Abstract
Background and Aims
Endoplasmic reticulum (ER) stress is associated with liver inflammation and hepatocellular carcinoma (HCC). However, how ER stress links inflammation and HCC remains obscure. Mesencephalic astrocyte‐derived neurotrophic factor (MANF) is an ER stress‐inducible secretion protein that inhibits inflammation by interacting with the key subunit of nuclear factor kappa light chain enhancer of activated B cells (NF‐κB) p65. We hypothesized that MANF may play a key role in linking ER stress and inflammation in HCC.
Approach and Results
Here, we found that MANF mRNA and protein levels were lower in HCC tissues versus adjacent noncancer tissues. Patients with high levels of MANF had better relapse‐free survival and overall survival rates than those with low levels. MANF levels were also associated with the status of liver cirrhosis, advanced tumor‐node‐metastasis (TNM) stage, and tumor size. In vitro experiments revealed that MANF suppressed the migration and invasion of hepatoma cells. Hepatocyte‐specific deletion of MANF accelerated N‐nitrosodiethylamine (DEN)‐induced HCC by up‐regulating Snail1+2 levels and promoting epithelial‐mesenchymal transition (EMT). MANF appeared in the nuclei and was colocalized with p65 in HCC tissues and in tumor necrosis factor alpha (TNF‐α)‐treated hepatoma cells. The interaction of p65 and MANF was also confirmed by coimmunoprecipitation experiments. Consistently, knockdown of MANF up‐regulated NF‐κB downstream target genes TNF‐α, interleukin (IL)‐6 and IL‐1α expression in vitro and in vivo. Finally, small ubiquitin‐related modifier 1 (SUMO1) promoted MANF nuclear translocation and enhanced the interaction of MANF and p65. Mutation of p65 motifs for SUMOylation abolished the interaction of p65 and MANF.
Conclusions
MANF plays an important role in linking ER stress and liver inflammation by inhibiting the NF‐κB/Snail signal pathway in EMT and HCC progression. Therefore, MANF may be a cancer suppressor and a potential therapeutic target for HCC.
Abbreviations
- ATF6
activating transcription factor 6
- Co‐IP
coimmunoprecipitation
- DEN
N‐nitrosodiethylamine
- EMT
epithelial‐mesenchymal transition
- ER
endoplasmic reticulum
- ERSE
ER stress response element
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- IHC
immunohistochemistry
- IL
interleukin
- IOD
integral optical density
- KO
knockout
- MANF
mesencephalic astrocyte‐derived neurotrophic factor
- NC‐shRNA
Negative control shRNA
- NF‐κB
nuclear factor kappa light chain enhancer of activated B cells
- NLS
nuclear localization signal
- OGD
oxygen‐glucose deprivation
- OS
overall survival
- RFS
relapse‐free survival
- shRNA
short hairpin RNA
- SIM/SBM
SUMO‐interaction/binding motif
- SUMO1
small ubiquitin‐related modifier 1
- TNF‐α
tumor necrosis factor alpha
- TNM
tumor‐node‐metastasis
- WT
wild type
Continuous accumulation of misfolded or unfolded proteins in the endoplasmic reticulum (ER) disturbs the environment of the ER lumen, which induces ER stress and unfolded protein response (UPR).1 UPR is a signaling cascade initiated by three ER transmembrane sensors: inositol‐requiring enzyme 1; activating transcription factor 6 (ATF6); and double‐stranded RNA‐dependent protein kinase–like ER kinase. ER stress has also been implicated in the pathogenesis of many human diseases, such as Alzheimer’s disease, Parkinson’s disease, diabetes, atherosclerosis, and heart diseases, and viruses‐related diseases, including hepatitis C virus and hepatitis B virus (HBV).2, 3 It was reported that ER stress is involved in hepatocellular carcinoma (HCC) and chronic inflammation that contributes to HCC development.4, 5 However, how ER stress links chronic inflammation and HCC remains obscure.
Mesencephalic astrocyte‐derived neurotrophic factor (MANF), identified as a secreted protein, is up‐regulated by ER stress and protects against various ER stress‐induced damage.6 MANF protects neurons and improves the symptoms in a rat model of Parkinson’s disease.7 Our previous studies have shown that cerebral ischemia triggers ER stress and enhances MANF expression. Recombinant human MANF promotes neuron proliferation and prevents neurons from apoptosis.8, 9 ER stress‐related proteins are up‐regulated in pancreatic cell‐specific MANF deficiency mice, and recombinant MANF enhances pancreatic β cell proliferation in vivo and in vitro.10, 11 Recombinant MANF protects mice from heart tissue damage in a mice myocardial infarction model.12 Meanwhile, our studies have indicated that inflammation triggers ER stress and promotes MANF nuclear translocation, which facilitates the interaction of MANF and nuclear factor kappa light chain enhancer of activated B cells (NF‐κB) p65 and inhibits the transcriptional activity of NF‐κB.13, 14 As a result, MANF inhibits inflammatory response.15
In chronic hepatitis B, a persistent inflammation with impaired antiviral immune response promotes the initiation and progression of HCC. The inflammatory tumor microenvironment also induces ER stress in tumor cells. However, how MANF behaves in HCC is not clear. In the current study, we have detected MANF expression in liver tissues of HCC patients and analyzed the correlation of MANF level with patients’ survival ratios. We also identified MANF as a suppressor for HCC and explored the mechanisms. Our data have demonstrated that p65 SUMOylation is required for MANF recruitment and its interaction with p65, which suppresses the NF‐κB/Snail signal pathway and thereby inhibits epithelial‐mesenchymal transition (EMT) and HCC progression.
Materials and Methods
Human Liver Tissue Samples and Ethics Statement
The human liver tissues of HCC patients were collected from the First Affiliated Hospital of Anhui Medical University (Hefei, China) in this study. The use of clinical HCC specimens was in accord with the Declaration of Helsinki. This study was approved by the Ethics Committee of Anhui Medical University (NO. 20131359), and written informed consent was obtained from each participant enrolled in this study.
Hepatocyte‐Specific MANF Deficiency Mice
MANFflox/flox mice bearing loxP sites flanking exon 3 of the manf gene on the C57BL/6 background were kindly provided by Prof. Jia Luo of the University of Kentucky (Lexington, KY). MANFflox/flox mice were cross‐bred with Alb‐cre mice to specifically knock out the manf gene in hepatocytes (MANF‐KO [knockout] mice). The details of N‐nitrosodiethylamine (DEN)‐induced HCC mouse model were described in the Supporting Data. All the mice experiments were approved by the Animal Experimental Committee of Anhui Medical University.
Plasmids, Antibodies, and Reagents
The details of plasmids, antibodies, and reagents used are presented in the Supporting Data.
Tissue Microarray Construction, in Situ Hybridization, and Immunohistochemistry
Tissue microarray was constructed by using liver tissues of HCC patients, and then in situ hybridization and immunohistochemistry (IHC) were performed. The details are presented in the Supporting Data.
Gene Silencing of MANF by Lentiviral Vector‐Delivered Short Hairpin RNA and Stable Cell Line Establishment
Establishment of cell lines stably expressing MANF short hairpin (shRNA) or negative control shRNA (NC‐shRNA) in HepG2 cells is described in the Supporting Data.
Quantitative Real‐Time Polymerase Chain Reaction
Total RNA was extracted with TRIzol. The details of the qPCR assay are described in the Supporting Data.
Malignant Biological Behavior Assay
The methods of the wound‐healing and invasion assay are described in the Supporting Data.
Oxygen‐Glucose Deprivation
Cells were treated with oxygen‐glucose deprivation (OGD). The details are described in the Supporting Data.
Immunofluorescence, IHC, and Integral Optical Density
The details for immunofluorescence, IHC, and integral optical density (IOD) are described in the Supporting Data.
Coimmunoprecipitation
HepG2 cells and liver tissues of HCC patients were used to perform the coimmunoprecipitation (Co‐IP) assay. The details are presented in the Supporting Data.
Statistical Analysis
All the data are expressed as mean ± SD. The specific statistical methods are described in the Supporting Data.
