Abstract
Background
Brain-derived neurotrophic factor (BDNF) and microRNA (miRNA) play crucial roles in the etiology of depression. However, the molecular mechanisms underlying this disease are not fully understood. The primary objective of this study was to investigate the relationship between miR-202-3p and BDNF in a chronic unpredictable mild stress (CUMS) model.
Methods
Depression model was established with chronic mild unpredictable mild stimulation (CUMS) combined with solitary feeding. The expression levels of miR-202-3p and BDNF in rat hippocampus were measured by qRT-PCR. The novelty inhibition feeding test (NSFT), sucrose preference test (SPT), and forced swimming test (FST) were used to evaluate the functions of miR-202-3p and BDNF. Target gene prediction and screening and luciferase reporter assay were used to verify the target of miR-202-3p. The expression levels of BNDF, CREB1 and p-CREB1 were detected by Western blot.
Results
Upregulation of miR-202-3p was associated with decreased expression of BDNF in the hippocampus of the CUMS model. Antidepressant was observed when LV-BDNF or LV-si-miR-202-3p was injected into the hippocampus. In addition, in the rat hippocampus and cultured nerve cells, the expression levels of BDNF and cyclic AMP response element binding protein 1 (CREB1), which is a target gene of BDNF, were reduced after LV-miR-202-3p injection. Overexpression of miR-202-3p aggravated depressive behavior and decreased the expression levels of BDNF. Luciferase reporter assay also confirmed that BDNF was a target of miR-202-3p.
Conclusion
Silencing miR-202-3p can reduce the damage to hippocampal nerve in CUMS rats; the mechanism may be related to the upregulation of BNDF expression. miR-202-3p may be an effective target for the treatment of depression.
Keywords: miR-202-3p, BDNF, CREB1, depression, hippocampus
Introduction
Depression is one of the most common mental diseases, which is mainly manifested as depression (showing a lasting unpleasant and unsustainable feeling), loss of enjoyment or interest in usual activities, inability or inadequacy of self-feeling, as well as depressed psychological needs and social disconnection.1,2 It leads to sleep cycle disorders and appetite disorders, distraction and suicidal thoughts. These symptoms may persist in patients with depression for a long time or even relapse.3 It affects the quality of life of the patients and increases family burden if not treated or improved timely.4 Endocrine system disorder, which usually results in elevated cortisol levels and causes nerve cell damage, is an important pathological feature of patients with depression.5 Therefore, protecting nerve cells from injury is an important direction for depression treatment.
With the development of neurobiology and molecular biology, researchers have found that regulating neuroprotective-related genes can protect nerve cells from damage. Studies have shown that decreased expression levels of BDNF play a part in the development of depression.6,7 BDNF is one of the most important members of the neurotrophic factor family. During brain development, the initial expression level of BDNF is very low, and its expression level gradually increases with the development. BDNF is mainly synthesized in neurons, transported by anterograde ax plasm to axon terminals, and released to act on target tissues through specific receptors. Antidepressant treatment can increase the expression of BDNF; therefore, the regulation of BDNF might serve as a potential therapeutic target for antidepressants.8,9 BDNF is regulated by a variety of miRNAs.10 For example, overexpression of miR-206 attenuates neuropathic pain by targeting BDNF.11 And studies have found that miR-613 is involved in the regulation of BDNF in the hippocampus of mice and the occurrence of Alzheimer’s disease process.12 However, the roles of miRNA targeting BDNF in depression remain unknown.
miRNAs are involved in essential biological processes such as the regulation of physiological functions.13,14 Studies in recent years have identified miRNAs as critical regulators of gene expression.15 A large number of miRNAs have been shown to be specifically expressed in the brain.16,17 The specificity of miRNAs in some neurons is their location between synaptic dendrites, which is related to their ability to regulate the translation of target miRNAs.18 The turnover rate of miRNA in neurons appears to be faster than other cell types. MiRNAs play critical roles in the maturation of the nervous system, participate in various pathophysiological processes in the brain, and regulate the occurrence of central nervous system diseases.19,20 Studies have found that in the prefrontal cortex of patients with depression, the expression of various miRNAs, such as miR-145, miR-504 and miR-889, is altered. It suggests that miRNAs might be involved in the formation of depression by regulating the expression of optic genes related to neural activities.21 MiR-202-3p has been shown to be closely related to the occurrence and development of many diseases, and the expression pattern and functions in different diseases are different.22 At present, the expression, function and target of miR-202-3p in depression are not clear. It was speculated that miR-202-3p may regulate the progression of depression through BDNF. The main purpose of this study was to explore the regulation of miR-202-3p in depression. The findings in this study would provide a theoretical basis for developing new drug targets.
Materials and Methods
Animals
Male Wistar rats (180–200 g) were obtained from Shanghai Animal Center, China. Animals have free access to food and water during the study period. All animal protocols were available from the Qingdao Mental Health Center Animal Protection and Use Committee. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of “Qingdao Mental Health Center” and approved by the Animal Ethics Committee of “Animal Ethical and Welfare Committee (AEWC)”.
