Abstract
Sargassum is a cosmopolitan brown algal genus spanning the three ocean basins of the Atlantic, Pacific and Indian Oceans, inhabiting temperate, subtropical and tropical habitats. Sargassum has been postulated to have originated in the Oligocene epoch approximately 30 mya according to a broad phylogenetic analysis of brown macroalgae, but its diversification to become one of the most widespread and speciose macroalgal genera remains unclear. Here, we present a Bayesian molecular clock study, which analyzed data from the order Fucales of the brown algal crown radiation (BACR) group to reconstruct a time‐calibrated phylogeny of the Sargassum clade. Our phylogeny included a total of 120 taxa with 99 Sargassum species sampled for three molecular markers – ITS‐2, cox3 and rbcLS – calibrated with an unambiguous Sargassaceae fossil from between the lower and middle Miocene. The analysis revealed a much later origin of Sargassum than expected at about 6.7 mya, with the genus diversifying since approximately 4.3 mya. Current geographic distributions of Sargassum species were then analyzed in conjunction with the time‐calibrated phylogeny using the dispersal‐extinction‐cladogenesis (DEC) model to estimate ancestral ranges of clades in the genus. Results strongly support origination of Sargassum in the Central Indo‐Pacific (CIP) region with subsequent independent dispersal events into other marine realms. The longer history of diversification in the ancestral CIP range could explain the much greater diversity there relative to other marine areas today. Analyses of these dynamic processes, when fine‐tuned to a higher spatial resolution, enable the identification of evolutionary hotspots and provide insights into long‐term dispersal patterns.
Keywords: ancestral range, brown macroalgae, cladogenesis, global distribution, macroevolution, time‐calibrated phylogeny
Abbreviations
- BACR
brown algal crown radiation
- cox3
mitochondrial cytochrome coxidase subunit 3
- DEC
dispersal‐extinction‐cladogenesis
- HPD
highest posterior density
- ITS‐2
internal transcribed spacer 2 region of nuclear ribosomal RNA
- MCC
maximum clade credibility
- PP
posterior probability
- rbcLS
chloroplastic ribulose bisphosphate carboxylase large subunit
Species richness is high in many marine environments despite the scarcity of hard barriers isolating populations from gene flow (Cowman and Bellwood 2013). While there is strong interest in the study of diversification and evolution of marine organisms (Vieira et al. 2017), the interconnectedness of the oceans makes it challenging to study the macroevolutionary processes driving the distribution of species (Cowman and Bellwood 2013). Fundamentally, spatial variation in species richness over evolutionary timescales can be explained by biogeographic models, which make predictions about the relative contributions of the primary macroevolutionary processes, speciation, extinction and dispersal (Stebbins 1974, Jablonski 1993, Jablonski et al. 2006). Interestingly, marine taxa exhibiting concordant diversity distributions may have discordant and complex biogeographic histories (Bowen et al. 2013). For example, the high species diversity of reef fish in the Indo‐West Pacific region has been shown to be driven by rapid speciation (Mora et al. 2003, Tornabene et al. 2015) while the high coral diversity could have resulted from long‐term dispersal of species into this region (Connolly et al. 2003, Huang et al. 2018).
While many marine groups, including corals, cowries and shore fishes, exhibit high relative diversity in the tropical Indo‐West Pacific region (Hughes et al. 2002, Bellwood et al. 2005, 2012, Bellwood and Meyer 2009), marine algae are typically more genus rich in temperate regions (Kerswell 2006). Despite this, there are taxon‐specific patterns where high algal richness has been observed in the tropics, specifically in the central Indo‐Pacific region such as for the brown macroalga Lobophora (Dictyotales, Phaeophyceae; Vieira et al. 2017) and red macroalga Portieria (Gigartinales, Rhizophyllidaceae; Leliaert et al. 2018). Likewise, the cosmopolitan genus Sargassum, which comprises over 350 currently accepted species (Guiry and Guiry 2018), exhibits a similar pattern by dominating the tropical and subtropical regions (Mattio and Payri 2011). It is also especially species rich in the Pacific (Phillips 1995), although few studies on marine algae, apart from Fucus (Cánovas et al. 2011) and Lobophora (Vieira et al. 2017), have tested biogeographic hypotheses of the richness divergence between the Pacific and Atlantic Oceans. Despite the widespread and far‐ranging ecological associations Sargassum has with other marine organisms – from being a food source and providing key tropical habitats equivalent to the temperate kelp forests (Mattio and Payri 2011, Mattio et al. 2013), to being a potent competitor of corals on reef environments (McManus and Polsenberg 2004) – the contemporary distribution and historical biogeography of the genus remain poorly understood.
