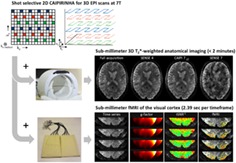
Abstract
To be able to examine dynamic and detailed brain functions, the spatial and temporal resolution of 7 T MRI needs to improve. In this study, it was investigated whether submillimeter multishot 3D EPI fMRI scans, acquired with high‐density receive arrays, can benefit from a 2D CAIPIRINHA sampling pattern, in terms of noise amplification (g‐factor), temporal SNR and fMRI sensitivity. High‐density receive arrays were combined with a shot‐selective 2D CAIPIRINHA implementation for multishot 3D EPI sequences at 7 T. In this implementation, in contrast to conventional inclusion of extra kz gradient blips, specific EPI shots are left out to create a CAIPIRINHA shift and reduction of scan time. First, the implementation of the CAIPIRINHA sequence was evaluated with a standard receive setup by acquiring submillimeter whole brain T2*‐weighted anatomy images. Second, the CAIPIRINHA sequence was combined with high‐density receive arrays to push the temporal resolution of submillimeter 3D EPI fMRI scans of the visual cortex. Results show that the shot‐selective 2D CAIPIRINHA sequence enables a reduction in scan time for 0.5 mm isotropic 3D EPI T2*‐weighted anatomy scans by a factor of 4 compared with earlier reports. The use of the 2D CAIPIRINHA implementation in combination with high‐density receive arrays, enhances the image quality of submillimeter 3D EPI scans of the visual cortex at high acceleration as compared to conventional SENSE. Both the g‐factor and temporal SNR improved, resulting in a method that is more sensitive to the fMRI signal. Using this method, it is possible to acquire submillimeter single volume 3D EPI scans of the visual cortex in a subsecond timeframe. Overall, high‐density receive arrays in combination with shot‐selective 2D CAIPIRINHA for 3D EPI scans prove to be valuable for reducing the scan time of submillimeter MRI acquisitions.
Keywords: 3D EPI, 7T, CAIPI, high resolution, receive arrays, shot selective, visual cortex
This study investigated whether submillimeter 3D EPI fMRI scans acquired with high‐density receive arrays can still benefit from a CAIPI sampling pattern, in terms of scan time per volume and (temporal) SNR. A shot‐selective 2D CAIPIRINHA acceleration scheme for multishot 3D EPI scans was implemented and evaluated with submillimeter 3D T2*‐weighted anatomical scans. When combining this implementation with high‐density receive coil arrays at 7 T, it enabled substantial reduction in scan time for submillimeter fMRI scans of the visual cortex.
Abbreviations used
- BOLD
blood oxygenation level‐dependent
- CAIPIRINHA
controlled aliasing in parallel imaging
- EPI
echo‐planar imaging
- FOV
field of view
- MB
multiband
- RF
radio frequency
- SENSE
sensitivity encoding
- tSNR
temporal signal‐to‐noise ratio
1. INTRODUCTION
The combination of both a high temporal resolution and a high spatial resolution is essential to examine dynamic and spatially detailed brain functions using functional MRI (fMRI). Within the cortex, detailed brain structures (columns, layers) play different roles with respect to different brain functions. To which brain function a detailed cortical structure contributes can vary on a small scale of less than 1 mm. Recent developments in fMRI acquisition methods and hardware technologies, particularly at high field (≥7 T), have enabled visualization of functional detail at a laminar or columnar level.1 However, in most studies, the required high spatial resolution comes at the cost of low temporal resolution or reduced coverage.2, 3 In general, fMRI measures neuronal function via associated changes in hemodynamics, most commonly using the blood oxygenation level‐dependent (BOLD) contrast. To reveal biologically relevant characteristics of the hemodynamic response at detailed spatial scale requires not only a high spatial resolution but also a high temporal resolution.4, 5, 6 Nowadays, despite the strong drive to image the cortex dynamically with high detail, fMRI with both a high temporal resolution (<1 second) and a high spatial resolution (<1 mm) combined, is rarely seen.
In pursuit of high‐resolution fMRI with a short scan time per volume, implementations of controlled aliasing in parallel imaging (CAIPIRINHA)7, 8, 9 in combination with simultaneous multislice methods (SMS) including multiband (MB), have been studied extensively in the recent literature.2, 10, 11 Most of these studies are aimed at 2D or multislice imaging. More recently, the fMRI field has advanced towards 3D imaging, because of the gains in SNR at small voxel sizes12, 13 and the time efficiency of the acquisition.14 Multishot 3D echo‐planar imaging (EPI) sequences are effective high‐resolution fMRI scans. Previous studies have been performed in which the benefits of 3D EPI sequences were combined with the benefits of 2D CAIPIRINHA.15, 16 By implementing additional gradient blips on the slice axis of the EPI sequence, the k‐space trajectory can be influenced, allowing the aliasing patterns caused by undersampling to be controlled. Overlapping aliasing patterns are avoided or moved outside the brain, which leads to reduced noise amplification and higher achievable acceleration factors compared with the traditional acceleration methods like SENSE17 and GRAPPA.18
Similarly, high‐density receive coil arrays also facilitate the use of high acceleration factors with reduced g‐factors at high resolution.19, 20, 21 For imaging the cortex, the benefits of using high‐density receive coil arrays are 2‐fold. Locally, at the periphery of the brain it results in an improved SNR. Second, it enhances the encoding performance, because the variation in spatial sensitivity is increased, which can be exploited by parallel imaging techniques to reduce scan time. Applying these high‐density receive coil arrays at ultrahigh field strengths results in an intrinsic SNR gain and an additional spatial variance of local B1 − fields.
A combination of high‐density receive coil arrays and a 2D CAIPIRINHA sampling pattern at a high field strength of 7 T could potentially increase the resolution even further. However, the use of high‐density receiver arrays often goes together with reduced field of view (FOV) acquisitions. Since there is less space around the brain in reduced FOV acquisitions, the difference between SENSE and CAIPIRINHA could become smaller. Furthermore, the high variation in spatial sensitivity of small coil elements in high‐density receive arrays is already sufficient to achieve high acceleration factors with the traditional parallel imaging methods such as SENSE.21 Can 2D CAIPIRINHA still significantly contribute to the already achievable high acceleration performance of high‐density receiver arrays?
This study investigates whether submillimeter multishot 3D EPI fMRI scans acquired with high‐density receive arrays can still benefit from a 2D CAIPIRINHA sampling pattern, in terms of noise amplification (g‐factor), temporal SNR and fMRI sensitivity. A straightforward implementation of shot‐selective 2D CAIPIRINHA for multishot 3D EPI sequences was developed, which, instead of adding extra gradients, leaves them out. First, the implementation of the sequence was evaluated by submillimeter T2*‐weighted 3D EPI anatomical imaging. Afterwards, the combination of high‐density receive arrays and 2D CAIPIRINHA was evaluated on its capability to accelerate submillimeter fMRI acquisitions.
