SUMMARY
Innate regeneration following digit tip amputation is one of the few examples of epimorphic regeneration in mammals. Digit tip regeneration is mediated by the blastema, the same structure invoked during limb regeneration in some lower vertebrates. By genetic lineage analyses, the digit tip blastema has been defined as a population of heterogeneous, lineage restricted progenitor cells. These previous studies, however, do not comprehensively evaluate blastema heterogeneity or address lineage restriction of closely related cell types. In this report we present single cell RNA sequencing of over 38,000 cells from mouse digit tip blastemas and unamputated control digit tips and generate an atlas of the cell types participating in digit tip regeneration. We computationally define differentiation trajectories of vascular, monocytic, and fibroblastic lineages over regeneration, and while our data confirm broad lineage restriction of progenitors, our analysis reveals 67 genes enriched in blastema fibroblasts including a novel regeneration-specific gene, Mest.
Keywords: blastema, digit tip regeneration, single cell RNAseq, fibroblast heterogeneity, Mest
eTOC BLURB
Johnson et al. use single-cell RNA sequencing to define the cell types of homeostatic and regenerating mouse digit tips. They find that all broad cell types present in the unamputated digit tip appear in the early blastema and that dynamic gene expression in fibroblast populations underlies the regenerative state.
INTRODUCTION
Many animals have the amazing ability to regenerate entire body parts such as the limb, tail, or spinal cord following amputation. This process has been termed epimorphic regeneration, where a complex structure comprised of multiple tissue types is regenerated from progenitor cells within a structure termed the blastema (Carlson, 1978; Hay and Fischman, 1961; Morgan, 1901). Examples of vertebrates that employ epimorphic regeneration include axolotl, newt, and juvenile xenopus which can regenerate many structures including limbs and the spinal cord (Dent, 1962; Overton, 1963; Spallanzani, 1768); and zebrafish, which can regenerate their fins (Johnson and Weston, 1995). In contrast to these species, mammals have limited epimorphic regeneration of complex tissues, though examples do exist: deer can repeatedly shed and regenerate antlers, and mice and human children can regenerate amputated digit tips (Goss, 1961; Illingworth, 1974; Neufeld and Zhao, 1995). Mouse is a well characterized model for studying digit tip regeneration. Following amputation in adult digit tips, there is an initial inflammation and wound healing phase (Fernando et al., 2012). When the wound epithelium has closed, the blastema, a proliferative and heterogeneous structure, forms and goes on to regenerate all non-epidermal structures of the digit tip by approximately 28 days post-amputation (dpa) (Fernando et al., 2012; Lehoczky et al., 2011; Rinkevich et al., 2011).
The blastema is the common structure that links together regeneration in species that seem disparate such as zebrafish, axolotl, and mouse. The blastema is a critical yet transient structure and much remains to be learned about how it mediates regeneration of complex tissues, particularly in mammals. Two hypotheses exist as to how blastema cells give rise to regenerated tissues. One posits that blastema cells are multipotent and can differentiate into any of the regenerating tissues. Another is that the blastema is a heterogeneous population of cells that are lineage restricted, and only contribute to their tissue of origin in the regenerate. While species-specific nuances likely exist, genetic lineage tracing studies in several regenerative models support that the blastema contains cells that are lineage restricted, not multipotent (Flowers et al., 2017; Gargioli and Slack, 2004; Tu and Johnson, 2011). In the mammalian digit tip specifically, mouse genetic lineage analyses have revealed that embryonic germ layer identities hold true during digit tip regeneration and there is no evidence for transdifferentiation (Lehoczky et al., 2011; Rinkevich et al., 2011). Progeny of epithelial progenitor cells traced using Krt5 or Krt14 inducible cre drivers remain restricted to the regenerated epithelium (Lehoczky et al., 2011; Rinkevich et al., 2011; Takeo et al., 2013). Similarly, Sp7 or Sox9 marked skeletal progenitors contribute solely to the regenerating bone and periosteum, and VE-cadherin- or Tie2-expressing endothelial cells only give rise to endothelium in the regenerate (Lehoczky et al., 2011; Rinkevich et al., 2011). In the neural lineage, Schwann cells marked by Sox2 contribute only to the regenerated glial lineage (Johnston et al., 2016). Fibroblasts are one of the most abundant cell types within the digit tip, and as found for the other cell types, lineage marked Prrx1-expressing fibroblasts remain fate restricted to the regenerated mesoderm (Rinkevich et al., 2011). In a similar experiment, Msx1-expressing cells in the mesenchyme and bone contribute highly to the blastema but do not transdifferentiate into tissues lineages derived from other germ layers (Lehoczky et al., 2011). Collectively, these studies support a heterogeneous blastema comprised of lineage restricted progenitor cells in the regenerating mouse digit tip. However, these previous analyses lack a precise description of all the cell types present in the blastema and an assessment of lineage restriction among closely related cell types.
To this point, previous genetic lineage analyses in axolotl found the regenerating limb blastema to be heterogenous and lineage restricted (Kragl et al., 2009), and recently single cell RNAseq and lineage tracing have been combined to elucidate a more detailed understanding of the axolotl limb blastema (Gerber et al., 2018; Leigh et al., 2018). For example, the presence of macrophages, muscle progenitors, and fibroblasts was confirmed while additional cell types were discovered in both regenerating and homeostatic limbs (Leigh et al., 2018). Moreover, supporting transdifferentiation of closely related lineages, a multipotent fibroblast-like progenitor competent to contribute to multiple regenerated lineages including tendon, skeleton, and fibroblasts was found in the blastema (Gerber et al., 2018). However other lineages, including muscle and wound epithelium, remained more restricted (Gerber et al., 2018; Leigh et al., 2018). These studies demonstrate that single cell transcriptome profiling can offer a more nuanced view of the blastema not possible with genetic lineage analyses alone. Addressing similar questions in the context of mouse digit tip regeneration is important, especially in the context of working towards regenerative therapies.
In this paper we build upon previous findings that the mouse digit tip blastema is heterogeneous and lineage restricted by generating single cell transcriptomes of four stages of regenerating mouse digit tip blastemas as well as unamputated control digit tips. We sequenced over 38,000 total cells, allowing us to comprehensively define the cell type heterogeneity of the blastema throughout regeneration. We analyze an integrated data set from all regenerative and control time points and find that a clear signature of the cell types found in the quiescent control digit tip already exists in the early blastema, supporting lineage restriction. We find that blastema population dynamics vary by cell type and we focus specifically on a population of fibroblasts enriched in early blastema stages as compared to unamputated control digit tips. Differential expression analyses concentrated on these blastema-enriched fibroblasts reveals ten highly significant genes. Of these, Mest is expressed broadly in the blastema by RNA in situ hybridization, but not in the unamputated digit tip. This finding supports the notion of regeneration-specific factors and opens the door to more subtle transdifferentiation relationships within the fibroblast lineage. Collectively, these data have important implications for regeneration of other musculoskeletal tissues and our broad understanding of epimorphic regeneration in mammals.
RESULTS
The early blastema is heterogeneous in cell type
Previous studies have used tissue/cell-type specific mouse genetic lineage analyses to characterize the regenerating digit tip blastema as both cellularly heterogeneous and lineage restricted (Lehoczky et al., 2011; Rinkevich et al., 2011). However, these studies leave room for additional insights into the origin of the blastema cells, the complexity of the cellular heterogeneity, and lineage relationships within germ layers. Toward these questions, we set out to characterize the adult mouse digit tip blastema as it first emerges from the stump tissue. By histology in outbred mice, the blastema is first detected at 10dpa (Fernando et al., 2012). While this timing is consistent with inbred FVB/NJ mice used in our study, we find gross microdissection of the blastema is not possible until 11dpa due to lack of tissue integrity at earlier stages. We amputated adult FVB/NJ mouse hindlimb digits midway through the terminal phalanx at a level permissive for innate regeneration (Figure 1A) (Fernando et al., 2012; Han et al., 2008; Neufeld and Zhao, 1995). At 11dpa we euthanized the mice and manually dissected the blastemas away from the surrounding epithelium and stump tissue. 12 blastemas dissected from two mice were pooled, dissociated, and subjected to single cell RNA sequencing using the 10X Genomics platform (Figure 1A). 7,830 cells were captured, with an average of 15,491 sequencing reads per cell. Quality control and filtering of reads was performed using a standard computational pipeline in Seurat, leaving RNA sequencing data for 7,610 high quality cells (Butler et al., 2018; Stuart et al., 2019), which was then used for unbiased cell clustering based on differential gene expression.
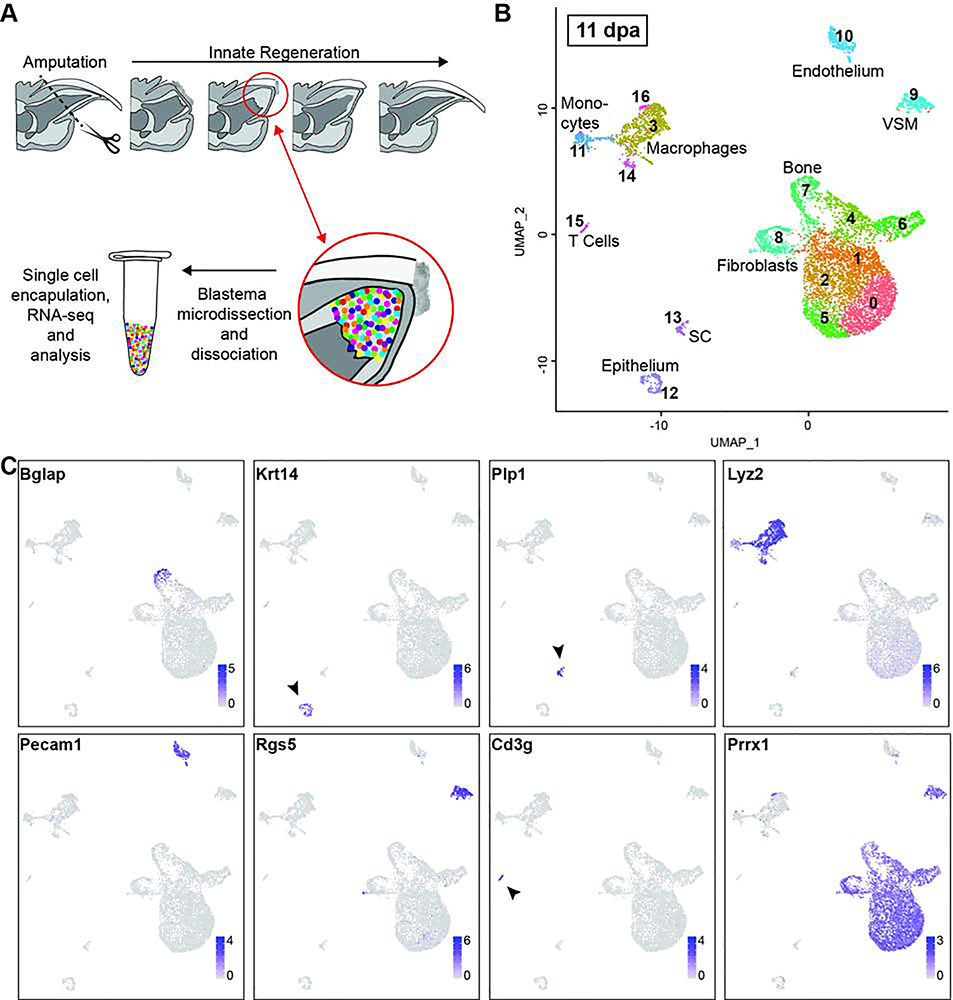
Figure 1. Cellular heterogeneity of the 11dpa blastema.
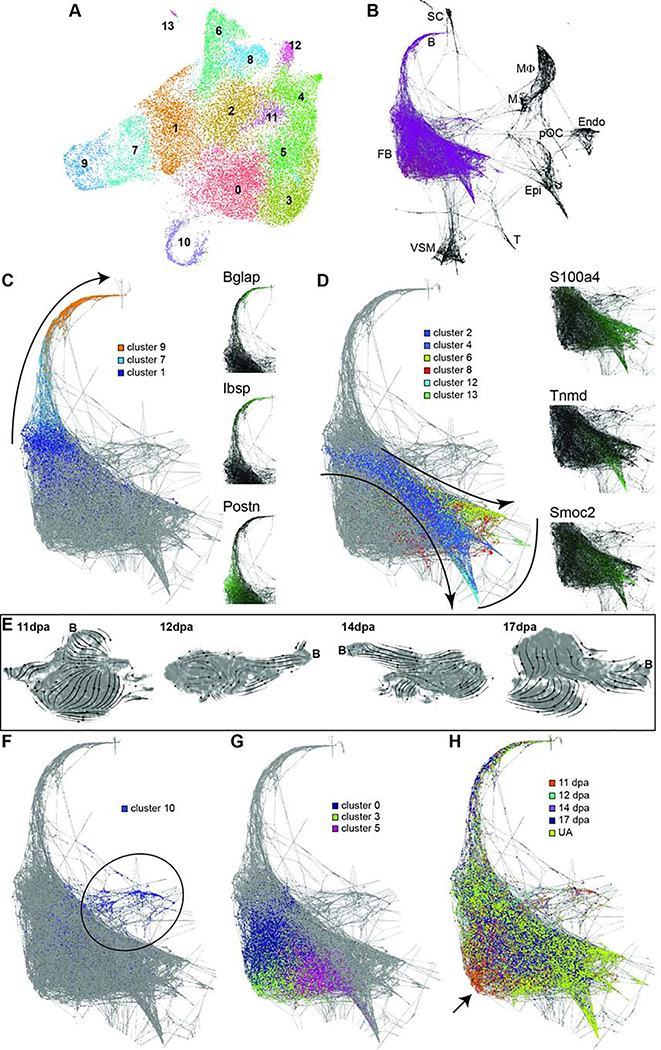
(A) Schematic overview of innate mouse digit tip regeneration following amputation mid-way through the terminal phalanx. Schematic of the experimental design whereby blastemas were dissected, dissociated, and single cells captured. Single cell RNA libraries were prepared and sequenced for computational analysis. (B) Unbiased single cell clustering of 7,610 high quality cells visualized by UMAP. Each dot represents a single cell and cells assigned to the same cluster are similarly colored. Cell type identities are assigned as follows: fibroblasts (clusters 0–2, 4–6, and 8), macrophages (clusters 3, 14, and 16), bone (cluster 7), vascular smooth muscle cells (VSM) (cluster 9), endothelial cells (cluster 10), monocytes (cluster 11), epithelial cells (cluster 12), Schwann cells (SC) (cluster 13), T cells (cluster 15). (C) Gene expression UMAP overlay with examples of highly expressed, cell type specific markers used to assign cluster cell identities: Bglap (bone), Krt14 (epithelial cells), Plp1 (SCs), Lyz2 (macrophages and monocytes), Pecam1 (endothelial cells), Rgs5 (vascular smooth muscle cells), Cd3g (T cells), Prrx1 (fibroblasts). Gray depicts low expression and purple depicts high expression as specified on the scale for each gene.
