across these organisms contrasted with the notion that many Paraburkholderia species lack such systems (Estrada-de los Santos et al. 2016). Moreover, the finding of differential presence/absence of the T4SS across P. hospita DSM 17164T, P. terrae DSM 17804T (presence in both), and P. caribensis DSM 13236T (absence) points at a differential legacy of interactive (i.e., horizontal gene transfer—HGT) events across the former two organisms versus the latter one.
Glycerol and Oxalate Transformations and Biofilms
Membrane transporters are tightly related to the nutrient and mineral fluxes that are necessary to maintain the biological processes in living cells. The glycerol and oxalic acid released by Lyophyllum sp. strain Karsten can serve as carbon sources as well as molecular signals for Paraburkholderia, much like shown for strain BS001 (Haq et al. 2016, 2017). Here, we addressed the potential of P. hospita DSM 17164T, P. terrae DSM 17804T, and P. caribensis DSM 13236T to capture and metabolize these compounds. Clearly, the OFA family transporters found might be important in a biochemical cycle consisting of: 1) oxalate influx, 2) oxalate decarboxylation, and 3) formate efflux. This energy generating system has been intensively studied in Oxalobacter formigenes (Anantharam et al. 1989).
Moreover, the five-gene cluster present across all Paraburkholderia strains studied here was previously reported to be significantly expressed in (strain BS001) cells that are in contact with fungal hyphae (Haq et al. 2017), as an energy generating system. Its conservation in P. hospita DSM 17164T (and other P hospita strains), P. terrae DSM 17804T (and other P. terrae strains), and the four fungiphiles may indicate its importance in the strains of the two-species cluster, in processes similar to those in selected fungal-derived Paraburkholderia strains (fig. 7).
In the light of the importance of biofilm formation by Paraburkholderia at fungal surfaces (Warmink et al. 2011; Haq et al. 2016), we analyzed the genomes of P. hospita DSM 17164T, P. terrae DSM 17804T, and P. caribensis DSM 13236T, and other strains of these three species, for the presence of biofilm formation genes (fig. 4 and supplementary fig. 3, Supplementary Material online). The fact that the pel, pga and alginate-related genetic systems were found across the three genomes indicates their importance in all three type strains. Here, the analyses of synteny showed high similarity within the P. terrae/P. hospita genomes versus lower similarity of this group with the P. caribensis genome. Hence, eco-evolutionary divergent paths were found across these two groups, again testifying that their lines of descent are different.
Mobile genetic elements
The lifestyles of the investigated Paraburkholderia strains in soil may be strongly determined by the RGPs they harbor, and hence HGT may be a main facet shaping both the fitness of the organisms and their genomes. Indeed, we found clear evidence for the presence of T4SS-like mobility systems that were linked to GIs in P. terrae DSM 17804T (RGP97) and P. hospita DSM 17164T (RGP99) (see supplementary tables 9 and 10, Supplementary Material online), versus the absence of such inserts from P. caribensis. The huge genome size of P. hospita DSM 17164T (11.5 Mb), next to its easy capture of the pEMT1 plasmid in soil, may indicate that 1) P. hospita DSM 17164T is an avid recipient of exogenous genetic material, enabling it to rapidly adjust its genomic complement to the vagaries of the environment (Dagan et al. 2008), and 2) P. hospita DSM 17164T has been subjected to frequent events of HGT and, presumably, positive selection of foreign DNA. Consistent with this tenet was the finding of high numbers of transposases and integrases in the genome of this organism. It is possible that P. terrae DSM 17804T (and P. caribensis DSM 13236T) has the same capabilities, but has been subjected to a lower number of HGT and selection events. However, the extent to which different types of MGEs have entered the genomes of the three type strains, are maintained or deleted there, and endow the host with fitness traits still needs further analysis (Pratama and van Elsas 2019).