2. SEQUENCE DESIGN
A shot‐selective 2D CAIPIRINHA sampling pattern was implemented for 3D EPI scans on a Philips 7 T platform. Several EPI shots of a multishot interleaved 3D EPI sequence were selectively skipped, resulting in a 2D CAIPIRINHA sampling pattern and a reduction in scan time (Figure 1). The EPI shots were selected in such a way that the acquired kz planes had varying ky start positions (a Δky offset), in correspondence with the targeted 2D CAIPIRINHA sampling pattern. Note that 2D CAIPIRINHA8 is typically used for volumetric 3D sequences, while CAIPIRINHA7 was originally introduced for slice‐selective 2D sequences. Blipped‐CAIPI22 applied the CAIPIRINHA concept to 2D EPI. It was later shown to be equivalent to 2D CAIPIRINHA in an “SMS 3D k‐space”.11, 23, 24 The concept of a shot‐selective 2D CAIPIRINHA for 3D EPI scans has been suggested before,16, 25 but here we implemented it for fMRI scans with high spatiotemporal resolution. The implementation is based on a multishot interleaved 3D EPI sequence,26 therefore no gradient blips on the kz axis during the EPI train were required, in contrast to other reports for single‐shot and kz‐segmented 3D EPI sequences, which also use gradient blips on the kz axis during the EPI train.15 Both the RF pulses and the kz gradients of the multishot 3D EPI sequence are left untouched, so there is no additional burden on the specific absorption rate (SAR) as can be the case when using MB RF pulses, or the use of extra kz‐gradients, as can be the case in blipped CAIPIRINHA acquisitions. The reconstruction was performed with an adapted SENSE reconstruction,17, 23 in contrast to the more commonly reported CAIPIRNIHA reconstructions in literature, such as GRAPPA22 and SENSE‐GRAPPA hybrid27, 28 reconstructions.
Figure 1.
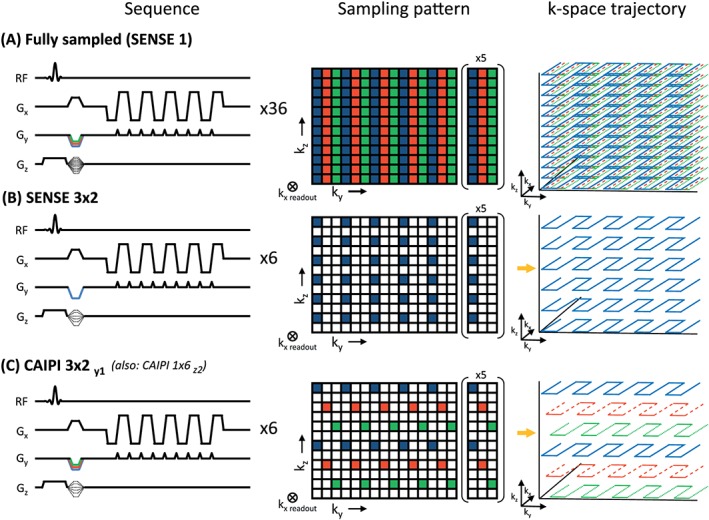
Sampling schemes of multishot 3D EPI sequences. The k‐space sampling patterns are displayed for (A) full acquisition, (B) SENSE 3 x 2 and (C) CAIPIRINHA 3 x 2 shift y1. Note that the fully sampled k‐space contains three shots (indicated in blue, red and green) per kz‐plane. When using SENSE 3 x 2, only one (blue) shot is acquired for every other kz‐plane. 2D CAIPIRINHA uses one shot per other kz‐plane as well, but alternates between the blue, red and green shots to reduce the g‐factor penalty
3. METHODS
Three healthy participants were scanned with a 7 T Achieva system (Philips, Best, the Netherlands). The participants gave informed consent, and the work was approved by the Medical Ethics Committee of the University Medical Center Utrecht. Before combining the implemented shot‐selective 2D CAIPIRINHA sequence with high‐density receive arrays, first the quality of the implementation was evaluated. This was done by performing T2*‐weighted 3D EPI anatomical imaging using a standard receive setup (details below). Afterwards, the combination of high‐density receive arrays and the 2D CAIPIRINHA was evaluated on its capability to accelerate submillimeter 3D EPI fMRI acquisitions, while maintaining low noise amplification (g‐factor) and high temporal SNR (tSNR). Prospectively undersampled datasets were acquired. Both receive setups (Figure 2) were connected to the same transmit setup, consisting of a dual channel transmit/receive birdcage coil (Nova Medical, USA).
Figure 2.
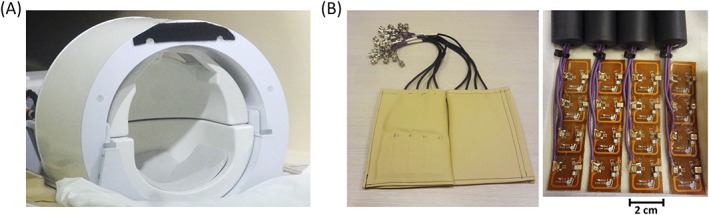
The two setups used for data acquisition. The 32‐channel headcoil (A) is used for the acquisition of whole brain of T2*‐weighted anatomy scans. The 2 x 16 channel high‐density surface coil arrays (B) are used for small FOV imaging of the visual cortex with a short scan time
3.1. Shimming and reference scan
B0 shimming of the brain was performed before acquiring the 3D EPI scans. A B0‐map was acquired, driving only the dual channel transmit/receive birdcage coil, and the following scan parameters: TE/TR = 1.54/4.0 ms, flip angle 10°, 3.75 x 3.75 x 3.75 mm3 voxels, 240 x 180 x 180 mm3 FOV, 48 slices and a total acquisition time of 18 seconds. Second‐order shimming parameters were calculated and applied to subsequent scans. After shimming, the same B0‐map was reacquired, this time including the calculated shimming values. As last preparation step, a (SENSE) coil sensitivity reference scan was acquired by successive signal reception with the volume T/R birdcage coil and the individual receive elements. The raw datasets of these scans were saved and used to calculate the coil sensitivity maps required for reconstruction. The coil sensitivity reference scan was a 3D gradient echo with the following scan parameters: TE/TR = 1.01/8.0 ms, flip angle 1°, 3 x 3 x 3 mm3 voxels, 240 x 240 x 240 mm3 FOV, 80 slices and a total acquisition time of 1.55 minutes.
3.2. Evaluation of sequence implementation, T2*‐weighted 3D EPI anatomical imaging
Whole brain T2*‐weighted multishot 3D EPI anatomical scans were acquired with a standard 32‐channel headcoil (Nova Medical, USA).29 The headcoil consists of 32 receive elements shaped in a dome‐like structure around the head (Figure 2A). The coil elements are large square loops of ~ 5 x 4 cm2. First, scans with 1 mm isotropic resolution were acquired to evaluate the implementation of the shot‐selective 2D CAIPIRINHA sequence. Fully sampled datasets were compared with prospectively undersampled datasets in transverse and coronal views. Second, scans with 0.5 mm isotropic resolution were acquired to compare the implementation in relation to the SENSE method. The individual scan parameters of the T2*‐weighted 3D EPI anatomical scans can be found in Table 1. The parameters of T2*‐weighted anatomy scans were based on Zwanenburg et al30 and consisted of the following: a transverse slice orientation (readout in anterior–posterior direction), fat suppression by spectral presaturation with inversion recovery (SPIR) using a frequency offset of 250 Hz, a sinc‐shaped RF excitation pulse of 0.65 ms (12 kHz bandwidth), no partial Fourier, a readout oversampling factor of 2, low peripheral nerve stimulation (dB/dt less than 60% of the threshold). In the figures displaying results, the CAIPIRINHA sampling patterns are denoted in the abbreviated format: for example, CAIPI 5y2, representing an acceleration factor of 5 and a shift of 2 points along the ky direction. The corresponding full specification can be found in Table 1, in the format CAIPI 5 x 1 y2.