Unbiased clustering of the 11dpa blastema cells revealed 17 discrete populations (Figure 1B). We assigned cell identities to each of these clusters based on the top 10–20 most differentially expressed genes associated with each cluster and the known expression of these genes based on the literature, as well as checking expression of broadly established cell type marker genes in each cluster (Table S1). The assigned cell types include: fibroblasts (clusters 0–2, 4–6, and 8; Prrx1, Msx1, Vim); bone (cluster 7; Bglap, Ibsp, Spp1); monocytes (cluster 11; Lyz2, Cd14, Cd86) and macrophages (clusters 3, 14, 16; Adgre1, C1qa, Lyz2); T cells (cluster 15; Cd3g, Icos, Trdc); endothelial cells (cluster 10; Pecam1, Cd93, Egfl7); vascular smooth muscle (VSM) cells (cluster 9; Rgs5, Notch3, Myh11), Schwann cells (cluster 13; Plp1, Mbp, and Scn7a); and epithelial cells (cluster 12; Krt14, Krt42, Perp) (Figure 1B and 1C). Our experiment was designed to capture the cells within the blastema, and because previous genetic lineage analyses demonstrate that epithelial cells do not contribute to the blastema (Lehoczky et al., 2011; Rinkevich et al., 2011), we interpret the presence of epithelial cells within our sample as a technical artifact secondary to dissection (Figure 1B, cluster 12). While the nail and wound epithelia are both important cell types necessary for digit tip regeneration (Fernando et al., 2012; Lehoczky and Tabin, 2015; Mohammad et al., 1999; Takeo et al., 2013) they were not intentionally captured in this study and have been excluded from all of our analyses. We also interpret the presence of a mature bone population at 11dpa as a dissection artifact consistent with inclusion of stump bone adjacent to the blastema (Figure 1B, cluster 7). We include this cluster in our analyses as these cells provide terminally differentiated tissue to facilitate analysis of osteoprogenitor differentiation in the blastema.
Of the non-epithelial 11dpa populations, we captured several cell types already described in mouse digit tip regeneration. We find most cells in our sample are fibroblasts marked in particular by Prrx1 and Msx1; previous genetic lineage analyses with these markers demonstrate that these cell types contribute broadly to the blastema (Lehoczky et al., 2011; Rinkevich et al., 2011). In addition, de-differentiated Schwann cells have been shown to secrete growth factors that may play a role in expansion of blastema cells during regeneration (Johnston et al., 2016), and in line with this we observe a population of Schwann cells (Figure 1B and 1C, cluster 13). Macrophages have been described in the post-amputation digit tip during wound closure and have been shown to be necessary for successful digit tip regeneration (Simkin et al., 2017). While this previous study finds peak numbers of macrophages prior to blastema formation by histology (Simkin et al., 2017), our blastema stage single cell analysis identifies three discrete macrophage populations (Figure 1B, clusters 3, 14, and 16), with one of the populations (cluster 14) likely representing mitotic macrophages based on expression of cell cycle genes such as Top2a and Cdk1. In addition, we find a population of endothelial cells (Figure 1B, cluster 10). Sca1/Ly6a positive endothelial cells have been characterized in the 10dpa blastema (Yu et al., 2014) and in line with previous data, 68% of the 11dpa endothelial cells in our dataset express Sca1/Ly6a (Table S1). Collectively, the presence of these previously described populations (Schwann cells, macrophages, endothelial cells, and fibroblasts) validates the robustness of our experimental approach. Importantly, unbiased single cell RNA sequencing also enabled us to identify cell populations that have not formally been described in the digit tip blastema. We isolated VSM cells and a small population of T cells (Figure 1B, clusters 9, and 15). We also isolated monocytes which likely contribute to the local macrophage population (Figure 1B, cluster 11).
To begin exploring the relationships among these populations, we constructed a cluster dendrogram and find the clusters fall into four main branches. The bone cells (cluster 7) make up their own branch of the dendrogram. Monocytes (cluster 11) and two macrophage populations (clusters 3 and 14) make up a separate branch (Figure S1A), which correlates with the known lineage relationship between monocytes and macrophages (van Furth and Cohn, 1968; Virolainen, 1968). A third branch of the dendrogram is comprised of the fibroblast populations (clusters 0–2, 4–6, and 8) and the remaining macrophage population (cluster 16). While we expected the fibroblast populations to be closely related, the presence of a macrophage population in this clade was unanticipated. The fourth branch is made up of the remaining unrelated populations: T-cells, endothelial cells, VSM cells, epithelial cells, and Schwann cells. As validation of the cell population relationships, we calculated the Pearson correlation between in silico bulk transcriptomes of each cluster (Figure S1B). Consistent with the dendrogram, the resultant correlation matrix shows that the fibroblast clusters are highly correlated with macrophage cluster 16. Many of the genes marking cluster 16 indicate that it is made up of macrophages (C1qa (83%), Adgre1 (74%)), while this cluster also expresses fibroblast marker genes (Prrx1 (100%), Fmod (100%)). While this could be a rare hybrid cell type, the mixed expression is more parsimonious with doublet cells (encapsulating two cells in one droplet before library preparation). To investigate this, we analyzed our dataset with DoubletFinder (McGinnis et al., 2019) and found 419 cells (5.5 %) classified as potential doublets, including 29 out of 53 cells (54.7%) in macrophage cluster 16 (Figure S1C). This finding prompted us to remove all putative doublet cells from subsequent integrated analyses, though the existence of hybrid cell types has not been formally ruled out.
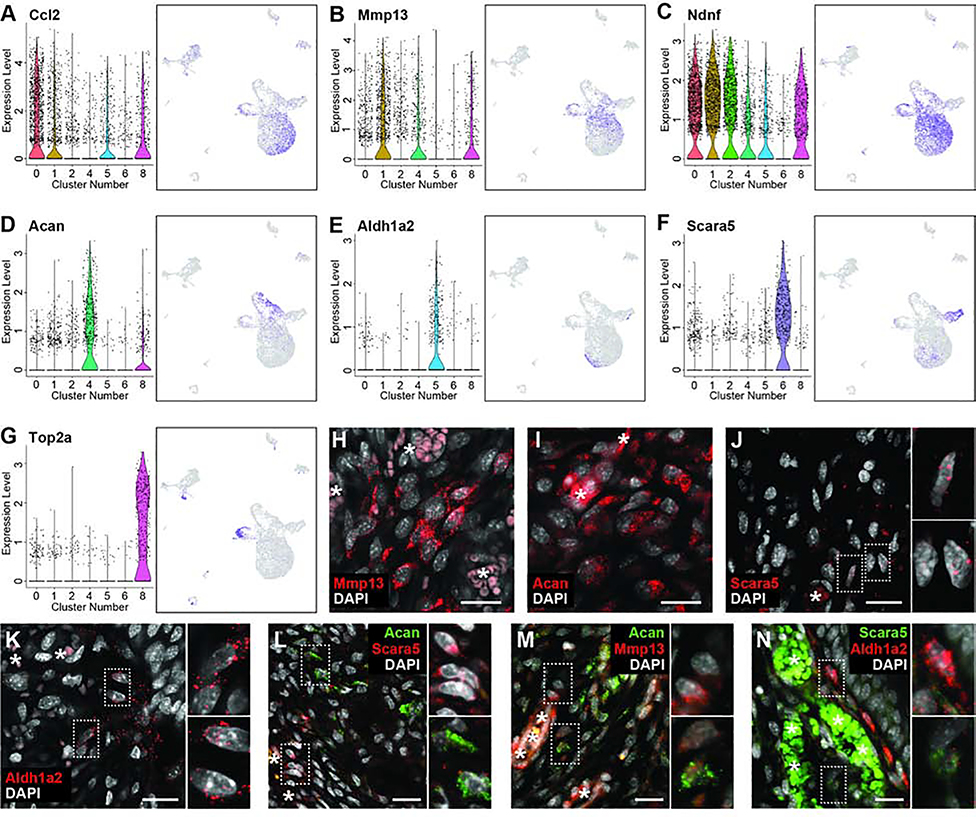
Although fibroblasts are known to participate in digit tip regeneration (Lehoczky et al., 2011; Marrero et al., 2017; Rinkevich et al., 2011; Wu et al., 2013b), the high proportion of the 11dpa blastema comprised of fibroblasts and the heterogeneity of these cells is striking, and has not been described previously. To begin to understand the biological significance of the seven discrete 11dpa fibroblast populations, we investigated the cluster marker genes that specifically mark these populations (Table S1). While all fibroblast clusters have gene expression in common, such as Prrx1, Msx1, and Pdgfra (Carr et al., 2018; Lehoczky et al., 2011; Rinkevich et al., 2011), these broad fibroblast markers ultimately mask the underlying heterogeneity of fibroblasts in the blastema. In line with the dendrogram and correlation matrix, clusters 0, 1, and 2 show common expression of many genes like Ndnf (Figure 2C). However, Ccl2 expression is more specific to cluster 0, and Mmp13 expression to cluster 1, both perhaps consistent with a role in cytokine signaling or immune response (Figure 2A and 2B, Table S2). Cells in clusters 4 and 6 are not closely related and have distinct expression profiles, for example Acan and Scara5 respectively (Figure 2D and 2F), and GO analysis predicts involvement in the different biological processes of skeletal development and ECM organization for cluster 4 and iron ion import and transmembrane receptor protein tyrosine kinase signaling pathway for cluster 6 (Table S2). Cluster 5 is also predicted to be involved in ECM organization, though these cells also express different genes than cluster 4, including Aldh1a2 (Figure 2E). Cluster 8, which expressed markers of proliferation such as Top2a, is comprised of mitotic fibroblasts (Figure 2G). Single and double labeled RNA FISH for Acan, Mmp13, Aldh1a2, and Scara5 on 11dpa sections confirms the heterogeneity of fibroblastic cells in the blastema (Figure 2H–N), which suggests that blastema fibroblasts may participate in a diverse set of functions and lineages in the regenerating digit tip.
Figure 2. 11dpa fibroblast heterogeneity.
Representative genes differentially expressed among fibroblast clusters. (A-H) Gene expression is shown by violin plot (left) where black dots represent individual cells and the colored curve shows the distribution of cells at a given expression level, and feature plot (right) where purple is high expression and grey is low expression. Cluster numbers and UMAP plots relate to data in Figure 1B. (A) Ccl2 is expressed by cells in clusters 0, 1, 5, and 8; (B) Mmp13 is expressed in clusters 1, 4, and 8; (C) Ndnf is expressed in clusters 0, 1, 2, 4, 5, and 8; (D) Acan is expressed in cluster 4; (E) Aldh1a2 is expressed in cluster 5; (F) Scara5 is expressed in cluster 6; (G) Top2a is expressed in cluster 8. (H-N) HCR RNA FISH for fibroblast cluster markers. Mid-blastemal region of 11dpa section probed for (H) Mmp13, (I) Acan, (J) Scara5, (K) Aldh1a2, or co-probed for (L) Acan (green) and Scara5 (red), (M) Acan (green) and Mmp13 (red), or (N) Scara5 (green) and Aldh1a2 (red). DAPI is shown in white. Scale bars = 20μm. Asterisks (*) denote blood vessel/RBC autofluorescence. Dashed boxes show magnified panels on right.
