Summary
MicroRNAs (miRNAs) are a class of small noncoding RNAs that play important roles in plant growth and development as well as in stress responses. However, little is known about their regulatory functions affecting rice grain yield.
We functionally characterized a novel miRNA in rice, OsmiR530, its target OsPL3, and its upstream regulator phytochrome‐interacting factor‐like 15 (OsPIL15). Their effects on rice yield were dissected comprehensively.
We determined that OsmiR530 negatively regulates grain yield. Blocking OsmiR530 increases grain yield, whereas OsmiR530 overexpression significantly decreases grain size and panicle branching, leading to yield loss. Additionally, OsPL3, which encodes a PLUS3 domain‐containing protein, is targeted directly by OsmiR530. Knocking out OsPL3 decreases the grain yield. In‐depth analyses indicated that OsPIL15 activates OsMIR530 expression by directly binding to the G‐box elements in the promoter. Analyses of genetic variations suggested that the OsMIR530 locus has likely been subjected to artificial selection during rice breeding.
The results presented herein reveal a novel OsPIL15–OsmiR530 module controlling rice grain yield, thus providing researchers with a new target for the breeding of high‐yielding rice.
Keywords: grain yield, miR530, OsPIL15, panicle branching, PLUS3 domain‐containing protein, seed size
Introduction
Rice (Oryza sativa), a model monocotyledonous plant species, is the staple food for most of the global population. Consequently, breeding high‐yielding rice varieties is urgently required to cope with food shortages worldwide. The number of panicles per plant, number of effective grains per panicle, grain weight and seed‐setting rate are generally considered to be the decisive factors affecting rice yield. Recent studies have revealed that several genes involved in transcriptional regulation and hormone signal transduction are important for regulating rice yield, such as Growth‐regulating factor (OsGRF4 (Duan et al., 2015), OsGRF6 (Gao et al., 2015)), SOUAMOSA promoter binding protein‐like (OsSPL13 (Si et al., 2016), OsSPL16 (Wang et al., 2015)), MADS‐domain transcription factor (OsMADS1, Liu et al., 2018) and GAGA‐binding protein 3 (OsGBP3; Gong et al., 2018). However, the molecular mechanisms underlying the rice yield regulatory network remain elusive.
Phytochrome‐interacting factors (PIFs) form a class of basic helix‐loop‐helix transcription factors influencing a series of biological processes by regulating the expression of their target genes which contain G‐box and/or PIF‐binding E‐box elements (Hornitschek et al., 2012; Y. Zhang et al., 2013). During the past 20 yr, PIF functions have been well‐studied in the model plant Arabidopsis thaliana, including their effects on phytochrome‐mediated photomorphogenesis, hormone signaling, and responses to biotic and abiotic stresses (Leivar & Quail, 2011; Oh et al., 2012; Quint et al., 2016; Paik et al., 2017; Shor et al., 2017). The rice genome contains six phytochrome‐interacting factor‐like genes (OsPILs), designated as OsPIL11 to OsPIL16 (Nakamura et al., 2007). Compared with the Arabidopsis PIFs, there is relatively little available information regarding rice PIF functions related to growth and development, especially in terms of rice yield regulation. A recent study revealed that one of the rice PIL genes, OsPIL15, negatively regulates grain size (Ji et al., 2019). Thus, OsPILs may contribute to unique signaling networks associated with rice seed development.
MicroRNAs (miRNAs) are a class of short noncoding RNAs that regulate the expression of target genes, with important implications for plant development and stress responses (Bartel, 2004). To date, only a few miRNAs, including OsmiR156 (Jiao et al., 2010), OsmiR397 (Y. C. Zhang et al., 2013), OsmiR396 (Duan et al., 2015; Gao et al., 2015) and miR408 (Zhang et al., 2017), have been confirmed as important regulators of rice yield via their effects on grain size or panicle architecture. We previously determined that a miRNA, OsmiR530‐5p_R+1 (OsmiR530), is significantly more abundant in the phytochrome B (phyB) mutant than in the wild‐type (WT) control (Sun et al., 2015). To elucidate its roles in rice growth and development, we generated transgenic lines overexpressing OsmiR530 (miR530‐OE) or the OsmiR530 target mimic (MIM530). Functional analyses revealed that OsmiR530 negatively regulates rice yield by altering the grain size and panicle architecture. Subsequent experiments demonstrated that OsmiR530 functions directly downstream of OsPIL15. Our results establish a new OsPIL15–OsmiR530 signaling pathway that regulates rice yield.
Materials and Methods
Plant materials and growth conditions
All of the rice plants used in this study were Oryza sativa L. cv Nipponbare (Nip). The phyB mutant and phytochrome‐interacting factor‐like gene (OsPIL)15‐overexpressing (PIL15‐OE) transgenic lines have been described previously (Takano et al., 2005; Zhou et al., 2014).
Seeds were surface‐sterilized in 70% (v/v) ethanol for 30 s and then in 5% NaClO (v/v) for 20 min, after which they were rinsed six times in sterile double‐distilled water. Seeds were incubated in darkness at 28°C for 3 d to induce germination.
In order to analyze the agronomic traits of the transgenic plants, we sowed seeds in mid‐May. The resulting seedlings were transplanted to an irrigated field in Jinan, China (lat 36°40′N, long 117°00′E) in late June. The plants were grown in the field with routine management practices. The following agronomic traits were measured before seeds were harvested: plant height, number of primary and secondary branches, seed‐setting rate and number of effective grains per main panicle. The grain length and width were measured when the seeds were harvested. For each line, c. 20 plants were used for statistical analyses.
Generation of transgenic rice plants
In order to generate the microRNA OsmiR530‐overexpression construct, the genomic DNA sequence surrounding the pre‐miR530 was amplified by PCR. The amplicon was sequenced and subcloned into the BamHI and SpeI sites downstream of the Ubi promoter in the p1390‐Ubi vector (Li et al., 2011). To prepare the OsmiR530 target mimic construct, a 24‐nucleotide motif complementary to the miR399 in IPS1 (Franco‐Zorrilla et al., 2007) was replaced with a sequence complementary to miR530. The IPS1 fragment with the miR530‐complementary motif was inserted into the pENTR/D‐TOPO vector (Invitrogen) and then transferred to the destination vector PC186 via LR Clonase reactions (Thermo Fisher Scientific, Waltham, MA, USA). The OsPIL15‐overexpressing (PIL15‐OE) transgenic plants were generated in our laboratory during a previous study (Zhou et al., 2014). For the OsPIL15‐eGFP construct, the full‐length eGFP cDNA was amplified by PCR and then inserted at the BstEII restriction site of the PIL15‐OE vector (Zhou et al., 2014) (GFP, green fluorescent protein). To construct the PHYB‐GFP vector, the rice PHYB cDNA sequence was amplified by PCR with a nucleotide substitution that replaced the PHYB translation termination codon (TAG) with an oligonucleotide sequence containing a KpnI site. The PHYB moiety encoded in the Arabidopsis PHYB‐GFP fusion construct (Yamaguchi et al., 1999) was replaced with rice PHYB to obtain the PHYB‐GFP construct in which the rice PHYB‐GFP fusion sequence was inserted between the constitutive cauliflower mosaic virus 35S promoter and the Nos terminator. The PLUS3 domain containing protein (OsPL3)‐knockout (PL3‐KO) and OsPIL15‐knockout (PIL15‐KO) lines were generated by the Biogle company (Hangzhou, China) using CRISPR/Cas9 technology. We designed two single‐guide RNAs (sgRNAs) targeting OsPL3 (sg1: 5′‐GTGAAGCCAACGGATTGCAG‐3′; sg2: 5′‐GCCCAGTGTTAGCCTGTTGG‐3′), and two sgRNAs targeting OsPIL15 (sg3679: 5′‐GACCACCAGGGAACCCTCCA‐3′; sg3680: 5′‐GGAGTCGACGGTCGTGCAGA‐3′) to minimize the off‐target effects. All sgRNAs were generated in the BGK03 vector, which contains the Cas9 gene. All constructs were introduced into Agrobacterium tumefaciens strain EHA105 cells for the subsequent transformation of Nip rice plants as described previously (Toki et al., 2006). The homozygous T3 generation plants were used for a phenotypic analysis. Details regarding the primers used for constructing vectors are provided in Supporting Information Table S1.
Field plot analysis
For the field plot experiment, the WT, miR530‐OE and MIM530 plants were grown under natural conditions in Jinan, China (lat 36°40′N, long 117°00′E) in 2018. Each plot was 3 m2 and comprised a mixture of three transgenic lines, except for the WT plots, with a planting density of 20 cm × 25 cm. The miR530‐OE plots included miR530‐OE #a1, #b5 and #c2 transgenic plants, whereas the MIM530 plots consisted of MIM530 #2, #3 and #6 transgenic plants. Plot yields were determined when the seeds were harvested. The field trial was completed according to a randomized complete block design with four replicates. Data are presented herein as the mean ± SD.