Chronic Unpredictable Mild Stress (CUMS) Protocol
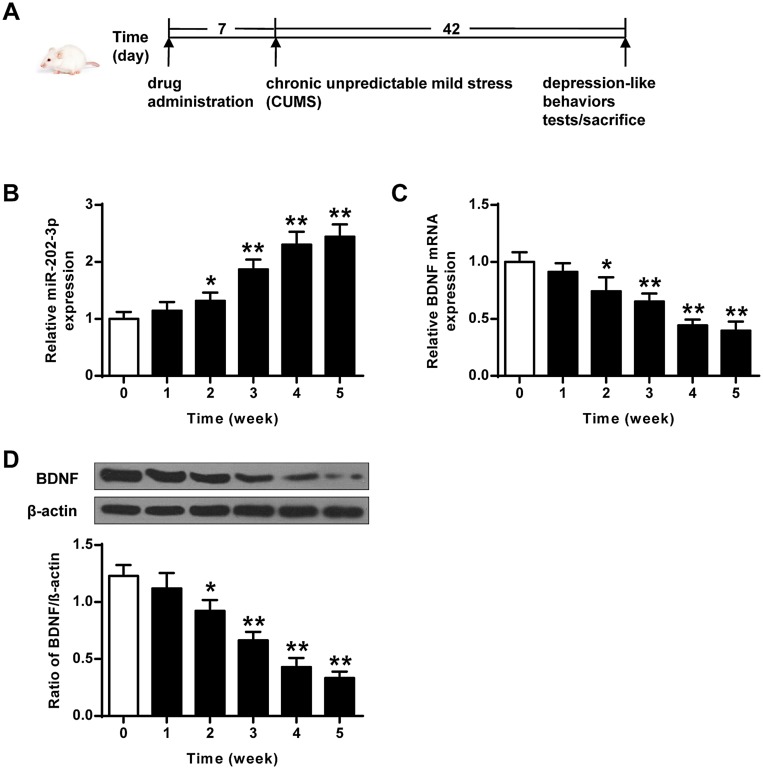
The CMS rat depression model was a classic model that induces behavioral abnormalities in the body by simulating the body’s long-term pressure and unpredictable adverse changes in the surrounding environment. The stress modes were 10 min tail clamping, 24 h food deprivation, 20 min low-temperature swimming (8°C), 24 hours drinking water deprivation, one group of six animals, each shaking for 1 hour with the frequency of 240 Hz, restriction of freedom (60 min), 20 min warm water swimming (40°C), and 24 h high-density space (one cage for every 5–6). Unpredictable stimulation was given once a day for 42 d. The specific flow chart of the experiment is shown in Figure 1A.
Figure 1.
CUMS-induced expression of miR-202-3p and BDNF in rat hippocampus. (A) The specific flow chart of the experiment. (B) CUMS-induced mRNA expression of miR-202-3p in rat hippocampus. (C) CUMS-induced hippocampus in rat mRNA expression level of BDNF. (D) CUMS-induced protein levels of BDNF in rat hippocampus. β-Actin was used as an internal control. *P < 0.05, **P < 0.01 compared with 0 group.
Lentivirus Vectors and Stereotaxic Injection
The primer sequences of BDNF in the plasmid (ptk431) derived from lentivirus driven by CMV promoter were 5ʹ-CGCGGGATCCATGACCATCCTTTTCCTT-3ʹ (forward) and 5ʹ-GCCGCTCGAGCTACAGGTCCTCCTCTGAGATCAGCTTCTGTCTCCCTTTTTAA-3ʹ (reverse). LV-simulation acted as a negative control. LV-miR-202 -3p (108 TU/mL) and LV-SI-miR-202-3p (108 TU/mL) were obtained from Gene Pharmaceuticals (Shanghai, China). Virus injection was performed according to the literature.23
For stereotactic surgery, rats were anesthetized with a mixture of ketamine/xylazine and fixed in a stereotactic frame. Using a precision Hamilton microinjector with a 26 G needle, 1 mL of virus solution was injected into both sides of the rat according to the following coordinates: hippocampus with the first injection: 4.8 mm behind the anterior terior, 2.5 mm inside the suture, and 3.5 mm ventral surface of the skull. Second injection: 4.8 mm behind the front leg, 5 mm inside suture, and 6 mm ventral of the skull.
Depression-Like Behavioral Tests
In the novelty-suppressed feeding test (NSFT), a square box (52 × 42 × 30 cm, length × width × height) was used, and a pellet of food was placed in the center. After 24 h of fasting, the mice were put into the box, with each time from the same direction and position. The first incubation period of feeding was recorded within 5 min (based on the standard of starting to bite the food pellet), that is, the time from the animals being put into the cage to eating the food for the first time.24
In the sucrose preference test (SPT), rats were trained to drink water with sugar in a quiet room. Two water bottles were placed in each cage at the same time. In the first 24 h, both bottles were filled with 1% sucrose water. In the second 24 h, one bottle had 1% sucrose water, one bottle had pure water, and two bottles were randomly placed. After 23 h of water ban, the rat sucrose water consumption experiment was started. After 1 h, two bottles of water were taken to weigh, the sucrose water consumption, pure water consumption and total liquid consumption of the rats were recorded.
In the forced swimming test (FST), the rats were placed in a glass circular cylinder with an inner diameter of 18 cm and a height of 40 cm. The water depth in the cylinder was 20 cm, the water temperature was 21°C to 23°C, and the time of test was 6 min. The time that rats in the water was recorded (stop struggling, floating state was not moving). The immobility time was recorded during the final 4 min.
Primary Hippocampal Neuronal Cultures
The Wistar newborn rats were taken out within 24 h of birth. The hippocampus was isolated under sterile conditions, digested with 0.125% trypsin, dispersed and made into a cell suspension of 1×106/mL density. On the 3rd to 5th day after inoculation, 4 μg/mL of cytarabine, the cell division inhibitor, was added to the culture dish to inhibit the excessive proliferation of non-neuronal cells. Fresh culture medium was changed twice a week. Cells were treated with LV-miR-202-3p for 48 h, and protein expression levels of BDNF and CREB1 were detected by Western blot.
Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNAs were extracted from cells using TRIzol reagent (Kehaojia, Wuhan, China). RNA was reverse transcribed into cDNA using the BeyoRTTM First Strand cDNA Synthesis Kit (Beyotime Institute of Biotechnology). The qRT-PCR reactions were prepared using the ViiATM 7 real-time PCR system (Life Technologies, Grand Island, NY). β-Actin and U6 were used as internal references. Gene expression levels were detected using the SYBR Premix Ex Taq II (Takara Biotechnology). The qRT-PCR was performed following the methods described in the literature.25 Primer sequences were listed in Table 1.
Table 1.
Sequences of Primers Used in qRT-PCR
| Gene | Forward Primer (5ʹ-3ʹ) | Reversed Primer (5ʹ-3ʹ) |
|---|---|---|
| miR-202-3p | ACACTCCAGCTGGGAGAGGTATAGGGCA | CTCAACTGGTGTCGTGGAGTCGGCAATTCAG |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| BDNF | GGTTCGAGAGGTCTGACGAC | CAAAGGCACTTGACTGCTGA |
| β-actin | GCTTCTCCTTAATGTCACGC | CCCACACTGTGCCCATCTAC |
Western Blot Analysis
The transfected cells were collected and total proteins were extracted. Protein concentration was quantified using the BCA Protein Assay Kit. Protein samples were incubated with BDNF (1:500, sc-546), CREB1 (1:500, sc-25785), p-CREB1 (1:500, sc-7978, Abcam, Cambridge, UK), andβ-actin (1:1000, A9044, Sigma, St. Louis, MO) primary antibodies at 4°C for overnight. And then followed by incubation with horseradish peroxidase-conjugated secondary antibody (1:5000; Abcam) at room temperature for 1 h. Western blot analysis was referred to the literature.26
Luciferase Reporter Gene Assay
Wild-type BDNF-3ʹ-UTR (WT) and mutant BDNF-3ʹ-UTR (MT) containing the putative binding site of miR-202-3p were constructed. The report vector containing WT or MT BDNF 3 ‘- UTR and mir-202-3p mimic or NC was co-transfected into cells by Lipofectamine 2000. The luciferase activity was measured by the dual luciferase assay system (Promega) at 48 h post-transfection.
Statistical Methods
The monitoring data were analyzed by SPSS19.0 statistical software. The data were presented as mean ± standard deviation (mean ± SD). Multigroup comparison was done based on one-way ANOVA. LSD test was used for subsequent analysis. P < 0.05 indicated significant difference.
Results
Upregulation of mR-202-3p and Downregulation of BDNF in the Hippocampus of Rats Exposed to CUMS
The expression levels of miR-202-3p were detected in hippocampus of rats exposed to CUMS. As the duration of CUMS was prolonged, the expression levels of miR-202-3p were gradually increased (P < 0.05, P < 0.01) (Figure 1B). As shown in Figure 1C and D, the expression levels of BDNF mRNA and protein decreased gradually with the prolongation of CUMS (P < 0.05, P < 0.01). These results indicated that CUMS induced upregulation of miR-202-3p and downregulation of BDNF in the hippocampus of rats.
Effects of Overexpression and Silencing of miR-202-3p on the Behavioral Dysfunctions Induced by CUMS
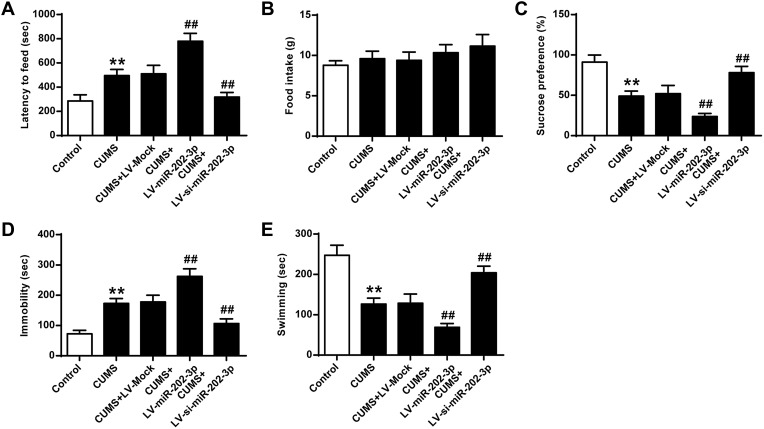
The functional role of miR-202-3p was assessed in CUMS-induced depression in rats. Compared to the Control group, sucrose preference (Figure 2C) and swimming time (Figure 2E) were significantly decreased (P < 0.01), latency to feed (Figure 2A) and the immobility time were significantly increased (Figure 2D) (P < 0.01) in CUMS rats. The sucrose preference of Lv-miR-202-3p in CUMS rats (Figure 2C) was significantly prolonged (P < 0.01), the latency to feed (Figure 2A) and immobility time were significantly shorter (Figure 2D), indicating that depression behavior was more pronounced. The role of LV-si-miR- 202-3p group was reversed. Overexpression or inhibition of miR-202-3p did not cause significant changes in food intake (Figure 2B). These evidence suggested that miR-202-3p can promote CUMS-induced depression-like behaviors.
Figure 2.
Effect of miR-202-3p on CUMS-induced depression-like behavior. (A) Delay in food intake in mice. (B) Determination of feed consumption. (C) Determination of sucrose preference. (D) Measurement of immobility time in FST of rats. (E) Measurement of swimming time in FST of rats. n = 6. **P<0.01 vs control group; ##P<0.01 vs CUMS+LV-MOCK group.