Signature of the terminally differentiated digit tip already exists in the early blastema
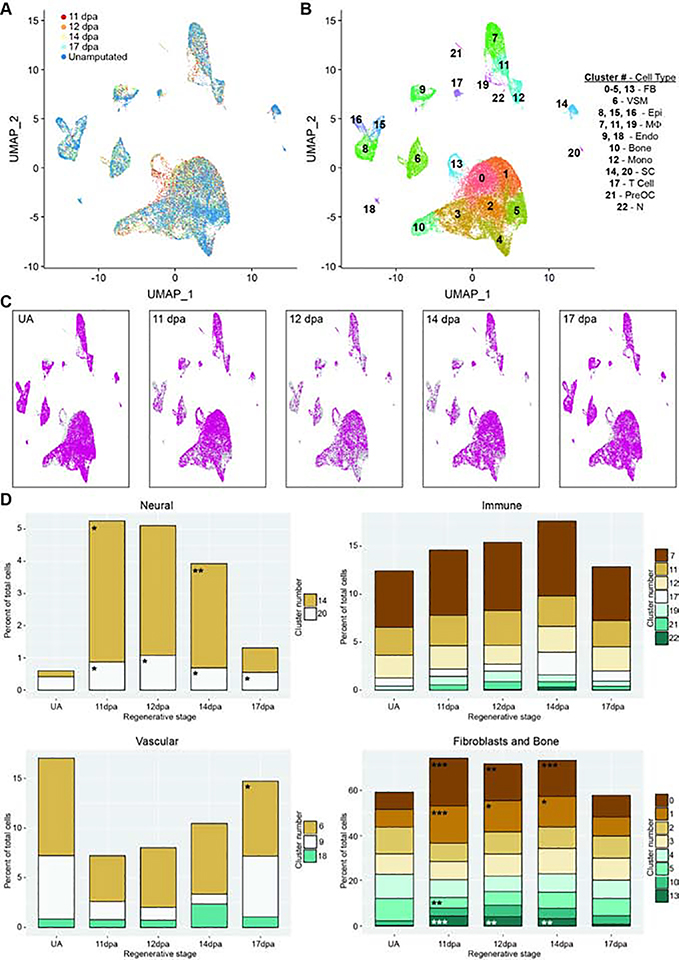
The diversity of cell types we find by single cell RNAseq in the 11dpa blastema is supported by previous studies which demonstrate the mouse digit tip blastema is heterogeneous through genetic lineage analyses and staining for cell type specific markers (Carr et al., 2018; Johnston et al., 2016; Lehoczky and Tabin, 2015; Lehoczky et al., 2011; Rinkevich et al., 2011). However, digit tip regeneration is a prolonged, dynamic process, and little is known about how the heterogeneous blastema resolves into regenerated tissues or how the blastema cells relate to the cells of the original digit tip. Toward addressing these questions, we generated single cell RNAseq data from progressive blastema stages, as well as from the mesenchyme of unamputated digit tips. As with our 11dpa experiment, we amputated adult FVB/NJ mouse hindlimb digits (Figure 1A) and manually dissected blastemas at 12, 14, or 17dpa. For unamputated samples, mice were euthanized and non-epithelial tissues distal to our standardized amputation plane were dissected from hindlimb digits. All four samples were separately dissociated and subjected to single cell RNA sequencing as above. Likely due to variation in cell dissociation, encapsulation, and library preparation we captured a range of cell numbers and reads for our samples: 12dpa (3,433 cells/27,628 average reads per cell), 14dpa (6,065 cells/27,026 reads), 17dpa (9,112 cells/21,416 reads), unamputated (UA) (13,750 cells/9,831 reads). Samples and reads were processed as with the 11dpa sample, and quality control and filtering left 3,309, 5,896, 8,778, and 12,871 cells in each data set respectively. We first analyzed each sample separately to determine which cell types were present at each regenerative stage (Figures S2–5 and Table S1). Intriguingly, all the cell types identified in the 11dpa blastema are also present in all four more mature blastema stages, as well as in the unamputated digit tip. Moreover, there are only a few additional cell types that appear in any sample and are limited to pre-osteoclasts (12dpa and 14dpa), neutrophils (14dpa), and a second population of Schwann cells (UA), though the emergence of these cell types in only certain regenerative stages could be explained by the differing number of sequenced cells.
The finding that the majority of cell types identified in the unamputated digit tip are already present in the 11dpa blastema presents at least two scenarios: 1) these are the same cells in terms of gene expression and only differ in quantity and perhaps spatial organization, or 2) these are cells within the same tissue-specific lineage that differ in gene expression and differentiation state at the time points sampled. Importantly, these possibilities need not be mutually exclusive given many of our assigned cell types have multiple discrete populations of cells (for example macrophages or fibroblasts) which could have separate roles in digit tip regeneration. To begin to address these questions, we removed all predicted doublet cells from the 11dpa, 12dpa, 14dpa, 17dpa, and UA single cell RNAseq data sets (Figures S1C, S2C, S3C, S4C, and S5C), and combined and normalized the data for the remaining cells using the Integration workflow in Seurat (Stuart et al., 2019). Unbiased clustering of this combined data set revealed that cells from all stages were qualitatively well-mixed among 23 clusters (Figures 3A and 3B, Table S3). This combined data set also allowed for increased resolution of cell types that might be rare in each stage; we now observed defined populations for myelinating (cluster 20, marked by Mbp and Plp1) and non-myelinating Schwann cells (cluster 14, marked by C4b and Scn7a), and lymphatic endothelium (cluster 18, marked by Pdpn and Lyve1) (Figure 3B). Overlaying each regenerative stage individually over the total integrated data set reveals that no cluster is comprised of cells from a single time point (Figure 3C), reinforcing the conclusion that unamputated digit cell types exist at all blastema stages and ruling out a broadly multipotent cell in the blastema.
Figure 3. Integrated analysis of single cell populations through a regenerative time course.
All analyses use combined and normalized 11dpa, 12dpa, 14dpa, 17dpa, and unamputated (UA) scRNAseq data sets. (A) UMAP plot of integrated data sets colored by regenerative stage: 11dpa (red), 12dpa (orange), 14dpa (yellow), 17dpa (light blue), and unamputated (blue). (B) UMAP plot of integrated data sets showing clusters and cluster cell type annotations. Assigned cell types are: fibroblasts (FB; clusters 0–5, and 13), vascular smooth muscle cells (VSM; cluster 6), epithelial cells (Epi; clusters 8, 15, and 16), macrophages (Mф; clusters 7, 11, and 19), endothelial cells (Endo; clusters 9 and 18), bone (cluster 10), monocytes (Mono; cluster 12), Schwann cells (SC; clusters 14 and 20), T cells (cluster 17), pre-osteoclasts (PreOC; cluster 21), and neutrophils (N; cluster 22). (C) UMAP plot of integrated data set (gray) showing the cluster distribution of cells from each regenerative stage (pink). (D) The percentage of total cells represented by each cluster for the given regenerative stage. Each stage has been compared to the proportion of cells in UA, and significant changes were determined by differential proportion analysis (marked with asterisk); all p-values reported in Table S4. Clusters are categorized by overarching cell types (fibroblast or bone, immune, vasculature, or neural). Significance values are as follows: * denotes p<0.05, ** denotes p<0.01, *** denotes p<0.001.
Blastema population dynamics during regeneration vary by cell type
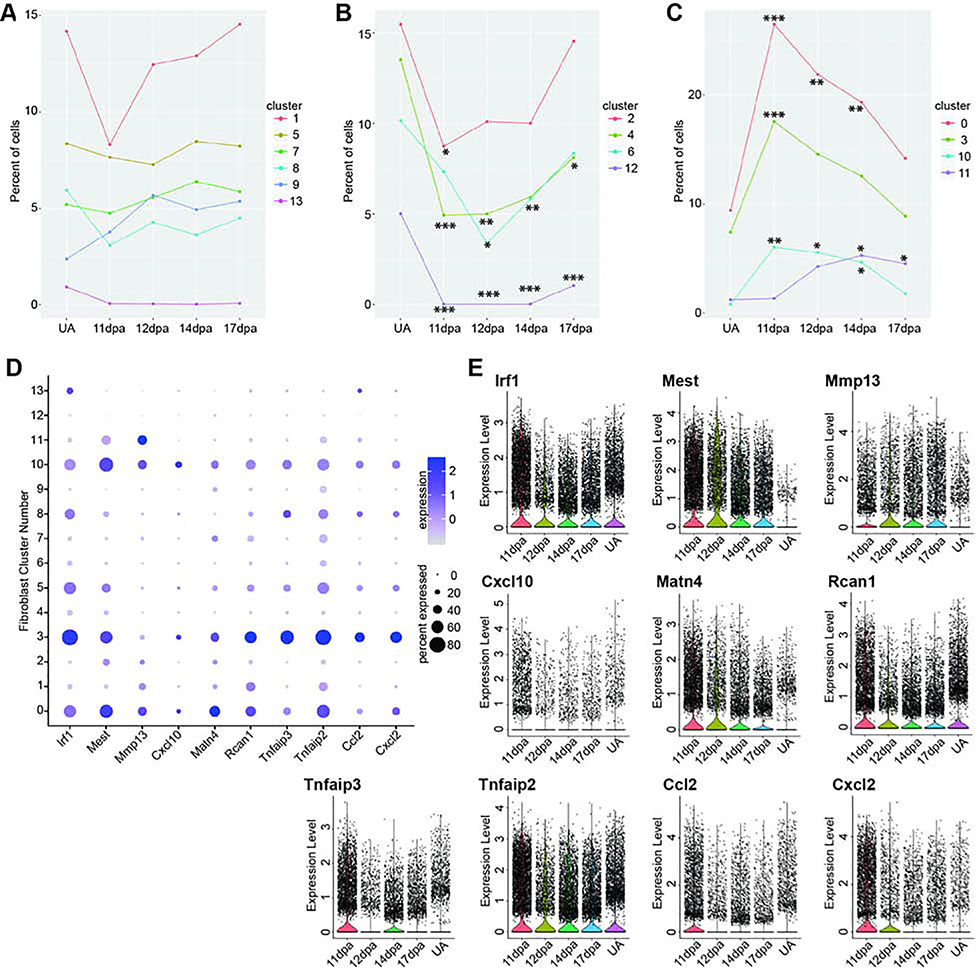
To examine the dynamics of individual blastema cell populations throughout regeneration, we sought to assess 1) the proportion of cells in each cluster present at each regenerative stage and whether it changes over time, and 2) cell type specific gene expression changes through regeneration and whether this reflects tissue specific differentiation states. To compare cluster membership over time, we performed differential proportion analysis (Farbehi et al., 2019) on our integrated data set segregated by stage. This statistical test detects changes in population proportions across conditions while controlling for false positives. We used this analysis to build hypotheses about the timing and function of blastema populations and whether they are regeneration or homeostasis specific. As a first pass analysis we looked for changes in relative population size as compared to the unamputated digit and found significant regenerative population dynamics for Schwann cells (clusters 14 and 20), VSM cells (cluster 6), and several fibroblast populations (clusters 0, 1, 5, and 13). Notably, no significant population dynamics were found in the immune-related clusters (Figure 3D, Table S4).
Little is known about the influence of immune cells in digit tip regeneration. Only macrophages have been characterized and found to be necessary (Simkin et al., 2017), but it is important to understand whether additional immune cells play a role in the blastema as well as the initial inflammation response. In our data, differential proportion analysis finds no significant differences in proportion of monocytes, macrophages, pre-osteoclasts, T cells, or neutrophils between any two stages in our data set (Figure 3D, Table S4). Given the small number of cells (551 total) in the pre-osteoclast, T cell, and neutrophil clusters, we are likely statistically underpowered to make meaningful conclusions for these cell types. That said, we have relatively large populations of macrophages and monocytes at all stages of our data (4,534 cells total), thus the absence of significant population dynamics for these cell types is likely reflective of the biology of the digit tip regeneration immune response. To understand the lineage relationship of these cells, and if there is a differentiation trend during regeneration, we subjected the cells in these clusters to SPRING, an algorithm to detect cell lineage relationships in single-cell gene expression datasets (Weinreb et al., 2018). The data predict a major differentiation trajectory from monocytes to macrophages, with no skewing in differentiation state based on regenerative/unamputated stage (Figure S6A–D). This finding suggests that the production of macrophages from monocytes in the digit tip is at a homeostatic rate once the blastema is formed. RNA velocity data support this prediction, showing that monocytes and macrophages in both regenerative and non-regenerative stages are increasing expression of Adgre1 (Figure S6E and S6F). No specific lineage relationships are revealed for the population of ECM producing macrophages, T-cells, or neutrophils, though we have a minimal sampling of these populations (Figure S6A). However, there is a qualitative increase in differentiation of monocytes to pre-osteoclasts and an increase in proliferative macrophages marked by Adgre1 and Top2a in the blastema ( Figure S6B, S6D, and S6H–I). The presence of proliferative macrophages could reflect a lingering response to the initial wound or a physiological role in the blastema itself that is not satisfied by recruited monocytes.
Previous studies demonstrate that innervation and neural associated cell types such as Schwann cell precursors are necessary for digit tip regeneration (Carr et al., 2018; Dolan et al., 2019; Johnston et al., 2016; Mohammad and Neufeld, 2000; Takeo et al., 2013). Both sensory and sympathetic axons innervate the connective tissue of the unamputated digit tip, and they are accompanied by both myelinating and non-myelinating Schwann cells (Dolan et al., 2019). In the unamputated digit tip, we find that myelinating and non-myelinating Schwann cells make up 0.40% and 1.9% of the captured cells, respectively. Despite small numbers of cells, differential proportion analysis reveals a significant increase of both populations in the 11dpa blastema compared to the unamputated digit tip (Figure 3D, Table S4). This conflicts with previous work, which showed that Schwann cells are present in the blastema but are qualitatively less abundant than in the quiescent digit tip, and only non-myelinating Schwann cells appear to recover to pre-amputation levels by 4 weeks post amputation (Dolan et al., 2019; Johnston et al., 2016). While it is important to understand whether these dynamics correlate with the differentiation trajectory of these cell types, these clusters (clusters 14 and 20) contained too few cells for a meaningful SPRING trajectory analysis, thus these questions remain for future experiments designed to specifically enrich these populations.