Histological analysis
Internodes just below the neck node and young spikelet hulls were fixed in 4% (w/v) glutaraldehyde for ≥ 16 h. The samples were dehydrated with a series of increasing ethanol concentrations, and then embedded in Paraplast Plus (Sigma). After preparing 8‐µm sections, the samples were counter‐stained and observed with the BX53 microscope (Olympus, Tokyo, Japan). For scanning electron microscopy (SEM), the lemmas from mature seeds were fixed in 2.5% (w/v) glutaraldehyde. After dehydrating with a series of increasing ethanol concentrations, the samples were dried to a critical point and mounted on stubs. The inner surfaces of the lemmas were observed with the S‐3000N scanning electron microscope (Hitachi High‐Technologies Corp., Tokyo, Japan). The cell length, width and area were measured with the imagej program (US National Institutes of Health, Bethesda, MD, USA).
Quantitative real‐time (qRT)‐PCR analysis
Total RNA (including miRNA) was extracted using the miRNeasy Mini kit (Qiagen). Residual genomic DNA was eliminated with RNase‐free DNase I (Promega). To analyze miR530 expression, miRNA first‐strand cDNA was synthesized with the miRcute miRNA First‐Strand cDNA Synthesis kit (Tiangen, Beijing, China). The expression of miR530 was then quantified by qRT‐PCR with the SYBR PrimeScript™ miRNA RT‐PCR kit (Tiangen) with 5.8S rRNA as the internal control. To analyze the expression of the OsmiR530 precursor, the target mimic and OsPL3, c. 2 μg total RNA for each sample was reverse‐transcribed with the M‐MLV RTase cDNA synthesis kit (TaKaRa, Dalian, China). The resulting cDNA was used as the template for a qRT‐PCR assay, which was completed with the SYBR Green PCR master mix (TaKaRa). The OsEF‐1α gene was used as the internal control. The qRT‐PCR analysis involved three biological replicates for each miRNA and gene. The relative expression ratios of OsmiR530, its precursor OsMIR530 and OsPL3 were calculated according to the delta‐delta threshold cycle relative quantification method. Details regarding the qRT‐PCR primers are provided in Table S1.
In situ hybridization
The miRNA in situ hybridization was completed as described previously (Y. C. Zhang et al., 2013). Digoxigenin‐labeled antisense (5′‐AGTGTTAGCCTGTTGGCGGTGTAA‐3′) and sense (5′‐TTACACCGCCAACAGGCTAACACT‐3′) probes for OsPL3 were synthesized by Beijing AuGCT Biotech Company, China. The 5′‐ and 3′‐digoxin‐labeled LNA™ probe for OsmiR530 (5DiGN/TAGGTGCAGGTGCAAATGCA/3DiG_N) was purchased from Qiagen (http://www.exiqon.com). A scrambled miRNA probe was used as a negative control.
RNA ligase‐mediated 5′‐rapid amplification of cDNA ends (RLM 5′‐RACE)
The RLM 5′‐RACE assay was performed with the RLM‐RACE kit (Takara) according to the manufacturer′s instruction. Approximately 2 µg WT seedling total RNA was ligated to the RNA Oligo adaptor without a calf intestinal phosphatase treatment. Two rounds of nested PCR were performed, after which the PCR products were inserted into a cloning vector for sequencing. The primers used in this assay are listed in Table S1.
Subcellular localization
In order to generate the 35S::OsPL3‐GFP plasmid, the full‐length OsPL3 coding sequence without the stop codon was inserted into the Cam35S‐GFP vector between the BamHI and XbaI sites. Rice protoplasts were transformed with the 35S::OsPL3‐GFP construct as described previously (Zhang et al., 2012). GFP signals were observed with the FV10‐ASW confocal microscope (Olympus). The far‐red fluorescent protein mKate with an N‐terminal nuclear localization sequence (NLS), NLS‐mKate, was used for an analysis of the co‐localization. The primers used in this assay are listed in Table S1.
Yeast‐one‐hybrid assay (Y1H)
A Y1H assay was completed with the Matchmaker™ Gold Yeast One‐Hybrid Library Screening System (Clontech Laboratories, Mountain View, CA, USA). Two DNA fragments containing two adjacent G‐box (5′‐CACGTG‐3′) and one normal G‐box element in the OsMIR530 promoter were synthesized separately (Table S1) and inserted into the reporter vector pAbAi to obtain the pMIR530A‐AbAi and pMIR530B‐AbAi plasmids, respectively. To test the specificity of the binding sites, two fragments carrying the same flanking regions but with mutated G‐box elements (i.e. replaced with 5′‐TGACCT‐3′) (Table S1), were synthesized and inserted into the pAbAi reporter vector to generate the pMIR530Am‐AbAi and pMIR530Bm‐AbAi plasmids. The full‐length OsPIL15 coding sequence was amplified by PCR and inserted into the pGADT7 vector. The recombinant pGADT7‐OsPIL15 plasmid was used to transform Y1HGold yeast strain cells carrying the linearized pMIR530A‐AbAi, pMIR530B‐AbAi, pMIR530Am‐AbAi and pMIR530Bm‐AbAi. Transformed yeast cells were detected by spotting serial dilutions (1 : 1, 1 : 10, 1 : 100 and 1 : 1000) of yeast onto agar‐solidified synthetic dextrose (SD)/−Leu medium supplemented with 600 ng ml−1 aureobasidin A (AbA). The pGADT7‐Rec‐p53 and p53‐AbAi plasmids were used as positive controls. Details regarding the primers used for this assay are listed in Table S1.
Electrophoretic mobility shift assay (EMSA)
An EMSA was conducted as described previously (Ma et al., 2009). The full‐length OsPIL15 coding sequence was synthesized and cloned into the expression vector pET28a. The resulting recombinant plasmid was transformed into Escherichia coli BL21 (DE3) cells to obtain positive clones. The His‐tagged fusion proteins produced in the transformed E. coli cells were purified as described previously (Ji et al., 2019).
The G‐box oligonucleotide sequences were synthesized and separately labeled with biotin by using the Biotin 3′ End DNA Labeling Kit (Beyotime, Shanghai, China). DNA probes were obtained by annealing two complementary oligonucleotides (Table S1), and then used in the assay completed with the EMSA kit (Beyotime).
Chromatin immunoprecipitation and qRT‐PCR (ChIP‐qPCR) analysis
A ChIP‐qPCR assay was performed as described previously (Lee et al., 2007). Briefly, the aboveground parts of OsPIL15:eGFP transgenic plants at the four‐leaf stage were treated with 1% formaldehyde (i.e. cross‐linking treatment). The chromatin complexes were sonicated at 4°C to generate 200–500‐bp fragments. The sheared chromatin was immunoprecipitated, washed and reverse cross‐linked. A polyclonal anti‐GFP antibody (Abcam, Cambridge, UK) was used, with IgG as a negative control. The purified precipitated DNA was dissolved in water for a qRT‐PCR analysis. Details regarding the primers used for this assay are listed in Table S1.
Transcriptional activity assay
The transcriptional activity in Nicotiana benthamiana leaves was examined as described previously (Sun et al., 2012). The 2‐kb OsMIR530 promoter sequence was ligated to the luciferase reporter gene LUC in the plant binary vector pGWB35 (Nakagawa et al., 2007) via Gateway reactions (Invitrogen) to generate the reporter construct. To prepare the effector construct, the OsPIL15 coding sequence was cloned into the pCAMBIA 1300‐FLAG vector between the BamHI and SpeI restriction sites. The reporter and effector constructs were inserted separately into A. tumefaciens strain GV3101 cells for the subsequent co‐infiltration of N. Benthamiana leaves. The LUC signals were detected and quantified with the NightSHADE LB 985 Plant Imaging System (Berthold, Bad Wildbad, Germany) at 48 h after infiltrations. Ten independent N. benthamiana leaves were infiltrated and analyzed for each of three biological replicates. Details regarding the primers used for this assay are listed in Table S1.
Yeast‐two‐hybrid (Y2H) assay
The GAL4‐AD and GAL4‐BD derivatives were generated by fusing the full‐length OsPIL15 coding sequence and the C‐terminal of PHYB sequence (i.e. 1891–3513 bp), respectively. The AD‐OsPIL15 and BD‐PHYB‐C constructs were used to co‐transform the Saccharomyces cerevisiae strain AH109 cells. The transformed yeast colonies were transferred to agar‐solidified SD/−Leu/−Trp/−His medium to detect interactions. Details regarding the primers used for this assay are listed in Table S1.
Firefly luciferase complementation imaging (LCI) assay
An LCI assay was performed in N. benthamiana leaves as described previously (Sun et al., 2013). The full‐length PHYB and OsPIL15 coding sequences were separately fused to the fragments encoding the N‐terminal and C‐terminal parts of LUC. The N. benthamiana leaves were co‐infiltrated with the A. tumefaciens cells harboring the nLUC and cLUC constructs. After 48 h, the leaves were analyzed for LUC activity. The primers used in this assay are listed in Table S1.