Results
Expression of MANF in the Cancer and Adjacent Noncancer Tissues of HCC Patients
We collected cancer tissues from 150 HCC patients and adjacent noncancer tissues from 136 HCC patients. MANF mRNA level in cancer tissues was much lower than that in adjacent noncancer tissues (Fig. 1A,B). Meanwhile, a high level of MANF mRNA was found in 38.7% of 150 HCC specimens, with 72.8% of 136 adjacent noncancer tissues; and a high level of MANF protein was found in 25.3% of 150 HCC specimens, with 73.5% of 136 adjacent noncancer tissues (P < 0.001; Supporting Table S1). Similarly, MANF protein level in cancer tissues was much lower than that in noncancer tissues of HCC patients (Fig. 1C,D). Furthermore, high expression of MANF protein was detected only in 25.3% of HCC tissues, but in 73.5% of noncancer tissues (P < 0.001), as shown in Supporting Table S1. Additionally, we also found that MANF level was down‐regulated in liver cancer tissues of DEN‐induced HCC mice, compared with that in para‐cancer tissues (Fig. 2F). These results suggest that MANF expression was decreased in cancer tissues of HCC patients and mice.
Figure 1.
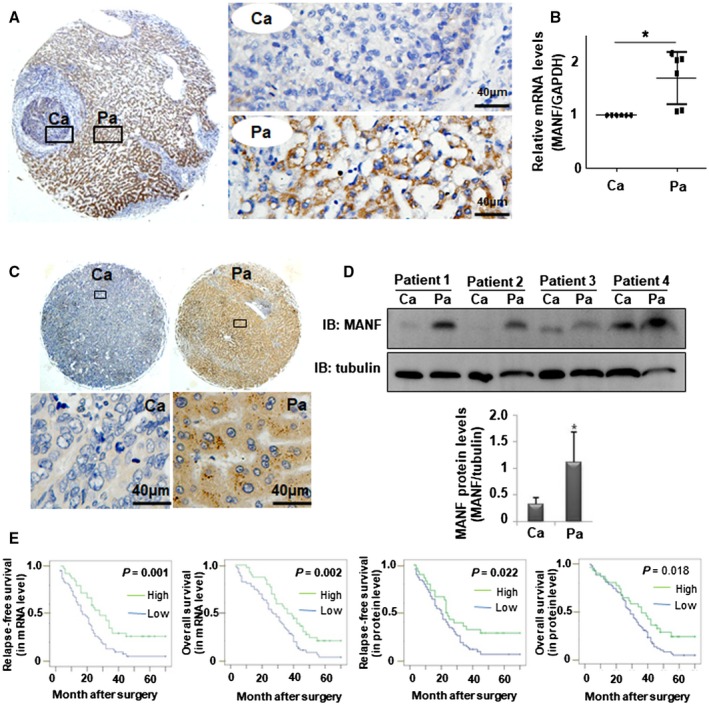
MANF level is associated with the survival rates of HCC patients. (A,B) MANF mRNA levels were detected in cancer (Ca) and para‐cancer (Pa) tissues of HCC patients by using in situ hybridization (A) and qPCR (B), respectively. *P < 0.05, compared with Ca. (C,D) MANF protein levels in Ca and Pa tissues of HCC patients were detected by IHC (C) and western blotting (D), respectively. *P < 0.05, compared with Ca. (E) Kaplan‐Meier analysis of the relationship between MANF mRNA or protein levels and RFS or OS of HCC patients (as indicated). Abbreviation: GAPDH, glyceraldehyde 3‐phosphate dehydrogenase.
Figure 2.
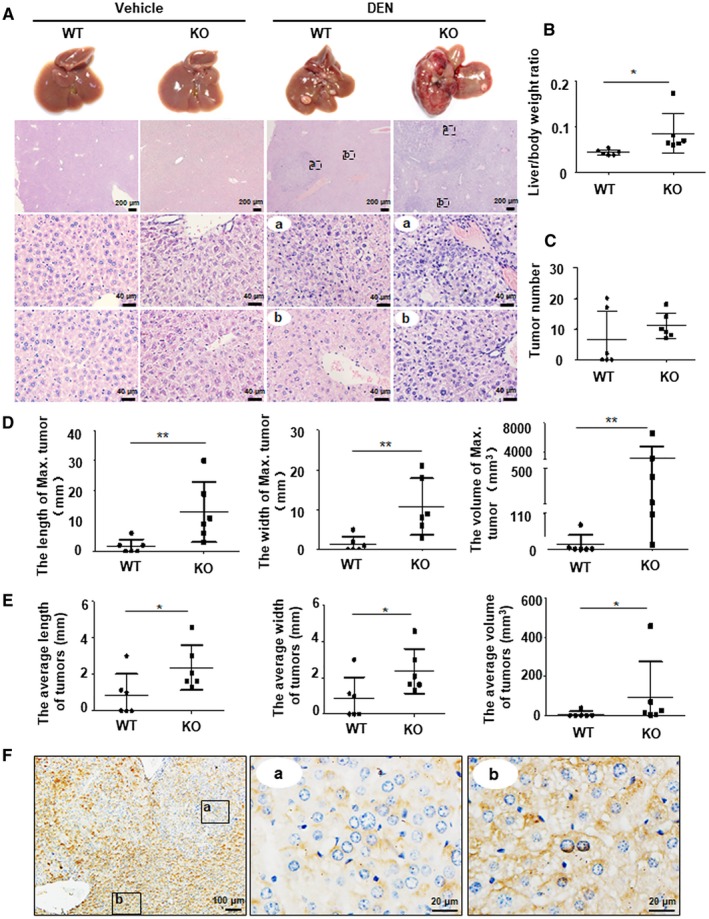
Hepatocyte‐specific MANF deficiency promotes mice HCC growth. (A) Liver morphology and H&E staining. WT and MANF‐KO mice were treated with DEN (30 mg/kg) by intraperitoneal injection at postnatal day 15, and livers were harvested at 8 months after DEN injection. a: cancer foci, b: para‐cancer. (B‐E) Effects of MANF knockout on body weight and tumor growth. Mice were sacrificed at 8 months after DEN treatment. n = 6; *P < 0.05; **P < 0.01, compared to WT. (B) The liver/body weight ratio. (C) The number of tumors (diameter ≥1 mm) on the surface of livers. (D) The length (left), width (middle), and volume (right) of maximal tumor on the surface of livers. (E) The average length (left), width (middle), and volume (right) of tumors on the surface of livers. (F) MANF expression in liver tissues of WT mice treated with DEN. a: cancer foci, b: para‐cancer. Abbreviation: H&E, hematoxylin and eosin.
Correlation Between MANF Expression and Clinicopathological Features of HCC
As mentioned above, that MANF level in cancer tissues was decreased, we further analyzed the correlation between MANF level and HCC patients’ survival by using Kaplan‐Meier curves. As expected, patients in the MANF‐high group had better relapse‐free (RFS) and overall survival (OS) rates than those in the MANF‐low group (Fig. 1E). Expressions of MANF in mRNA and protein levels were significantly associated with the status of liver cirrhosis (P = 0.008 and 0.004, respectively), tumor size (P = 0.010 and 0.016, respectively), and tumor‐node‐metastasis (TNM) stage (P = 0.001 and 0.031, respectively; Supporting Table S2). When the 150 HCC patients were divided into two groups (hepatitis B virus [HBV] positive and HBV negative), we found that MANF level was significantly associated with the status of liver cirrhosis (P = 0.005) and tumor size (P = 0.010) in HBV‐positive patients, not HBV‐negative patients (Supporting Tables S3 and S4). A low level of MANF was also found in a pulmonary metastasis tumor originally derived from HCC (Supporting Fig. S1). These results suggest that a low level of MANF in cancer tissues of HCC patients predicts poor prognosis.