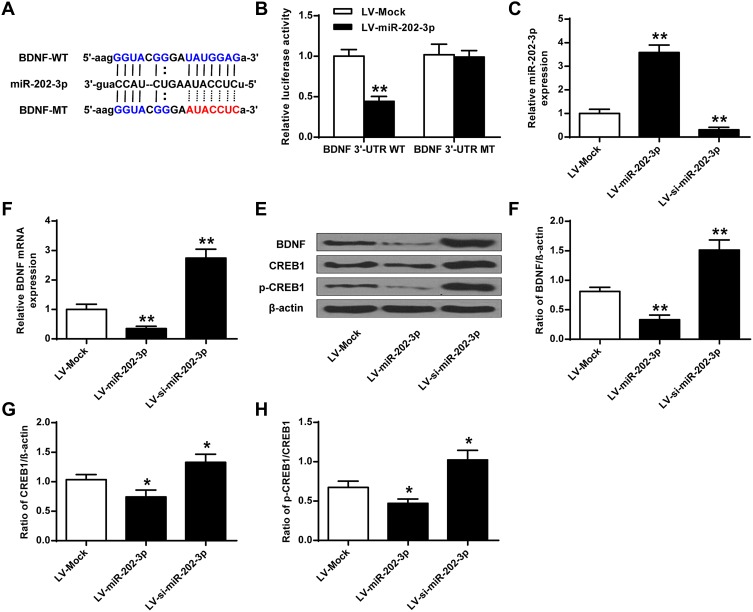
BDNF Was a Functional Target of miR-202-3p
Bioinformatics analysis performed using Targetscan predicted that BDNF was a potential target for miR-202-3p (Figure 3A). The results of luciferase reporter gene assay showed that the luciferase activity in cells co-transfected with Lv-miR-202-3p and BDNF-WT decreased significantly (P < 0.05), but the luciferase activity of BDNF-MUT did not change (Figure 3B). Compared with LV-MOCK group, the expression level of miR-202-3p was significantly increased in nerve cells of LV-miR-202-3p group (P < 0.05, P < 0.01), and the expression level of miR-202-3p was significantly decreased in LV-si-miR-202-3p group (P < 0.01, Figure 3C). As shown in Figure 3D–H, compared to the LV-MOCK group, the expression levels of BDNF, CREB1 and p-CREB1 were significantly decreased in the LV-miR-202-3p group (P < 0.05, P < 0.01), while the expression levels of BDNF, CREB1 and p-CREB1 in the LV-si-miR-202-3p group were significantly increased (P < 0.05, P < 0.01). In summary, miR-202-3p may directly target BDNF in neuronal cells.
Figure 3.
BDNF was a functional target of miR-202-3p. (A) Putative targeting sites for miR-202-3p and BDNF. (B) Fluorescence in cells co-transfected with BDNF-WT or BDNF-Mut vector analysis of the activity of the enzyme. (C) Expression level of miR-202-3p in hippocampal neurons treated with LV-miR-202-3p or LV-si-miR-202-3p. (D) The mRNA expression level of BDNF in hippocampal neurons after treatment with LV-miR-202-3p or LV-si-miR-202-3p. (E) Effect of miR-202-3p on BDNF, CREB1 and pCREB1 protein expression levels. (F–H) Quantitative analysis of BDNF, CREB1 and pCREB1 protein levels. *P <0.05, **P <0.01 vs LV-MOCK.
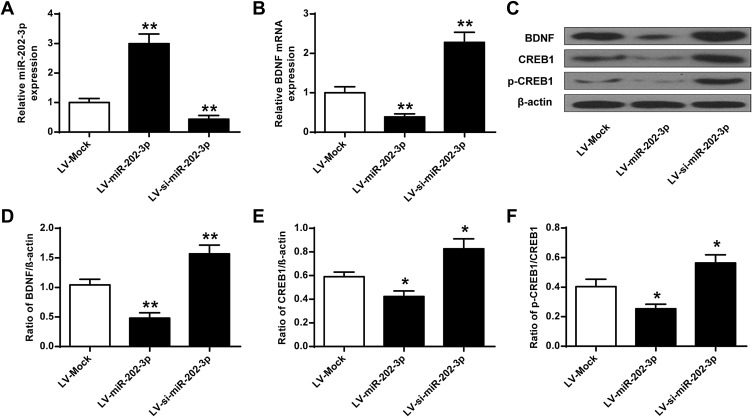
miR-202-3p Down-Regulated the Expression of BDNF in the Hippocampus
The effect of BDNF on the expression of miR-202-3p in the hippocampus of rats was assessed. Compared with the LV-MOCK group, the expression level of miR-202-3p was significantly increased in the hippocampus of the LV-miR-202-3p group (P < 0.05, P < 0.01), and the expression level of miR-202-3p was significantly decreased in the LV-si-miR-202-3p group (P < 0.01) (Figure 4A). As shown in Figure 4B–F, the expression levels of BDNF, CREB1 and p-CREB1 in the hippocampus of the LV-miR-202-3p group were significantly reduced (P < 0.05, P < 0.01), while the expression levels of BDNF, CREB1 and p-CREB1 were significantly increased in the LV -si-miR-202-3p group (P < 0.05, P < 0.01). In summary, miR-202-3p may directly target the BDNF in the hippocampus of rats.
Figure 4.
miR-202-3p down-regulated BDNF expression in rat hippocampus. (A) miR-202-3p of LV- or miR-202-3p of LV-SI- after treatment with miR-202-3p in rat hippocampus expression level. (B) mRNA expression level of BDNF in rat hippocampus after treatment with LV-miR-202-3p or LV-si-miR-202-3p. (C) miR-202-3p in BDNF in rat hippocampus, CREB1 and pCREB1 expression levels were affected. (D–F) BDNF, CREB1 and pCREB1 protein levels were quantified. N = 6. *P <0.05, **P <0.01 vs LV-MOCK.