Table 1.
Acquisition parameters of the T2*‐weighted 3D EPI anatomical scans
| Sampling pattern | Resolution isotropic (mm) | FOV (mm3) | Slices | Slice ovs factor | TR/TE (ms) | Flip angle | Matrix size | EPI factor | ESP (ms) | Readout BW (kHz) | Total scan time (min) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SENSE 1 x 1 | 1 | 240 x 196 x 150 | 150 | 1 | 72/27 | 16° | 240 x 195 | 13 | 1.3 | 313 | 2.43 |
| CAIPI 5 x 1 y2 | 1 | 240 x 196 x 150 | 150 | 1 | 72/27 | 16° | 240 x 195 | 13 | 1.3 | 313 | 0.33 |
| SENSE 1 x 1 | 0.5 | 240 x 186 x 150 | 300 | 1 | 72/27 | 19° | 480 x 364 | 13 | 2.0 | 315 | 10.05 |
| SENSE 4 x 1 | 0.5 | 240 x 186 x 150 | 300 | 1 | 72/27 | 19° | 480 x 364 | 13 | 2.0 | 315 | 2.31 |
| SENSE 7 x 1 | 0.5 | 240 x 186 x 150 | 300 | 1 | 72/27 | 19° | 480 x 364 | 13 | 2.0 | 315 | 1.27 |
| CAIPI 7 x 1 y3 | 0.5 | 240 x 186 x 150 | 300 | 1 | 72/27 | 19° | 480 x 364 | 13 | 2.0 | 315 | 1.27 |
Abbreviations: BW, bandwidth; ESP, echo spacing; FOV, field‐of‐view (AP, anterior–posterior, RL; left–right, FH, feet–head); slice ovs factor, slice oversampling factor; TE, echo time; TR, repetition time.
3.3. Combination with high‐density receive arrays, 3D EPI functional imaging
3D EPI scans of the visual cortex were acquired with 2 x 16 channel high‐density surface coil receive arrays (MR Coils BV, Zaltbommel, the Netherlands).21 The high‐density surface coil array consists of 32 receive channels distributed over two patches of 16 receive elements, which mainly cover the back of the head (Figure 2B). The receive elements are rectangular‐shaped with a size of ~ 1.5 x 2 cm.
Submillimeter 3D EPI scans were acquired at rest using the shot‐selective 2D CAIPINHA sequence and different scan parameters to explore the possibilities and the balance between a high spatial and a high temporal resolution. Several scans were tested with variable spatial resolution, temporal resolution and acceleration factors, as shown in Table 2. For comparison, a number of scans were also acquired using SENSE (Table 2).
Table 2.
Acquisition parameters of the high‐resolution 3D EPI scans of the visual cortex
| Sampling pattern | Resolution isotropic (mm) | FOV (mm3) | Slices | Slice ovs factor | TR/TE (ms) | Flip angle | Matrix size | EPI factor | ESP (ms) | Readout BW (kHz) | Scan time per vol. (s) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SENSE 1 x 1 | 0.99 | 64 x 164 x 11.9 | 12 | 1.25 | 54/27 | 20° | 64 x 165 | 33 | 1.4 | 57 | 4.3 |
| SENSE 5 x 1 | 0.99 | 64 x 164 x 11.9 | 12 | 1.25 | 54/27 | 20° | 64 x 165 | 33 | 1.4 | 57 | 0.86 |
| CAIPI 5 x 1 y2 | 0.99 | 64 x 164 x 11.9 | 12 | 1.25 | 54/27 | 20° | 64 x 165 | 33 | 1.4 | 57 | 0.86 |
| CAIPI 7 x 1 y3 | 0.95 | 65 x 180 x 13 | 14 | 1 | 54/27 | 20° | 68 x 189 | 27 | 1.4 | 58 | 0.79 |
| CAIPI 8 x 1 y3 | 0.95 | 65 x 176 x 15 | 16 | 1 | 54/27 | 20° | 68 x 184 | 23 | 1.4 | 58 | 0.89 |
| SENSE 1 x 1 | 0.80 | 50 x 175 x 26 | 32 | 1.25 | 54/27 | 20° | 64 x 216 | 27 | 1.7 | 44 | 17.5 |
| SENSE 7 x 1* | 0.80 | 51 x 163 x 28 | 35 | 1.2 | 57/28 | 20° | 64 x 203 | 29 | 1.7 | 45 | 2.39 |
| CAIPI 7 x 1 y4* | 0.80 | 51 x 163 x 28 | 35 | 1.2 | 57/28 | 20° | 64 x 203 | 29 | 1.7 | 45 | 2.39 |
| CAIPI 8 x 1 y3 | 0.80 | 50 x 175 x 26 | 32 | 1 | 54/27 | 20° | 64 x 216 | 27 | 1.7 | 44 | 1.75 |
| CAIPI 5 x 1 y2 | 0.70 | 50 x 164 x 21 | 30 | 1 | 54/27 | 20° | 72 x 230 | 23 | 1.9 | 44 | 3.28 |
| CAIPI 10 x 1 y2 | 0.70 | 50 x 164 x 21 | 30 | 1 | 54/27 | 20° | 72 x 230 | 23 | 1.9 | 44 | 1.64 |
Abbreviations: BW, bandwidth; ESP, echo spacing; FOV, field‐of‐view (AP, anterior–posterior, RL; left–right, FH, feet–head); slice ovs factor, slice oversampling factor; TE, echo time; TR, repetition time; vol., volume.
fMRI time series.
fMRI time series of the visual cortex were acquired during visual stimulation and at rest, using both SENSE undersampling and 2D CAIPIRINHA undersampling. The scan parameters of the fMRI datasets are noted in Table 2. The scans were acquired with a transverse slice orientation (readout in anterior–posterior direction), a nominal resolution of 0.8 mm isotropic, no fat suppression, a sinc‐shaped RF excitation pulse of 0.69 ms (12 kHz bandwidth), no partial Fourier, low peripheral nerve stimulation (dB/dt less than 60% of the threshold) and a readout oversampling factor of 2. Due to the rapidly decaying receive sensitivity profile along the anterior–posterior direction, a readout oversampling factor of 2 was sufficient to avoid aliasing in the readout direction. Each fMRI time series had a volume acquisition time of 2.39 seconds, two preceding startups, 100 timeframes and a total acquisition time of 4 minutes. The CAIPI acceleration scheme used during the fMRI scan (CAIPI 7 x 1 y4) had a minimum distance dmin of 2.24 between the aliased points in the elementary aliasing cell, where greater values of dmin were favored, since they correspond to a greater minimum distance between aliases before parallel imaging reconstruction.8 The SENSE fMRI acquisition (SENSE 7 x 1) had a dmin of 1. The visual stimulus consisted of a black and white checkerboard that reversed contrast at 8 Hz, alternated with a gray screen (15 seconds on/15 seconds off). The stimulus is projected onto a screen placed at the base of the transmit coil, which the subject viewed through prism glasses.