MANF Suppresses HCC Progression in a DEN‐Induced Cancer Model
The above clinical data suggest that MANF may be a suppressor for HCC. To confirm this, we investigated the suppressive effect of MANF on HCC progression in a DEN‐induced mouse HCC model. Solid tumor nodules developed more quickly in DEN‐treated MANF‐KO mice, compared to wild‐type (WT) controls (Fig. 2A,C‐E). Meanwhile, the liver/body weight ratio in MANF‐KO mice was higher than that in WT controls (Fig. 2B). Histological images showed that liver tissues in MANF‐KO mice had a higher‐grade malignancy than that in WT controls (Fig. 2A). Serum aspartate aminotransferase was increased and albumin was decreased in MANF‐KO mice, compared to WT controls (Supporting Fig. S2B,D). There was no significant difference in serum alanine aminotransferase levels between WT and MANF‐KO mice (Supporting Fig. S2C). Furthermore, much more serious pulmonary metastasis was found in MANF‐KO mice 20 months after DEN treatment, compared to WT controls (Supporting Fig. S3F).
MANF Suppresses Hepatocyte EMT
The above in vivo data suggest that MANF suppresses pulmonary metastases of HCC. To confirm this, we investigated the effects of MANF on migration and invasion of hepatoma cells. The wound healing assay and transwell assay demonstrated that knockdown of endogenous MANF promoted the migration and invasion of hepatoma cells (Fig. 3B,C). To test whether or not MANF affects hepatocyte EMT, we detected mRNA and protein levels of E‐cadherin, β‐catenin, and vimentin in liver tissues of a DEN‐induced HCC model. We found that the levels of epithelial marker E‐cadherin in liver tissues of MANF‐KO mice were much lower than those in WT mice (Fig. 3D and Supporting Fig. S4). In contrast, the levels of mesenchymal markers β‐catenin and vimentin were up‐regulated in liver tissues of MANF‐KO mice, compared to WT mice (Fig. 3E,F and Supporting Figs. S5 and S6). These data suggest that MANF prevents EMT in HCC. Moreover, we found that hepatocyte‐specific MANF deficiency increased the number of inflammatory cells in liver cancer tissues of a DEN‐treated mice HCC model (Supporting Figs. S7 and S8). These findings indicate that MANF suppresses inflammation and EMT in HCC.
Figure 3.
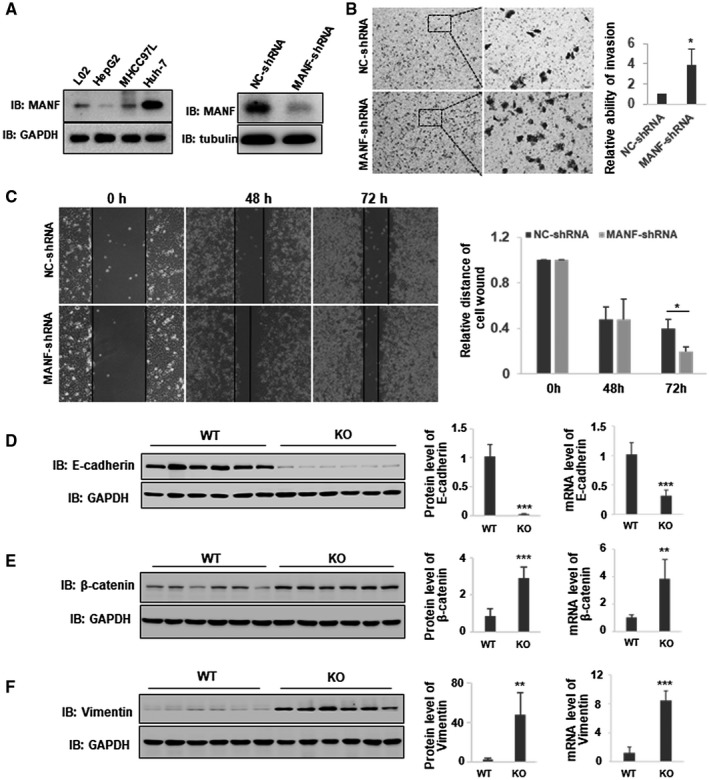
MANF suppresses hepatocyte EMT. (A) MANF expression in hepatoma cell lines (left panel) and the efficacy of MANF knockdown in HepG2 cells stably transfected with NC‐shRNA or MANF‐shRNA plasmid (right panel). (B,C) Knockdown of MANF promotes the migration and invasion of HepG2 cells. Transwell assay (B) and wound healing assay (C) were used to detect the invasion and migration of HepG2 stable cells, respectively. *P < 0.05, compared with NC‐shRNA. (D‐F) Effects of MANF knockout on hepatocyte EMT. WT and MANF‐KO mice were treated with DEN (30 mg/kg) by intraperitoneal injection, and liver tissues were harvested at 8 months after DEN treatment. The mRNA and protein levels of E‐cadherin (D), β‐catenin (E), and vimentin (F) in liver cancer tissues were detected by qPCR and western blotting, respectively. n = 6; **P < 0.01; ***P < 0.001, compared to WT. Abbreviations: GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; IB, immunoblotting.
MANF Interacts With p65 and Inhibits the NF‐κB Signal Pathway in HCC
We have verified that MANF suppresses inflammation and EMT and thereby inhibits invasion and metastases in HCC. It was reported that activation of the NF‐κB signal pathway enhances inflammation and EMT of hepatocytes.16, 17 Our previous study also showed that MANF inhibits NF‐κB transcriptional activity by binding to the DNA‐binding domain of p65 in nuclei.14 Therefore, we speculated that the underlying mechanism by which MANF suppresses hepatocyte EMT and inflammation may be associated with inhibition of the NF‐κB signal pathway. In HCC tissue, we found that MANF appeared in the nuclei of hepatocytes and colocalized with p65 (Fig. 4A,B, as indicated by the arrows). Co‐IP assay verified the interaction of MANF and p65 in cancer tissues of HCC (Fig. 4C) and HepG2 cells that were overexpressing MANF‐FLAG after being treated with tumor necrosis factor alpha (TNF‐α; Fig. 4D). To investigate the subcellular localization where MANF interacts with p65, cytoplasmic and nuclear proteins were isolated for Co‐IP assay. The major interaction of MANF and p65 was localized in the nuclei of hepatoma cells (Fig. 4E). Additionally, TNF‐α treatment promoted the nuclear translocation and colocalization of p65 and MANF in the nuclei of hepatoma cells (Fig. 4F). These data demonstrate that MANF interacts with p65 in the nuclei in HCC.
Figure 4.