Effects of the Overexpression of BDNF on CUMS-Induced Depression-Like Behavior in Rats
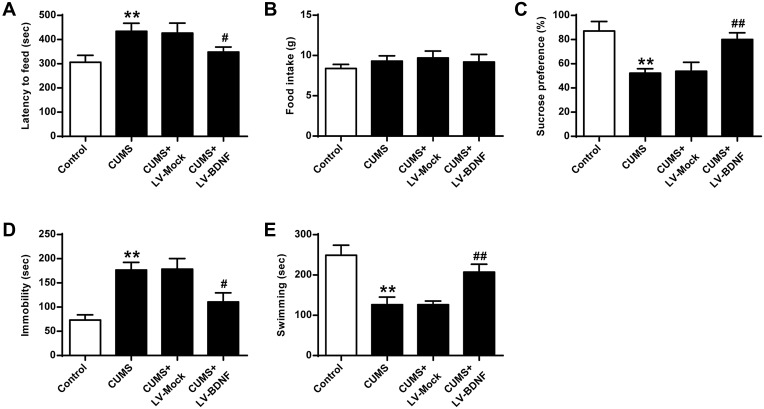
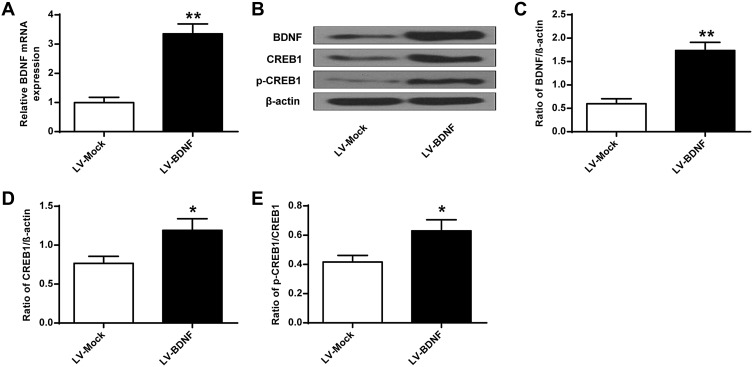
As shown in Figure 5, compared with the Control group, sucrose preference in CUMS rats (Figure 5C) and swimming time (Figure 5E) was significantly lower (P < 0.01), and the feeding latency (Figure 5A) and the immobility time were significantly prolonged (Figure 5D) (P < 0.01). In addition, the sucrose preference in CUMS rats in the LV-BDNF group (Figure 5C) and the swimming time (Figure 5E) was significantly prolonged (P < 0.01). The feeding latency (Figure 5A) and the immobility time were significantly reduced (Figure 5D). There was no significant difference in food intake between groups (Figure 5B).These results indicated that overexpression of BDNF could alleviate depression. Furthermore, as shown in Figure 6A, the expression level of BDNF in the LV-BDNF group was significantly increased compared with that in the LV-MOCK group, indicating that the transfection was successful. In addition, the expression levels of BDNF, CREB1 and p-CREB1 in the LV-BDNF group were significantly increased (P < 0.05, P < 0.01) (Figure 6B–E).
Figure 5.
Effect of BDNF overexpression on CUMS-induced depression in NSFT, SPT, and FST. (A) Delayed feeding in mice. (B) Determination of feed consumption. (C) Determination of sucrose preference. (D) Determination of the amount of immobility in rat. (E) Determination of swimming time in rats. n = 6. **P <0.01 vs control group; #P <0.05, ##p <0.01, vs CUMS + LV-MOCK.
Figure 6.
BDNF, CREB1 and p-CREB1 protein expression levels in rat hippocampus after LV-BDNF treatment. (A) BDNF mRNA expression level in rat hippocampus treated with LV-BDNF. (B) The effects of LV-BDNF regulation on BDNF, CREB1 and pCREB1 protein expression levels in the hippocampus of rats. (C–E) Quantitative analysis of BDNF, CREB1 and pCREB1 protein levels. n = 6. *P<0.05, **P<0.01 vs LV-MOCK group.
Discussion
Depression is a chronic neuropsychiatric disorder that seriously affects the life of patients and increases the burden on patients’ families.27 At present, the treatment effect of clinical depression is still not satisfactory. The treatment with drugs instead of electroconvulsive therapy brings good news to patients, but the side effects of drugs have adverse consequences.28,29 Therefore, it is particularly important to find new therapeutic targets for antidepressants based on the underlying mechanisms of depression. It has been studied to improve the symptoms of depression from the perspective of reversing neuronal damage, and it is expected to be safer and more effective.30 The development of molecular biology and bioinformatics provides a new research direction for disease treatment. It is of potential significance for disease treatment to explore the mechanisms of disease occurrence and find effective therapeutic gene targets. The existing point of view associates the generation of new neurons with the execution of hippocampal-dependent functions, such as learning, memory, and emotions. In addition, the high excitability of young neurons that has been demonstrated in some studies suggests that they may be involved in the recognition of novel stimuli. Chronic stress and elevated level of glucocorticoids, which play a role in the development of depression disorders, decrease the proliferation and survival rate of new cells in the hippocampus.31 On the other hand, antidepressant drugs enhance the proliferation of hippocampal cells in experimental animals.32 In this study, the effects of miR-202-3p and its targeted BDNF on depression in rats were investigated. Our results showed that the silencing of miR-202-3p could reduce the CUMS damage to hippocampal neurons in rats.