3.4. Reconstruction
The scans were reconstructed offline in a modified Philips Recon 2.0 environment (Philips Healthcare, Best, the Netherlands). Due to undersampling, the total number of acquired k‐space points in the raw dataset was smaller than the fully sampled acquisition expected by the reconstructor. Therefore, in the current implementation, zeros were placed on the k‐space positions that were not acquired, to preserve information on matrix size and the position of the acquired k‐space points. In the reconstruction pipeline, coil sensitivity maps were shifted, reordered and then provided to the traditional SENSE reconstruction algorithm to be able to reconstruct CAIPIRINHA undersampled data.
The reconstruction time was fast, in general not more than 2 minutes, depending on the size of the dataset. The reconstruction time of the data acquired with CAIPIRINHA was in principle equal to the reconstruction time of SENSE data, of which the latter is nowadays also commonly used in clinic.
3.5. Data analysis (g‐factor, tSNR and fMRI activation)
The g‐factor maps were calculated using the exported coil sensitivity maps. The coil sensitivity maps were masked to include the brain region only. The g‐factor maps were calculated according to the equations of Pruessmann et al.17
The time series datasets were analyzed using AFNI.31 The datasets were corrected for motion and drift. The maps of temporal fluctuation (1/tSNR) were derived from the 4‐minute time series acquired at rest (no visual stimulus). The temporal fluctuation, expressed as a percentage, was calculated by dividing the standard deviation (SD) of the signal over time by the mean of the signal over time. This was a measure of the temporal instability of the time series.
fMRI activation maps were constructed from the time series acquired during on/off visual stimulus. Activated voxels were identified with linear regression (GLM) of the time series data with the stimulus waveform convolved with a canonical hemodynamic response, using the function 3dDeconvolve in AFNI. Activation maps were obtained by thresholding the resulting parametric maps of T‐statistics at P < 0.01 FDR‐corrected.
4. RESULTS
The results are presented in two steps. First, the implementation of the shot‐selective 2D CAIPIRINHA sequence for 3D EPI scans was evaluated using T2*‐weighted anatomical imaging. Second, the combination of high‐density receive arrays and the 2D CAIPIRINHA sequence was evaluated based on the acceleration performance for submillimeter fMRI acquisitions.
4.1. Evaluation of sequence implementation, T2*‐weighted 3D EPI anatomical imaging
To evaluate whether the shot‐selective 2D CAIPIRINHA sequence was implemented correctly, fully sampled 1 mm 3D‐EPI whole brain T2* anatomy‐weighted images were compared with CAIPI undersampled images in transverse and coronal views, as shown in Figure 3. The full non‐undersampled SENSE 1 dataset was acquired in 2.43 minutes (Figure 3A,D), and the prospectively CAIPIRINHA undersampled dataset with an undersampling factor of 5 was acquired in 33 seconds. The resulting aliasing because of undersampling is displayed as a sum of the separate channels (Figure 3B,E), and, especially in the coronal view, the typical CAIPI aliasing patterns can be seen. The reconstructed images (Figure 3E,F) align well with the fully sampled images (Figure 3A,D). Pro‐ and retrospectively undersampled datasets were also compared and found to be similar.
Figure 3.
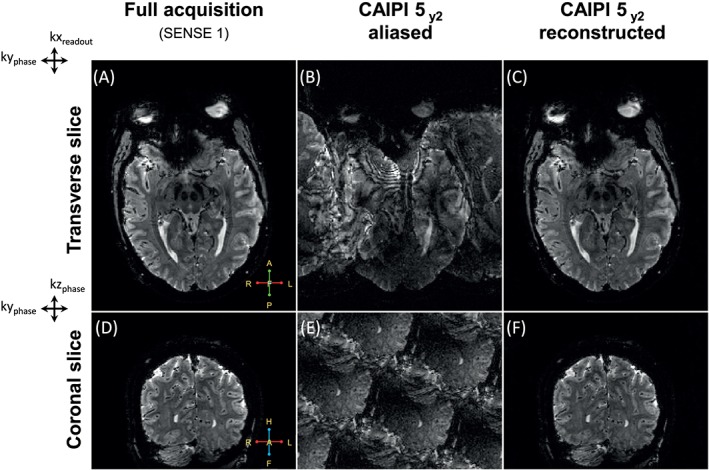
Overview of the shot‐selective 2D CAIPIRINHA implementation for 3D T2*‐weighted anatomical images. The images are shown in both transverse (A‐C) and coronal (D‐F) view. A full non‐undersampled SENSE 1 dataset (A, D) is acquired in 2.43 minutes, prospectively undersampled CAIPI 5 data is acquired in 33 seconds. The sum of the aliased channels is shown (B, E), and especially in the coronal view (two phase directions) the typical CAIPI pattern becomes apparent. The reconstructed images (C, F) are in close agreement with the fully sampled images (A, D), confirming an accurate implementation of the shot‐selective 2D CAIPIRINHA sequence
To evaluate the performance of the 2D CAIPIRINHA implementation in comparison with SENSE acceleration, the resolution of 3D EPI whole brain T2*‐weighted anatomical scans was pushed to 0.5 mm isotropic. These scans are displayed in Figure 4. A central axial slice is depicted, as well as a slice through the lower part of the brain that shows the deeper brain structures. Both a fully sampled dataset of 10.05 minutes (Figure 4A) and undersampled SENSE and CAIPIRINHA datasets are displayed (Figure 4B‐D). The undersampling patterns used are SENSE 4 acquired in 2.31 minutes (Figure 4B), CAIPI 7 acquired in 1.27 minutes (Figure 4C) and SENSE 7 acquired in 1.27 minutes (Figure 4D). Note that both SENSE and CAIPIRINHA can be used to shorten the total scan time substantially. However, at an acceleration factor of 7, the SENSE image is heavily spoiled by artifacts, while the CAIPIRINHA image still preserves the anatomy of the brain, allowing the scan time to be shortened even further.
Figure 4.