Semi‐in vivo pull‐down assay
In order to determine the interaction between PHYB and OsPIL15, leaves were collected from WT and PHYB‐GFP seedlings incubated for 14 d in a growth chamber with a 12‐h light (28°C) : 12‐h dark (24°C) cycle. Total proteins were extracted from the leaves with lysis buffer 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA (pH 8.0), 0.1% Triton X‐100 and 0.2% NP‐40, with freshly added 10 mM PMSF, 20 mM MG132 and a protease inhibitor cocktail (Roche). Approximately 5 µg OsPIL15‐His protein was mixed with the total protein extracts from WT or PHYB‐GFP plants and then incubated overnight with anti‐GFP (MBL, Nagoya, Japan) conjugated beads at 4°C under red‐light conditions. The beads were then washed five times with 2‐ml lysis buffer and eluted samples were analyzed by immunoblotting with anti‐GFP (1 : 2000; Roche Diagnostics) and anti‐His (1 : 3000; CWBIO, Beijing, China) antibodies.
Results
OsmiR530 regulates grain yield by altering grain size and panicle architecture
In order to investigate the functions of OsmiR530 related to rice growth and development, we generated miR530‐OE and MIM530 lines. The expression of OsmiR530 and the target mimic were under the control of a constitutively active ubiquitin promoter. The qRT‐PCR analyses indicated that the expression levels of OsmiR530 in the miR530‐OE lines and the OsmiR530 target mimic in the MIM530 lines were significantly elevated to varying degrees (Fig. S1a,b). The miR530‐OE plants exhibited dwarfism (c. 78.3% of the WT height; P < 0.01) (Figs 1a, S1c). Compared with the WT plants, the miR530‐OE plants produced shorter panicles (c. 77.8% of the WT panicle length; P < 0.01) and more aborted seeds (c. 64.4% of the WT seed‐setting rate; P < 0.01) (Figs 1b, S1d–f). Further analyses of the panicle indicated that compared with the WT plants, the miR530‐OE lines had considerably fewer primary (c. 93.8% of WT; P < 0.05) and secondary (c. 68.0% of WT; P < 0.001) branches and fewer effective grains per main panicle (c. 66.1% of WT; P < 0.001) (Fig. 1c). Moreover, the grain length and width and 1000‐grain weight decreased significantly in the miR530‐OE lines (Fig. 1d–f). In contrast to the miR530‐OE plants, the MIM530 plants were taller than the WT plants and produced longer panicles, more panicle branches and larger seeds (Figs 1a–f, S1c–g). The grain length, grain width and 1000‐grain weight also were significantly greater for the MIM530 lines than for the WT plants (Fig. 1d–f).
Figure 1.
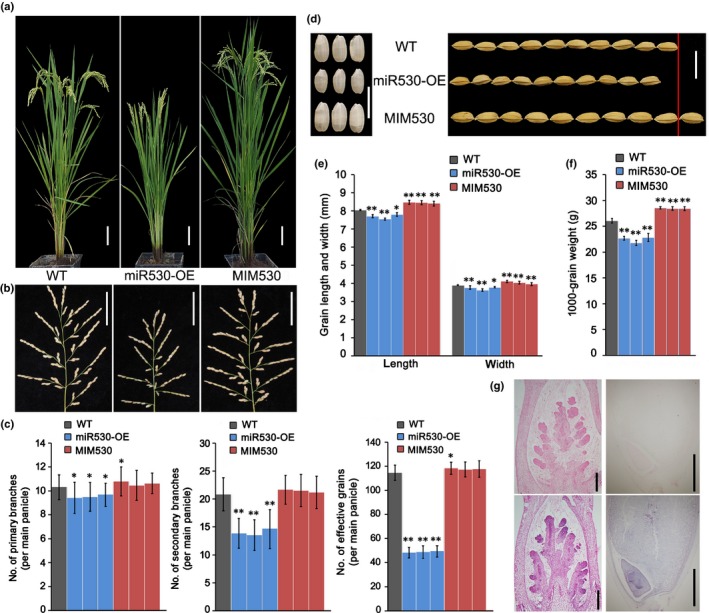
Phenotypes and agronomic traits of OsmiR530‐overexpressing (miR530‐OE) and target mimic (MIM530) lines (miRNA, microRNA; Os, Oryza sativa). (a) Morphologies of rice transgenic and wild‐type (WT) plants. Bars, 10 cm. (b) Panicle morphologies of the rice transgenic and WT plants. Bars, 5 cm. (c) Number of primary and secondary branches and number of effective grains per main panicle in rice transgenic and WT plants. (d) Brown rice grains (left) and grains with hulls (right) of rice transgenic and WT plants. Bars, 1 cm. (e) Grain length and width of rice transgenic and WT plants. (f) The 1000‐grain weight of rice transgenic and WT plants. (g) In situ hybridization of OsmiR530 in the panicle at the secondary branch formation stage (left column) and in the seed at 8 d after pollination (right column) in rice. The upper panels are the negative controls. Bars, 200 μm (left column), and 1 mm (right column). For all histograms, gray bars represent WT, blue bars represent miR530‐OE transgenic lines #a1, #b5 and #c2 (from left to right), and red bars represent MIM530 transgenic lines #2, #3 and #6 (from left to right). The data in (c), (e) and (f) are presented as the mean ± SD (c, n = 20 plants; e, n > 300 seeds; f, n = 3 replicates). Significant differences: *, P < 0.05; **, P < 0.01 (Student's t‐test).
The number of vascular bundles in the peduncle is reportedly related to the number of panicle branches and grains (Ikeda‐Kawakatsu et al., 2009; Terao et al., 2010). Correspondingly, compared with the peduncle diameter of the WT plants, those of the miR530‐OE and MIM530 lines were smaller and larger, respectively (Fig. S2a). Transverse sections of the internode just below the neck node revealed that there were considerably fewer large and small vascular bundles in the miR530‐OE lines than in the WT plants, whereas there were more vascular bundles in the MIM530 lines (Fig. S2b–d). These results suggest that OsmiR530 affects the formation of vascular bundles in the peduncle, which influences the peduncle diameter and panicle branching.
The grain yields of the WT plants and the miR530‐OE and MIM530 lines were determined in field plots in Jinan, China (lat 36°40′N, long 117°00′E) to evaluate the effect of OsmiR530. Compared with the grain yield of WT plants, those of the miR530‐OE and MIM530 lines were c. 43.29% lower and c. 7.93% higher, respectively (Table 1). Overall, these results indicate that OsmiR530 negatively regulates rice yield.
Table 1.
Rice grain yields of wild‐type (WT), OsmiR530‐overexpressing (miR530‐OE) and OsmiR530 target mimic (MIM530) rice lines, and knockout PLUS3‐domain‐containing protein (PL3‐KO) plants grown in a paddy field.
| Traits | WT | miR530‐OE | MIM530 | PL3‐KO |
|---|---|---|---|---|
| Yield per plot (kg) | 1.64 ± 0.06 | 0.93 ± 0.09 (P < 0.001) | 1.77 ± 0.08 (P = 0.018) | 1.31 ± 0.07 (P < 0.001) |
| Yield comparison with WT (%) | — | −43.29 | +7.93 | −20.12 |
Plants were arranged in plots according to a randomized complete block design with four replicates. Three transgenic lines were mixed and grown in one plot. Nipponbare (WT) was used as a control. Plants were grown under natural conditions in Jinan, China in 2018. The planting density was 20 cm × 25 cm, with one plant per hill. Data are presented as the mean ± SD (n = 4 plots, 3.0 m2 per plot). Significant differences were determined with Student's t‐test.
Spatial and temporal expression pattern of OsmiR530
MicroRNA expression patterns may provide clues regarding its functions. Thus, OsmiR530 expression was examined in the root, flag leaf, node, panicle, embryo and seed. The qRT‐PCR data indicated that OsmiR530 was expressed in all examined organs, especially in the flag leaf (Fig. S3a). Previous studies on rice revealed that the photosynthetic products related to grain‐filling are derived mainly from the top three leaves, especially the flag leaf (Al‐Sady et al., 2006; Yue et al., 2006). Therefore, the accumulation of OsmiR530 in the flag leaf implies that it may have an important effect on grain yield. Consistent with this possibility, overexpression or repressed expression of OsmiR530 altered the panicle architecture and grain size (Fig. 1). We performed an RNA in situ hybridization experiment to examine OsmiR530 expression in the inflorescence meristem and seeds at 8 d after pollination. OsmiR530 was strongly expressed in the secondary branch primordia (Fig. 1g, left column) at the late secondary branch formation stage. In seeds, intense and weak OsmiR530 signals were detected in the embryo and endosperm cells, respectively (Fig. 1g, right column).
OsmiR530 regulates grain size by controlling cell division and expansion
Because the spikelet hull restricts grain size, we measured the spikelet hulls of the WT, miR530‐OE and MIM530 plants. The spikelet hulls of the miR530‐OE lines were significantly shorter and thinner than those of the WT plants, whereas spikelet hulls of the MIM530 lines were longer and wider than those of the WT plants (Figs 2a, S1g). The enlargement of plant organ is generally due to cell division and expansion. To clarify the grain size changes in the transgenic lines, histological cross‐sections of the spikelet hulls just before anthesis were analyzed. Compared with the WT plants, the MIM530 and miR530‐OE lines had more and fewer outer parenchyma cells in the grain hull, respectively (Fig. 2b,c). These findings suggest that miR530 negatively regulates cell division in rice. Furthermore, the inner epidermal cells of the lemma in the miR530‐OE, MIM530 and WT plants were analyzed by SEM. The areas of the inner epidermal cells of the spikelet followed the same rank order as the number of parenchyma cells: miR530‐OE < WT < MIM530 (Fig. 2d,e). The increasing epidermal cell areas in the MIM530 lines were due mainly to increases in cell length, but not cell width (Fig. 2f,g). These results imply that repressing the expression of OsmiR530 sufficiently promotes the cell expansion and proliferation in the spikelet hulls to produce larger seeds.