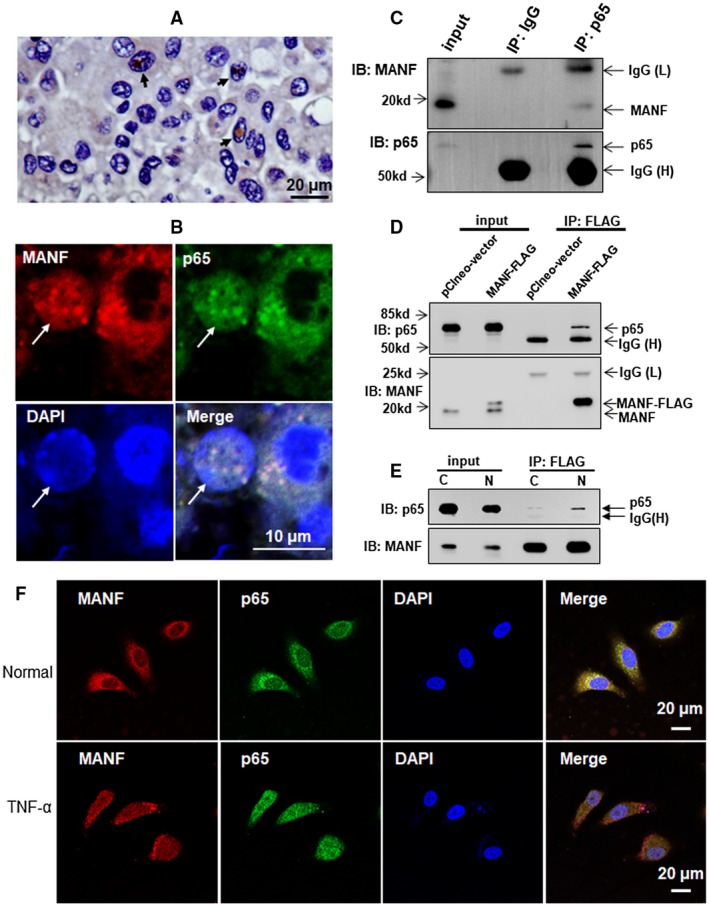
MANF interacts with p65 in HCC and hepatoma cells. (A) MANF was localized in the nuclei in cancer tissues of HCC patients. MANF was detected by using IHC. (B) Colocalization of p65 (green) and MANF (red) in the nucleus in liver tissues of HCC patients. DAPI was used to stain the nuclei (blue). (C) MANF interacts with p65 in liver tissues of HCC patients. Tissues were lysed for Co‐IP assay by using anti‐p65 antibody. The isotype, IgG, was used as a negative control. (D) MANF interacts with p65 in hepatoma cells. HepG2 cells were transfected with pClneo‐MANF‐FLAG or pClneo‐vector plasmid, and treated with TNF‐α (10 ng/ml) for 8 hours at 24 hours after transfection. The interaction was detected by Co‐IP assay by using anti‐FLAG M2 Affinity Gel. (E) MANF interacts with p65 in the nuclei of hepatoma cells. HepG2 cells were transfected with MANF‐FLAG plasmid. Twenty‐four hours later, the cells were treated with 10 ng/ml TNF‐α for 8 hours. Then, cytoplasmic (C) and nuclear (N) proteins were extracted, and Co‐IP assay was performed using anti‐FLAG M2 Affinity Gel. MANF and p65 was detected with anti‐MANF and anti‐p65 antibodies, respectively. (F) TNF‐α‐induced co‐localization of MANF and p65 in hepatoma cells. HepG2 cells were treated with 10 ng/ml TNF‐α (lower panel) or vehicle (upper panel) for 8 hours and then stained with immunofluorescent antibodies against MANF (red) and p65 (green). The nuclei were stained with DAPI (blue). Abbreviations: DAPI, 4′,6‐diamidino‐2‐phenylindole; IB, immunoblotting; IgG, immunoglobulin G; IP, immunoprecipitation.
Next, we investigated the effect of MANF on NF‐κB transcriptional activity. We found that MANF did not affect the total level of p65 (Supporting Fig. S9) and p65 phosphorylation (Supporting Fig. S10) in a DEN‐treated mice HCC model. However, MANF knockdown increased the mRNA levels of interleukin (IL)‐1α, TNF‐α, and IL‐6 in HepG2 cells stably silenced by MANF (Fig. 5B). Consistently, expressions of IL‐1α (Fig. 5C) and TNF‐α (Fig. 5D) were up‐regulated in mRNA and protein levels in liver tissues of MANF‐KO mice, compared to WT mice. These data suggest that MANF decreases the levels of NF‐κB target genes.
Figure 5.
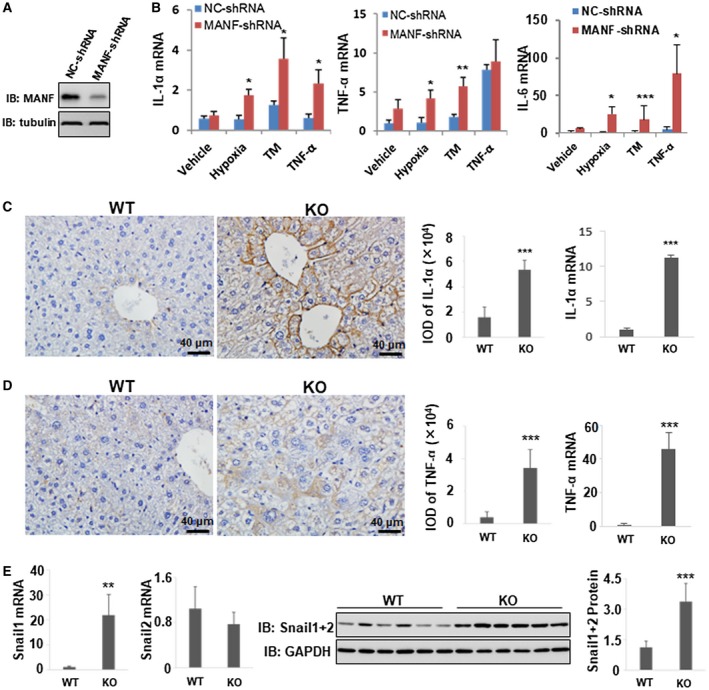
MANF down‐regulates the expressions of NF‐κB target genes. (A) Western blotting verifies the efficacy of MANF knockdown by MANF‐shRNA in HepG2 cells. (B) Effects of MANF knockdown on proinflammatory cytokine production in hepatoma cells. HepG2 cells stably transfected with NC‐shRNA or MANF‐shRNA plasmid were treated with hypoxia, tunicamycin (TM; 2.5 μg/mL), or TNF‐α (10 ng/mL) for 48 hours. Then, total RNA was extracted, and mRNA levels of IL‐1α, TNF‐α, and IL‐6 were detected by using the qPCR assay. GAPDH was used as an internal control. *P < 0.05; **P < 0.01; ***P < 0.001, compared with NC‐shRNA. (C‐E) Effects of MANF knockout on inflammatory factor level in HCC tissues. WT and MANF‐KO mice were treated with DEN (30 mg/kg) by intraperitoneal injection, and liver tissues were harvested at 8 months after DEN treatment. Levels of IL‐1α (C) and TNF‐α (D) were detected using IHC and qPCR. n = 6; ***P < 0.001. (E) Snail1 and Snail2 mRNA levels were detected using qPCR. Western blotting was performed to detect Snail1+2 protein levels. n = 6; **P < 0.01; ***P < 0.001, compared to WT. Abbreviations: GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; IB, immunoblotting.
It has been reported that Snail1 and Snail2 are EMT inducers.18 Meanwhile, p65 triggers Snail1 transcription and promotes hepatocyte EMT by binding to its promoter.17 Moreover, TNF‐α, the downstream target of the NF‐κB signaling pathway, stabilizes Snail2 protein by inhibiting its ubiquitination and degradation.19 In this study, we found that the Snail1 mRNA level in liver tissues of MANF‐KO mice was increased, compared to WT mice. However, there was no statistical difference in Snail2 mRNA levels. Additionally, both Snail1 and Snail2 protein levels in liver tissues of MANF‐KO mice were much higher than those in WT mice (Fig. 5E and Supporting Figs. S11‐S13), suggesting that MANF decreases Snail1+2 expression. Similar data were obtained in the clinical specimens. MANF level was low in cancer tissues in most of the HCC patients, whereas Snail1+2 was high (Supporting Fig. S14, left panels). On the contrary, in a few cases, MANF level was high, whereas Snail1+2 level was low (right panels). Furthermore, we analyzed the correlation between MANF, Snail1+2, and p65 expression in liver tissues of 150 HCC patients. We found a negative correlation between MANF and Snail1+2 (rs = −0.238; P = 0.003) and a close positive correlation between p65 and Snail1+2 (rs = 0.405; P = 0.000; Supporting Table S5). However, there was no significant correlation between MANF and p65 levels. These data suggest that MANF suppresses the NF‐κB/Snail signal pathway and, subsequently, prevents inflammation and hepatocyte EMT.