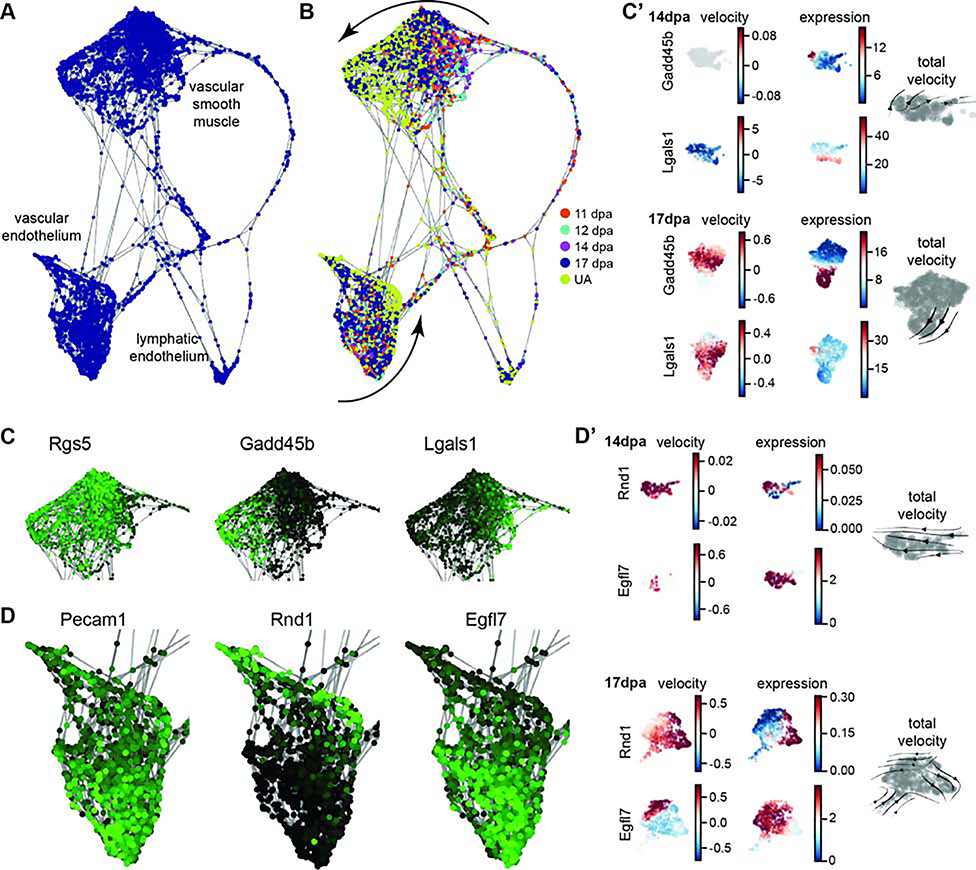
In previous studies, VE-cadherin-expressing endothelial cells have been shown to be lineage restricted during digit tip regeneration (Rinkevich et al., 2011) and individual endothelial cells are found in the blastema (Fernando et al., 2012). However, the overall dynamics of vascular-related cells in the blastema, including VSM cells, have not yet been characterized. Differential proportion analysis of our all stage integrated single cell RNAseq data reveals no significant change in the relative population sizes of endothelial cells or VSM cells between 11dpa or 14dpa compared to UA. However, at 17dpa, VSM is significantly expanded compared to UA, and endothelial cells are significantly expanded compared to 14dpa (Figure 3D, Table S4). The low relative percentage of vascular cell types in the early regenerative stages is consistent with previous work that describes minimal angiogenesis in the early blastema (Yu et al., 2014), and the spike in vascular cell types at 17dpa could be indicative of over-sprouting of blood vessels before they are pruned (reviewed in Korn and Augustin, 2015). To explore these cell types further, we computationally predicted the differentiation trajectory using SPRING. The three vascular-related cell clusters (Figure 3B clusters 6, 9, and 18) appear separate in the SPRING visualization (Figure 4A). No major lineage relationship is predicted between vascular and lymphatic endothelial cells (Figure 4A). Lymphatic endothelial cells have not yet been described in digit tip regeneration so it is an important advance to have captured them, and their associated markers; that said, this is an extremely small population and we are underpowered to make predictions on a differentiation trajectory. When analyzing the VSM cells and the vascular endothelial populations by regeneration stage, there is qualitative spatial variation between the different time points on the SPRING plot (Figure 4B). Closer evaluation of the VSM cells confirms that all of the cells express the tissue-specific marker Rgs5 (Figure 4C) (Li et al., 2004). However, early blastema cells (11, 12, and 14dpa) are concentrated on one side of the cluster, while UA cells are on the other and 17dpa cells appear throughout. This observation is consistent with the differentiation of VSM cells from a progenitor state to terminally differentiated cells and is supported by the differential expression of Gadd45b and Lgals1 as well as the gene-specific and total RNA velocity data at 14dpa and 17dpa for these clusters (Figures 4C and 4C’) (Gizard et al., 2008; Kim et al., 2014; Moiseeva et al., 2000). Similarly, the cluster of vascular endothelial cells all express the broad marker Pecam1 (Figure 4D) (Albelda et al., 1990; Müller et al., 2002), though the computationally defined trajectory shows UA cells concentrated on one edge of the SPRING plot and blastema cells throughout the rest of the cluster, consistent with differentiation from vascular endothelial progenitors (expression of Egfl7 (Campagnolo et al., 2005)) to terminally differentiated cells (expression of Rnd1 (Suehiro et al., 2014)) (Figure 4D). RNA velocity analysis of the vascular endothelium supports this predicted trajectory and shows that endothelial cells are specifically increasing expression of Rnd1 and, at 17dpa, terminating expression of Egfl7 (Figure 4D’). The total RNA stream plots from 14dpa and 17dpa also show that endothelial cell velocity moves towards the more differentiated state (Figure 4D’). Overall, the different vascular populations have recovered by 17dpa, and there are differences in differentiation state between the blastema stages and the unamputated digit tip. The 17dpa vascular cells span both mature and immature states.
Figure 4. Vasculature differentiation trajectory of integrated data set.
Computationally defined SPRING trajectory analysis of cells from the integrated data set vascular clusters 6, 9, and 18. (A) Force-directed plot of cells showing clusters of VSMs, vascular endothelial cells, and lymphatic endothelial cells. (B) SPRING plot as in (A) with regenerative stages of each cell colored coded: 11dpa (orange), 12dpa (light blue), 14dpa (purple), 17dpa (dark blue), unamputated (yellow). Differential clustering of blastema cells and unamputated cells suggests tissue specific differentiation of VSMs and the vascular endothelium (curved arrows). (C) Gene expression overlay on VSMs. Rgs5 is expressed in all cells, Gadd45b is more highly expressed in UA cells, and Lgals1 is more highly expressed in blastema cells. High expression is in green and low expression is black. (C’) UMAP plots of RNA velocity and expression data for individual genes Gadd45b and Lgals1 at 14dpa and 17dpa; total RNA velocity stream plot for each stage in gray (right). (D) Gene expression overlay on vascular endothelial cells. Pecam1 is expressed in all cells, Rnd1 is more highly expressed in UA cells, and Egfl7 is more highly expressed in blastema cells. (D’) UMAP plots of RNA velocity and expression data for individual genes Rnd1 and Egfl7 at 14dpa and 17dpa; total RNA velocity stream plot for each stage in gray (right).
Diversity and dynamics of fibroblasts during regeneration reveal regeneration-specific markers
As fibroblasts make up the majority of the blastema and are more heterogeneous than previously described (Figure 2) (Lehoczky et al., 2011; Rinkevich et al., 2011; Wu et al., 2013b), we analyzed all fibroblast and bone cells separately from the rest of the cell types. This unbiased clustering resulted in 14 populations that were broadly concordant with the original all-cell-type clustering, yet more refined (Figure 5A compared to Figure 3B). We performed SPRING analysis on the all-cell-integrated data set and found no populations predicted to transdifferentiate from fibroblasts or bone into any other cell type within the blastema or unamputated digit tip (Figure 5B), supporting the lineage restriction found in previous genetic lineage studies (Lehoczky et al., 2011; Rinkevich et al., 2011). Among the fibroblast clusters there is a distinct differentiation trajectory predicted from clusters 1 and 7 into bone (cluster 9; Bglap and Ibsp expression) (Figure 5C); total RNA velocity stream plots from 11dpa to 17dpa support this lineage (Figure 5E). While the presence of mature bone cells at all regenerative stages (Figure 5H) is an artifact of our microdissection, these cells facilitate trajectory mapping and allow for cell type identification of clusters 1 and 7 as osteoprogenitors and differentiating osteoblasts, respectively (Figure 5C, Postn expression; Table S5). Another possible group of trajectories originate in cluster 2, then branch and terminate in clusters 4, 6, 8, 12, and 13 (Figure 5D). It is unclear if these trajectories reflect the differentiation of resident fibroblast subtypes within the digit tip, or whether they reflect skeletal tissue lineages (ex. tenocytes or adipocytes). Clusters 6 and 12 express several tendon-specific genes, such as Fibin and Tnmd (Table S5) (Brandau et al., 2001; Pearse et al., 2009), and we hypothesize that cluster 2 contains mesenchymal stem cells (MSCs) or minimally, tendon progenitor cells because of Scx expression (Schweitzer et al., 2001). However, no discrete skeletal lineage can be assigned to clusters 4, 8, and 13 based on marker gene expression, for example S100a4 and Smoc2, thus they may be incompletely differentiated MSCs that also reside in the unamputated digit, or resident fibroblast subtypes that have not been characterized. Clusters 0, 3, and 5 make up a third major concentration of cells. They do not appear to differentiate into a specific lineage and remain centrally located on the trajectory map (Figure 5G). Intriguingly, this analysis reveals that these clusters are enriched for cells from early blastema stages and this is not a function of proliferation (Figures 5F and 5H). This finding could be consistent with the dedifferentiation of fibroblasts (lineage contribution), or regenerative-specific fibroblasts (non-lineage, providing signals).
Figure 5. Fibroblast differentiation trajectory of integrated data set.
(A) UMAP plot of unbiased re-clustering of fibroblast and bone cells from integrated data set (Figure 3: clusters 0–5, 10, and 13), reveals 14 refined clusters. (B) Computationally defined SPRING trajectory analysis of cells from the integrated data set showing fibroblasts (FB) or bone (B) are not predicted to transdifferentiate into Schwann cells (SC), monocytes (M), macrophages (MΦ), pre-osteoclasts (pOC), endothelium (Endo), epithelium (Epi), T cells (T), or vascular smooth muscle (VSM). Fibroblast SPRING lineage trajectory overlaid with (C) predicted bone lineage from cluster 1 to cluster 9 (curved arrow). Marker gene expression for each cluster shown with Bglap, Ibsp, and Postn. High expression is in green and low expression is black. (D) Computationally proposed mesenchymal stem cell lineage from cluster 2 to clusters 4, 6, 8, 12, and 13 (curved arrows), with distinct lineages marked by Tnmd, S100a4, and Smoc2. Curved line depicts proposed terminally differentiated cells. (E) UMAP plots of total RNA velocity stream data for 11dpa, 12dpa, 14dpa, and 17dpa individual stage datasets; the position of bone cells in each plot is marked with a ‘B’. (F) Cluster 10 marks mitotic cells (majority highlighted by black circle) and (G) clusters 0, 3, and 5 may not contribute to a lineage, but are (H) enriched for early stage blastema cells (arrow pointing to orange).
Differential proportion analyses of the re-clustered fibroblasts support our qualitative findings from the trajectory analysis. Clusters 1, 5, 7, 8, 9, and 13 do not have significant changes in population size through regeneration, though for clusters 1, 7, and 9, this can be attributed to inclusion of bone in all dissections (Figure 6A and Table S6). Cell populations in clusters 2, 4, 6, and 12 all are significantly depleted at 11dpa, and are restored to unamputated levels by 17dpa, with the exception of clusters 12 and 4 (Figure 6B and Table S6). This profile may be consistent with amputated tissue lineages being restored through regeneration. Unexpectedly, we found the cell populations in clusters 0, 3, and 10 to be significantly increased at 11dpa and cluster 11 at 14dpa; by 17dpa clusters 0, 10, and 11 have still not returned to unamputated levels (Figure 6C and Table S6). Cluster 10 population dynamics can be attributed to cellular proliferation (Figure 5F and Table S5), however for clusters 0, 3, and 11 this suggests a regeneration specific function. Gene expression analysis between cells from blastema-enriched clusters (Figure 6C) and blastema-depleted clusters (Figure 6B) results in 370 significantly differentially expressed genes, 67 of which are upregulated in blastema-enriched clusters (Table S7). We prioritized genes with an average log fold-change ≥ 0.75 and with the percent of cells in other clusters expressing the gene ≤ 0.25, leaving 10 genes (Figure 6D). Of these, several had distinct regeneration-specific expression profiles. Ccl2 and Cxcl2 both showed increased expression at early blastema stages, with low expression in late regeneration as well as the unamputated digit tip (Figure 6E). Mmp13, Mest, and Matn4 both showed expression at all blastema stages, with negligible expression in the unamputated digit tip (Figure 6E). While Mmp13 has already been implicated in regeneration in other species as a necessary mediator of ECM remodeling (Calve et al., 2010; Miyazaki et al., 1996; Vinarsky et al., 2005; Wu et al., 2013a), the other 9 genes are novel markers of the blastema and epimorphic regeneration (Figure 7A–7E).
Figure 6. Analysis of blastema fibroblast population dynamics.