Figure 2.
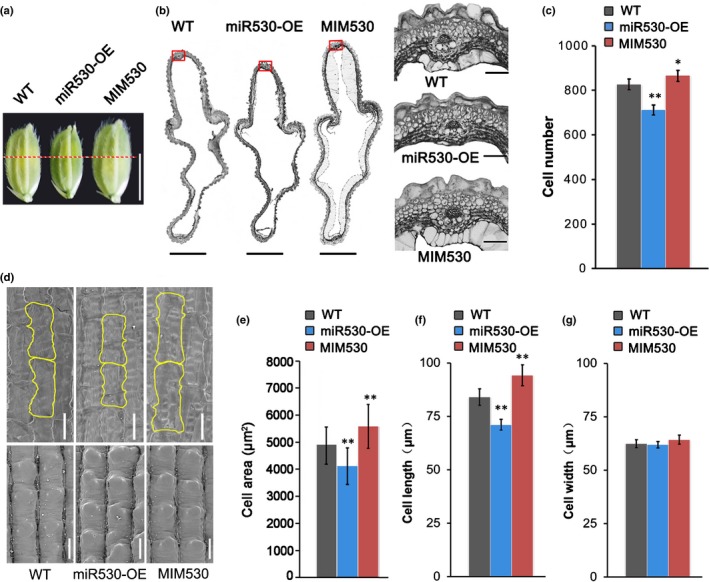
Histological analysis of spikelet hulls in OsmiR530 transgenic lines and wild‐type (WT) plants (miRNA, microRNA; Os, Oryza sativa). (a) Spikelet hulls of rice WT and transgenic plants just before anthesis. Bar, 5 mm. (b) Cross‐sections of the middle parts of spikelet hulls (marked by red dashed lines in a) of rice WT and transgenic plants just before anthesis (left). Bars, 500 μm. Magnified images of the boxed cross‐section areas are provided on the right. Bars, 50 μm. (c) Number of cells in the outer parenchyma layer of the spikelet hulls of rice WT and transgenic plants (n = 5 spikelets). (d) Scanning electron micrographs of the inner epidermal cells of lemmas in the mature seeds of rice WT and transgenic plants. Bars, 50 μm. (e) Cell areas in the middle part of the inner epidermis of lemmas in fully mature rice seeds (n = 5 seeds). (f) Cell lengths in the middle part of the inner epidermis of lemmas in fully mature rice seeds (n = 5 seeds). (g) Cell widths in the middle part of the inner epidermis of lemmas in fully mature rice seeds (n = 5 seeds). The data in (c), (e), (f) and (g) are presented as the mean ± SD. Significant differences: *, P < 0.05; **, P < 0.01 (Student's t‐test).
OsPL3 is a direct target of OsmiR530
In plants, miRNAs exert their functions by inhibiting the expression of their target genes. To uncover the molecular mechanism underlying the regulatory function of OsmiR530 influencing rice yield, we identified its downstream targets by two methods. A previous study predicted that one of the genes targeted by OsmiR530 is OsPL3 (LOC_Os01g56780), which encodes a PLUS3 domain‐containing protein. OsPL3 mRNAs were spliced between the 10th and 11th base pair of the OsmiR530 target region based on degradome sequencing result (Sun et al., 2015). To further validate the target, we performed RLM5′‐RACE to map the OsmiR530‐directed cleavage sites in OsPL3 mRNA. Of 15 randomly selected clones, 13 possessed the 5′ ends of the cleaved fragments at the same site of the OsmiR530 target sequence as that detected by degradome sequencing (Fig. 3a), indicating that OsPL3 can be precisely cleaved by OsmiR530 in vivo.
Figure 3.
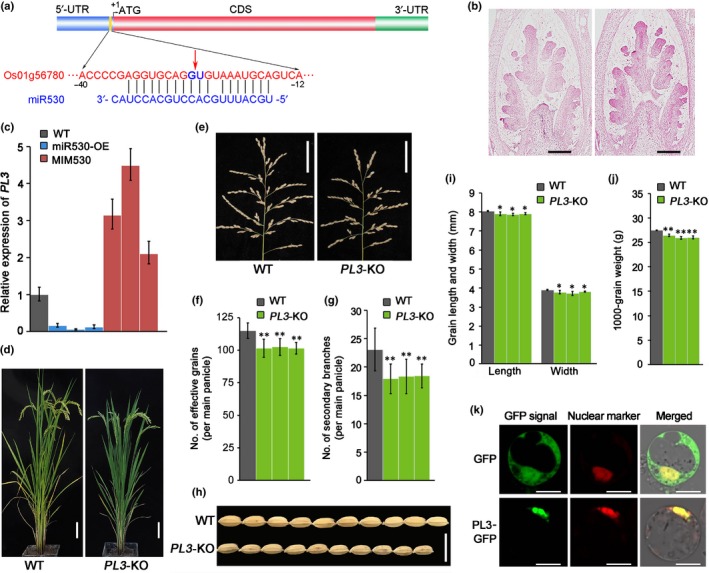
Phenotypes and agronomic traits of OsPL3‐knockout (PL3‐KO) lines (Os, Oryza sativa; PL3, PLUS3‐domain‐containing protein). (a) Illustration of the OsmiR530 cleavage site in the OsPL3 mRNA. (b) In situ hybridization of OsPL3 in the panicle at the secondary branch formation stage in rice. The left graph presents the data for the negative control (sense probe). Bars, 200 μm. (c) The expression of OsPL3 in OsmiR530‐overexpressing (miR530‐OE) and OsmiR530 target mimic (MIM530) rice lines. The OsEF‐1α gene was used as an internal reference for quantitative real‐time (qRT)‐PCR. Data are presented as the means ± SD of three biological replicates. (d) Morphologies of rice PL3‐KO and wild‐type (WT) plants. Bars, 10 cm. (e) Panicle morphologies of the rice PL3‐KO and WT plants. Bars, 5 cm. (f) Number of effective grains per main panicle in rice PL3‐KO and WT plants. (g) Number of secondary branches in rice PL3‐KO and WT plants. (h) Grains with hulls from rice WT and PL3‐KO plants. Bar, 1 cm. (i) Grain length and width of rice transgenic and WT plants. (j) The 1000‐grain weight of rice transgenic and WT plants. (k) Subcellular localization of OsPL3 in rice protoplasts. Bars, 10 μm. For all histograms, gray bars represent WT, green bars represent PL3‐KO transgenic lines #12, #13 and #16 (from left to right). The data in (c), (f), (g), (i) and (j) are presented as the mean ± SD (f and g, n = 20 plants; i, n > 300 seeds; j, n = 3 replicates). Significant differences: *, P < 0.05; **, P < 0.01 (Student's t‐test).
We then investigated the spatial expression patterns of OsPL3 in various rice organs using qRT‐PCR. Similar to OsmiR530, OsPL3 was detected in most of the tissues analyzed, especially in the flag leaf (Fig. S3b). The results of an RNA in situ hybridization revealed that OsPL3 also was expressed in the primordia of the secondary panicle branches in the inflorescence meristem (Fig. 3b). Strikingly, the OsPL3 expression patterns were anti‐correlated with OsmiR530 in the examined floral organs and seeds (Fig. S3). Additionally, OsPL3 expression was significantly downregulated in the miR530‐OE lines and upregulated in the MIM530 lines (Fig. 3c), suggesting that OsmiR530 negatively regulates OsPL3 expression. These results imply that the OsPL3 expression level is dependent on the level of OsmiR530 through an OsmiR530‐directed cleavage mode in vivo.
In order to elucidate the function of OsPL3 related to rice panicle and seed development, we generated three independent OsPL3‐knockout (PL3‐KO) lines in the Nip background using CRISPR/Cas9 technology. The PL3‐KO1 (#12) plants had a 10‐bp deletion in the third exon of OsPL3, whereas the PL3‐KO2 (#13) and PL3‐KO3 (#16) plants had a single base (A) inserted in the third exon at 1693 and 1797 bp from the translation start site, respectively (Fig. S4). The deletion or insertions created new stop codons that result in a truncated OsPL3 lacking the PLUS3 domain (Fig. S5). As expected, the three PL3‐KO lines showed similar phenotypes as those of miR530‐OE plants, including dwarfism, smaller panicles, decreased effective grains per main panicle, low seed‐setting rate, fewer secondary branches per main panicle and smaller seeds (Figs 3d–j, S6). A field trial showed that knocking out OsPL3 led to c. 20.12% yield losses (Table 1). These results confirmed that OsPL3 is the major target of OsmiR530 related to the regulation of panicle and seed development in rice.