Recent availability of DNA sequence data for a large number of Sargassum species has improved our understanding of phylogenetic relationships within the group (Mattio et al. 2008, 2009, 2015, Mattio and Payri 2009, Yip et al. 2018). When calibrated using relevant paleontological data, the phylogeny can be used to infer species ancestral ranges and the origination of the genus (Moore and Donoghue 2007). Paleontological data on brown macroalgae (Phaeophyceae), such as Sargassum, are sparse due to their general inability to fossilize (Silberfeld et al. 2010). Nevertheless, the diversification of Sargassum has been estimated to be relatively recent – no earlier than the Neogene period (Silberfeld et al. 2010). Therefore, like for many successful marine taxa, late geological events in the Pliocene may have been the main drivers of its diversification and distribution (Hallam 1985).
Biogeographic studies of brown macroalgal groups have focused on the macroevolution of common and widespread genera like Macrocystis (Rothman et al. 2017), Lobophora (Vieira et al. 2017), and Fucus (Cánovas et al. 2011). Those of Sargassum are largely limited to the use of hydrodynamic and ocean current models. For example, Mattio et al. (2015) utilized hydrodynamic models to characterize dispersal patterns between isolated islands of the western Indian Ocean, while Phillips (1995) invoked ocean circulation to explain why the western Pacific is more diverse than eastern Pacific despite higher endemism in the latter. By leveraging on a more robust understanding of the Sargassum phylogeny, biogeographic analyses of species ancestral ranges could be performed to reconstruct macroevolutionary processes that have shaped present species distributions and diversity gradients.
Congruent diversity patterns between macroalgae and other major coral reef taxa such as fishes and corals have generally been assumed to result from associations that drive the diversification of reef organisms as a whole (Hughes et al. 2002, Cowman and Bellwood 2011). For example, ecological interactions like herbivory have been known to play key roles in macroalgal evolution (Hay 1997), suggesting that diversification trajectories of macroalgae and their herbivores may be correlated. Furthermore, as the foundation of reef habitats, corals provide opportunities for new niche colonization that may promote reef algal speciation (Cowman and Bellwood 2011). As such, the importance of investigating diversity and distribution patterns of Sargassum are underlain by its interactions with modern corals. Macroalgae compete directly with hard corals via several means, including shading, physical abrasion, competition for space, and impacting coral larval settlement (McCook et al. 2001). Thus, phase shift to macroalgal dominance by this ubiquitous reef genus is a serious threat to corals and an indication of poor reef health (Azevedo et al. 2011, Smith et al. 2016).
Apart from co‐diversification of reef organisms, there are other hypotheses and growing evidence suggesting that large‐scale geological events and tectonic activities could account for concordant biogeographic processes and diversification patterns, particularly between reef fishes and corals (Keith et al. 2013, Leprieur et al. 2016). In addition, availability of habitats and empty niches promotes speciation, which could also result in similar patterns across taxa (Moura et al. 2013, Sanciangco et al. 2013). Therefore, understanding the processes that drive diversification and diversity patterns involves integrating evolutionary models with paleoclimatic and paleoceanographic data. In uncovering the macroevolutionary patterns of Sargassum that have led to the contemporary global distribution and species richness gradients (Okamura 1932, Yoshida 1989, Phillips 1995), biodiversity drivers on habitats in which Sargassum thrives could eventually be elucidated (Etti and Schils 2016).