The establishment of animal models is the key for studying the pathogenesis and treatment of depression. The chronic unpredictable mild stimulation (CUMS) model can simulate all kinds of stress including stress in daily life and work, which is very closed to the cause of human depression.33 It is widely recognized by researchers as the animal model for depression.34 In this study, the sucrose intake of rats with depression was significantly reduced in the animal models of depression. The lack of pleasure was the main symptom of patients with depression. In the forced swimming experiment, the behavior changes of rats were judged by recording the time of swimming, struggling and immobility, while the immobility of rats was considered as behavior despair, which was also the main criterion of depression judgment.35 In the unfamiliar environment feeding test, the incubation period of the model rats was significantly prolonged, and the unit of food intake was decreased. It indicated that the depression model rats were accompanied by an anxiety-like phenotype.36 The above results were consistent with previous studies, indicating that the depression model was successfully established.
MiRNAs are a group of endogenous highly conserved non-coding single-stranded RNAs discovered in recent years.37 MiRNAs are widely distributed in various tissues and organs, and participate in the regulation of various physiological functions of the body including development, immune response, hematopoiesis, metabolism, and so on.38 Recent studies have found that miRNAs widely exist in peripheral blood, and abnormal expression of miRNAs was observed in patients with diseases. It indicates that miRNAs in peripheral blood can be used as potential biomarkers for early diagnosis and differential diagnosis of disease.39 Previous studies have demonstrated the involvement of miRNAs in the occurrences of depression or resilience relevant to chronic stress.40 Although one or a few miRNAs are presumably involved in depression or resilience in response to chronic stress, these studies indicate primary data about the role of miRNAs in major depression and resilience. Some of them are granted by our analysis with high throughput sequencing.41 miR-202-3p is a recently discovered miRNA, and it was found to be abnormally expressed in various diseases.42 For example, studies have found that miR-202-3p can inhibit the occurrence and development of gastric cancer.43 The roles of miR-202-3p in depression remain unknown. Our study found that CUMS induced the expression of miR-202-3p. In Lv-miR-202-3p CUMS rats, sucrose preference and swimming time were prolonged, and the feeding latency and immobility time became shorter, indicating that overexpression of miR-202-3p makes the depression behavior of rats more obvious.
Depression is associated with reduced expression of BDNF, which regulates synaptic structure and morphological plasticity. BDNF is also involved in the regulation of cell proliferation, migration, and phenotypic differentiation.44 It is required for neuronal production and function in adulthood.45 Studies suggest that miRNA and BDNF participate in the onset of depression, and the feedback regulation network of miR-134 and BDNF on neural plasticity is a typical example.46 Some studies have detected the expression of BDNF, miR-132 and miR-182 in patients with depression, and the expression of BDNF in the serum of patients is decreased, and the expression of miR-132 and miR-182 is increased. Therefore, it is speculated that over-expressed miR-132 and miR-182 participate in the pathogenesis of depression by inhibiting the expression of BDNF.41 Studies have shown that inhibition of miR-15a can promote the maturation of methylated Cp G-binding protein 2-deficient neurons, which directly targets BDNF.47 We screened BDNF as a target gene of miR-202-3p by analyzing the database. CUMS inhibits the expression of BDNF in a time-dependent manner. CREB is a junction of several signal transduction pathways.48 It is activated by phosphorylation of its residue Ser133, which can regulate the transcription of related genes, affect the expression of many cytokines, and start cell, endocrine and behavioral responses through these cytokines.49,50 The present study found that in the LV-miR-202-3p group, the expression levels of pCREB1, BDNF and CREB1 in neuronal cells and hippocampus of rats were significantly decreased, while their expression levels were significantly increased in the LV-si-miR-202-3p group. In addition, sucrose preference and swimming time were significantly prolonged in CUMS rats in the LV-BDNF group, and the feeding latency and immobility time were significantly shorter. Furthermore, the expression levels of BDNF, CREB1 and p-CREB1 were significantly increased in the LV-BDNF group, indicating that overexpression of BDNF can alleviate depression. In future research, we would try to assess whether BDNF can negatively regulate the expression of miR-202-3p to expand our findings.
Conclusion
Silencing of miR-202-3p attenuated CUMS injury in rat hippocampal neurons, which may be related to the up-regulated expression of BNDF. miR-202-3p might serve as an effective target for depression therapy.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.O’Keane V, Marsh MS. Depression during pregnancy. J Midwifery Womens Health. 2017;62(6):157–179. [DOI] [PubMed] [Google Scholar]
- 2.Cocker F, Sanderson K, LaMontagne AD. Estimating the economic benefits of eliminating job strain as a risk factor for depression. J Occup Environ Med. 2017;59(1):12. doi: 10.1097/JOM.0000000000000908 [DOI] [PubMed] [Google Scholar]