Small Ubiquitin‐Related Modifier 1 Promotes the Interaction of MANF and p65
As mentioned above, we found nuclear MANF in HCC and hepatoma cells, because MANF is a secreted protein and usually localized in the cytoplasm under normal conditions.20 How does MANF enter the nucleus in HCC? To answer this question, we searched for the nuclear localization signal (NLS) in the MANF sequence by using PredictProtein software. The result showed that MANF lacks an NLS domain. Next, we screened the MANF‐interacting proteins using yeast two‐hybrid experiment. The result showed that small ubiquitin‐related modifier (SUMO) proteins may interact with MANF. It is reported that RanBP2/RanGAP1*SUMO1/UBC9 was a E3 ligase complex for SUMOylation in the nuclear membrane and promoted the nuclear import of SUMO1‐related SUMOylated target proteins.21 Therefore, we investigated the effect of SUMO1 on MANF. In HepG2 cells overexpressing MANF‐FLAG, we detected the high‐molecular‐weight SUMO1 bands after being immunoprecipitated with anti‐FLAG antibody, suggesting that MANF may be SUMOylated by SUMO1 protein (Fig. 6A). Next, we investigated the effect of SUMO1 on MANF nuclear import. The results showed that SUMO1 overexpression increased OGD‐induced MANF nuclear import, and SUMO1 knockdown decreased the nuclear import of MANF (Fig. 6B,C and Supporting Fig. S15). Because of the colocalization of p65 and MANF in the nuclei in HCC, we wondered whether or not SUMO1 regulates the interaction of MANF and p65 in the nuclei. The data showed that knockdown of SUMO1 reduced the interaction of p65 and MANF in the nuclei (Fig. 6D). These data indicate that SUMO1 regulates the interaction of p65 and MANF through promoting MANF nuclear import.
Figure 6.
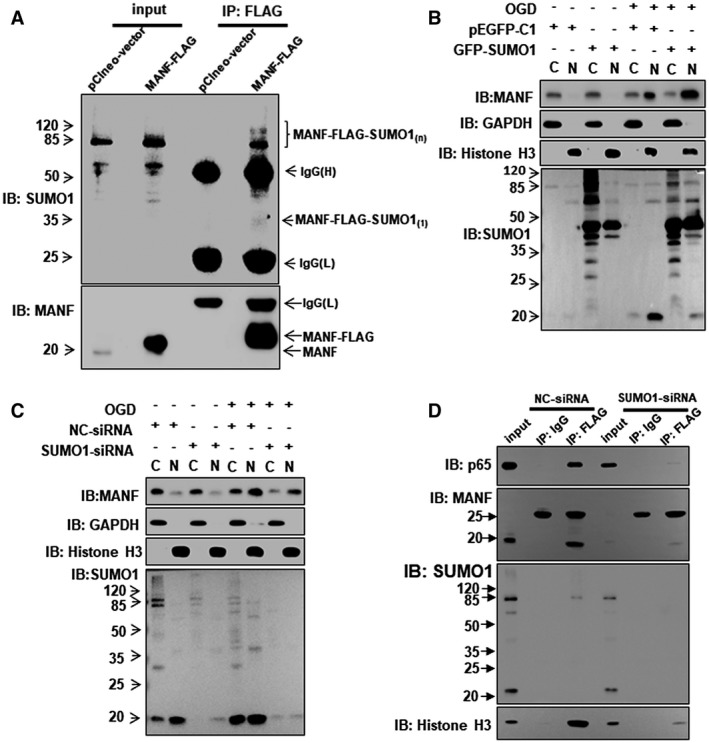
SUMO1 promotes the nuclear import of MANF. (A) MANF interacts with SUMO1. HepG2 cells were transfected with pClneo‐vector and pClneo‐MANF‐FLAG plasmid, respectively. Twenty‐four hours later, cells were treated with TNF‐α (10 ng/mL) for 8 hours. Co‐IP assay was performed to detect the interaction of MANF and SUMO1 by using anti‐FLAG M2 Affinity Gel. (B,C) SUMO1 promotes MANF nuclear translocation. HepG2 cells were transfected with GFP‐SUMO1 (B), SUMO1‐siRNA (C), or corresponding controls and treated with or without OGD for 150 minutes at 48 hours posttransfection. The cytoplasmic and nuclear proteins were extracted and western blotting was performed. GAPDH and histone H3 were used as cytoplasmic and nuclear markers, respectively. C, cytoplasm; N, nucleus. (D) SUMO1 knockdown inhibits the interaction of MANF and p65 in the nuclei. HepG2 cells were transfected with MANF‐FLAG plus NC‐siRNA or SUMO1‐siRNA, respectively. Forty‐eight hours later, cells were treated with OGD for 150 minutes and then harvested for the extraction of cytoplasmic and nuclear proteins. The nuclear proteins were used for Co‐IP assays by using anti‐FLAG M2 Affinity Gel. The isotype, IgG, was used as a negative control, and 5% nuclear protein lysate was loaded as input. Abbreviations: GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; GFP, green fluorescent protein; IB, immunoblotting; IgG, immunoglobulin G; IP, immunoprecipitation; pEGFP, phosphorylated enhanced green fluorescent protein; siRNA, small interfering RNA.
p65 SUMOylation is Essential for the Interaction of p65 and MANF in Hepatocytes
It was thought that low‐level transcription factor SUMOylation can lead to repression of transcriptional activity.22 Our previous data showed that p65, as a transcription factor, can be SUMOylated by SUMO1 and SUMO2/3.23, 24 We searched for the SUMO‐interaction/binding motif (SIM/SBM) of p65 by using SUMOsp2.0 software. The result indicates that p65 is a protein containing SIM/SBM at Lys37, Lys122, Lys123, and Lys221 (Fig. 7A). The target proteins of SUMOylation are recruited to SUMOylated transcription factors and then a repressor complex is assembled.22 Interestingly, MANF also has SIM/SBM according to the prediction results of SUMOsp2.0 software. Therefore, we assumed that SUMOylated p65 can recruit MANF for the assembly of a repressor complex to inhibit NF‐κB activation. To test this hypothesis, we constructed p65 mutates at the SIM motif and performed Co‐IP. As predicted, the levels of SUMOylated p65 modified by SUMO1 and SUMO2/3 were reduced after SIM motifs of p65 were mutated (Fig. 7B). Meanwhile, the mutates did not interact with MANF (Fig. 7C). These findings suggest that interaction of p65 and MANF depends on p65 SUMOylation. Moreover, we detected SUMO1‐mediated MANF SUMOylation and p65 SUMOylation in liver tissues of HCC patients. The SUMOylated MANF was higher in the para‐cancer tissues than that in the cancer tissues (Supporting Fig. S16A). However, the SUMOylated p65 was lower in the para‐cancer tissues than in cancer tissues (Supporting Fig. S16B).
Figure 7.
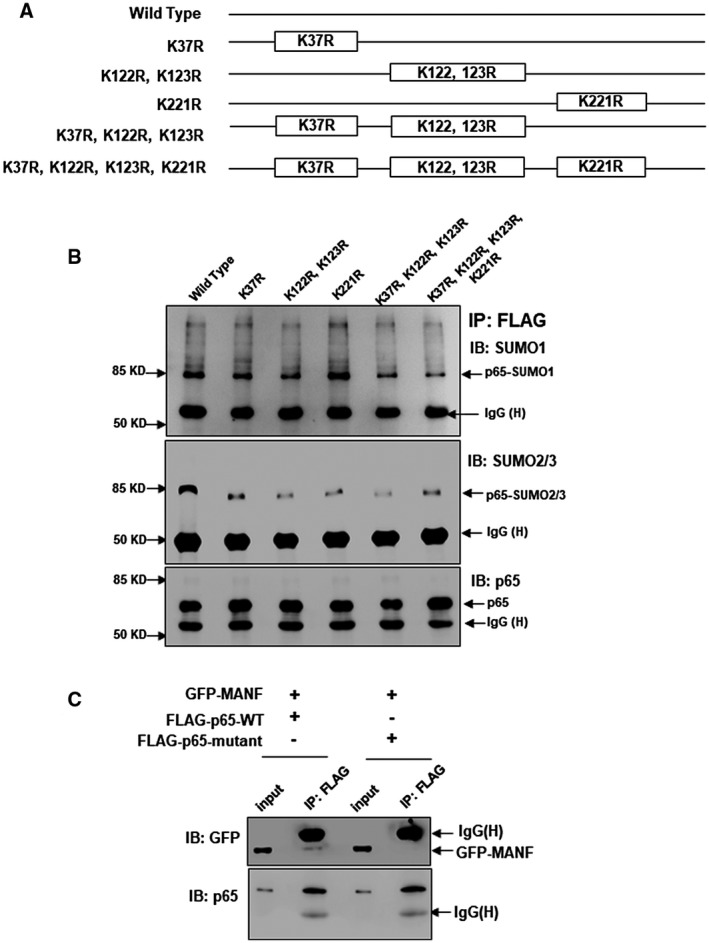
SUMOylation is essential for the interaction of p65 and MANF. (A,B) Identifying the SIM motif of p65 for SUMOylation. (A) Schematic diagram of p65 mutants. (B) Hela cells were transfected with the plasmids of FLAG‐p65‐WT or FLAG‐p65‐mutants, respectively. Twenty‐four hours later, cells were treated with TNF‐α (10 ng/mL) for 30 minutes, and Co‐IP assay was performed with anti‐FLAG M2 Affinity Gel. SUMOylated p65 was detected with anti‐SUMO1 and anti‐SUMO2/3 antibodies, respectively. (C) p65 SUMOylation is essential for the interaction of p65 and MANF. HepG2 cells were transfected with GFP‐MANF and FLAG‐p65‐WT or FLAG‐p65 mutated at the sites of K37R, K122, 123R, and K221R, respectively. Twenty‐four hours later, cells were treated with TNF‐α (10 ng/mL) for 8 hours, and Co‐IP assay was performed. MANF and p65 was detected with anti‐GFP and anti‐p65 antibodies, respectively. Abbreviations: GFP, green fluorescent protein; IB, immunoblotting; IgG, immunoglobulin G; IP, immunoprecipitation.