Overall Conclusions
The genomes of P. terrae DSM 17804T, P. hospita DSM 17164T and P. caribensis DSM 13236T (9–11.5 Mb) are, as previously reported for, for example, fungiphilic strain BS001, among the largest bacterial genomes described. Clearly, these genome sizes fall within the range of sizes previously reported for the four fungiphilic strains (selectively included in the analyses presented here). The evaluation of the ANIm (and TETRA) values across our strain set revealed the existence of a tight species cluster typified by P. hospita DSM 17164T (and named the P. hospita species cluster), which includes P. hospita DSM 17164T, P. terrae DSM 17804T and the four fungiphilic strains BS001, BS007, BS110, and BS437, but excludes P. caribensis DSM 13236T. Further supporting evidence for this relatedness was found in the phylogenetic analyses (extended with additional genomes) as well as the shared orthologous genes between these strains.
Based on the collective data, we propose to: 1) reassign the formerly named P. terrae strains BS001, BS007, BS110, and BS437 to P. hospita and 2) group the P. terrae/P. hospita-like strains within one species cluster named the P. hospita species cluster. To reach this conclusion, we took into account our own ANIm and TETRA analyses, the phenotypic analyses, DDH results reported previously (Goris et al. 2002; Yang et al. 2006) and the ANI cutoff values for Burkholderia species (Ciufo et al. 2018). Genomic analyses (comparative and CAZyme) of the two type strains, next to the four fungiphiles, revealed these all share traits conferring interactivity with soil organisms such as fungi, which is consistent with a presumed common ancestry and ecophysiology. The traits that were used as “reporters” of fungal interactivity capacities included secretion systems, type-4 pili, biofilm formation, flagellar motility and chemotaxis, the five-gene metabolic response cluster and glycerol and oxalate uptake systems. By the criteria applied, P. caribensis was not strongly fungal-interactive and it was also genomically divergent. Overall, our results confirm the tight relatedness of P. hospita DSM 17164T, P. terrae DSM 17804T and the fungiphiles BS001, BS007, BS110, and BS437. We extended our analyses to other currently available genomes of the three species, and suggest that this analysis indeed indicates that a cluster of soil organisms with fungal-associative behavior exists around the species P. hospita and P. terrae.
Supplementary Material
Acknowledgments
We thank Simone Severitt and Nicole Heyer for excellent technical assistance regarding SMRTbell template preparation and sequencing and Jolantha Swiderski for bioinformatics assistance concerning genome assemblies. This study was supported by a scholarship of the Indonesia Endowment Fund for Education (Lembaga Pengelola Dana Pendidikan—LPDP, Departemen Keuangan, Republik Indonesia) to A.A.P. and the China Scholarship Council (CSC) to Q.C.
Author Contributions
Performed bacterial growth and genomic DNA preparation: A.A.P. Performed whole-genome sequencing and assembly: C.S., B.B., and J.O. Comparative genome analyses: A.A.P., D.J.J.A., and J.D.v.E. Ecological feature genomic analysis: A.A.P. A.A.P. and Q.C. did the experiments. Wrote the manuscript: A.A.P., D.J.J.A., and J.D.v.E. All authors read and approved of the manuscript.