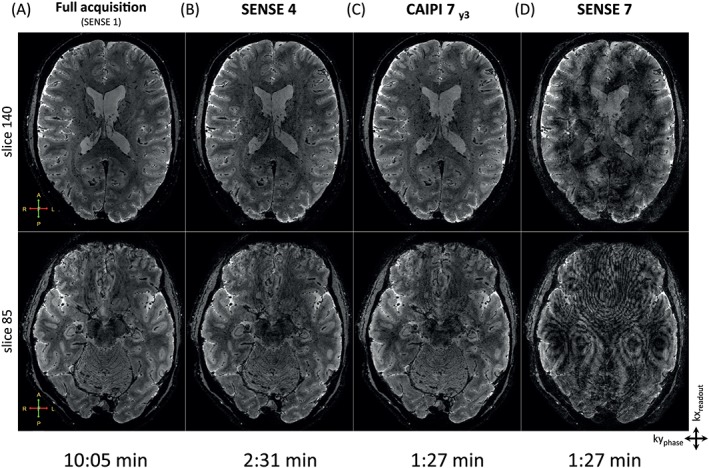
Whole brain T2* anatomy scans acquired with multishot 3D EPI sequences. Example slices are displayed for (A) fully sampled data, (B) SENSE 4, (C) CAIPI 7 and (D) SENSE 7. The resolution of the scans is 0.5 mm isotropic. The fully sampled data takes 10.05 minutes to acquire, whereas the SENSE and CAIPIRINHA methods are able to significantly shorten the scan time to 2.31 and 1.27 minutes, respectively. Note that even with the shortened scan time, the detailed brain structures are still visible for the CAIPIRINHA scan, in contrast to the SENSE 7 scan, which is spoiled by artifacts. The complete 3D datasets are available as animation in the supporting information
4.2. Combination with high‐density receive arrays, 3D EPI functional imaging
3D EPI scans of the visual cortex, acquired with a combination of high‐density receive arrays and a 2D CAIPIRINHA sequence, are shown in detail in Figure 5. To illustrate the different possibilities for imaging with a high spatial and high temporal resolution, different combinations of submillimeter imaging of the visual cortex are displayed. A fully sampled (SENSE 1) acquisition is displayed on the left (Figure 5A,F). The accelerated CAIPIRINHA acquisitions (Figure 5B‐D) have varying scan parameters (as indicated in the figure). The range of the acquired resolutions is 0.70–0.99 mm isotropic, the acquisition time per volume is 0.82–3.28 seconds, and the coverage is 12–32 slices. As the resolution increases (i.e. smaller voxels), the scan time becomes longer. Note that it is possible to perform submillimeter and subsecond EPI imaging of the visual cortex without losing substantial image quality (Figure 5B,C), in contrast to using SENSE (Figure 5E).
Figure 5.
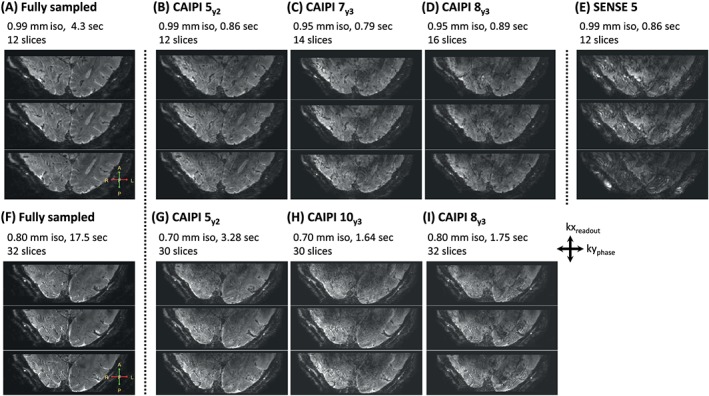
3D EPI scans of the visual cortex acquired with different settings for resolution, scan time and imaging coverage (scaled differently for visualization). The same three slices are displayed for both the fully sampled SENSE 1 dataset (A, F) and the accelerated datasets, respectively (B‐E, G‐I). Note that it is possible to scan with both a high spatial resolution (<1 mm) and a high temporal resolution (<1 second) combined
The fMRI results are shown in Figure 6 for both CAIPIRINHA (Figure 6A,C,E,G) and SENSE undersampling (Figure 6B,D,F,G). For each dataset, eight consecutive axial slices of the visual cortex are illustrated. The mean signal intensity over the time series is displayed (A,B), as well as g‐factor maps (C,D), maps of temporal fluctuation (E,F) and fMRI activation maps (G,H). The mean signal intensity is comparable between the CAIPIRINHA and SENSE datasets, although there is a slight drop of signal visible deeper in the center of the visual cortex for the dataset acquired with SENSE. The g‐factor maps show a reduction of the noise amplification factor for the CAIPIRINHA data compared with the SENSE data, especially in the center of the visual cortex. A mean g‐factor of 1.5 was found for the CAIPIRINHA dataset and 2.52 for the SENSE dataset. Likewise, the maps of temporal fluctuation show that the CAIPIRINHA method is more stable over time. The mean and SD of the tSNR−1 values are 6.57% ± 3.8% for the CAIPIRINHA dataset and 12.48% ± 6.6% for the SENSE dataset. In the fMRI maps, it is seen that the extent of fMRI activation measured with CAIPIRINHA is much larger in comparison with SENSE. In this example, the CAIPIRINHA measurement counts 26 135 activated voxels with a mean and SD of the T‐statistic of 5.01 ± 3.68, whereas the SENSE measurement counts 2446 activated voxels with a mean and SD of the T‐statistic of 4.38 ± 2.72. An overview of the values from the quantitative time series comparison can be found in Table 3.
Figure 6.
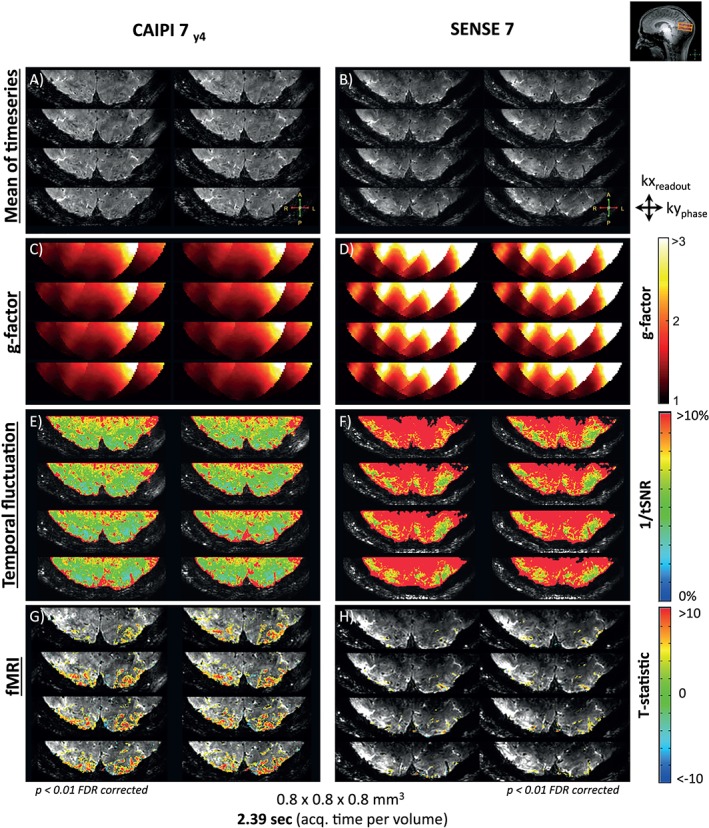
3D EPI fMRI time series of the visual cortex, acquired with the high‐density receive arrays in combination with 2D CAIPIRINHA (A, C, E, G) and SENSE (B, D, F, H). For eight slices of the visual cortex, the mean image of the time series is displayed (A, B), as well as the g‐factor maps (C, D), tSNR−1 maps (E, F) and fMRI maps (G, H). Both datasets have an acceleration factor of 7, a resolution of 0.8 mm isotropic, a volume acquisition time of 2.39 seconds and a total acquisition time of 4 minutes. Note that the g‐factor is lower when using 2D CAIPIRINHA compared with SENSE. This results in an improved temporal stability, making it possible to detect more fMRI activation
Table 3.