Discussion
In this study, our data indicated that MANF level in cancer tissues of HCC was lower than that in the adjacent noncancer tissues. We also found that low‐level MANF is correlated with poor RFS and OS. We further verified that MANF was a suppressor for HCC in vitro and in vivo through inhibiting inflammation and preventing hepatocyte EMT. We also demonstrated the mechanisms involved in HCC development, including SUMO1‐mediated MANF SUMOylation and nuclear translocation. SUMOylation is essential for the interaction of MANF and p65 in HCC.
MANF is also named as arginine‐rich, mutated in early‐stage tumors or arginine‐rich protein, and it is considered to be a highly conserved mutation in early‐stage tumors.25 A subsequent report showed that this variation was not a tumor‐specific mutation, but a normal polymorphism.26 In this study, we found that MANF level in liver cancer tissues of HCC patients was lower than that in the adjacent noncancer tissues. A previous study has shown that the MANF gene is an ER stress‐sensitive gene. Our previous study showed that X‐box binding protein 1 promotes MANF transcription through binding to ER stress response element (ERSE)‐I.27 It was also reported that MANF is induced through ERSE‐II in its 5′‐flanking region.28 Recent data have shown that ATF6 was the strongest ER stress response transcription factor to increase the promoter activity of MANF through ERSE‐II.29 The tissue‐based map of the human proteome has shown that ATF6 is decreased in liver cancer tissues, compared to that in normal liver tissues.30 ATF6 knockdown suppressed ER stress‐induced MANF expression.28 These data suggest that a low level of ATF6 in liver tumor tissues may inhibit MANF transcription through ERSE‐II and down‐regulate MANF protein level. Of course, it is possible that there are other elements involved in the regulation of MANF level in HCC, including MANF transcriptional regulation by other ER stress‐related transcription factors, MANF gene modification, alternative RNA splicing, and MANF degradation. This hypothesis needs to be proven.
Previous studies have demonstrated that MANF protects dopaminergic neuron and has therapeutic potential for Parkinson’s disease.7 We also have demonstrated that the ER stress inducer, tunicamycin, up‐regulates MANF expression in neurons and recombinant human MANF promotes the proliferation of neurons and prevents neurons from cerebral ischemia‐induced apoptosis.8, 9 Meanwhile, MANF promotes the survival and proliferation of pancreatic β cells.10, 11 Furthermore, MANF as an immune modulator mediates tissue repair in the mammalian retina and maintains liver metabolic homeostasis.15, 31 Additionally, MANF plays key roles in facilitating the survival of cardiac cells from ischemic injury, modulating myocardial hypertrophy and heart failure as well.32 These findings demonstrate that MANF protects normal cells against the apoptosis induced by various stimuli and promotes the survival and proliferation of normal cells. However, in the present study, our data have indicated that MANF suppresses the migration and invasion of a liver cancer cell line in vitro, as well as inflammation and hepatocyte EMT in a DEN‐induced mouse HCC model in vivo. These results suggest that MANF may have protective effects on the survival of normal cells and suppressive effects on the malignant biological behavior of tumor cells.
The mechanism by which MANF suppresses HCC progression attracts our interests. An increasing number of references indicate that hepatocellular EMT plays a key role in the progression of malignant hepatocytes.33 In our present study, the epithelial marker, E‐cadherin, was down‐regulated and the mesenchymal markers, β‐catenin and vimentin, were up‐regulated in DEN‐treated MANF‐KO mice, compared to WT controls. The results indicate that MANF can suppress hepatocyte EMT during HCC progression. Recently, it was reported that p65 overexpression in the hepatoma cell lines decreases E‐cadherin and increases vimentin level.16 These findings suggest that activation of the NF‐κB pathway promotes hepatocyte EMT. Our previous study demonstrated that nuclear MANF could bind to the p65 DNA‐binding domain, which blocks p65 binding to the promoter regions of its target genes and suppresses NF‐κB activation in fibroblast‐like synoviocytes and 293T cells.14 In the present study, we also verified the interaction of MANF and p65 in the nuclei of HCC. In the stable cell line with MANF knockdown, we found that MANF knockdown increased the mRNA levels of IL‐1α, IL‐6, and TNF‐α. Similar data were obtained in a mice HCC model. We also found that MANF knockdown up‐regulated the number of CD68+ and CD3+ cells, whereas the mRNA levels of CD68 and CD3 were not changed, even down‐regulated in the liver tissues. The explanations for the inconsistency may include two aspects; on the one hand, MANF knockdown recruits the circulating inflammatory/immune cells to the liver (data not shown). On the other hand, the protein synthesis was attenuated in the liver because of liver injury. It was reported that liver injury can recruit macrophages and T cells in the liver.34, 35 However, MANF deficiency did not affect the total level of p65 and its phosphorylation in a mice HCC model, suggesting that MANF suppresses the levels of p65 target genes by blocking p65 binding to its target gene promoters in HCC.
It has been proven that NF‐κB binds to the promoter of Zinc‐finger transcription factor Snail1 and triggers its transcription.17 Meanwhile, TNF‐α, the target gene of NF‐κB, stabilizes Snail2 protein by inhibiting its ubiquitination.19 Snail1 and Snail2 can bind to the E‐boxes of the E‐cadherin promoter to inhibit its transcriptional activity and act as epithelial repressors and mesenchymal promoters.36, 37 Our data indicate that MANF decreases the levels of TNF‐α and Snail1; the underlying mechanism may be related to suppressive regulation in the promoter region of NF‐κB target genes. Reduction of TNF‐α leads to the increase of Snail2 degradation. It is reported that the chronic inflammation microenvironment of cancer activates the NF‐κB pathway, which is a decisive factor in the induction of cancer cell EMT. Therefore, NF‐κB is an important connection between inflammation, cancer, and EMT.38 Our previous data indicate that liver inflammation up‐regulates MANF expression.39 The present study has demonstrated that MANF inhibits the NF‐κB/Snail pathway and prevents the EMT of hepatocytes, thereby suppressing HCC occurrence. Once hepatocytes become malignant from benign, the cellular MANF level is dramatically down‐regulated. The low level of MANF further exacerbates the activation of the NF‐κB/Snail pathway and promotes the EMT of HCC. Therefore, MANF inhibits inflammation and EMT of HCC by suppressing the NF‐κB/Snail pathway.