- 3.Goodyer IM, Herbert J, Tamplin A, Altham PME. Recent life events, cortisol, dehydroepiandrosterone and the onset of major depression in high-risk adolescents. Br J Psychiatry. 2018;177(4):499. [DOI] [PubMed] [Google Scholar]
- 4.Smolderen KG, Spertus JA, Gosch K, et al. Depression treatment and health status outcomes in young patients with acute myocardial infarction: insights from the VIRGO study (variation in recovery: role of gender on outcomes of young AMI patients). Circulation. 2017;135(18):1762. doi: 10.1161/CIRCULATIONAHA.116.027042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey R, Kaskutas V, Fox I, Baum CM, Mackinnon SE. Effect of upper extremity nerve damage on activity participation, pain, depression, and quality of life. J Hand Surg. 2009;34(9):1682–1688. [DOI] [PubMed] [Google Scholar]
- 6.Erickson KI, Miller DL, Roecklein KA. The aging hippocampus: interactions between exercise, depression, and BDNF. Neuroscientist. 2012;18(1):82. doi: 10.1177/1073858410397054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Z, Lu T, Xu G, et al. Decreased serum brain-derived neurotrophic factor (BDNF) is associated with post-stroke depression but not with BDNF gene Val66Met polymorphism. Clin Chem Lab Med. 2011;49(2):185–189. [DOI] [PubMed] [Google Scholar]
- 8.Miyakawa K, Yamazaki A, Sakano C, et al. GPR39 (zinc receptor) knockout mice exhibit depression-like behavior and CREB/BDNF down-regulation in the hippocampus. Int J Neuropsychopharmacol. 2015;18(3):337–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian X, Wang M. BDNF gene and depression. Adv Psychol Sci. 2016;24(10):1583. doi: 10.3724/SP.J.1042.2016.01583 [DOI] [Google Scholar]
- 10.Sharma RP, Schuhmacher M, Kumar V. Developing integrated PBPK/PD coupled mechanistic pathway model (miRNA-BDNF): an approach towards system toxicology. Toxicol Lett. 2017;280:S037842741731161X. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Yang Q, Wang X, et al. MicroRNA expression profile and functional analysis reveal that miR-206 is a critical novel gene for the expression of BDNF induced by ketamine. Neuromolecular Med. 2014;16(3):594–605. doi: 10.1007/s12017-014-8312-z [DOI] [PubMed] [Google Scholar]
- 12.Li W, Li X, Xin X, Kan P-C, Yan Y. MicroRNA-613 regulates the expression of brain-derived neurotrophic factor in Alzheimer\”s disease. Biosci Trends. 2016;10(5):372–377. doi: 10.5582/bst.2016.01127 [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Zhang X, Huang S, Wang Q, Liu W. miRNA-101-3p inhibits proliferation and migration of gastric cancer cells by targeting EZH2. Chin J Pathophysiol. 2017;33:2143–2150. [Google Scholar]
- 14.Bian B, Yu XF, Wang GQ, Teng TM. Role of miRNA-1 in regulating connexin 43 in ischemia-reperfusion heart injury: a rat model. Cardiovasc Pathol. 2017;27:37–42. [DOI] [PubMed] [Google Scholar]
- 15.Kim JH, Lee DK, Kim J, et al. A miRNA-101-3p/Bim axis as a determinant of serum deprivation-induced endothelial cell apoptosis. Cell Death Dis. 2017;8:e2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hohjoh H, Fukushima T. Expression profile analysis of microRNA (miRNA) in mouse central nervous system using a new miRNA detection system that examines hybridization signals at every step of washing. Gene. 2007;391(1–2):39–44. [DOI] [PubMed] [Google Scholar]
- 17.Jaitner C, Reddy C, Abentung A, et al. Satb2 determines miRNA expression and long-term memory in the adult central nervous system. Elife. 2016;5:e17361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner RW, Kruskic M, Teves M, Scheidl-Yee T, Hameed S, Zamponi GW. Neuronal expression of the intermediate conductance calcium-activated potassium channel KCa3.1 in the mammalian central nervous system. Pflügers Archiv -Eur J Physiol. 2015;467(2):311–328. doi: 10.1007/s00424-014-1523-1 [DOI] [PubMed] [Google Scholar]
- 19.Kynast KL, Russe OQ, Möser CV, Geisslinger G, Niederberger E. Modulation of central nervous system–specific microRNA-124a alters the inflammatory response in the formalin test in mice. Pain®. 2013;154(3):368–376. doi: 10.1016/j.pain.2012.11.010 [DOI] [PubMed] [Google Scholar]
- 20.Peckham-Gregory EC, Thapa DR, Martinson J, et al. MicroRNA-related polymorphisms and non-Hodgkin lymphoma susceptibility in the Multicenter AIDS Cohort Study. Cancer Epidemiol. 2016;45:47–57. doi: 10.1016/j.canep.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma K, Guo L, Xu A, Cui S, Wang JH. Molecular mechanism for stress-induced depression assessed by sequencing miRNA and mRNA in medial prefrontal cortex. PLoS One. 2016;11(7):e0159093. doi: 10.1371/journal.pone.0159093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farahani M, Rubbi C, Liu L, Slupsky J.R, Kalakonda N. CLL exosomes modulate the transcriptome and behaviour of recipient stromal cells and are selectively enriched in miR-202-3p. PLoS One. 2015;10(10):e0141429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Favre D, Provost N, Blouin V, Favre. Immediate and long-term safety of recombinant adeno-associated virus injection into the nonhuman primate muscle. Mol Ther. 2001;4(6):559–566. doi: 10.1006/mthe.2001.0494 [DOI] [PubMed] [Google Scholar]
- 24.Yi L-T, Liu -B-B, Li J, et al. BDNF signaling is necessary for the antidepressant-like effect of naringenin. Progress Neuro-Psychopharmacol Biol Psychiatr. 2014;48:135–141. doi: 10.1016/j.pnpbp.2013.10.002 [DOI] [PubMed] [Google Scholar]
- 25.Kong Q, Yuan J, Gao L, et al. Identification of suitable reference genes for gene expression normalization in qRT-PCR analysis in watermelon. PLoS One. 2014;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Campillo M, Bini L, Comanducci M, et al. Identification of immunoreactive proteins of Chlamydia trachomatis by Western blot analysis of a two-dimensional electrophoresis map with patient sera. Electrophor Int J. 2015;20(11):2269–2279. [DOI] [PubMed] [Google Scholar]