Quantitative case comparison of the acquired time series
| CAIPI 7 x 1 y4 (mean ± SD) | SENSE 7 x 1 (mean ± SD) | |
|---|---|---|
| g‐factor | 1.50 ± 0.52 | 2.52 ± 1.71 |
| Temporal fluctuation (tSNR−1) | 6.57% ± 3.8% | 12.48% ± 6.6% |
| fMRI, number of activated voxels | 26 135 | 2446 |
| fMRI, T‐statistic | 5.01 ± 3.68 | 4.38 ± 2.72 |
5. DISCUSSION
This study investigated whether submillimeter multishot 3D EPI fMRI scans acquired with high‐density receive arrays can benefit from a 2D CAIPIRINHA sampling pattern, in terms of noise amplification (g‐factor), temporal SNR and fMRI sensitivity. A shot‐selective 2D CAIPIRINHA sequence for multishot 3D EPI scans was implemented, which, instead of adding extra gradients, leaves them out. With the 2D CAIPIRINHA sequence it was possible to acquire high‐resolution T2*‐weighted anatomical scans at high acceleration factors while maintaining sufficient image quality, confirming an accurate sequence implementation. When combining high‐density receive arrays with the shot‐selective 2D CAIPIRINHA sequence it was possible to acquire submillimeter, subsecond 3D EPI scans of the visual cortex. The image quality and fMRI performance of the CAIPIRINHA acquisitions were improved compared with SENSE acquisitions with the same resolution and scan time.
5.1. Evaluation of sequence implementation, T2*‐weighted 3D EPI anatomical imaging
The T2*‐weighted 3D EPI anatomical scans acquired with the standard receive setup show that the proposed shot‐selective 2D CAIPIRINHA sequence for multishot 3D EPI scans allows for artifact‐free images at high acceleration. Several EPI shots of a multishot interleaved 3D EPI sequence were selectively skipped, resulting in a CAIPIRINHA sampling pattern and a reduction in scan time. The implementation resulted in the expected image quality, as evaluated by comparing fully sampled and undersampled datasets. These datasets are in agreement, as can be seen by the similarities of the reconstructed images. When the sequence is used to acquire anatomical scans with an isotropic resolution of 0.5 mm, the total scan time can be shortened significantly. The total scan time can be reduced by a factor of 4 compared with an earlier optimized implementation of 6 minutes for whole brain T2*‐weighted 3D EPI anatomy scans at 7 T.30 In general, undersampling schemes shorten scan times at the cost of the overall SNR, which is also the case for the currently implemented acceleration method. Nonetheless, the accelerated scans in this study still maintain sufficient SNR to depict the deeper structures of the brain in high detail. The current results are acquired at a field strength of 7 T; however, the applied method can be easily translated to more clinically available 3 T scanners. This holds promise for future applications in routine examinations among patients.
5.2. Combination with high‐density receive arrays, 3D EPI functional imaging
The combination of high‐density receive arrays and a 2D CAIPIRINHA sequence made it possible to acquire submillimeter 3D EPI scans of the visual cortex with a temporal resolution of less than 1 second. Multiple scans with different settings for a high resolution and a short scan time were acquired. In the literature, submillimeter functional imaging is often characterized by a relatively slow repetition time. Previous work from Fracasso et al32 showed that when using high‐density receive arrays, high‐resolution fMRI data of 0.7 mm isotropic could be acquired in 4 seconds. In this study, when using a similar scan protocol, high‐density receive arrays, and the proposed CAIPIRINHA sequence, we were able to push down the scan time of 0.7 mm isotropic data to 1.64 seconds. However, note that some slices contain aliasing artifacts due to the high acceleration factor.
For the fMRI time series, where CAIPIRINHA and SENSE acquisitions with identical acquisition parameters are compared (Figure 6), it is seen that the CAIPIRINHA method outperforms SENSE. The 0.8 mm isotropic 3D EPI scans of the visual cortex with a scan time of 2.39 seconds per volume have less noise amplification, especially in the center of the visual cortex. This is visible both in the g‐factor maps (Figure 6C,D) as well as in the maps for temporal fluctuation (Figure 6D,E). The fMRI maps show that the capability to distinguish the fMRI signal from background noise was improved, as the detected number of activated voxels in the visual cortex was larger for the CAIPIRINHA dataset compared with the SENSE dataset.
For both methods, unfolding artifacts due to the high acceleration can still be seen at the outer slices of the datasets with high acceleration. However, these artifacts are less dominant in the CAIPIRINHA dataset compared with the SENSE dataset. Overall, despite the already high achievable acceleration factor when using high‐density receive arrays, it is possible to further increase the acceleration factor by combining the high‐density receive arrays with a shot‐selective 2D CAIPIRINHA sequence.
5.3. Position in the literature
The novelty of this study is the combination of high‐density receive arrays and a 2D CAIPIRINHA sequence for 3D EPI scans to push MRI resolutions to a subsecond and submillimeter scale. The CAIPIRINHA acquisition was implemented in a shot‐selective manner, and the reconstruction was performed with an adapted SENSE reconstruction. The concept of a shot‐selective 2D CAIPIRINHA for 3D EPI scans has been suggested before.16, 25 Here, we show that it enables high spatiotemporal resolution imaging with high temporal SNR, low g‐factors, and high fMRI sensitivity. The sequence is simple and straightforward to implement, since it does not require SAR‐demanding (MB) RF pulses or additional kz gradient blips during the EPI readout train. Additionally, the odd‐even EPI phase corrections (necessary because of alternating positive and negative readout gradients) are identical to the corrections used in a traditional multishot interleaved 3D EPI sequence and do not require modification. The combination with high‐density receive arrays enabled us to push the acceleration further compared with using high‐density receive arrays or CAIPI alone.
When compared with blipped‐CAIPIRINHA, the shot‐selective CAIPIRINHA implementation uses shorter EPI gradient trains. Therefore, the EPI factor is smaller, and a shorter TRshot and TE can be used. Consequently, shot‐selective CAIPIRINHA has a number of advantages in comparison with blipped CAIPIRINHA in terms of image quality. These include reduced imaging blurring (due to T2* decay over the EPI gradient train), lower ghost intensity (due to reduced phase error accumulation throughout the echo train),33 reduced image distortion and chemical shift displacement (due to increased effective phase‐encoding bandwidth). The major drawback of the improved image quality is that the shot‐selective method requires more EPI shots and is therefore generally slower than its blipped counterpart.