Protein structure analysis revealed that OsPL3 contains a PLUS3 domain in the C‐terminal, which is essential for nucleic acid binding. Subcellular localization in rice protoplasts detected OsPL3 in the nucleus (Fig. 3k). However, the molecular mechanism by which OsPL3 affects panicle branching and seed development in rice remains to be characterized.
OsPIL15 directly activates OsMIR530 transcription to regulate grain size and panicle architecture
In order to identify the upstream regulators of OsmiR530, the 2‐kb upstream sequence of OsMIR530 promoter was analyzed, and three G‐box elements were identified. Because the G‐box is the core element for the targets of PIFs and PILs (Martínez‐García et al., 2000; Castillon et al., 2007; Hornitschek et al., 2009; Leivar & Quail, 2011), we assumed that the rice PIL family members might be able to bind to the OsMIR530 promoter.
Among the six PIL genes (OsPIL11–16) in the rice genome (Nakamura et al., 2007), OsPIL15 was identified to play important roles in grain size regulation (Ji et al., 2019). We also analyzed the PIL15‐OE (Zhou et al., 2014) and PIL15‐KO transgenic lines (Figs 4, S7, S8). Consistent with the results of a recent study (Ji et al., 2019), the PIL15‐OE lines produced smaller seeds than the WT plants, whereas the PIL15‐KO lines produced larger seeds (Fig. 4a–c). Additionally, we observed that OsPIL15 negatively regulates secondary branching. The OsPIL15‐OE plants had significantly fewer secondary branches compared with the WT plants, whereas the OsPIL15‐KO plants had considerably more secondary branches (Fig. 4d). The phenotypic similarities between the OsPIL15‐OE and miR530‐OE plants, and between the OsPIL15‐KO and MIM530 plants suggest that OsPIL15 may directly regulate OsMIR530 expression.
Figure 4.
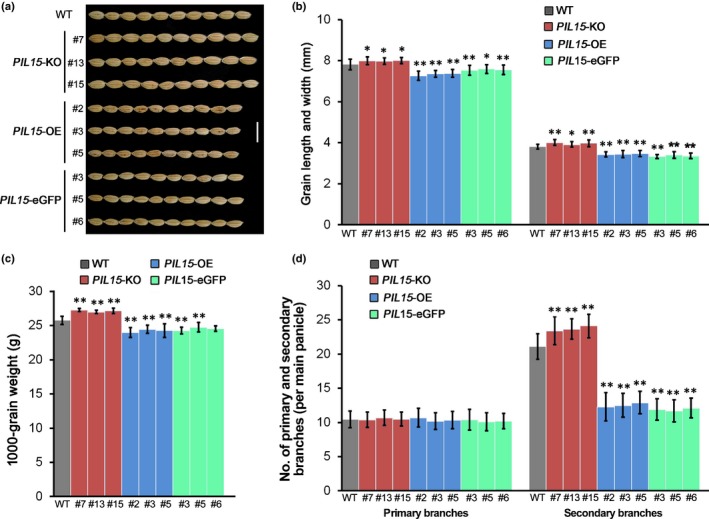
Agronomic traits of OsPIL15‐overexpressing (PIL15‐OE and PIL15‐eGFP) and knockout (PIL15‐KO) transgenic lines (GFP, green fluorescent protein; Os, Oryza sativa: PIL, phytochrome‐interacting factor (PIF)‐like gene). (a) Grains with hulls (right) from rice wild‐type (WT) and transgenic plants. Bar, 1 cm. (b) Grain length and width of rice WT and transgenic plants. (c) The 1000‐grain weight of rice WT and transgenic lines. (d) Number of primary and secondary branches in rice WT and OsPIL15 transgenic plants. The data in (b), (c) and (d) are presented as the mean ± SD (b, n > 300 seeds; c, n = 3 replicates; d, n = 20 panicles). Significant differences: *, P < 0.05; **, P < 0.01 (Student's t‐test).
The three G‐box elements identified in the OsMIR530 promoter were located < 600 bp from the OsMIR530 transcription start site and were divided artificially into fragments A and B (Fig. 5a). The interaction between OsPIL15 and the OsMIR530 promoter was first assessed in a Y1H assay, which revealed that OsPIL15 can bind to both fragments A and B with normal G‐box elements (Fig. 5b). However, mutations to the G‐box elements of these two fragments in the OsMIR530 promoter abolished this binding (Fig. 5b). To further determine whether the G‐box element in the OsMIR530 promoter is the direct binding site for OsPIL15, we conducted EMSA with the purified OsPIL15‐His recombinant protein and OsMIR530 probes with normal and mutated G‐box elements. As presented in Fig. 5(c), OsPIL15 specifically bound to OsMIR530 promoter fragments A and B with the normal G‐box sequence. The binding abilities to the two fragments decreased significantly in the presence of unlabeled competitor probes A and B, but not with the unlabeled probes with mutated G‐box sequences (Fig. 5c). These results suggest the G‐box motif is critical for the binding of OsPIL15 to the OsMIR530 promoter. A ChIP‐qPCR analysis was performed with the OsPIL15‐eGFP lines, which have phenotypes similar to those of PIL15‐OE lines (Fig. 4). The enrichment of fragments A and B containing one or two G‐box motifs in the 2‐kb sequence upstream of the OsMIR530 promoter was analyzed, with a fragment C located 1950 bp upstream of the OsMIR530 transcription start site used as a negative control (Fig. 5a). As shown in Fig. 5(d), fragments A and B were obviously enriched in the PIL15‐eGFP plants, whereas fragment C was not. These results further indicate that OsPIL15 binds to the G‐box elements in the OsMIR530 promoter region.
Figure 5.
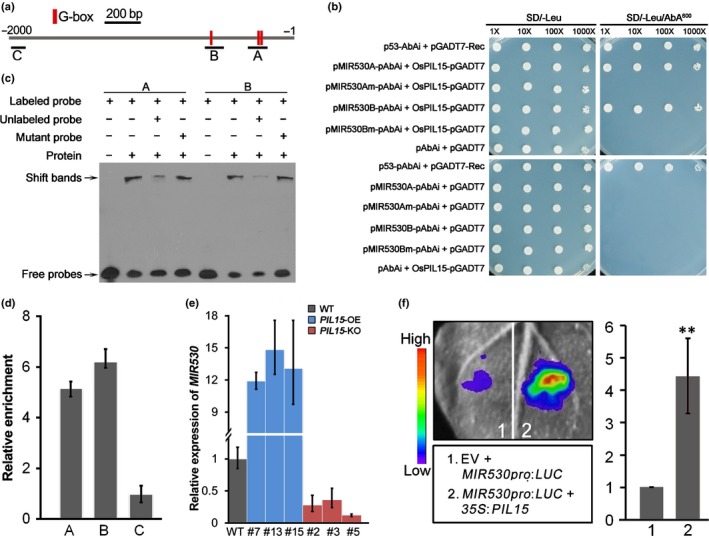
Identification of the OsMIR530 gene as a direct target of OsPIL15 (miRNA, microRNA; Os, Oryza sativa; PIL, phytochrome‐interacting factor (PIF)‐like gene). (a) Schematic representation of G‐box elements in the OsMIR530 promoter. (b) Yeast‐one‐hybrid assay of the binding of OsPIL15 to the OsMIR530 promoter. Two fragments containing two adjacent G‐box and one single G‐box elements were synthesized, and then were separately inserted into the reporter vector pAbAi to obtain the pMIR530A‐AbAi and pMIR530B‐AbAi construct, respectively. Meanwhile, the pMIR530Am‐AbAi and pMIR530Bm‐AbAi constructs were obtained using the same sequences with mutated G‐box elements. pGADT7‐Rec‐p53 and p53‐AbAi were used as positive controls. (c) Electrophoretic mobility shift assay. Unlabeled probes containing G‐box elements at 50× the amount of the labeled probe competed for the OsPIL15 binding sites. Unlabeled probes with mutations in G‐box elements were also used in this assay. (d) Chromatin immunoprecipitation and quantitative real‐time (qRT)‐PCR analysis of OsPIL15 binding to the OsMIR530 promoter. The DNA samples acquired before immunoprecipitation were used as the input, and IgG was set as a negative control. The qRT‐PCR was completed with three replicates. Signal intensities were first normalized relative to the input, then the enrichment of each fragment was calculated using the Input% of IgG as a baseline. (e) Expression of OsMIR530 (OsmiR530 precursor) in rice PIL15‐OE and PIL15‐KO transgenic plants. The OsEF‐1α gene was used as an internal reference for the qRT‐PCR. Data are presented as the mean ± SD of three biological replicates. (f) Transient expression assays indicated that OsMIR530 transcription was activated by OsPIL15. Representative images of Nicotiana benthamiana at 48 h post‐infiltration (left). More than 10 independent N. benthamiana leaves were infiltrated in the experiment. The histogram (right) presents the intensities of the luminescence indicated in the photo (left). Significant differences: *, P < 0.05; **, P < 0.01 (Student's t‐test).