- 27.Semkovska M, Ahern E. Online neurocognitive remediation therapy to improve cognition in community-living individuals with a history of depression: a pilot study. Internet Interv. 2017;9(1):7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazell P, Mirzaie M. Tricyclic drugs for depression in children and adolescents. Cochrane Database Syst Rev. 2013;6(6):CD002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergner CL, Smolinsky AN, Hart PC, et al. Mouse Models for Studying Depression-Like States and Antidepressant Drugs. 2016;602:267–282. [DOI] [PubMed] [Google Scholar]
- 30.Omata N, Mizuno T, Matsumoto H, et al. Mania is an extension of depression from the perspective of neuronal plasticity – evaluation of our hypothesis through the affective spectrum. Med Hypotheses. 2017;102:87–88. doi: 10.1016/j.mehy.2017.03.018 [DOI] [PubMed] [Google Scholar]
- 31.Papatriantafyllou JP, Gotsis EG, Toulas P, et al. P.1.125 Study of the left hippocampus with in vivo NMR spectroscopy in patients with depressive disorders and subjective complaints about their memory efficiency. Eur Neuropsychopharmacol. 2004;14(4):S229. [Google Scholar]
- 32.Aniol VA, Gulyaeva NV. Changes in adult neurogenesis in the hippocampus during depressive disorders in humans. Neurochem J. 2015;9(1):8–12. doi: 10.1134/S181971241501002X [DOI] [Google Scholar]
- 33.Tian JS, Zuo YM, Sun HF, Qin XM. Analysis on endogenous metabolites in cecal tissue of CUMS-induced depression rats after Xiaoyao San intervention by GC-MS metabolomics. Chinese Traditional and Herbal Drugs 2015. [Google Scholar]
- 34.Li B, Han L, Cao B, Yang X, Qiao W. Use of magnoflorine-phospholipid complex to permeate blood-brain barrier and treat depression in the CUMS animal model. Drug Deliv. 2019;26(1):566–574. doi: 10.1080/10717544.2019.1616236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu L, An SC, Lian T. [Involvement of hippocampal NMDA receptor and neuropeptide Y in depression induced by chronic unpredictable mild stress]. Sheng Li Xue Bao:[Acta physiologica Sinica]. 2010;62(1):14–22. [PubMed] [Google Scholar]
- 36.Liu W, Xue X, Xia J, Liu J, Qi Z. Swimming exercise reverses CUMS-induced changes in depression-like behaviors and hippocampal plasticity-related proteins. J Affect Disord. 2018;227:126–135. doi: 10.1016/j.jad.2017.10.019 [DOI] [PubMed] [Google Scholar]
- 37.Aghaeebakhtiari SH, Arefian E, Lau P. miRandb: a resource of online services for miRNA research. Brief Bioinform. 2018;18(5):904. [DOI] [PubMed] [Google Scholar]
- 38.Oka S, Furukawa H, Shimada K, Hashimoto A, Tohma S. Plasma miRNA expression profiles in rheumatoid arthritis associated interstitial lung disease. BMC Musculoskelet Disord. 2017;18(1). doi: 10.1186/s12891-017-1389-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hua FF, Liu SS, Zhu LH, et al. MiRNA-338-3p regulates cervical cancer cells proliferation by targeting MACC1 through MAPK signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(23):5342–5352. [DOI] [PubMed] [Google Scholar]
- 40.Tavakolizadeh J, Roshanaei K, Salmaninejad A, Yari R, Mirzaei H. MicroRNAs and exosomes in depression: potential diagnostic biomarkers. J Cell Biochem. 2017;119(5):3783–3797. [DOI] [PubMed] [Google Scholar]
- 41.Li Y-J, Xu M, Gao Z-H, et al. Alterations of serum levels of BDNF-related miRNAs in patients with depression. PLoS One. 2013;8(5):e63648. doi: 10.1371/journal.pone.0063648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu F, Li L, Wen Q, Yang J, Cheng L. A functional variant in ST2 gene is associated with risk of hypertension via interfering MiR-202-3p. J Cell Mol Med. 2017;21(7):1292–1299. doi: 10.1111/jcmm.13058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Z, Chenglong L, Ming W, et al. Decrease of miR-202-3p expression, a novel tumor suppressor, in gastric cancer. PLoS One. 2013. 8(7):e69756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monteiro BC, Monteiro S, Candida M, Adler N, Machado S. Relationship between brain-derived neurotrofic factor (Bdnf) and sleep on depression: a critical review. Clin Pract Epidemiol Ment Health. 2017;13(1):213–219. doi: 10.2174/1745017901713010213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H, Zhang Y, Helou LI, Zeng W, Qiao M. Shuyu capsules relieve liver‑qi depression by regulating ERK‑CREB‑BDNF signal pathway in central nervous system of rat. Exp Ther Med. 2017;14(5):4831–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan C, Zhu X, Song Q, Wang P, Liu Z, Yu SY. MiR-134 modulates chronic stress-induced structural plasticity and depression-like behaviors via downregulation of Limk1/cofilin signaling in rats. Neuropharmacology. 2018;131:364–376. doi: 10.1016/j.neuropharm.2018.01.009 [DOI] [PubMed] [Google Scholar]
- 47.Gao Y, Su J, Guo W, et al. Inhibition of miR-15a promotes BDNF expression and rescues dendritic maturation deficits in MeCP2-deficient neurons. Stem Cells. 2015;33(5):1618–1629. doi: 10.1002/stem.1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chrivia JC, Kwok RPS, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365(6449):855–859. doi: 10.1038/365855a0 [DOI] [PubMed] [Google Scholar]
- 49.Sanna MD, Mello T, Masini E, Galeotti N. Activation of ERK/CREB pathway in noradrenergic neurons contributes to hypernociceptive phenotype in H4 receptor knockout mice after nerve injury. Neuropharmacology. 2018;128:340–350. [DOI] [PubMed] [Google Scholar]
- 50.Zhang G, Tao Z, Ning L, et al. Tetramethylpyrazine nitrone activates the BDNF/Akt/CREB pathway to promote post-ischaemic neuroregeneration and recovery of neurological functions in rats. Br J Pharmacol. 2018;175:517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]