Literature Cited
- Achouak W, Christen R, Barakat M, Marte M, Heulin T.. 1999. Burkholderia caribensis sp. nov, an exopolysaccharide-producing bacterium isolated from vertisol microaggregates in Martinique. Int J Syst Bacteriol. 49(2):787–794. [DOI] [PubMed] [Google Scholar]
- Anantharam V, Allison MJ, Maloney PC.. 1989. Oxalate: formate exchange. The basis for energy coupling in Oxalobacter. J Biol Chem. 264(13):7244–7250. [PubMed] [Google Scholar]
- Beukes CW, et al. 2017. Genome data provides high support for generic boundaries in Burkholderia sensu lato. Front Microbiol. 8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Kunert G, Heckel DG, Pauchet Y.. 2017. Evolution and functional characterization of CAZymes belonging to subfamily 10 of glycoside hydrolase family 5 (GH5_10) in two species of phytophagous beetles. PLoS One 12(8):e0184305–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J, et al. 2018. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 68(1):461–466. [DOI] [PubMed] [Google Scholar]
- Ciufo S, et al. 2018. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int J Syst Evol Microbiol. 68(7):2386–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costaglioli P, et al. 2012. Evidence for the involvement of the anthranilate degradation pathway in Pseudomonas aeruginosa biofilm formation. Microbiol Open. 1(3):326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Artzy-Randrup Y, Martin W.. 2008. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci USA 105(29):10039–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J, Schmalhorst PS, Routier FH.. 2012. Biosynthesis of the fungal cell wall polysaccharide galactomannan requires intraluminal GDP-mannose. J Biol Chem. 287(53):44418–44424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-de los Santos P, et al. 2018. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 9(8):389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-de los Santos P, Rojas-Rojas FU, Tapia-García EY, Vásquez-Murrieta MS, Hirsch AM.. 2016. To split or not to split: an opinion on dividing the genus Burkholderia. Ann Microbiol. 66(3):1303–1314. [Google Scholar]
- Estrada-de los Santos P, Vinuesa P, Martínez-Aguilar L, Hirsch AM, Caballero-Mellado J.. 2013. Phylogenetic analysis of Burkholderia species by multilocus sequence analysis. Curr Microbiol. 67(1):51–60. [DOI] [PubMed] [Google Scholar]
- Goris J, et al. 2002. Diversity of transconjugants that acquired plasmid pJP4 or pEMT1 after inoculation of a donor strain in the A- and B-horizon of an agricultural soil and description of Burkholderia hospita sp. nov. and Burkholderia terricola sp. nov. Syst Appl Microbiol. 25(3):340–352. [DOI] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C.. 2008. CRISPRFinder: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 36:52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq IU, et al. 2016. Chemotaxis and adherence to fungal surfaces are key components of the behavioral response of Burkholderia terrae BS001 to two selected soil fungi. FEMS Microbiol Ecol. 92:1–14. [DOI] [PubMed] [Google Scholar]
- Haq IU, Dini-Andreote F, van Elsas JD.. 2017. Transcriptional responses of the bacterium Burkholderia terrae BS001 to the fungal host Lyophyllum sp. strain Karsten under soil-mimicking conditions. Microb Ecol. 73(1):236–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq IU, Graupner K, Nazir R, Van Elsas JD.. 2014. The genome of the fungal-interactive soil bacterium Burkholderia terrae BS001-a plethora of outstanding interactive capabilities unveiled. Genome Biol Evol. 6(7):1652–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq IU, Zwahlen RD, Yang P, van Elsas JD, .. 2018. The response of Paraburkholderia terrae strains to two soil fungi and the potential role of oxalate. Front Microbiol. 9:989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, et al. 2016. A new view of the tree of life. Nat Microbiol. 1:1–6. [DOI] [PubMed] [Google Scholar]
- Kim S-K, Park H-Y, Lee J-H.. 2015. Anthranilate deteriorates the structure of Pseudomonas aeruginosa biofilms and antagonizes the biofilm-enhancing indole effect. Appl Environ Microbiol. 81(7):2328–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, et al. 2009. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25(17):2283–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaoke H, Shinya S, Fukamizo T, Kimoto H.. 2017. Biochemical and biotechnological trends in chitin, chitosan, and related enzymes produced by Paenibacillus IK-5 Strain. Int J Biol Macromol. 104:1633–1640. [DOI] [PubMed] [Google Scholar]
- Latgé JP. 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol. 66(2):279–290. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. 2009. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:254–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Göker M.. 2019. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 10(1):2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M.. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35:182–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazir R, Zhang M, de Boer W, van Elsas JD.. 2012. The capacity to comigrate with Lyophyllum sp. strain Karsten through different soils is spread among several phylogenetic groups within the genus Burkholderia. Soil Biol Biochem. 50:221–233. [Google Scholar]