In this study, we inferred the most comprehensive time‐calibrated phylogeny of Sargassum yet, based on all available sequences of three commonly analyzed markers to estimate the age and origin of Sargassum. Species distribution data were then consolidated from isolated diversity studies over the last decade (Mattio et al. 2008, 2009, 2015, Mattio and Payri 2009, Nguyen 2014) and analyzed in conjunction with the phylogeny using a biogeographic model to estimate ancestral ranges and macroevolutionary processes (dispersal‐extinction‐cladogenesis; Ree and Smith 2008). Our reconstruction supports a Central Indo‐Pacific origination and initial diversification of Sargassum with very recent (<1.5 mya) independent dispersals into the Atlantic, resulting in high species richness in the Pacific but limited diversity and endemism in the Atlantic.
Materials and Methods
Taxon sampling and alignment
Molecular sequences of nuclear ITS‐2, chloroplastic partial RuBisCO operon rbcLS, and mitochondrial cox3 for Sargassum species were downloaded from the NCBI GenBank database (Table S1 in the Supporting Information). Sequences were selected based on a set of criteria to maximize taxonomic accuracy. Specifically, we preferred sequences generated in the recent Sargassum‐focused taxonomic work by Mattio et al. (2008, 2009, 2010, 2013, 2015, 2010), Mattio and Payri (2009), followed by sequences from other published studies (Camacho et al. 2015) and, lastly, unpublished sequences uploaded by taxonomists. After this screening process, data were found to be available for 99 species, representing 28% of all valid Sargassum species (Guiry and Guiry 2018). Gene sequences used for each species may not have been sampled from the same individual so as to increase gene coverage and reduce missing data. Sequences from Silberfeld et al. (2010) representing outgroup species from the Fucales order and Turbinaria sequences from Stiger et al. (2003) were also downloaded and included in the analysis. The 120‐taxon data matrices were each aligned with MAFFT 7.311 (Katoh and Standley 2013) using the L‐INS‐I alignment algorithm and trimmed to 710, 434, and 755 sites for ITS‐2, cox3, and rbcLS respectively in Mesquite 3.2 (Maddison and Maddison 2017), before being concatenated for phylogenetic analyses.
Bayesian divergence time‐estimated phylogeny
The best‐fit substitution model for each gene alignment was estimated using jModelTest2 (Guindon and Gascuel 2003, Darriba et al. 2012), comparing the fit of various models under the Akaike information criterion (ITS‐2: GTR + Γ; cox3 and rbcLS: GTR + I + Γ). Substitution models were included as priors in a Bayesian analysis to infer the time‐calibrated phylogeny of Fucales focusing on the genus Sargassum using a relaxed molecular clock implemented in BEAST 2.4.6 (Huelsenbeck et al. 2001, Drummond and Rambaut 2007, Bouckaert et al. 2014). The three gene partitions were linked with an uncorrelated lognormal clock model and a birth–death tree prior.
The lack of a continuous and well‐characterized fossil record stemmed from the nature of brown algal groups, which comprised only soft tissues (Silberfeld et al. 2010). The only geological formation that yielded fossils for calibrating the Fucales phylogeny was associated with the Sargassaceae family. The monophyletic group of the order Fucales, excluding the deepest branch Notheia anomala (Notheiceae), was the only node used as a calibration point by applying Sargassaceae fossils previously implemented in Silberfeld et al. (2010). The structurally complex fossils found in the Miocene deposits (13–17 mya) of the Monterey formation (Parker and Dawson 1965) belonged to the extinct genera Paleocystophora (P. subopposita) and Paleohalidrys (P. californica, P. superba, and P. occidentalis) and were comparable to the Fucalean family Sargassaceae based on their sympodial bifurcating patterns (Silberfeld et al. 2010). Since it was ambiguous to assign these extinct fossil genera to any extant genus, and the origin of Sargassaceae must predate the origin of the earliest fossil assigned to the family, the mid‐point of the Miocene deposits at 15 mya was set as a hard minimum bound for the stem node of Sargassaceae. A lognormal distribution (mean ± SE = 2.0 ± 0.85) with 95% highest posterior density (HPD) spanning 16.8 to 44.9 mya (minimum bound + 30 Ma; Wood et al. 2013) was applied to calibrate the node.