Differential proportion analysis of fibroblast clusters parsed by regeneration profile where clusters in (A) have no significant population dynamics between blastema stages and unamputated. (B) Cells in clusters 2, 4, 6, and 12 are significantly depleted as compared to unamputated and (C) cells in clusters 0, 3, 10, and 11 are enriched during regeneration as compared to unamputated. Significance values are as follows: * denotes p<0.05, ** denotes p<0.01, *** denotes p<0.001. All p-values are presented in Table S6. (D) Subset of genes enriched in blastema stages as compared to unamputated. Gray depicts low expression and dark purple is high expression; small circles depict a low percentage of cells and large circles depict a high percentage. (E) Violin plots of genes in (D) at four blastema stages as compared to unamputated. Black points represent individual cells and the colored curve shows the distribution of cells at a given expression level.
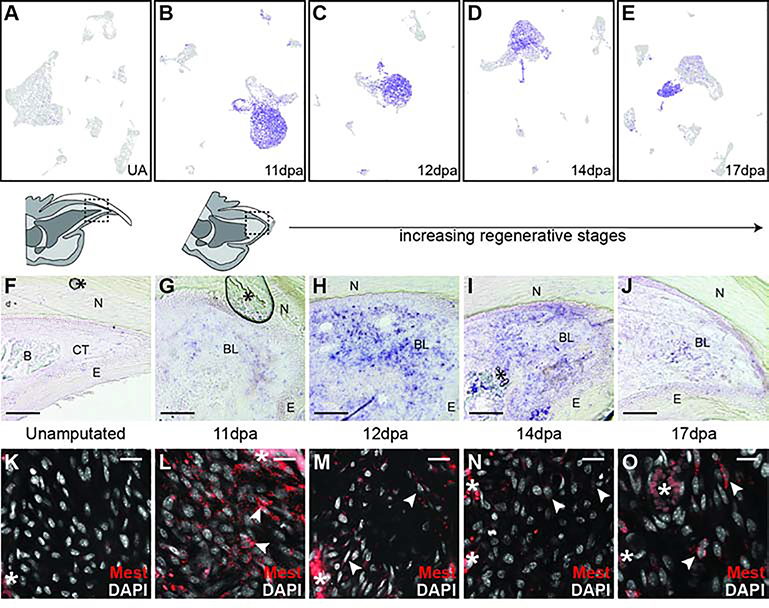
To validate these regeneration specific markers we focused on Mest, a gene expressed in mesenchymal tissues during embryonic development (Kaneko-Ishino et al., 1995). To determine the distribution of Mest expressing cells within the regenerating mouse digit tip we utilized RNA in situ hybridization. The Mest antisense RNA probe revealed the expected expression domains, including tongue and vertebrae, on control E12.5 embryonic mouse sections (Figure S7A–C). No significant Mest expression was found on unamputated digit tip sections (Figure 7F) and appeared comparable to Mest sense RNA control probe hybridization on unamputated digits (Figure S7E). In contrast, at 11dpa Mest expression is found scattered throughout blastema cells which is not seen for sense RNA probe 11dpa controls (Figure 7G and Figure S7D). Heterogeneous Mest blastema expression becomes even more pronounced at 12dpa and 14 dpa, then begins to decrease and become centrally restricted at 17dpa (Figure 7H–J). This expression was confirmed by Mest RNA FISH (Figure 7K–O). These in situs validate our computational analysis and establish Mest as a novel regeneration-specific marker of mouse digit tip blastema.
Figure 7. Mest expression during digit tip regeneration.
(A-E) UMAP feature plots of Mest expression in all digit tip cells in UA, 11dpa, 12dpa, 14dpa, and 17dpa, respectively. Purple marks high expression and gray marks low expression; plots refer to Figures S5, S4, S3, S2, and 1, respectively. (F-J) DIG-labeled RNA section in situ hybridization for Mest on unamputated and regenerating digit tips. (F) Unamputated digit tip with orientation shown by schematic above. (G) 11dpa with region of the blastema depicted in the schematic above. Additional regenerative stages include (H) 12dpa, (I) 14dpa, and (J) 17dpa. Asterisks (*) denote artifacts from coverslipping. Abbreviations: (N) nail, (CT) connective tissue, (E) epithelium, (B) bone, (BL) blastema. Scale bar = 100μm. (K-O) HCR RNA FISH for Mest. (K) Mesenchymal region in the unamputated digit tip. (L-O) Mid blastemal region in (L) 11dpa, (M) 12dpa, (N) 14dpa, and (O) 17dpa blastemas. Arrows denote examples of Mest positive cells. Asterisks (*) denote blood vessel/RBC autofluorescence. Scale bar = 20μm.
DISCUSSION
Historically, the blastema has been described as a collection of proliferative and homogeneous cells that give rise to the regenerated tissue (Hay and Fischman, 1961). Based on this description, we would expect there to be a high proportion of actively dividing cells in the blastema. We find a dividing fibroblast cluster in 11, 12, and 14dpa blastema stages that is depleted by 17dpa and not found at all in the unamputated digit tip. These dividing fibroblasts account for less than 10% of the total cells captured, which is consistent with previous results using EdU (Johnston et al., 2016). This challenges the idea of the blastema as a highly proliferative structure and suggests that relatively few proliferative cells are needed to support regeneration once the blastema is formed.
The classical assessment of the blastema as homogeneous was based on cell morphology in the regenerating newt limb (Hay and Fischman, 1961), however recent studies of digit tip regeneration use genetic lineage tracing to collectively conclude that the digit tip blastema contains progenitors that are heterogeneous in cell-type and lineage restricted (Johnston et al., 2016; Lehoczky et al., 2011; Rinkevich et al., 2011). While these studies clearly demonstrate the blastema is not pluripotent across tissue germ layers, multipotency within germ layers was never formally addressed likely due to the tissue-specificity limitations of the available genetic alleles. Our single cell RNAseq analysis reveals that all of the defined cell populations of the unamputated digit tip are already present in the 11dpa blastema, validating both the lineage restriction and heterogeneity of the blastema as defined by genetic lineage analyses (Lehoczky et al., 2011; Rinkevich et al., 2011). This regenerative dataset allows for an unbiased view of the cell types within the mouse digit tip blastema, and includes several cell types that have already been described during digit tip regeneration including Schwann cells, macrophages, neutrophils, endothelial cells, osteoblasts, and fibroblasts (Johnston et al., 2016; Lehoczky et al., 2011; Rinkevich et al., 2011; Simkin et al., 2017). Importantly, our clustered data reveals subpopulations and more detailed gene expression associated with these previously reported populations, including two Schwann cell populations, three macrophage populations, and 14 distinct fibroblast populations (Figures 3B and 5A). Our data also provide insight into cell types that have not previously been described during digit tip regeneration, including T cells, monocytes, pre-osteoclasts, VSM cells, and lymphatic endothelium. From the standpoint of epimorphic regeneration, this adds considerable information to our understanding of the number of unique cell types participating in digit tip regeneration. From an experimental standpoint, we now have access to specific genetic markers for each of these individual cell types to refine future in vivo experimentation.
Integration of our longitudinal regenerative data set reveals that a signature of unamputated digit tip cell types exists in the early blastema. Importantly, these are not necessarily identical cell populations and can instead be related cells in distinct cell states (Morris, 2019). With trajectory analysis, we predict differentiation from monocytes to macrophages equally at all regenerative stages, however we do find more blastema cells in the pre-osteoclast lineage than are found in the homeostatic digit tip (Figure S7G). This likely indicates that our analysis missed the post-amputation macrophage response which occurs prior to the emergence of the blastema (Simkin et al., 2017). Conversely, our data find discrete clusters of vascular related cells (VSM, vascular endothelium, and lymphatic endothelium) taking on distinct cell states throughout digit tip regeneration (Figure 4). Our analysis provides a refined view of these tissue-specific differentiating cells; for example, a canonical cell-type specific marker such as Pecam1 would label all vascular endothelial cells, whereby our data details genes and timing of emergence of different populations potentially useful for experimental access to vascular endothelial progenitors (Figure 4, Egfl7) or terminally differentiated cells (Figure 4, Rnd1).
A similar analysis with the digit tip fibroblast and bone populations enriches our previous understanding of heterogeneity and lineage restriction within the connective tissue and skeletal lineages of the regenerating digit tip. The extensive fibroblastic heterogeneity seemed unprecedented given the limited number of mesenchymally-derived tissues within the digit tip regenerate, which includes bone and tendon but not cartilage or muscle. This may suggest that only a portion of the fibroblast populations are progenitors (mesenchymal stem cells (MSCs)) differentiating into tissue-specific lineages, whereby the remaining populations might function as niche fibroblasts for ECM production, chemotaxis, etc. Trajectory analysis with these cells indeed suggests multiple tissue-specific lineages, including osteoprogenitors into bone, as well as MSCs into tendon (Figure 5). From this analysis it is not clear if these progenitors can transdifferentiate between skeletal lineages (Figure 5, clusters 1 and 2). This analysis also underscores the importance of re-visiting conclusions from previous fibroblastic genetic lineage analyses, as it is likely that these cre alleles (ex. Msx1 or Prrx1) mark the majority of our newly defined fibroblastic clusters, ultimately limiting the conclusions about transdifferentiation that can be drawn (Lehoczky et al., 2011; Rinkevich et al., 2011).
Beyond lineage restriction and heterogeneity, our data offer new insight into the molecular biology of digit tip regeneration. Differential gene expression analysis between blastema cells and homeostatic digit cells enabled us to identify markers of regenerating fibroblasts (Figure 6 and Table S7). We found several such markers of blastemal fibroblasts that are upregulated in clusters associated with regeneration and not the quiescent digit tip, including some associated with inflammation (Ccl2, Cxcl2) and some that regulate extracellular matrix (Matn4, Mmp13). The gene with the most dramatic change in expression from unamputated digit tip to blastema is Mest. The molecular function of Mest is not known, but it bears resemblance to the α/β hydrolase family of enzymes and is important for embryonic growth (Lefebvre et al., 1998). Intriguingly, Mest has been associated with other regenerative models, in particular the regeneration of adipocytes and hair follicles following skin wounding, where it is thought to be a marker of de-differentiated fibroblasts that differentiate into myofibroblasts (Guerrero-Juarez et al., 2019). The role of Mest in digit tip regeneration needs to be explored in vivo. It will be important to determine whether Mest-expressing cells are MSCs or de-differentiated fibroblasts that can transdifferentiate into multiple mesenchymal lineages or whether these cells are regeneration-specific fibroblasts that do not contribute to a tissue lineage, but instead provide niche factors. These findings can give insight into inducing epimorphic regeneration in other mammalian tissues.
This work presents extensive new and refined data for the regenerating mouse digit tip. Moving forward, much experimental work is required to determine which of these cell types and genes are necessary for regeneration and what molecular role they play. Studies on the necessity and role of Schwann cells and macrophages exemplify the types of focused experiments needed to put this comprehensive digit tip cell atlas into biological context (Johnston et al., 2016; Simkin et al., 2017). Importantly, our study cannot conclusively define the origin of blastema cells and whether they arise via de-differentiation of terminally differentiated cells or whether they are derived from tissue-resident progenitor cells. Our data suggest that both could be true, depending on the lineage. For instance, macrophages in the blastema appear to originate from resident monocytes (Figure S7B), whereby vascular cells and at least a subset of fibroblasts may de-differentiate to form the blastema (Figures 4B and 5H). Future experiments, taking advantage of the markers defined in this work, are needed to formally distinguish between these mechanisms for each cell-type.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jessica Lehoczky (jlehoczky@bwh.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mice were housed in the Hale BTM specific pathogen free vivarium at Brigham and Women’s Hospital. All mouse breeding and surgery was performed in accordance with BWH IACUC approved protocols. All single cell RNA sequencing experiments used inbred wild-type FVB/NJ mice (JAX 001800), maintained in our colony. 6-week-old adult male mice were used for unamputated controls and digit tip amputation surgeries and subsequent blastema collection; 2 mice were used for each RNAseq time point (12 total hindlimb digits). The 11 days post amputation digits for all fluorescent RNA in situ experiments were derived from adult FVB/NJ mice of both sexes (JAX 001800). Colorimetric RNA in situ experiments utilized adult digit and embryo tissues of both sexes, derived from wild-type CD-1(ICR) mice (Charles River 022), and stored in frozen lab tissue stocks.
METHOD DETAILS
Mouse digit tip amputation surgery
Six-week-old adult male mice were used for unamputated controls and digit tip amputation surgeries and subsequent blastema collection; 2 mice were used for each time point (12 total hindlimb digits). Mice were anesthetized with inhaled isoflurane (1–2% in oxygen) and digits were visualized with a Leica MZ6 stereomicroscope. For each mouse, digits 2, 3, and 4 of both hindlimbs were amputated midway through the distal digit segment using a #11 disposable scalpel. Subcutaneous buprenorphine (0.05 mg/kg) was given peri- and post-operatively as analgesia. Post-surgical animals were housed individually. Mice were euthanized and digits were collected at 11, 12, 14, and 17 days post amputation for blastema collection.
Digit tip single cell isolation
For all regenerating digits, blastemas were microdissected from the digit tip while being visualized under a Leica M165FC stereomicroscope. To minimize collection of epithelial cells, the nail and associated epithelium was reflected and removed, leaving direct access to the blastema. The blastema was removed intact with super-fine forceps and placed into ice-cold PBS. Control unamputated digit tip samples were collected in a similar manner whereby the nail and associated epithelium was removed and the exposed digit tip bone and attached connective tissues were amputated with a #11 scalpel at a position comparable to all other digit tip amputations. These control digit tips were collected into ice-cold PBS and processed in parallel with the blastema samples. All tissues were enzymatically dissociated with trypsin (Thermo Fisher) (0.25%, 37°C for 1 hour), then with collagenase type I (Thermo Fisher) (0.65%, 37°C for 20 minutes), followed by manual trituration with a pipette. Red blood cells were lysed using ACK lysis buffer. Dissociated cells were washed, filtered, and resuspended in 0.4% BSA in PBS for cell counting on. All samples were adjusted to a concentration of 1,000 cells/uL for the single cell RNAseq pipeline.