In order to determine how OsPIL15 regulates OsMIR530 expression, we detected the expression of the OsmiR530 precursor (OsMIR530) and the mature OsmiR530 in PIL15‐OE and PIL15‐KO lines. The OsMIR530 transcripts obviously accumulated in the PIL15‐OE lines, but decreased in PIL15‐KO lines (Fig. 5e). The mature OsmiR530 and its precursor OsMIR530 exhibited similar expression patterns in OsPIL15 transgenic lines (Fig. S9). These results imply that the expression of OsMIR530 is positively correlated with that of OsPIL15. We subsequently conducted transient expression assays to confirm the in vivo result. The A. tumefaciens strains harboring the p35S:OsPIL15 effector and pOsMIR530:LUC reporter plasmids were used to infiltrate N. benthamiana leaf epidermal cells. The LUC activity was examined after a 2‐d incubation in darkness. The co‐expression of p35S:OsPIL15 and pOsMIR530:LUC sharply increased the LUC reporter activity, in contrast to the effects of the empty vector control (Fig. 5f). Overall, these results demonstrate that OsPIL15 can activate OsMIR530 expression in vivo by directly binding to the promoter G‐box elements.
Evolution and genetic variations in OsmiR530
By screening the miR530 family in the miRBase database, we determined that miR530 is specific to plants. Most plant species, including rice, have only one miR530 member in their genomes, but the Glycine max genome has the most miR530 family members (Gma‐miR530a, b, c, d and e) (Fig. S10). Compared with the mature osa‐miR530‐5p sequence in miRBase, the OsmiR530 identified in the phyB mutant and WT (Nip) plants contains an additional base (cytosine) at the 3′ end. Both osa‐miR530‐5p and OsmiR530 were derived from the same precursor and osa‐miR530‐5p was not detected in the phyB mutant and Nip small RNA libraries. These observations suggest that OsmiR530 is the main miR530 family member in our plant materials. Additionally, OsmiR530 homologs were not identified in the genomes of the widely used model plants Arabidopsis and maize. These findings imply that miR530 has unique functions during plant divergence and speciation.
Considering that OsmiR530 plays an important role in mediating rice grain yield, we explored the variations in the mature miR530 sequence as well as in its precursor sequence and its promoter sequences among 3024 rice varieties in the 3000 Rice Genome Project (Zheng et al., 2015). No polymorphisms were identified in the mature sequences of all rice varieties, whereas three single nucleotide polymorphisms (SNPs), G/A‐C/T‐G/A, were identified in the miR530 precursor sequences (Fig. 6a). Most of the rice varieties (97.7%, 2953 of 3024) were classified into two haplotypes (HAP1 with G‐C‐G SNPs and HAP2 with A‐T‐A SNPs in the miR530 precursor sequences, respectively) (Fig. 6a,b). The majority of indica varieties (98.6%, 1733 of 1758) were HAP1 haplotype (Fig. 6b). By contrast, the occurrence of the HAP2 haplotype was observed mainly in the temperate japonica (92.3%, 250 of 271) and subtropical japonica (75.9%, 85 of 112) varieties (Fig. 6b). However, the functional significance of these three SNPs remains elusive. In the OsMIR530 promoter regions, 13 SNPs and three insertion–deletion polymorphisms (InDels) were identified in all examined rice accessions (Fig. 6a). Given that InDels cause more dramatic alterations to the genome structure than SNPs, subsequent analyses were focused on the InDels in the OsMIR530 promoter regions (Fig. 6c). Most of the rice accessions (99.1%, 2998 of 3024) were classified into three haplotypes (MIR530NIP, MIR530MH63 and MIR530GL4) represented by Nip, Minghui63 (MH63) and Guangluai4 (GL4), respectively. The MIR530NIP and MIR530MH63 haplotypes have a single 6‐bp (5′‐AGATGG‐3′) and 4‐bp (5′‐CATA‐3′) insertion at different positions, respectively (Fig. 6a). The MIR530GL4 haplotype has both insertions in the promoter region (Fig. 6a). The majority of japonica varieties were the MIR530NIP haplotype, as indicated by the 93.4% (268 of 287), 76.1% (86 of 113) and 58.5% (48 of 82) occurrence in the temperate japonica, subtropical japonica and admixed japonica varieties, respectively (Fig. 6c). Only a small portion of the tropical japonica varieties (12.6%, 47 of 372) were classified as the MIR530NIP haplotype. Additionally, occurrences of MIR530MH63 haplotype in centrum‐Aus and indica varieties were 41.4% (82 of 198) and 34.5% (611 of 1771), respectively. However, the greater frequency of the MIR530GL4 haplotype was observed not only in the centrum‐Aus (54.0%, 107 of 198) and indica (61.0%, 1080 of 1771) varieties, but also in the tropical japonica varieties (86.3%, 321 of 372). Interestingly, HAP1 based on three SNPs in the miR530 precursor sequences was well correlated with MIR530MH63 and MIR530GL4, whereas HAP2 was correlated with MIR530NIP (Fig. 6d). These results suggest that the OsMIR530 locus was subjected to artificial selection during rice breeding.
Figure 6.
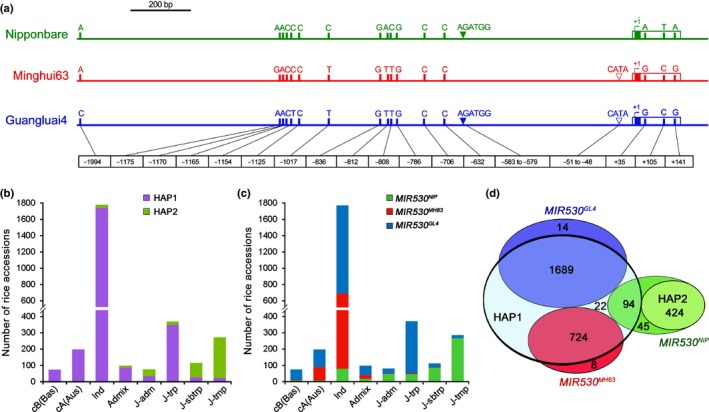
Analysis of genetic variations in OsMIR530 (miRNA, microRNA; Os, Oryza sativa). (a) Schematic representation of the genetic variations in OsMIR530 in a japonica variety (Nipponbare) and two indica varieties (Minghui63 and Guangluai4). The white boxes at the 3′ terminal represent the pre‐OsmiR530, which contains the mature OsmiR530 sequences of indicated by solid boxes marked in green, red and blue, respectively. The promoter sequence is presented with lines. The numbers at the bottom indicate the positions of the variations (The 5′ terminal nucleotide of the mature OsmiR530 is referred to as +1). (b) Distribution of three single nucleotide polymorphisms (SNPs) in the miR530 precursor sequences in 2953 rice varieties. The classification of the rice varieties is based on the 3000 Rice Genome Project database. (c) Distribution of three haplotypes of insertion–deletion polymorphisms (InDels) in the OsMIR530 promoter in 2998 rice varieties. (d) Schematic representation of the relationship between the three SNPs in the OsmiR530 precursor sequence and the three InDel polymorphisms in the OsMIR530 promoter sequence. NIP, Nipponbare; MH63, Minghui63, GL4, Guangluai4; cB (Bas), centrum‐Basmati population; cA(AUS), centrum‐Aus population; Ind, indica rice; Admix, admixed between any two or more of indica, japonica, cA(AUS) and cB (Bas); J‐adm, admixed japonica with two or more japonica subpopulations; J‐trp, tropical japonica; J‐sbtrp, subtropical japonica; J‐tmp, temperate japonica.
Discussion
Grain size and panicle branching, which are controlled by complex genetic networks, are two key factors influencing grain yield. Although a number of genes regulating grain yield have been identified, considerably fewer microRNAs (miRNAs) have been confirmed as important regulators of grain yield.
In this study, OsmiR530 was confirmed as a negative regulator of rice grain yield via affecting cell division and expansion in spikelet hulls. The overexpression of OsmiR530 decreased the seed size and inhibited panicle branching, whereas the OsmiR530 target mimic had the opposite effects (Fig. 1). To the best of our knowledge, this is the first report describing miR530 as a potential regulator of grain yield. Four other miRNAs have been confirmed as important regulators of rice grain yield, namely OsmiR156 (Jiao et al., 2010; Wang et al., 2012; Si et al., 2016), OsmiR396 (Duan et al., 2015; Gao et al., 2015), OsmiR397 (Y. C. Zhang et al., 2013) and OsmiR408 (Zhang et al., 2017). Although the effects of the previously reported miRNAs on grain yield are clear, their upstream factors were uncharacterized. In this study, we determine that OsmiR530 expression can be activated by phytochrome‐interacting factor (PIF)‐like gene OsPIL15 (Fig. 5), which is one of the homologs of Arabidopsis PIFs. Knocking out OsPIL15 enhances cell division in spikelet hulls, resulting in enlarged rice grains (i.e. longer and wider) and increased grain yield (Ji et al., 2019). Moreover, in this study, we observed that similar to OsmiR530, OsPIL15 negatively regulates grain yield by controlling grain size as well as panicle branching. The PIL15‐OE lines produced smaller seeds and fewer panicle branches compared with the wild‐type (WT) plants, whereas the knockout (PIL15‐KO) lines exhibited the opposite phenotypes (Fig. 4). These results suggest that OsPIL15 has important regulatory roles influencing grain size and panicle branching.