Eight independent Monte Carlo Markov Chain (MCMC; Altekar et al. 2004) runs were performed using BEAST 2.4.6 on the molecular data with fossil constraints, implemented at the Cyberinfrastructure for Phylogenetic Research Science Gateway 3.3 (Miller et al. 2010). Markov chains were run for 60 million generations and sampled every 1,000th generation. Convergence of all parameters was monitored using Tracer 1.6 (Rambaut et al. 2014), and a burn‐in of 10 million generations was determined. The post‐burn‐in output tree files from the eight runs were combined using LogCombiner 2.4.7 (Bouckaert et al. 2014), resampled at a lower frequency of one per 20,000 trees, and summarized as a maximum clade credibility (MCC) chronogram with TreeAnnotator 2.4.7 based on 20,000 posterior trees.
Ancestral range estimation
The maximum likelihood‐based dispersal‐extinction‐cladogenesis (DEC) biogeographic model (Ree and Smith 2008) was applied to infer the evolution of geographic ranges and estimate the dispersal and local extinction rates using R 3.4.2 (R Core Team 2013) package BioGeoBEARS (Matzke 2013). DEC accounts for subset inheritance of sister species ranges from sympatric speciation events, which other biogeographic models like DIVA and BAYAREA do not (Matzke 2013). An additional model accounting for founder‐event speciation (+J) in island clades was assessed and compared with the DEC model (Matzke 2013, 2014). The rare jump dispersal event was reflected in the ancestral range estimation through a genetically diverging isolated lineage. The model was fitted to the MCC tree and species distribution data, with geographic ranges demarcated according to Vieira et al. (2017) and bioregionalization of coastal areas by Spalding et al. (2007). A total of five areas were defined: Central Indo‐Pacific (A), Western Indo‐Pacific (B), Eastern Indo‐Pacific (C), Tropical Eastern Pacific (D), and Atlantic (E). Geographic ranges of the 99 Sargassum species were obtained from relevant publications (Tseng et al. 1985, Tseng and Lu 1988, Yoshida 1988, Lee and Yoo 1992, Trono 1992, Ajisaka et al. 1994, 1995, 1999, Phillips 1995, Silva et al. 1996, Ajisaka 2002, Yoshida et al. 2002, Noiraksar and Ajisaka 2008) compiled at AlgaeBase (http://algaebase.org) and coded for these areas (Table S2 in the Supporting Information). Dispersal rates for area combinations of ocean basins were relaxed to allow for equal dispersal probabilities, and all possible dispersal directions were permitted considering the possibility of high connectivity among marine regions.
Results
Time‐calibrated phylogeny of Sargassum
Following the removal of burn‐in samples, the effective sample sizes (ESS) were above 200 for all divergence time parameters. The Bayesian posterior probabilities (PP) of selected clades along with the inferred ages and 95% HPDs are presented in Table 1, with the corresponding labelled nodes shown in Figure 1. The fossil calibration point at the origin of the Sargassaceae family was estimated to have a mean age of 20.6 mya, and the origin of Sargassum was dated to be Late Miocene with mean age of 6.7 mya. The internal deep nodes of the Sargassum lineages from 4.3 to 1.6 mya representing taxonomic sections were moderately to strongly supported (PP > 0.9), while shallower nodes representing interspecific relationships were less strongly supported.
Table 1.