There are other promising approaches to achieve similar or potentially even higher spatial and temporal resolution for MRI. These approaches include sequence design as well as hardware developments. Recent efforts aim to increase the number of channels for whole brain coverage at high fields34, 35, 36 similar to lower fields.37, 38 Combined with advanced acceleration schemes such as the recent wave‐CAIPI,39 these can further improve fMRI sensitivity and specificity by enabling high spatiotemporal resolution. Another approach is to use insert gradients for head imaging. Insert gradients allow fast switching of strong gradients, which can reduce the length of the EPI readout train, thereby enhancing temporal resolution.40 The current study used 2D CAIPIRINHA for 3D EPI sequences, although recently major advances have been made with methods that use simultaneous multislice approaches,19 such as MB41 and blipped‐CAIPI.22 Other approaches focus more on the excitation side of the sequence. Selective RF excitation can be used to either reduce the FOV, or to reduce parallel imaging noise amplification,42 which has already been combined with the benefits of a 3D CAIPI EPI sequence.43
5.4. Considerations
In this study, the achieved gains from high‐density receive arrays and 2D CAIPIRINHA were used to shorten the scan time of high‐resolution 3D EPI scans. However, the gains can be used in different ways, not only to reduce scan time, but also to increase the number of slices or imaging coverage (FOV). The results are promising, but the methods can be further optimized in the future, because artifacts are present in some CAIPIRINHA datasets, in particular in the top and bottom slices. In fact, these may suggest imperfect slab excitations, which could possibly also explain the artifacts of SENSE 7 (Figure 4). In this study, a shot‐selective 2D CAIPIRINHA sequence for multishot 3D EPI scans was used. The sequence is practical to implement, since it mainly involves removing selected EPI shots from a multishot 3D EPI sequence. A disadvantage of the current implementation is that the undersampling factor is directly connected to the EPI factor, which reduces the freedom of choice in the sequence settings. For example, the acquisition of a 3D volume with a single EPI shot is not possible with the current implementation. Offline reconstruction times of both methods were shorter than those of state‐of‐the‐art iterative minimization approaches such as compressed sensing.44, 45 Reconstruction times can be reduced further when the calculations are performed on the dedicated reconstruction computer at the scanner. Despite the high acceleration factors achieved in this study, there might be more advantageous applications for CAIPIRINHA. In our study we used reduced FOV acquisitions (as commonly acquired with surface coils); this can be considered as a nonideal case for combination with CAIPIRINHA, since CAIPIRINHA exploits the ability to shuffle sensitivity variations over a (large) FOV relative to the coil sensitivity profiles. On the other hand, this study does use a large channel count of 32 receivers, which further enhances the benefits of CAIPIRINHA.
6. CONCLUSIONS
The combination of high‐density receive arrays and shot‐selective 2D CAIPIRINHA for 3D EPI pushes the temporal resolution of submillimeter fMRI scans. The implemented shot‐selective sequence makes it possible to reduce the scan time of high‐resolution whole brain T2*‐weighted anatomical 3D EPI scans. When high‐density receive arrays are combined with the shot‐selective 2D CAIPIRINHA sequence, the noise amplification is low compared with the SENSE scans, resulting in enhanced image quality for submillimeter 3D EPI scans of the visual cortex. Time series show that the temporal SNR of the CAIPIRINHA scans is higher than the temporal SNR of the SENSE scans. Consequently, the sensitivity to the fMRI signal is improved. The gains in temporal SNR and fMRI sensitivity demonstrate that the benefit of CAIPIRINHA with the high‐density arrays can be used to complement both techniques for maximizing spatial and temporal resolution. Overall, high‐density receive arrays in combination with shot‐selective 2D CAIPIRINHA for 3D EPI scans prove to be valuable for reducing the scan time of submillimeter fMRI acquisitions.
Supporting information
Data S1. Supporting Information
Movie S1. Supporting Information
ACKNOWLEDGEMENTS
This work was supported by the Dutch Research Council (NWO), grant number: 040.11.581, ALW‐834.14.004 and Vidi Grant 13339 (Petridou).
Hendriks AD, D'Agata F, Raimondo L, et al. Pushing functional MRI spatial and temporal resolution further: High‐density receive arrays combined with shot‐selective 2D CAIPIRINHA for 3D echo‐planar imaging at 7 T. NMR in Biomedicine. 2020;33:e4281 10.1002/nbm.4281
REFERENCES
- 1. Dumoulin SO, Fracasso A, van der Zwaag W, Siero JCW, Petridou N. Ultra‐high field MRI: advancing systems neuroscience towards mesoscopic human brain function. Neuroimage. 2018;168:345‐357. [DOI] [PubMed] [Google Scholar]
- 2. Setsompop K, Feinberg DA, Polimeni JR. Rapid brain MRI acquisition techniques at ultra‐high fields. NMR Biomed. 2016;29:1198‐1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van der Zwaag W, Schafer A, Marques JP, Turner R, Trampel R. Recent applications of UHF‐MRI in the study of human brain function and structure: a review. NMR Biomed. 2016;29:1274‐1288. [DOI] [PubMed] [Google Scholar]
- 4. Lin FH, Polimeni JR, Lin JL, et al. Relative latency and temporal variability of hemodynamic responses at the human primary visual cortex. Neuroimage. 2018;164:194‐201. [DOI] [PubMed] [Google Scholar]
- 5. Petridou N, Siero JCW. Laminar fMRI: what can the time domain tell us? Neuroimage. 2019;197:761‐771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoo PE, John SE, Farquharson S, et al. 7T‐fMRI: faster temporal resolution yields optimal BOLD sensitivity for functional network imaging specifically at high spatial resolution. Neuroimage. 2018;164:214‐229. [DOI] [PubMed] [Google Scholar]
- 7. Breuer FA, Blaimer M, Heidemann RM, Mueller MF, Griswold MA, Jakob PM. Controlled aliasing in parallel imaging results in higher acceleration (CAIPIRINHA) for multi‐slice imaging. Magn Reson Med. 2005;53:684‐691. [DOI] [PubMed] [Google Scholar]
- 8. Breuer FA, Blaimer M, Mueller MF, et al. Controlled aliasing in volumetric parallel imaging (2D CAIPIRINHA). Magn Reson Med. 2006;55:549‐556. [DOI] [PubMed] [Google Scholar]
- 9. Jurrissen M, Fuderer M, van den Brink J. Diamond‐SENSE: undersampling on a crystallographic grid In: Proceedings of the 12th Annual Meeting of the ISMRM. Kyoto, Japan; 2004:2643. [Google Scholar]