PIFs are believed to be one of the major targets of photoactivated phytochromes in Arabidopsis (Nakamura et al., 2007; Leivar & Quail, 2011). In the present study, we observed that OsMIR530 expression can be activated by OsPIL15 (Fig. 5). Additionally, the OsmiR530 expression is significantly upregulated in the phytochrome B (phyB) mutant (Sun et al., 2015). Thus, clarifying the relationship between OsPIL15 and PHYB is warranted. Accordingly, in the current study, we proved that OsPIL15 interacts physically with PHYB in vitro and in vivo (Fig. S11), which is consistent with the result from a recent study involving a bimolecular fluorescence complementation system (Xie et al., 2019). However, it was unclear how OsPIL15 and PHYB coordinately control rice grain size. Xie et al. (2019) revealed that the light‐induced degradation of OsPIL15 is partially dependent on PHYB in rice. In this context, there should be more OsPIL15 in the phyB mutant than in the WT control. We observed that the phyB mutant had smaller seeds (Fig. S12), resembling those of OsPIL15‐OE (Fig. 4). Thus, the direct interaction between PHYB and OsPIL15 regulates grain size by inducing the degradation of OsPIL15 and decreasing OsmiR530 expression.
The degradation of PIFs or PILs is essential for activating or repressing the expression of their downstream genes (Nakamura et al., 2007; Leivar & Quail, 2011). A recent study indicated that OsPIL15 upregulates the expression of a purine permease gene, OsPUP7, by binding to the N‐box (5′‐CACGCG‐3′) motifs in the promoter (Ji et al., 2019). The rice grains of the loss‐of‐function mutant ospup7, are significantly larger than those of the control (Zhonghua 11) (Qi & Xiong, 2013), which is consistent with the effects of a loss‐of‐function mutation to OsPIL15 (Ji et al., 2019). Although the 50‐seed weight of the ospup7 mutant is c. 125% greater than that of Zhonghua 11, the mutant produces c. 50% fewer tillers and spikelets per panicle than the control (Qi & Xiong, 2013). Thus, it is obvious that the reported increased grain yield of OsPIL15‐knockout plants (Ji et al., 2019) is not due to OsPUP7. In the present study, we identified OsMIR530 as a critical target of OsPIL15. Moreover, OsPIL15 can bind directly to the G‐box in the OsMIR530 promoter region to activate its expression (Fig. 5b–f). Most importantly, the phenotypes of the PIL15‐KO and PIL15‐OE lines were highly similar to those of the MIM530 and miR530‐OE lines, respectively (Fig. 4). Thus, we conclude that OsPIL15 regulates grain yield mainly by controlling OsmiR530 expression.
In this study, we identified OsPL3, which encodes a PLUS3‐domain‐containing protein, is directly targeted by OsmiR530 to regulate rice branching and grain size. Knocking out OsPL3 decreased the overall grain yield by ≤ 20.12% in a field trial (Table 1), but it cannot completely explain the 42.29% decrease in the grain yield of miR530‐OE lines (Table 1). In this context, we speculate that OsmiR530 decreases the grain yield partially by downregulating OsPL3 expression and that other targets probably also are involved in OsmiR530‐regulated grain yield. The PLUS3 domain is involved in nucleotide binding, especially for RNA and single‐stranded DNA (de Jong et al., 2008). Previous studies on human and yeast revealed that the PLUS3 domain affects transcriptional elongation, chromatin re‐organization and co‐transcriptional splicing (Ciftci‐Yilmaz & Mittler, 2008; de Jong et al., 2008; Wier et al., 2013). The only functionally characterized PLUS3 domain containing protein in plant is AtTZP in Arabidopsis, wherein it is important for regulating morning‐specific growth (Loudet et al., 2008) and initiating the flowering process (Kaiserli et al., 2015). Unlike AtTZP, which contains a zinc finger domain and a PLUS3 domain at its C‐terminal, OsPL3 has only a PLUS3 domain at the C‐terminal, implying that its function may differ from that of AtTZP. Thus the molecular mechanism underlying the OsPL3‐regulated rice grain yield should be explored. Considering that OsPL3 was localized to the nucleus in rice (Fig. 3k), we predict that OsPL3 is crucial for transcriptional control.
On the basis of our findings, we propose a working model for the regulatory roles of OsmiR530 in grain yield (Fig. 7). When OsPIL15 accumulates, it binds to G‐box elements in the OsMIR530 promoter to activate its expression in the nucleus. The increased production of OsmiR530 inhibits seed enlargement and panicle branching via downregulating the expression of OsPL3 and other targets, causing substantial yield losses.
Figure 7.

Proposed working model of the regulation of rice grain yield by the OsPIL15‐OsmiR530 module (miRNA, microRNA; Os, Oryza sativa; PIL, phytochrome‐interacting factor (PIF)‐like gene). The accumulation of OsPIL15 promotes OsMIR530 transcription. The overexpression of OsMIR530 leads to the accumulation of mature OsmiR530, which downregulates OsPL3 expression, thereby inhibiting grain size enlargement and panicle branching.
Genetic variations were identified in the OsMIR530 promoter among 2998 rice varieties (Fig. 6). Two insertions (5′‐AGATGG‐3′ and 5′‐CATA‐3′) were detected mainly in the promoter of indica, centrum‐Aus and tropical japonica rice, but were infrequently detected in temperate japonica and subtropical japonica rice varieties (Fig. 6a,c). However, a single insertion (5′‐AGATGG‐3′) in the OsMIR530 promoter was observed mainly in temperate japonica and subtropical japonica, but rarely in indica and centrum‐Aus rice (Fig. 6a,c). By contrast, another single insertion 5′‐CATA‐3′ in the OsMIR530 promoter was identified relatively frequently in indica and centrum‐Aus (Fig. 6a,c). It seems that two InDels were selected in the indica and japonica varieties under certain circumstances. These results imply that the OsMIR530 locus has been artificially selected during breeding, although the functional significance of the genetic variation in OsMIR530 remains elusive at present. Additionally, two haplotypes were identified based on three single nucleotide polymorphisms (SNPs) in the miR530 precursor (Fig. 6a,b), which are well‐correlated with MIR530GL4, MIR530MH63 and MIR530NIP (Fig. 6d). Whether and how these three SNPs affect OsMIR530 functions is one question that remains open. Moreover, the relationship between the genetic variation at the OsMIR530 locus and the functions of the encoded miRNA, especially regarding the regulation of grain yield, is worthy of being explored comprehensively in the future.
Author contributions
XX and XHX designed the experiments, wrote and revised the paper; WS and YL performed most of the experiments with the assistance of LX, YH, XL and HS; and WL and XHX completed the bioinformatics analysis. WS and XHX contributed equally to this work.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Agronomic traits of OsmiR530‐OE and target mimic (MIM530) lines.
Fig. S2 Comparison of the peduncle in miR530‐OE, target mimic (MIM530) and WT plants.
Fig. S3 Expression of OsmiR530 and OsPL3 in various rice tissues.
Fig. S4 Diagram of two sgRNAs targeting OsPL3 in the rice genome.
Fig. S5 Analysis of the OsPL3 protein sequence in the WT and OsPL3‐KO lines.
Fig. S6 Agronomic traits of OsPL3‐KO lines.
Fig. S7 Diagram of two sgRNAs targeting OsPIL15 in the rice genome.
Fig. S8 Analysis of the OsPIL15 protein sequence in the rice WT and OsPIL15‐KO lines.
Fig. S9 Expression of OsmiR530 in rice OsPIL15 transgenic lines.
Fig. S10 Alignment of mature miR530 sequences identified in miRBase.
Fig. S11 PHYB physically interacts with OsPIL15.
Fig. S12 Knockout of PHYB in rice results in smaller seed.
Table S1 Details regarding the primers and DNA fragments used in this study.