- Pratama AA, Chaib De Mares M, van Elsas JD.. 2018. Evolutionary history of bacteriophages in the genus Paraburkholderia. Front Microbiol. 9:853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratama AA, van Elsas JD.. 2019. Gene mobility in microbiomes of the mycosphere and mycorrhizosphere—role of plasmids and bacteriophages. FEMS Microbiol Ecol. 95:fiz053. [DOI] [PubMed] [Google Scholar]
- Pratama AA, Haq IU, Nazir R, Chaib de Mares M, van Elsas JD.. 2017. Draft genome sequences of three fungal-interactive Paraburkholderia terrae strains, BS007, BS110 and BS437. Stand Genomic Sci. 12:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rossello-Mora R.. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 106(45):19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J.. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32(6):929–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl JW, et al. 2015. Genomic characterization of Burkholderia pseudomallei isolates selected for medical countermeasures testing: comparative genomics associated with differential virulence. PLoS One 10(3):e0121052–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles JF, de Souza FA, van Elsas JD.. 2002. Molecular method to assess the diversity of Burkholderia species in environmental samples. Appl Environ Microbiol. 68(4):1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawana A, Adeolu M, Gupta RS.. 2014. Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet. 5:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya S, et al. 2016. Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase. Biochem J. 473(8):1085–1095. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- Stoyanova M, Pavlina I, Moncheva P, Bogatzevska N.. 2007. Biodiversity and incidence of Burkholderia species. Biotechnol Biotechnol Equip. 21(3):306–310. [Google Scholar]
- Talavera G, Castresana J, Kjer K, Page R, Sullivan J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. [DOI] [PubMed] [Google Scholar]
- Vallenet D, et al. 2017. MicroScope in 2017: an expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res. 45(D1):D517–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Straube MJ, et al. 2016. Genetic and functional analysis of the biosynthesis of a non-ribosomal peptide siderophore in Burkholderia xenovorans LB400. PLoS One 11(3):e0151273–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos GS, Parkhill J.. 2006. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22(18):2196–2203. [DOI] [PubMed] [Google Scholar]
- Waack S, et al. 2006. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics 7(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Coleman-Derr D, Chen G, Gu YQ.. 2015. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 43(W1):W78–W84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmink JA, van Elsas JD.. 2008. Selection of bacterial populations in the mycosphere of Laccaria proxima: is type III secretion involved? ISME J. 2(8):887–900. [DOI] [PubMed] [Google Scholar]
- Warmink JA, van Elsas JD.. 2009. Migratory response of soil bacteria to Lyophyllum sp. strain Karsten in soil microcosms. Appl Environ Microbiol. 75(9):2820–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmink JA, Nazir R, Corten B, van Elsas JD.. 2011. Hitchhikers on the fungal highway: the helper effect for bacterial migration via fungal hyphae. Soil Biol Biochem. 43(4):760–765. [Google Scholar]
- Warmink JA, Nazir R, van Elsas JD.. 2009. Universal and species-specific bacterial ‘fungiphiles’ in the mycospheres of different Basidiomycetous fungi. Environ Microbiol. 11(2):300–312. [DOI] [PubMed] [Google Scholar]
- Yang HC, Im WT, Kim KK, An DS, Lee ST.. 2006. Burkholderia terrae sp. nov., isolated from a forest soil. Int J Syst Evol Microbiol. 56(2):453–457. [DOI] [PubMed] [Google Scholar]
- Yang P, Zhang M, van Elsas JD.. 2017. Role of flagella and type four pili in the co-migration of Burkholderia terrae BS001 with fungal hyphae through soil. Sci Rep. 7(1):2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Zhang M, Warmink JA, Wang M, van Elsas JD.. 2016. The type three secretion system facilitates migration of Burkholderia terrae BS001 in the mycosphere of two soil-borne fungi. Biol Fertil Soils. 52(7):1037–1046. [Google Scholar]
- Yin Y, et al. 2012. DbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40:445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Brons JK, van Elsas JD.. 2016. The complete sequences and ecological roles of two IncP-1ß plasmids, pHB44 and pBS64, isolated from the mycosphere of Laccaria proxima. Front Microbiol. 7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Pereira e Silva M de C, Chaib de Mares M, van Elsas JD.. 2014. The mycosphere constitutes an arena for horizontal gene transfer with strong evolutionary implications for bacterial–fungal interactions. FEMS Microbiol Ecol. 89(3):516–526. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Liu H, Wang C, Xu JJ-R.. 2013. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics 14(1):274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS.. 2011. PHAST: a fast phage search tool. Nucleic Acids Res. 39:347–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.