- 10. Barth M, Breuer F, Koopmans PJ, Norris DG, Poser BA. Simultaneous multislice (SMS) imaging techniques. Magn Reson Med. 2016;75:63‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poser BA, Setsompop K. Pulse sequences and parallel imaging for high spatiotemporal resolution MRI at ultra‐high field. Neuroimage. 2018;168:101‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poser BA, Koopmans PJ, Witzel T, Wald LL, Barth M. Three dimensional echo‐planar imaging at 7 tesla. Neuroimage. 2010;51:261‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van der Zwaag W, Marques JP, Kober T, Glover G, Gruetter R, Krueger G. Temporal SNR characteristics in segmented 3D‐EPI at 7T. Magn Reson Med. 2012;67:344‐352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stirnberg R, Huijbers W, Brenner D, Poser BA, Breteler M, Stocker T. Rapid whole‐brain resting‐state fMRI at 3 T: efficiency‐optimized three‐dimensional EPI versus repetition time‐matched simultaneous‐multi‐slice EPI. Neuroimage. 2017;163:81‐92. [DOI] [PubMed] [Google Scholar]
- 15. Narsude M, Gallichan D, van der Zwaag W, Gruetter R, Marques JP. Three‐dimensional echo planar imaging with controlled aliasing: a sequence for high temporal resolution functional MRI. Magn Reson Med. 2016;75:2350‐2361. [DOI] [PubMed] [Google Scholar]
- 16. Poser BA, Ivanov D, Kannengiesser SA, Uludag K, Barth M. Accelerated 3D EPI using 2D blipped‐CAIPI for high temporal and/or spatial resolution In: Proceedings of the 22th Annual Meeting of the ISMRM. Milan, Italy; 2014:1506. [Google Scholar]
- 17. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42:952‐962. [PubMed] [Google Scholar]
- 18. Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med. 2002;47:1202‐1210. [DOI] [PubMed] [Google Scholar]
- 19. Feinberg DA, Vu AT, Beckett A. Pushing the limits of ultra‐high resolution human brain imaging with SMS‐EPI demonstrated for columnar level fMRI. Neuroimage. 2018;164:155‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fracasso A, Luijten PR, Dumoulin SO, Petridou N. Laminar imaging of positive and negative BOLD in human visual cortex at 7T. Neuroimage. 2018;164:100‐111. [DOI] [PubMed] [Google Scholar]
- 21. Petridou N, Italiaander M, van de Bank BL, Siero JC, Luijten PR, Klomp DW. Pushing the limits of high‐resolution functional MRI using a simple high‐density multi‐element coil design. NMR Biomed. 2013;26:65‐73. [DOI] [PubMed] [Google Scholar]
- 22. Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL. Blipped‐controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g‐factor penalty. Magn Reson Med. 2012;67:1210‐1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zahneisen B, Ernst T, Poser BA. SENSE and simultaneous multislice imaging. Magn Reson Med. 2015;74:1356‐1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zahneisen B, Poser BA, Ernst T, Stenger VA. Three‐dimensional Fourier encoding of simultaneously excited slices: generalized acquisition and reconstruction framework. Magn Reson Med. 2014;71:2071‐2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Poser BA, Kemper VG, Ivanov D, Kannengiesser SA, Uludag K, Barth M. CAIPIRINHA‐accelerated 3D EPI for high temporal and/or spatial resolution EPI acquisitions. In: Proceedings of the 30th Scientific Annual Meeting of the ESMRMB Toulouse, France, 2013, 287.
- 26. McKinnon GC. Ultrafast interleaved gradient‐echo‐planar imaging on a standard scanner. Magn Reson Med. 1993;30:609‐616. [DOI] [PubMed] [Google Scholar]
- 27. Blaimer M, Breuer FA, Seiberlich N, et al. Accelerated volumetric MRI with a SENSE/GRAPPA combination. J Magn Reson Imaging. 2006;24:444‐450. [DOI] [PubMed] [Google Scholar]
- 28. Koopmans PJ. Two‐dimensional‐NGC‐SENSE‐GRAPPA for fast, ghosting‐robust reconstruction of in‐plane and slice‐accelerated blipped‐CAIPI echo planar imaging. Magn Reson Med. 2017;77:998‐1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ledden PJ, Mareyam A, Wang S, van Gelderen P, Duyn J. 32 channel receive‐only SENSE array for brain imaging at 7T In: Proceedings of the 15th Annual Meeting of the ISMRM. Berlin, Germany; 2007:242. [Google Scholar]
- 30. Zwanenburg JJ, Versluis MJ, Luijten PR, Petridou N. Fast high resolution whole brain T2* weighted imaging using echo planar imaging at 7T. Neuroimage. 2011;56:1902‐1907. [DOI] [PubMed] [Google Scholar]
- 31. Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162‐173. [DOI] [PubMed] [Google Scholar]
- 32. Fracasso A, Petridou N, Dumoulin SO. Systematic variation of population receptive field properties across cortical depth in human visual cortex. Neuroimage. 2016;139:427‐438. [DOI] [PubMed] [Google Scholar]
- 33. Bernstein MA, King KF, Zhou XJ. Chapter 16.1: Echo Planar Imaging. Handbook of MRI Pulse Sequences . Burlington: Academic Press. 2004;722‐725. [Google Scholar]
- 34. Auerbach EJ, DelaBarre L, Van de Moortele PF, et al. An integrated 32‐channel transmit and 64‐channel receive 7 Tesla MRI system In: Proceedings of the 25th Annual Meeting of the ISMRM. Hawaii, USA; 2017:1218. [Google Scholar]
- 35. Beckett AJS, Vu AT, Schillak S, Wald LL, Feinberg DA. A high density 24 channel array coil extendable to 48 channels for human cortical MRI at 7T In: Proceedings of the 25th Annual Meeting of the ISMRM. Hawaii, USA; 2017:2654. [Google Scholar]
- 36. Hendriks AD, Luijten PR, Klomp DWJ, Petridou N. Potential acceleration performance of a 256‐channel whole‐brain receive array at 7 T. Magn Reson Med. 2019;81:1659‐1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Keil B, Blau JN, Biber S, et al. A 64‐channel 3T array coil for accelerated brain MRI. Magn Reson Med. 2013;70:248‐258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wiggins GC, Polimeni JR, Potthast A, Schmitt M, Alagappan V, Wald LL. 96‐channel receive‐only head coil for 3 tesla: design optimization and evaluation. Magn Reson Med. 2009;62:754‐762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bilgic B, Gagoski BA, Cauley SF, et al. Wave‐CAIPI for highly accelerated 3D imaging. Magn Reson Med. 2015;73:2152‐2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weiger M, Overweg J, Rosler MB, et al. A high‐performance gradient insert for rapid and short‐T2 imaging at full duty cycle. Magn Reson Med. 2018;79:3256‐3266. [DOI] [PubMed] [Google Scholar]
- 41. Larkman DJ, Hajnal JV, Herlihy AH, Coutts GA, Young IR, Ehnholm G. Use of multicoil arrays for separation of signal from multiple slices simultaneously excited. J Magn Reson Imaging. 2001;13:313‐317. [DOI] [PubMed] [Google Scholar]
- 42. Mooiweer R, Sbrizzi A, Raaijmakers AJE, van den Berg CAT, Luijten PR, Hoogduin H. Combining a reduced field of excitation with SENSE‐based parallel imaging for maximum imaging efficiency. Magn Reson Med. 2017;78:88‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van der Zwaag W, Reynaud O, Narsude M, Gallichan D, Marques JP. High spatio‐temporal resolution in functional MRI with 3D echo planar imaging using cylindrical excitation and a CAIPIRINHA undersampling pattern. Magn Reson Med. 2018;79:2589‐2596. [DOI] [PubMed] [Google Scholar]
- 44. Jaspan ON, Fleysher R, Lipton ML. Compressed sensing MRI: a review of the clinical literature. Br J Radiol. 2015;88:20150487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58:1182‐1195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting Information
Movie S1. Supporting Information