Acknowledgements
We thank Daolin Fu for providing vector PC186. This study was funded by the National Natural Science Foundation of China (31801343), the Natural Science Foundation of Shandong Province (ZR2018ZC08N2 and ZR2018LC005), the Young Talents Training Program of Shandong Academy of Agricultural Sciences, and the Science and Technology Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2018E16). We thank LiwenBianji, Edanz Editing China (http://www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
References
- Al‐Sady B, Ni W, Kircher S, Schäfer E, Quail PH. 2006. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome‐mediated degradation. Molecular Cell 23: 439–446. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. 2007. Phytochrome Interacting Factors: central players in phytochrome‐mediated light signaling networks. Trends in Plant Science 12: 514–521. [DOI] [PubMed] [Google Scholar]
- Ciftci‐Yilmaz S, Mittler R. 2008. The zinc finger network of plants. Cellular and Molecular Life Sciences 65: 1150–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan P, Ni S, Wang J, Zhang B, Xu R, Wang Y, Chen H, Zhu X, Li Y. 2015. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nature Plants 2: 15203. [DOI] [PubMed] [Google Scholar]
- Franco‐Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio‐Somoza I, Leyva A, Weigel D, Garcia JA, Paz‐Ares J. 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Gao F, Wang K, Liu Y, Chen Y, Chen P, Shi Z, Luo J, Jiang D, Fan F, Zhu Y et al 2015. Blocking miR396 increases rice yield by shaping inflorescence architecture. Nature Plants 2: 15196. [DOI] [PubMed] [Google Scholar]
- Gong R, Cao H, Zhang J, Xie K, Wang D, Yu S. 2018. Divergent functions of the GAGA‐binding transcription factor family in rice. The Plant Journal 94: 32–47. [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López‐Vidriero I, Franco‐Zorrilla JM, Solano R, Trevisan M, Pradervand S et al 2012. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. The Plant Journal 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. 2009. Inhibition of the shade avoidance response by formation of non‐DNA binding bHLH heterodimers. EMBO Journal 28: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda‐Kawakatsu K, Yasuno N, Oikawa T, Iida S, Nagato Y, Maekawa M, Kyozuka J. 2009. Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem. Plant Physiology 150: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Du Y, Li F, Sun H, Zhang J, Li J, Peng T, Xin Z, Zhao Q. 2019. The basic helix‐loop‐helix transcription factor, OsPIL15, regulates grain size via directly targeting a purine permease gene OsPUP7 in rice. Plant Biotechnology Journal 17: 1527–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X et al 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genetics 42: 541–544. [DOI] [PubMed] [Google Scholar]
- de Jong RN, Truffault V, Diercks T, Ab E, Daniels MA, Kaptein R, Folkers GE. 2008. Structure and DNA binding of the human Rtf1 Plus3 domain. Structure 16: 149–159. [DOI] [PubMed] [Google Scholar]
- Kaiserli E, Páldi K, O'Donnell L, Batalov O, Pedmale UV, Nusinow DA, Kay SA, Chory J. 2015. Integration of light and photoperiodic signaling in transcriptional nuclear foci. Developmental Cell 35: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. 2007. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH. 2011. PIFs: pivotal components in a cellular signaling hub. Trends in Plant Science 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Li H, Hu X, Pan X, Wu G. 2011. Genetic transformation and overexpression of a rice Hd3a induces early flowering in Saussurea involucrata Kar. et Kir. ex Maxim. Plant Cell Tissue and Organ Culture 106: 363–371. [Google Scholar]
- Liu Q, Han R, Wu K, Zhang J, Ye Y, Wang S, Chen J, Pan Y, Li Q, Xu X et al 2018. G‐protein βγ subunits determine grain size through interaction with MADS‐domain transcription factors in rice. Nature Communications 9: 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Michael TP, Burger BT, Le Metté C, Mockler TC, Weigel D, Chory J. 2008. A zinc knuckle protein that negatively controls morning‐specific growth in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 105: 17193–17198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X et al 2009. Enhanced tolerance to chilling stress in OsMYB3R‐2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiology 150: 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐García JF, Huq E, Quail PH. 2000. Direct targeting of light signals to a promoter element‐bound transcription factor. Science 288: 859–863. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. 2007. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of bioscience and bioengineering 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kato T, Yamashino T, Murakami M, Mizuno T. 2007. Characterization of a set of phytochrome‐interacting factor‐like bHLH proteins in Oryza sativa . Bioscience Biotechnology and Biochemistry 71: 1183–1191. [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY. 2012. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nature Cell Biology 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Kathare PK, Kim JI, Huq E. 2017. Expanding roles of PIFs in signal integration from multiple processes. Molecular Plant 10: 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Xiong L. 2013. Characterization of a purine permease family gene OsPUP7 involved in growth and development control in rice. Journal of Integrative Plant Biology 55: 1119–1135. [DOI] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M. 2016. Molecular and genetic control of plant thermomorphogenesis. Nature Plants 2: 15190. [DOI] [PubMed] [Google Scholar]
- Shor E, Paik I, Kangisser S, Green R, Huq E. 2017. PHYTOCHROME INTERACTING FACTORS mediate metabolic control of the circadian system in Arabidopsis. New Phytologist 215: 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si L, Chen J, Huang X, Gong H, Luo J, Hou Q, Zhou T, Lu T, Zhu J, Shangguan Y et al 2016. OsSPL13 controls grain size in cultivated rice. Nature Genetics 48: 447–456. [DOI] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C. 2012. PIF4‐mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genetics 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Zhai Q, Li C. 2013. PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis . Plant Cell 25: 2102–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Xu XH, Wu X, Wang Y, Lu X, Sun H, Xie X. 2015. Genome‐wide identification of microRNAs and their targets in wild type and phyB mutant provides a key link between microRNAs and the phyB‐mediated light signaling pathway in rice. Frontiers in Plant Science 6: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, Ishizuka T, Yano M, Nishimura M, Miyao A, Hirochika H et al 2005. Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17: 3311–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao T, Nagata K, Morino K, Hirose T. 2010. A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice. Theoretical Applied Genetics 120: 875–893. [DOI] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. 2006. Early infection of scutellum tissue with Agrobacterium allows high‐speed transformation of rice. The Plant Journal 47: 969–976. [DOI] [PubMed] [Google Scholar]
- Wang S, Li S, Liu Q, Wu K, Zhang J, Wang S, Wang Y, Chen X, Zhang Y, Gao C et al 2015. The OsSPL16‐GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nature Genetics 47: 949–954. [DOI] [PubMed] [Google Scholar]
- Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q et al 2012. Control of grain size, shape and quality by OsSPL16 in rice. Nature Genetics 44: 950–954. [DOI] [PubMed] [Google Scholar]
- Wier AD, Mayekar MK, Heroux A, Arndt KM, VanDemark AP. 2013. Structural basis for Spt5‐mediated recruitment of the Paf1 complex to chromatin. Proceedings of the National Academy of Sciences, USA 110: 17290–17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhang G, An L, Chen X, Fang R. 2019. Phytochrome‐interacting factor‐like protein OsPIL15 integrates light and gravitropism to regulate tiller angle in rice. Planta 250: 105–114. [DOI] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. 1999. Light‐dependent translocation of a phytochrome B‐GFP fusion protein to the nucleusin transgenic Arabidopsis . Journal of Cell Biology 145: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue B, Xue WY, Luo LJ, Xing YZ. 2006. QTL Analysis for flag leaf characteristics and their relationship with yield and yield traits in rice. Yi Chuan Xue Bao 33: 824–832. [DOI] [PubMed] [Google Scholar]
- Zhang JP, Yu Y, Feng YZ, Zhou YF, Zhang F, Yang YW, Lei MQ, Zhang YC, Chen YQ. 2017. MiR408 regulates grain yield and photosynthesis via a phytocyanin protein. Plant Physiology 175: 1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH. 2013. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression‐patterning of shared target genes in Arabidopsis . PLoS Genetics 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YC, Yu Y, Wang CY, Li ZY, Liu Q, Xu J, Liao JY, Wang XJ, Qu LH, Chen F et al 2013. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nature Biotechnology 31: 848–852. [DOI] [PubMed] [Google Scholar]
- Zhang YM, Yan YS, Wang LN, Yang K, Xiao N, Liu YF, Fu YP, Sun ZX, Fang RX, Chen XY. 2012. A novel rice gene, NRR responds to macronutrient deficiency and regulates root growth. Moecular Plant 5: 63–72. [DOI] [PubMed] [Google Scholar]
- Zheng T, Yu H, Zhang H, Wu Z, Wang W, Tai S, Chi L, Ruan Y, Wei Z, Shi J et al 2015. Rice functional genomics and breeding database (RFGB)‐3K rice SNP and InDel sub‐database. Chinese Science Bulletin 60: 367–371. [Google Scholar]
- Zhou J, Liu Q, Zhang F, Wang Y, Zhang S, Cheng H, Yan L, Li L, Chen F, Xie X. 2014. Overexpression of OsPIL15, a phytochrome‐interacting factor‐like protein gene, represses etiolated seedling growth in rice. Journal of Integrative Plant Biology 56: 373–387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Agronomic traits of OsmiR530‐OE and target mimic (MIM530) lines.
Fig. S2 Comparison of the peduncle in miR530‐OE, target mimic (MIM530) and WT plants.
Fig. S3 Expression of OsmiR530 and OsPL3 in various rice tissues.
Fig. S4 Diagram of two sgRNAs targeting OsPL3 in the rice genome.
Fig. S5 Analysis of the OsPL3 protein sequence in the WT and OsPL3‐KO lines.
Fig. S6 Agronomic traits of OsPL3‐KO lines.
Fig. S7 Diagram of two sgRNAs targeting OsPIL15 in the rice genome.
Fig. S8 Analysis of the OsPIL15 protein sequence in the rice WT and OsPIL15‐KO lines.
Fig. S9 Expression of OsmiR530 in rice OsPIL15 transgenic lines.
Fig. S10 Alignment of mature miR530 sequences identified in miRBase.
Fig. S11 PHYB physically interacts with OsPIL15.
Fig. S12 Knockout of PHYB in rice results in smaller seed.
Table S1 Details regarding the primers and DNA fragments used in this study.