Single cell capture, library construction and next generation sequencing
All single cell RNAseq experiments used the 10x Chromium commercially available transcriptomics platform (10x Genomics) implemented by the Brigham and Women’s Hospital Single Cell Genomics Core. Single cells were captured using the 10X system; the 12dpa blastema sample cDNA library was made with Single Cell 3’ v2 chemistry, and all other libraries (11dpa, 14dpa, 17dpa, and UA) were made with Single Cell 3’ v3 chemistry. Libraries were sequenced at the Dana Farber Cancer Institute Molecular Biology Core Facilities on the Illumina NextSeq 500 sequencing system.
Single cell clustering and differential expression analysis
Computationally intensive portions of this research were conducted on the O2 High Performance Computing Cluster, supported by the Research Computing Group at Harvard Medical School (http://rc.hms.harvard.edu) using R version 3.5.1 (R Core Team, 2018). 10x Genomics Cell Ranger software (version 3.0.0 for 12dpa, version 3.0.2 for all other samples) was used to convert raw BCL files to FASTQ files, align reads to the mouse mm10 transcriptome, filter low quality cells, and count barcodes and unique molecular identifiers (UMIs). The cell by gene matrices for each of the five datasets generated by Cell Ranger were individually imported to Seurat version 3.1.1 (Stuart et al., 2019), and cells with unusually high numbers of UMIs (possible doublets) or mitochondrial gene percent (possible dying cells) were filtered out. Gene counts were normalized using the LogNormalize method and highly variable genes selected for downstream analysis (variable feature selection described in Stuart et al., 2019). Data was scaled and principal components selected and adjusted for each experimental group of cells for dimensional reduction. Thresholding parameters were: percent of mitochondrial genes upper bound (%Mito), number of unique molecular identifiers (nUMI) lower and upper bounds, number of principal components (#pc), and resolution (res). Parameters for individual samples were as follows: 11dpa (%Mito = 25; nUMI_lower = 200; nUMI_upper = 5500; #pc = 16; res = 0.6), 12dpa (%Mito = 20; nUMI_lower = 200; nUMI upper = 4000; #pc = 16; res = 0.6), 14dpa (%Mito = 20; nUMI_lower = 200; nUMI_upper = 6500; #pc = 20; res = 0.6), 17dpa (%Mito = 25; nUMI_lower = 200; nUMI_upper = 6500; #pc = 20; res = 0.6), and UA (%Mito = 25; nUMI_lower = 200; nUMI_upper = 5000; #pc = 20; res = 0.6).
Cells were clustered using the standard Seurat workflow and visualized using uniform manifold approximation and projection (UMAP) (McInnes et al., 2018). Cluster markers were found using FindAllMarkers with the Wilcoxon rank sum test, with only.pos = TRUE, min.pct = 0.25, logfc.threshold = 0.25. For the blastema-enriched vs. blastema-depleted differential expression analysis, FindMarkers was run on the fibroblast only Seurat object with clusters determined to be expanded in the blastema (0, 3, 10, and 11) as ident.1 and clusters determined to be depleted in the blastema (2, 4, 6, and 12) as ident.2. All other parameters were default.
Cell doublet identification
Initial broad screening for doublets in each data set was performed via quality control processing in Seurat by UMI thresholding (see above). For specific detection of putative doublet cells, we implemented the DoubletFinder (McGinnis et al., 2019) package in R version 3.5.1 (R Core Team, 2018) as described in detail at https://github.com/chris-mcginnis-ucsf/DoubletFinder. The doublet rate used (the nExp parameter in DoubletFinder) was estimated from the 10x Chromium users guide and the number of cells captured, and is as follows: UA, 7.6%. 11dpa, 5.5%. 12dpa, 2.5%. 14dpa, 4.6%. 17dpa, 6.9%. The pK value was estimated using the mean-variance normalized bimodality coefficient for each data set and is as follows: UA, 0.02. 11dpa, 0.09. 12dpa, 0.3. 14dpa, 0.13. 17dpa, 0.26. All identified putative doublets were removed from the integrated data set prior to clustering.
Batch correction, dataset integration and sub-clustering
The cells for our five experimental samples were collected and processed on multiple days, potentially contributing to batch effects in the data. To minimize this, we used the integrate function in Seurat version 3.1.1 to cluster all cells from all samples together with 11dpa as the anchor data set with dims = 1:20 and all other parameters set to default (Butler et al., 2018; Stuart et al., 2019). The integrated dataset was then scaled and 30 principal components used for clustering with a resolution of 0.6 and visualized with UMAP. For sub-clustering of fibroblast and bone populations, fibroblast and bone clusters were subsetted from the integrated data set as Seurat objects and re-normalized. These objects were re-integrated in Seurat, again using 11dpa as the anchor data set and dims = 1:20, scaled, and clustered with 20 principal components and resolution 0.6 to reveal any subpopulations.
Cell trajectory analyses
The SPRING web interface (https://kleintools.hms.harvard.edu/tools/spring.html) (Weinreb et al., 2018) was used to generate reproducible, continuous k nearest neighbors force-directed graphs of cells in gene expression space. A gene by cell expression matrix, a file containing time point and Seurat cluster metadata for each cell, and a list of gene names was the input to the web interface. All parameters were left at default values. Blastema datasets (11dpa, 12dpa, 14dpa, and 17dpa) were projected onto the unamputated dataset to avoid batch effects. Qualitative analysis of trajectories was facilitated by overlaying Seurat cluster information, regenerative stage, or gene expression. Differential gene expression associated with lineage trajectory (Figure 4, 5, and S6) was assessed in SPRING. Only genes with Z-score >1.96 were analyzed.
RNA velocity analyses
The velocyto command line tool (version 0.17.17) (La Manno et al., 2018) was used to generate a loom file for each sample (11dpa, 12dpa, 14dpa, 17dpa, and UA) from the BAM file outputted by Cell Ranger (version 3.0.0 for 12dpa, version 3.0.2 for all other samples). The loom file contained spliced and unspliced read counts for each gene and was used as input to scVelo (version 0.1.20) (Bergen et al., 2019) which was implemented in Python (version 3.7.5). The data was filtered using default parameters and normalized. The dynamical model was used to estimate RNA velocities for each cell. Velocity stream plots for each sample were generated in scVelo and use UMAP (McInnes et al., 2018) to visualize cell position in gene space.
Section RNA in situ hybridization
Blastema stage regenerating mouse digit tips, and contralateral unamputated controls, were collected and fixed in 4% paraformaldehyde at 4°C overnight, followed by washing and decalcification in decalcifying solution lite (Sigma Aldrich) (40 minutes at room temperature). Digits were prepared for embedding with a 5% to 30% sucrose gradient over 3 days, embedded in OCT (Tissue-Tek), and sectioned at 20μm on a Leica CM3050S cryostat. E12.5 embryos used for probe controls were collected from CD1(ICR) timed pregnant females, followed by PFA fixation and sucrose/OCT embedding as above, with solution change times of 30 minutes. A Mest cDNA for in situ probe template was PCR amplified from E10.5 mouse limb bud random-primed cDNA library with primers 5’GCTCCAGAACCGCAGAATCA and 5’GGGAGGTAATACAGGGAGGC (Mesman et al., 2018). The cDNA was cloned into the pGEM-T easy vector (Promega) and sequenced to confirm identity. Antisense RNA probe and sense negative control probe were generated by SP6 or T7 in vitro transcription with digoxigenin-UTP (Sigma Aldrich). Section RNA in situ hybridization was performed as previously reported (Murtaugh et al., 1999), with proteinase K used at 3ug/mL (room temp, 10 minutes). All colorimetric digit tip in situ hybridized sections were developed for the same amount of time and were imaged on a Leica DM2000 LED microscope.
For HCR fluorescent in situ hybridization, sections were prepared for probe hybridization in the same manner as described above. Probes targeted Acan (NM_001361500.1), Aldh1a2 (NM_009022.4), Mest (NM_001252292.1), Mmp13 (NM_008607.2), and Scara5 (NM_028903.2). Probe hybridization and signal amplification using amplifier hairpins were performed in accordance with the HCR v3.0 protocol for sections (https://www.molecularinstruments.com/protocols). The TrueVIEW autofluorescence quenching kit (Vector Laboratories) was used to quench endogenous fluorescence, then sections were counterstained with DAPI at a concentration of 1 ng/μL. No probe control stains were produced by omitting addition of the probe but still incubating with the probe amplifier hairpin (Figure S7F–I). Sections were imaged as 5–7μm thick z-stacks with a 0.5μm interval on either a Zeiss LSM 800 or Zeiss LSM 880 confocal microscope. Maximum intensity projections of z-stacks were generated in ImageJ (version 1.51v 9) where red, green, and blue channels were adjusted independently for brightness and contrast.
QUANTIFICATION AND STATISTICAL ANALYSIS
Hierarchical and correlation analyses
The dendrogram of 11dpa cell populations was built in R version 3.5.1 (R Core Team, 2018) using Seurat version 3.1.1 (Butler et al., 2018; Stuart et al., 2019) command BuildClusterTree on the 11dpa Seurat object with default parameters. The dendrogram was visualized using PlotClusterTree in Seurat. For the correlation analysis, bulk transcriptomes for each cluster were calculated using AverageExpression in Seurat. Pearson correlations were calculated from the resulting gene x cluster expression matrix using R base function cor with method = “pearson”. The correlation matrix was visualized using the corrplot function from the corrplot library (Wei and Simko, 2017).
GO biological process category analysis
For GO category analysis of 11dpa fibroblast populations, cluster marker genes with adjusted p-value ≤ 0.05 and average log fold-change ≥ 0.5 were used as input to the PANTHER classification system web interface (http://pantherdb.org) (Mi et al., 2010; Thomas et al., 2003). The statistical overrepresentation test was used with the slim biological processes category, fisher’s exact test, and the Bonferroni correction for multiple hypothesis testing. All genes in the gene by cell matrix from the 11dpa Seurat object were used as the background set for the overrepresentation test.
Differential proportion analysis
Differential proportion analysis (Farbehi et al., 2019) was performed in R to statistically test for significant changes in cluster membership over regenerative time. Cluster membership tables were calculated in Seurat and the resulting table of cells in each cluster by time point was used in differential proportion analysis. In the first step, generateNull was used with n = 100,000 and p = 0.1 as in the original reference. Significance values were calculated for pairwise comparisons of each time point with every other time point and were corrected for multiple hypothesis testing with the Benjamini-Hochberg method in R (Benjamini and Hochberg, 1995). Significance values reported in figures are: p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
DATA AND CODE AVAILABILITY
All raw single cell RNAseq FASTQ files and cell by gene expression matrices from this project are available in the NCBI Gene Expression Omnibus, Dataset Accession GSE143888. No new computational tools were developed in this project, however the code for the usage of existing tools as detailed above is available upon request.
Supplementary Material
Table S1 [Spreadsheet]
Cell cluster differential gene expression by regenerative stage (Relates to Figures 1 and S2–S5)
Differential gene expression analysis output from the FindAllMarkers function in Seurat. Each tab contains data from discrete regenerative stages: (A) 11dpa, (B) 12 dpa, (C) 14dpa, (D) 17dpa, and (E) unamputated. Column headers are: gene (NCBI gene ID), p-val (unadjusted p-value), avg logFC (average log fold-change among all cell clusters at this stage), pct.1 (percentage of cells in this cluster with this gene expression), pct.2 (percentage of cells in all other clusters with this gene expression), adj p-val (Bonferroni corrected p-value), cluster (cell cluster number on associated UMAP plot), cell type (cell cluster associated cell type assigned by literature review of most significant genes).
Table S3 [Spreadsheet]
Integrated data set cell cluster differential gene expression (Relates to Figure 3)
Differential gene expression analysis of 11dpa, 12dpa, 14dpa, 17dpa, and unamputated integrated data set. Output is from the FindAllMarkers function in Seurat. Column headers are: gene (NCBI gene ID), p-val (unadjusted p-value), avg logFC (average log fold-change among all cell clusters at this stage), pct.1 (percentage of cells in this cluster with this gene expression), pct.2 (percentage of cells in all other clusters with this gene expression), adj p-val (Bonferroni corrected p-value), cluster (cell cluster number on associated UMAP plot), cell type (cell cluster associated cell type assigned by literature review of most significant genes).
Table S5 [Spreadsheet]
Differential gene expression from all-stage integrated and re-clustered fibroblasts and bone (Relates to Figure 5)
Differential gene expression analysis from clustering of only fibroblast and bone cells of 11dpa, 12dpa, 14dpa, 17dpa, and unamputated digit tip data. Output is from the FindAllMarkers function in Seurat. Column headers are: gene (NCBI gene ID), p-val (unadjusted p-value), avg logFC (average log fold-change among all cell clusters at this stage), pct.1 (percentage of cells in this cluster with this gene expression), pct.2 (percentage of cells in all other clusters with this gene expression), adj p-val (Bonferroni corrected p-value), cluster (cell cluster number on associated UMAP plot (Figure 5A)), cell type (cell cluster associated cell type assigned by literature review of most significant genes).