MANF is a small secretory protein and normally localizes in the cytoplasm of cells. Unexpectedly, we observed that MANF was imported into the nuclei in the cancer cells of HCC in the present study. Given that MANF protein lacks an NLS domain, how does MANF move into the nuclei? To answer this question, we performed a yeast two‐hybrid experiment and found that SUMO proteins may interact with MANF. It was reported that SUMO1 protein mainly localizes in the nuclear membrane and mediates the nuclear translocation of proteins,21 especially the proteins that lack an NLS.40 In the present study, we have verified that SUMO1 mediates MANF SUMOylation and nuclear transport.
Our previous data showed that SUMO1 overexpression up‐regulated the nuclear transport of p65, and SUMO1 knockdown down‐regulated p65 nuclear translocation in hepatoma cells.23 However, our data also showed that SUMO1, either by overexpression or knockdown, led to inhibition of the NF‐κB transcriptional activity.23 The underlying mechanism is unclear. p65, as a transcriptional factor, can be SUMOylated by SUMO1 and SUMO2/3 proteins in hepatoma cells.23, 24 Interestingly, we found that SUMO1 knockdown inhibited the interaction of MANF and p65 in the nucleus. We also found p65 mutation at the SIM motifs cancelled the interaction of p65 and MANF, suggesting that p65 SUMOylation was essential for the interaction of MANF and p65. Frauke Melchior thought that SUMOylated transcription factor could lead to transcriptional repression and proposed a model for SUMOylation‐related transcriptional regulation, in which the proteins containing a SIM/SBM would be recruited to SUMOylated transcription factors and then assembled a repressor complex,22 such as corepressor of RE1‐silencing transcription factor and Sharp‐1.41, 42 MANF is a protein that contains SIM motifs, and we have demonstrated that it can be imported by SUMO1 in the present study. Therefore, we speculate that MANF can be recruited to SUMOylated p65 as a part of a repressor complex in NF‐κB transcriptional activation. Moreover, we found that in liver cancer tissues, the level of SUMO1‐related p65 SUMOylation was increased, which may lead to p65 nuclear import and subsequent NF‐κB signal pathway activation during HCC development. In the adjacent para‐cancer tissues, the level of SUMO1‐related MANF SUMOylation was increased, which may lead to MANF nuclear import and subsequent regulation on the NF‐κB /Snail signal pathway.
Taken together, our data demonstrate that SUMOylated p65 recruits MANF as a component of a repressor complex to inhibit NF‐κB activation and, subsequently, shuts down NF‐κB/Snail signaling, thereby limiting liver inflammation and preventing hepatocyte EMT. These findings suggest that MANF acts as an HCC suppressor (Fig. 8).
Figure 8.
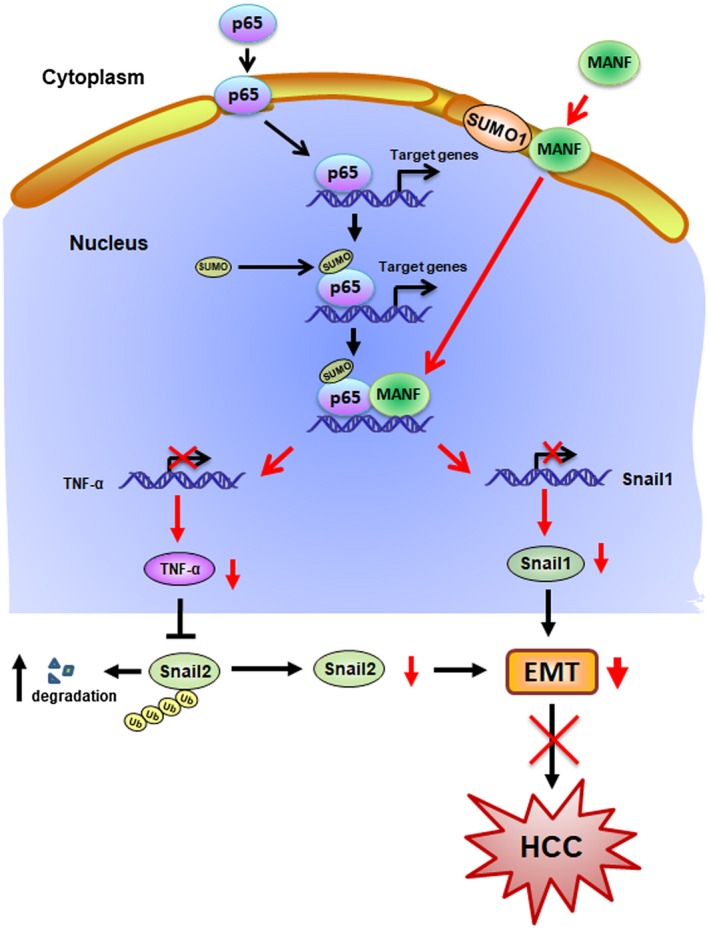
Schematic diagram of MANF inhibiting HCC. The nuclear import of p65 triggers the transcription of NF‐κB target genes. SUMO1 mediates MANF SUMOylation and nuclear import. SUMOylation of p65 recruits MANF to form a repressor complex to shut down NF‐κB signaling. As a result, the downstream genes of the NF‐κB signal pathway, such as Snail1 and TNF‐α, were inhibited. Subsequently, hepatocyte EMT and HCC were suppressed.
Author Contributions
Conceptualization: Y.S. Investigation: J.L., Z.W., D.H., C.W., Y.L., T.J., L.C., M.S., Y.C., F.H., X.G., J.Y., Y.S., H.W., L.F., D.W., S.F., S.W. Writing: J.L., Y.S. Funding acquisition: J.L., Y.S.
Supporting information
Acknowledgment
We thank Dr. Hiderou Yoshida and Dr. Steve Jackson for supplying myc‐UBC9 and GFP‐SUMO1 plasmids. We also thank Dr. Jia Luo for supplying MANF conditional knockout mice. We are grateful to Dr. Bin Gao for critical reading.
Supported by grants from the Natural Science Foundation of China (81372576 and 81673438 to Yuxian Shen and 81602120 to Jun Liu) and the Natural Science Foundation of Anhui Province (1708085QH176 to Jun Liu).
Potential conflict of interest: Nothing to report.
[Correction added on February 17, 2020, after first online publication: The corresponding author email was corrected.]
Contributor Information
Siying Wang, Email: sywang@ahmu.edu.cn.
Yuxian Shen, Email: shenyx@ahmu.edu.cn.
References
Author names in bold designate shared co‐first authorship.