Table S7 [Spreadsheet]
Differential gene expression of blastema-enriched versus blastema-depleted cell clusters (Relates to Figure 6)
Differential gene expression analysis from defined blastema-enriched clusters (Figure 6C) versus blastema-depleted clusters (Figure 6B). Output is from the FindAllMarkers function in Seurat. Column headers are: gene (NCBI gene ID), p-val (unadjusted p-value), avg logFC (average log fold-change among all cell clusters at this stage), pct.1 (percentage of cells in blastema-enriched clusters with detected expression of the gene), pct.2 (percentage of cells in blastema-depleted clusters with detected expression of the gene), and adj p-val (Bonferroni corrected p-value).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Digoxigenin-AP, Fab fragments | Sigma Aldrich | Cat# 11093274910; RRID:AB_2734716 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Embryos and adult digit tips from CD1 (ICR) mice, and adult digit tips from FVB/NJ mice | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Trypsin | Thermo Fisher | Cat# 15090046 |
| Collagenase Type I | Thermo Fisher | Cat# 17018029 |
| ACK Lysing Buffer | Thermo Fisher | Cat# A1049201 |
| DIG RNA 10x labeling mix | Sigma Aldrich | Cat# 11277073910 |
| Proteinase K | Sigma Aldrich | Cat# 3115879001 |
| SP6 RNA polymerase | Sigma Aldrich | Cat# 10810274001 |
| T7 RNA polymerase | Sigma Aldrich | Cat# 10881767001 |
| Critical Commercial Assays | ||
| Chromium Single Cell 3′ Library Construction Kit, v2 chemistry | 10X | N/A (No longer available) |
| Chromium Single Cell 3′ Library Construction Kit, v3 chemistry | 10X | Cat# 1000078 |
| Hybridization chain reaction v 3.0 probe hybridization buffer, probe wash buffer, and amplification buffer | Molecular Instruments | https://www.molecularinstruments.com/ |
| HCR Probe Set: Mouse Acan (Genbank: NM_001361500.1) B3 hairpin compatible, 20 probe sets | Molecular Instruments | https://www.molecularinstruments.com/ |
| HCR Probe Set: Mouse Aldh1a2 (Genbank: NM_009022.4) B5 hairpin compatible, 20 probe sets | Molecular Instruments | https://www.molecularinstruments.com/ |
| HCR Probe Set: Mouse Mest (Genbank: NM_001252292.1) B4 hairpin compatible, 20 probe sets | Molecular Instruments | https://www.molecularinstruments.com/ |
| HCR Probe Set: Mouse Mmp13 (Genbank: NM_008607.2) B2 hairpin compatible, 20 probe sets | Molecular Instruments | https://www.molecularinstruments.com/ |
| HCR Probe Set: Mouse Scara5 (Genbank: NM_028903.2) B4 hairpin compatible, 20 probe sets | Molecular Instruments | https://www.molecularinstruments.com/ |
| HCR amplifier B2, Alexa Fluor 647 label | Molecular Instruments | https://www.molecularinstruments.com/ |
| HCR amplifier B3, Alexa Fluor 594 label | Molecular Instruments | https://www.molecularinstruments.com/ |
| HCR amplifier B4, Alexa Fluor 594 label | Molecular Instruments | https://www.molecularinstruments.com/ |
| HCR amplifier B4, Alexa Fluor 647 label | Molecular Instruments | https://www.molecularinstruments.com/ |
| HCR amplifier B5, Alexa Fluor 647 label | Molecular Instruments | https://www.molecularinstruments.com/ |
| TrueVIEW Autofluorescence Quenching Kit | Vector Laboratories | Cat# SP-8400 |
| Deposited Data | ||
| Raw scRNA-seq data sets for adult digit tip blastemas at 11dpa, 12dpa, 14dpa, or 17dpa and the unamputated digit tip | This paper | GEO: GSE143888 |
| Mouse reference genome NCBI build 38, GRCm38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/genome/52 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Mouse: CD1(ICR) | Charles River Laboratories | Cat# 022 |
| Mouse: FVB/NJ | The Jackson Laboratory | Cat# JAX:001800 |
| Oligonucleotides | ||
| Primers for Mest cDNA amplification: 5’GCTCCAGAACCGCAGAATCA (Forward) and 5’GGGAGGTAATACAGGGAGGC (Reverse) | Mesman et al., 2018 | N/A |
| Recombinant DNA | ||
| Software and Algorithms | ||
| Cell Ranger (Multiple versions) | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation; RRID:SCR_017344 |
| R (Version 3.5.1) | R Core Team, 2018 | https://www.r-project.org/; RRID:SCR_001905 |
| Seurat (Version 3.1.1) | Stuart, Butler et al. 2018; Butler et al. 2018 | https://satijalab.org/seurat/; RRID:SCR_016341 |
| velocyto (Version 0.17.17) | La Manno et al. 2018 | https://velocyto.org/ |
| scVelo (Version 0.1.20) | Bergen et al. 2019 | https://scvelo-notebooks.readthedocs.io/index.html |
| Doublet finder (Version 2.0) | McGinnis et al. 2018 | https://github.com/chris-mcginnis-ucsf/DoubletFinder |
| SPRING | Weinreb et al. 2018 | https://kleintools.hms.harvard.edu/tools/spring.html |
| PANTHER | Mi et al., 2010; Thomas et al., 2003 | http://pantherdb.org; RRID:SCR_004869 |
| Python (Version 3.7.5) | Python Software Foundation | https://www.python.org/; RRID:SCR_008394 |
| Zen Image Acquisition and Processing software (Multiple versions) | Zeiss Microscope | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html; RRID:SCR_013672 |
| Adobe Illustrator CC | Adobe Systems | https://www.adobe.com/products/illustrator.html; RRID:SCR_010279 |
| Adobe Photoshop CC | Adobe Systems | https://www.adobe.com/products/photoshop.html; RRID:SCR_014199 |
| ImageJ (Version 1.51v 9) | NIH | https://imagej.net/; RRID:SCR_003070 |
| Differential proportion analysis | Farbehi et al. 2019 | N/A |
| Other | ||
TABLE WITH EXAMPLES FOR AUTHOR REFERENCE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Snail | Cell Signaling Technology | Cat#3879S; RRID: AB_2255011 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T9026; RRID: AB_477593 |
| Rabbit polyclonal anti-BMAL1 | This paper | N/A |
| Bacterial and Virus Strains | ||
| pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene AAV5; 44361-AAV5 |
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | Hope Center Viral Vectors Core | N/A |
| Cowpox virus Brighton Red | BEI Resources | NR-88 |
| Zika-SMGC-1, GENBANK: KX266255 | Isolated from patient (Wang et al., 2016) | N/A |
| Staphylococcus aureus | ATCC | ATCC 29213 |
| Streptococcus pyogenes: M1 serotype strain: strain SF370; M1 GAS | ATCC | ATCC 700294 |
| Biological Samples | ||
| Healthy adult BA9 brain tissue | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat#UMB1455 |
| Human hippocampal brain blocks | New York Brain Bank | http://nybb.hs.columbia.edu/ |
| Patient-derived xenografts (PDX) | Children’s Oncology Group Cell Culture and Xenograft Repository | http://cogcell.org/ |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MK-2206 AKT inhibitor | Selleck Chemicals | S1078; CAS: 1032350-13-2 |
| SB-505124 | Sigma-Aldrich | S4696; CAS: 694433-59-5 (free base) |
| Picrotoxin | Sigma-Aldrich | P1675; CAS: 124-87-8 |
| Human TGF-β | R&D | 240-B; GenPept: P01137 |
| Activated S6K1 | Millipore | Cat#14-486 |
| GST-BMAL1 | Novus | Cat#H00000406-P01 |
| Critical Commercial Assays | ||
| EasyTag EXPRESS 35S Protein Labeling Kit | Perkin-Elmer | NEG772014MC |
| CaspaseGlo 3/7 | Promega | G8090 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE63473 |
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Nanog STILT inference | This paper; Mendeley Data | http://dx.doi.org/10.17632/wx6s4mj7s8.2 |
| Affinity-based mass spectrometry performed with 57 genes | This paper; and Mendeley Data | Table S8; http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Experimental Models: Cell Lines | ||
| Hamster: CHO cells | ATCC | CRL-11268 |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Norbert Perrimon | FlyBase: FBtc0000181 |
| Human: Passage 40 H9 ES cells | MSKCC stem cell core facility | N/A |
| Human: HUES 8 hESC line (NIH approval number NIHhESC-09-0021) | HSCI iPS Core | hES Cell Line: HUES-8 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: Strain BC4011: srl-1(s2500) II; dpy-18(e364) III; unc-46(e177)rol-3(s1040) V. | Caenorhabditis Genetics Center | WB Strain: BC4011; WormBase: WBVar00241916 |
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1]; P{TRiP.HMS00609}attP2 | Bloomington Drosophila Stock Center | BDSC:34393; FlyBase: FBtp0064874 |
| S. cerevisiae: Strain background: W303 | ATCC | ATTC: 208353 |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | JAX: 006494 |
| Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J | The Jackson Laboratory | RRID: IMSR_JAX:008471 |
| Zebrafish: Tg(Shha:GFP)t10: t10Tg | Neumann and Nuesslein-Volhard, 2000 | ZFIN: ZDB-GENO-060207-1 |
| Arabidopsis: 35S::PIF4-YFP, BZR1-CFP | Wang et al., 2012 | N/A |
| Arabidopsis: JYB1021.2: pS24(AT5G58010)::cS24:GFP(-G):NOS #1 | NASC | NASC ID: N70450 |
| Oligonucleotides | ||
| siRNA targeting sequence: PIP5K I alpha #1: ACACAGUACUCAGUUGAUA | This paper | N/A |
| Primers for XX, see Table SX | This paper | N/A |
| Primer: GFP/YFP/CFP Forward: GCACGACTTCTTCAAGTCCGCCATGCC | This paper | N/A |
| Morpholino: MO-pax2a GGTCTGCTTTGCAGTGAATATCCAT | Gene Tools | ZFIN: ZDB-MRPHLNO-061106-5 |
| ACTB (hs01060665_g1) | Life Technologies | Cat#4331182 |
| RNA sequence: hnRNPA1_ligand: UAGGGACUUAGGGUUCUCUCUAGGGACUUAGGGUUCUCUCUAGGGA | This paper | N/A |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clonetech | Cat#632162 |
| Plasmid: GFP-Nito | This paper | N/A |
| cDNA GH111110 | Drosophila Genomics Resource Center | DGRC:5666; FlyBase:FBcl0130415 |
| AAV2/1-hsyn-GCaMP6- WPRE | Chen et al., 2013 | N/A |
| Mouse raptor: pLKO mouse shRNA 1 raptor | Thoreen et al., 2009 | Addgene Plasmid #21339 |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| ICS algorithm | This paper; Mendeley Data | http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Other | ||
| Sequence data, analyses, and resources related to the ultra-deep sequencing of the AML31 tumor, relapse, and matched normal. | This paper | http://aml31.genome.wustl.edu |
| Resource website for the AML31 publication | This paper | https://github.com/chrisamiller/aml31SuppSite |
HIGHLIGHTS.
Unbiased atlas of cell types throughout mouse digit tip regeneration
The early blastema contains all broad cell lineages found in the unamputated digit tip
Fibroblasts are the most abundant and heterogeneous population
Dynamics of distinct fibroblast populations reveal regeneration specific markers
ACKNOWLEDGEMENTS
We appreciate helpful conversations with Michael Brenner and Soumya Raychaudhuri. We thank Caleb Weinreb and Allon Klein for help with SPRING data input. We acknowledge the BWH confocal microscopy core. We are grateful for imaging technical assistance from Anne Golding, as well as constructive comments from all members of the Lehoczky Lab. This work was supported by the Eunice Kennedy Shriver National Institute of Child and Human Development (R03HD093922 and R21HD097405 to J.A.L), and the Osher Center for Integrative Medicine; J.A.L is also supported by the American Foundation for Surgery of the Hand, the William F. Milton Fund, the Stepping Strong Center for Trauma Innovation, and funds from BWH Department of Orthopedic Surgery.