- 1. Kaufman RJ, Scheuner D, Schroder M, Shen X, Lee K, Liu CY, et al. The unfolded protein response in nutrient sensing and differentiation. Nat Rev Mol Cell Biol 2002;3:411‐421. [DOI] [PubMed] [Google Scholar]
- 2. Yoshida H. ER stress and diseases. FEBS J 2007;274:630‐658. [DOI] [PubMed] [Google Scholar]
- 3. Malhi H, Kaufman RJ. Endoplasmic reticulum stress in liver disease. J Hepatol 2011;54:795‐809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Levrero M, Zucman‐Rossi J. Mechanisms of HBV‐induced hepatocellular carcinoma. J Hepatol 2016;64(1 Suppl):S84‐S101. [DOI] [PubMed] [Google Scholar]
- 5. Shuda M, Kondoh N, Imazeki N, Tanaka K, Okada T, Mori K, et al. Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. J Hepatol 2003;38:605‐614. [DOI] [PubMed] [Google Scholar]
- 6. Apostolou A, Shen Y, Liang Y, Luo J, Fang S. Armet, a UPR‐upregulated protein, inhibits cell proliferation and ER stress‐induced cell death. Exp Cell Res 2008;314:2454‐2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cordero‐Llana O, Houghton BC, Rinaldi F, Taylor H, Yanez‐Munoz RJ, Uney JB, et al. Enhanced efficacy of the CDNF/MANF family by combined intranigral overexpression in the 6‐OHDA rat model of Parkinson’s disease. Mol Ther 2015;23:244‐254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu YQ, Liu LC, Wang FC, Liang Y, Cha DQ, Zhang JJ, et al. Induction profile of MANF/ARMET by cerebral ischemia and its implication for neuron protection. J Cereb Blood Flow Metab 2010;30:79‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang W, Shen Y, Chen Y, Chen L, Wang L, Wang H, et al. Mesencephalic astrocyte‐derived neurotrophic factor prevents neuron loss via inhibiting ischemia‐induced apoptosis. J Neurol Sci 2014;344:129‐138. [DOI] [PubMed] [Google Scholar]
- 10. Lindahl M, Danilova T, Palm E, Lindholm P, Voikar V, Hakonen E, et al. MANF is indispensable for the proliferation and survival of pancreatic beta cells. Cell Rep 2014;7:366‐375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Danilova T, Belevich I, Li H, Palm E, Jokitalo E, Otonkoski T, et al. MANF is required for the postnatal expansion and maintenance of pancreatic beta‐cell mass in mice. Diabetes 2019;68:66‐80. [DOI] [PubMed] [Google Scholar]
- 12. Glembotski CC, Thuerauf DJ, Huang C, Vekich JA, Gottlieb RA, Doroudgar S. Mesencephalic astrocyte‐derived neurotrophic factor protects the heart from ischemic damage and is selectively secreted upon sarco/endoplasmic reticulum calcium depletion. J Biol Chem 2012;287:25893‐25904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang J, Cheng Q, Wang X, Zu B, Xu J, Xu Y, et al. Deficiency of IRE1 and PERK signal pathways in systemic lupus erythematosus. Am J Med Sci 2014;348:465‐473. [DOI] [PubMed] [Google Scholar]
- 14. Chen L, Feng L, Wang X, Du J, Chen Y, Yang W, et al. Mesencephalic astrocyte‐derived neurotrophic factor is involved in inflammation by negatively regulating the NF‐kappaB pathway. Sci Rep 2015;5:8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sousa‐Victor P, Neves J, Cedron‐Craft W, Ventura PB, Liao CY, Riley RR, et al. MANF regulates metabolic and immune homeostasis in ageing and protects against liver damage. Nat Metab 2019;1:276‐290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu TJ, Chang SS, Li CW, Hsu YH, Chen TC, Lee WC, et al. Severe hepatitis promotes hepatocellular carcinoma recurrence via NF‐kappaB pathway‐mediated epithelial‐mesenchymal transition after resection. Clin Cancer Res 2016;22:1800‐1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Song R, Song H, Liang Y, Yin D, Zhang H, Zheng T, et al. Reciprocal activation between ATPase inhibitory factor 1 and NF‐kappaB drives hepatocellular carcinoma angiogenesis and metastasis. Hepatology 2014;60:1659‐1673. [DOI] [PubMed] [Google Scholar]
- 18. Kudo‐Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail‐induced EMT of cancer cells. Cancer Cell 2009;15:195‐206. [DOI] [PubMed] [Google Scholar]
- 19. Liu S, Shi L, Wang Y, Ye D, Ju H, Ma H, et al. Stabilization of Slug by NF‐kappaB is essential for TNF‐alpha‐induced migration and epithelial‐mesenchymal transition in head and neck squamous cell carcinoma cells. Cell Physiol Biochem 2018;47:567‐578. [DOI] [PubMed] [Google Scholar]
- 20. Henderson MJ, Richie CT, Airavaara M, Wang Y, Harvey BK. Mesencephalic astrocyte‐derived neurotrophic factor (MANF) secretion and cell surface binding are modulated by KDEL receptors. J Biol Chem 2013;288:4209‐4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Werner A, Flotho A, Melchior F. The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol Cell 2012;46:287‐298. [DOI] [PubMed] [Google Scholar]
- 22. Geiss‐Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 2007;8:947‐956. [DOI] [PubMed] [Google Scholar]
- 23. Liu J, Tao X, Zhang J, Wang P, Sha M, Ma Y, et al. Small ubiquitin‐related modifier 1 is involved in hepatocellular carcinoma progression via mediating p65 nuclear translocation. Oncotarget 2016;7:22206‐22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu J, Sha M, Wang Q, Ma Y, Geng X, Gao Y, et al. Small ubiquitin‐related modifier 2/3 interacts with p65 and stabilizes it in the cytoplasm in HBV‐associated hepatocellular carcinoma. BMC Cancer 2015;15:675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shridhar R, Shridhar V, Rivard S, Siegfried JM, Pietraszkiewicz H, Ensley J, et al. Mutations in the arginine‐rich protein gene, in lung, breast, and prostate cancers, and in squamous cell carcinoma of the head and neck. Cancer Res 1996;56:5576‐5578. [PubMed] [Google Scholar]
- 26. Evron E, Cairns P, Halachmi N, Ahrendt SA, Reed AL, Sidransky D. Normal polymorphism in the incomplete trinucleotide repeat of the arginine‐rich protein gene. Cancer Res 1997;57:2888‐2889. [PubMed] [Google Scholar]
- 27. Wang D, Hou C, Cao Y, Cheng Q, Zhang L, Li H, et al. XBP1 activation enhances MANF expression via binding to endoplasmic reticulum stress response elements within MANF promoter region in hepatitis B. Int J Biochem Cell Biol 2018;99:140‐146. [DOI] [PubMed] [Google Scholar]
- 28. Mizobuchi N, Hoseki J, Kubota H, Toyokuni S, Nozaki J, Naitoh M, et al. ARMET is a soluble ER protein induced by the unfolded protein response via ERSE‐II element. Cell Struct Funct 2007;32:41‐50. [DOI] [PubMed] [Google Scholar]
- 29. Oh‐Hashi K, Hirata Y, Kiuchi K. Transcriptional regulation of mouse mesencephalic astrocyte‐derived neurotrophic factor in Neuro2a cells. Cell Mol Biol Lett 2013;18:398‐415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue‐based map of the human proteome. Science 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 31. Neves J, Zhu J, Sousa‐Victor P, Konjikusic M, Riley R, Chew S, et al. Immune modulation by MANF promotes tissue repair and regenerative success in the retina. Science 2016;353:aaf3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Glembotski CC. Functions for the cardiomyokine, MANF, in cardioprotection, hypertrophy and heart failure. J Mol Cell Cardiol 2011;51:512‐517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol 2016;65:798‐808. [DOI] [PubMed] [Google Scholar]
- 34. Tacke F, Zimmermann HW. Macrophage heterogeneity in liver injury and fibrosis. J Hepatol 2014;60:1090‐1096. [DOI] [PubMed] [Google Scholar]
- 35. Zwacka RM, Zhang Y, Halldorson J, Schlossberg H, Dudus L, Engelhardt JF. CD4(+) T‐lymphocytes mediate ischemia/reperfusion‐induced inflammatory responses in mouse liver. J Clin Invest 1997;100:279‐289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell 2016;166:21‐45. [DOI] [PubMed] [Google Scholar]
- 37. Casas E, Kim J, Bendesky A, Ohno‐Machado L, Wolfe CJ, Yang J. Snail2 is an essential mediator of Twist1‐induced epithelial mesenchymal transition and metastasis. Cancer Res 2011;71:245‐254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lopez‐Novoa JM, Nieto MA. Inflammation and EMT: an alliance towards organ fibrosis and cancer progression. EMBO Mol Med 2009;1:303‐314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pan G, Gao Y, Ye J, Jiang T, Shen Y, Shen Y. Study on correlation between MANF expression levels and the grade of liver fibrosis. Acta Universitatis Medicinalis Anhui 2015;50:78‐82. [Google Scholar]
- 40. Sehat B, Tofigh A, Lin Y, Trocme E, Liljedahl U, Lagergren J, et al. SUMOylation mediates the nuclear translocation and signaling of the IGF‐1 receptor. Sci Signal 2010;3:ra10. [DOI] [PubMed] [Google Scholar]
- 41. Ouyang J, Shi Y, Valin A, Xuan Y, Gill G. Direct binding of CoREST1 to SUMO‐2/3 contributes to gene‐specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell 2009;34:145‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Y, Shankar SR, Kher D, Ling BM, Taneja R. Sumoylation of the basic helix‐loop‐helix transcription factor sharp‐1 regulates recruitment of the histone methyltransferase G9a and function in myogenesis. J Biol Chem 2013;288:17654‐17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