Footnotes
DISCLOSURES:
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Albelda SM, Oliver PD, Romer LH, and Buck CA (1990). EndoCAM: a novel endothelial cell-cell adhesion molecule. J. Cell Biol 110, 1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2019). Generalizing RNA velocity to transient cell states through dynamical modeling. BioRxiv 820936. [DOI] [PubMed] [Google Scholar]
- Brandau O, Meindl A, Fässler R, and Aszódi A (2001). A novel gene, tendin, is strongly expressed in tendons and ligaments and shows high homology with chondromodulin-I. Dev. Dyn 221, 72–80. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calve S, Odelberg SJ, and Simon H-G (2010). A transitional extracellular matrix instructs cell behavior during muscle regeneration. Dev. Biol 344, 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagnolo L, Leahy A, Chitnis S, Koschnick S, Fitch MJ, Fallon JT, Loskutoff D, Taubman MB, and Stuhlmann H (2005). EGFL7 is a chemoattractant for endothelial cells and is up-regulated in angiogenesis and arterial injury. Am. J. Pathol 167, 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BM (1978). Types of morphogenetic phenomena in vertebrate regenerating systems. Integr. Comp. Biol 18, 869–882. [Google Scholar]
- Carr MJ, Toma JS, Johnston APW, Steadman PE, Yuzwa SA, Mahmud N, Frankland PW, Kaplan DR, and Miller FD (2018). Mesenchymal Precursor Cells in Adult Nerves Contribute to Mammalian Tissue Repair and Regeneration. Cell Stem Cell 1–17. [DOI] [PubMed] [Google Scholar]
- Dent JN (1962). Limb regeneration in larvae and metamorphosing individuals of the South African clawed toad. J. Morphol 110, 61–77. [DOI] [PubMed] [Google Scholar]
- Dolan CP, Yan M, Zimmel K, Yang T, Leininger E, Dawson LA, and Muneoka K (2019). Axonal regrowth is impaired during digit tip regeneration in mice. Dev. Biol 445, 237–244. [DOI] [PubMed] [Google Scholar]
- Farbehi N, Patrick R, Dorison A, Xaymardan M, Janbandhu V, Wystub-Lis K, Ho JWK, Nordon RE, and Harvey RP (2019). Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife 8, 1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando WA, Leininger E, Simkin J, Li N, Malcom CA, Sathyamoorthi S, Han M, and Muneoka K (2012). Wound healing and blastema formation in regenerating digit tips of adult mice. 350, 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers GP, Sanor LD, and Crews CM (2017). Lineage tracing of genome-edited alleles reveals high fidelity axolotl limb regeneration. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Furth R, and Cohn ZA (1968). The origin and kinetics of mononuclear phagocytes. J. Exp. Med 128, 415–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargioli C, and Slack JMW (2004). Cell lineage tracing during Xenopus tail regeneration. Development 131, 2669–2679. [DOI] [PubMed] [Google Scholar]
- Gerber T, Murawala P, Knapp D, Masselink W, Schuez M, Hermann S, Gac-Santel M, Nowoshilow S, Kageyama J, Khattak S, et al. (2018). Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science (80-. ). 0681, eaaq0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizard F, Nomiyama T, Zhao Y, Findeisen HM, Heywood EB, Jones KL, Staels B, and Bruemmer D (2008). The PPARalpha/p16INK4a pathway inhibits vascular smooth muscle cell proliferation by repressing cell cycle-dependent telomerase activation. Circ. Res 103, 1155–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss RJ (1961). Experimental investigation of morphogenesis in the growing antler. J. Embryol. Exp. Morphol 9, 342–354. [PubMed] [Google Scholar]
- Guerrero-Juarez CF, Dedhia PH, Jin S, Ruiz-Vega R, Ma D, Liu Y, Yamaga K, Shestova O, Gay DL, Yang Z, et al. (2019). Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun 10, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Yang X, Lee J, Allan CH, and Muneoka K (2008). Development and regeneration of the neonatal digit tip in mice. Dev. Biol 315, 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay ED, and Fischman DA (1961). Origin of the blastema in regenerating limbs of the newt Triturus viridescens. An autoradiographic study using tritiated thymidine to follow cell proliferation and migration. Dev Biol 3, 26–59. [DOI] [PubMed] [Google Scholar]
- Illingworth CM (1974). Trapped fingers and amputated finger tips in children. J. Pediatr. Surg 9, 853–858. [DOI] [PubMed] [Google Scholar]
- Johnson SL, and Weston JA (1995). Temperature-sensitive mutations that cause stage-specific defects in zebrafish fin regeneration. Genetics 141, 1583–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston APW, Yuzwa SA, Carr MJ, Mahmud N, Storer MA, Krause MP, Jones K, Paul S, Kaplan DR, and Miller FD (2016). Dedifferentiated Schwann Cell Precursors Secreting Paracrine Factors Are Required for Regeneration of the Mammalian Digit Tip. Cell Stem Cell 19, 433–448. [DOI] [PubMed] [Google Scholar]
- Kaneko-Ishino T, Kuroiwa Y, Miyoshi N, Kohda T, Suzuki R, Yokoyama M, Viville S, Barton SC, Ishino F, and Azim SuranP M (1995). Peg1 /Mest imprinted gene on chromosome 6 identified by eDNA subtraction hybridization. [DOI] [PubMed]
- Kim J-H, Qu A, Reddy JK, Gao B, and Gonzalez FJ (2014). Hepatic oxidative stress activates the Gadd45b gene by way of degradation of the transcriptional repressor STAT3. Hepatology 59, 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn C, and Augustin HG (2015). Mechanisms of Vessel Pruning and Regression. Dev. Cell 34, 5–17. [DOI] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, and Tanaka EM (2009). Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60–65. [DOI] [PubMed] [Google Scholar]
- Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, and Azim Surani M (1998). Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat. Genet 20, 163–169. [DOI] [PubMed] [Google Scholar]
- Lehoczky J, and Tabin C (2015). Lgr6 marks nail stem cells and is required for digit tip regeneration. Proc. Natl. Acad. Sci. U. S. A 112, 13249–13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehoczky J, Robert B, and Tabin C (2011). Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc. Natl. Acad. Sci. U. S. A 108, 20609–20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh ND, Dunlap GS, Johnson K, Mariano R, Oshiro R, Wong AY, Bryant DM, Miller BM, Ratner A, Chen A, et al. (2018). Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nat. Commun 9, 5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Adams LD, Wang X, Pabon L, Schwartz SM, Sane DC, and Geary RL (2004). Regulator of G protein signaling 5 marks peripheral arterial smooth muscle cells and is downregulated in atherosclerotic plaque. J. Vasc. Surg 40, 519–528. [DOI] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero L, Simkin J, Sammarco M, and Muneoka K (2017). Fibroblast reticular cells engineer a blastema extracellular network during digit tip regeneration in mice. Regen. (Oxford, England) 4, 69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis CS, Murrow LM, and Gartner ZJ (2019). DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 8, 329–337.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Saul N, and Groβberger L (2018). UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. [Google Scholar]
- Mesman S, Krüse SJ, and Smidt MP (2018). Expression analyzes of early factors in midbrain differentiation programs. Gene Expr. Patterns 27, 8–15. [DOI] [PubMed] [Google Scholar]
- Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, and Thomas PD (2010). PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38, D204–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Uchiyama K, Imokawa Y, and Yoshizato K (1996). Cloning and characterization of cDNAs for matrix metalloproteinases of regenerating newt limbs. Proc. Natl. Acad. Sci 93, 6819–6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad KS, and Neufeld DA (2000). Denervation retards but does not prevent toetip regeneration. Wound Repair Regen 8, 277–281. [DOI] [PubMed] [Google Scholar]
- Mohammad KS, Day FA, and Neufeld DA (1999). Bone Growth is Induced by Nail Transplantation in Amputated Proximal Phalanges. Calcif. Tissue Int 65, 408–410. [DOI] [PubMed] [Google Scholar]
- Moiseeva EP, Javed Q, Spring EL, and de Bono DP (2000). Galectin 1 is involved in vascular smooth muscle cell proliferation. Cardiovasc. Res 45, 493–502. [DOI] [PubMed] [Google Scholar]
- Morgan TH (1901). Regeneration and liability to injury. Science (80-. ). 14, 235–248. [DOI] [PubMed] [Google Scholar]
- Morris SA (2019). The evolving concept of cell identity in the single cell era. [DOI] [PubMed]
- Müller AM, Hermanns MI, Skrzynski C, Nesslinger M, Müller KM, and Kirkpatrick CJ (2002). Expression of the endothelial markers PECAM-1, vWF, and CD34 in Vivo and in Vitro. Exp. Mol. Pathol 72, 221–229. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Chyung JH, and Lassar AB (1999). Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 13, 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld DA, and Zhao W (1995). Bone regrowth after digit tip amputation in mice is equivalent in adults and neonates. Wound Repair Regen. 3, 461–466. [DOI] [PubMed] [Google Scholar]
- Overton J (1963). Patterns of limb regeneration inXenopus laevis. J. Exp. Zool 154, 153–161. [DOI] [PubMed] [Google Scholar]
- Pearse RV 2nd, Esshaki D, Tabin CJ, and Murray MM (2009). Genome-wide expression analysis of intra- and extraarticular connective tissue. J. Orthop. Res 27, 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2018). R: A Language and Environment for Statistical Computing.
- Rinkevich Y, Lindau P, Ueno H, Longaker MT, and Weissman IL (2011). Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature 476, 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer R, Chyung JH, Murtaugh LC, Brent AE, Rosen V, Olson EN, Lassar A, and Tabin CJ (2001). Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 128, 3855–3866. [DOI] [PubMed] [Google Scholar]
- Simkin J, Sammarco MC, Marrero L, Dawson LA, Yan M, Tucker C, Cammack A, and Muneoka K (2017). Macrophages are required to coordinate mouse digit tip regeneration. Development 144, 3907–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallanzani L (1768). Prodromo di un’ opera da imprimersi sopra la riproduzioni animali, Giovanni Montanari, Modena. An Essay Anim. Reprod [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suehiro J, Kanki Y, Makihara C, Schadler K, Miura M, Manabe Y, Aburatani H, Kodama T, and Minami T (2014). Genome-wide approaches reveal functional vascular endothelial growth factor (VEGF)-inducible nuclear factor of activated T cells (NFAT) c1 binding to angiogenesis-related genes in the endothelium. J. Biol. Chem 289, 29044–29059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo M, Chou WC, Sun Q, Lee W, Rabbani P, Loomis C, Taketo MM, and Ito M (2013). Wnt activation in nail epithelium couples nail growth to digit regeneration. Nature 499, 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, and Narechania A (2003). PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, and Johnson SL (2011). Fate restriction in the growing and regenerating zebrafish fin. Dev. Cell 20, 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinarsky V, Atkinson DL, Stevenson TJ, Keating MT, and Odelberg SJ (2005). Normal newt limb regeneration requires matrix metalloproteinase function. Dev. Biol 279, 86–98. [DOI] [PubMed] [Google Scholar]
- Virolainen M (1968). Hematopoietic origin of macrophages as studied by chromosome markers in mice. J. Exp. Med 127, 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T, and Simko V (2017). R package “corrplot”: Visualization of a Correlation Matrix.
- Weinreb C, Wolock S, and Klein AM (2018). SPRING: A kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34, 1246–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-H, Tsai M-H, Ho C-C, Chen C-Y, and Lee H-S (2013a). De novo transcriptome sequencing of axolotl blastema for identification of differentially expressed genes during limb regeneration. BMC Genomics 14, 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wang K, Karapetyan A, Fernando WA, Simkin J, Han M, Rugg EL, and Muneoka K (2013b). Connective Tissue Fibroblast Properties Are Position-Dependent during Mouse Digit Tip Regeneration. PLoS One 8, e54764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Yan M, Simkin J, Ketcham PD, Leininger E, Han M, and Muneoka K (2014). Angiogenesis is inhibitory for mammalian digit regeneration. 33–46. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 [Spreadsheet]
Cell cluster differential gene expression by regenerative stage (Relates to Figures 1 and S2–S5)
Differential gene expression analysis output from the FindAllMarkers function in Seurat. Each tab contains data from discrete regenerative stages: (A) 11dpa, (B) 12 dpa, (C) 14dpa, (D) 17dpa, and (E) unamputated. Column headers are: gene (NCBI gene ID), p-val (unadjusted p-value), avg logFC (average log fold-change among all cell clusters at this stage), pct.1 (percentage of cells in this cluster with this gene expression), pct.2 (percentage of cells in all other clusters with this gene expression), adj p-val (Bonferroni corrected p-value), cluster (cell cluster number on associated UMAP plot), cell type (cell cluster associated cell type assigned by literature review of most significant genes).
Table S3 [Spreadsheet]
Integrated data set cell cluster differential gene expression (Relates to Figure 3)
Differential gene expression analysis of 11dpa, 12dpa, 14dpa, 17dpa, and unamputated integrated data set. Output is from the FindAllMarkers function in Seurat. Column headers are: gene (NCBI gene ID), p-val (unadjusted p-value), avg logFC (average log fold-change among all cell clusters at this stage), pct.1 (percentage of cells in this cluster with this gene expression), pct.2 (percentage of cells in all other clusters with this gene expression), adj p-val (Bonferroni corrected p-value), cluster (cell cluster number on associated UMAP plot), cell type (cell cluster associated cell type assigned by literature review of most significant genes).
Table S5 [Spreadsheet]
Differential gene expression from all-stage integrated and re-clustered fibroblasts and bone (Relates to Figure 5)
Differential gene expression analysis from clustering of only fibroblast and bone cells of 11dpa, 12dpa, 14dpa, 17dpa, and unamputated digit tip data. Output is from the FindAllMarkers function in Seurat. Column headers are: gene (NCBI gene ID), p-val (unadjusted p-value), avg logFC (average log fold-change among all cell clusters at this stage), pct.1 (percentage of cells in this cluster with this gene expression), pct.2 (percentage of cells in all other clusters with this gene expression), adj p-val (Bonferroni corrected p-value), cluster (cell cluster number on associated UMAP plot (Figure 5A)), cell type (cell cluster associated cell type assigned by literature review of most significant genes).
Table S7 [Spreadsheet]
Differential gene expression of blastema-enriched versus blastema-depleted cell clusters (Relates to Figure 6)
Differential gene expression analysis from defined blastema-enriched clusters (Figure 6C) versus blastema-depleted clusters (Figure 6B). Output is from the FindAllMarkers function in Seurat. Column headers are: gene (NCBI gene ID), p-val (unadjusted p-value), avg logFC (average log fold-change among all cell clusters at this stage), pct.1 (percentage of cells in blastema-enriched clusters with detected expression of the gene), pct.2 (percentage of cells in blastema-depleted clusters with detected expression of the gene), and adj p-val (Bonferroni corrected p-value).
Data Availability Statement
All raw single cell RNAseq FASTQ files and cell by gene expression matrices from this project are available in the NCBI Gene Expression Omnibus, Dataset Accession GSE143888. No new computational tools were developed in this project, however the code for the usage of existing tools as detailed above is available upon request.