Bayesian estimates for selected nodes on the chronogram in Figure 1, showing posterior probability, mean age, and 95% highest posterior probability (HPD) interval for each node of interest
| Node | Description | Posterior probability | Mean age (mya) | 95% HPD Interval (mya) |
|---|---|---|---|---|
| 1 | Fucales | 1 | 25.1 | 16.4–39.4 |
| 2 | Diversification of Sargassaceae | 1 | 11.3 | 6.5–17.9 |
| 3 | Diversification of Sargassum | 1 | 4.3 | 2.2–6.8 |
| 4 | Subgen. Sargassum | 1 | 2.8 | 1.4–4.3 |
| 5 | Subgen. Bactrophycus | 1 | 2.9 | 1.5–4.8 |
| 6 | – | 0.93 | 2.6 | 1.3–4.1 |
| 7 | Sect. Teretia | 1 | 1.6 | 0.8–2.6 |
| 8 | Sect. Halochloa | 1 | 0.5 | 0.2–0.9 |
| 9 | – | 1 | 2.1 | 1.1–3.3 |
| 10 | Sect. Binderiana | 1 | 1.5 | 0.6–2.6 |
| 11 | Sect. Polycystae | 1 | 0.4 | 0.1–0.8 |
| 12 | Sect. Ilicifolia | 1 | 0.9 | 0.5–1.5 |
| 13 | Sect. Zygocarpicae | 0.99 | 0.6 | 0.2–1.1 |
| 14 | Sect. Sargassum | 0.95 | 1.0 | 0.5–1.6 |
| 15 | Undefined section | 0.83 | 1.5 | 0.6–2.6 |
| 16 | Origin of Sargassum | 1 | 6.7 | 3.4–11.0 |
Figure 1.
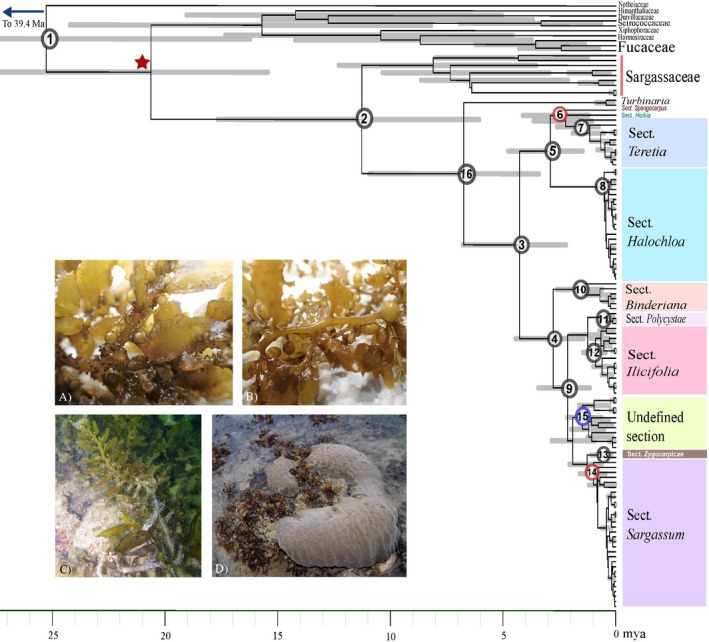
Maximum clade credibility chronogram from the BEAST analysis. Gray bars represent 95% highest posterior density (HPD) intervals of node ages. The calibrated node is denoted by a red star. Posterior probabilities of the numbered nodes (see Table 1) are denoted by color: PP > 95: black; 95 ≤ PP < 90: red; PP ≤ 90: blue. Each colored box on the right represents a section within subgenus Sargassum. The inset shows: A) Sargassum polycystum; B) Sargassum sp.; C) S. swartzii (photo credit: J.K.Y. Low); and D) coral‐Sargassum interaction (photo credit: J. Fong).
Sargassum was recovered as sister taxon to genus Turbinaria with strong support (PP = 1.00). The divergence of Sargassum into two clades – subgenera Sargassum (PP = 1.00) and Batrophycus (PP = 1.00) – was estimated at 4.3 mya in the early Pliocene before closure of the Isthmus of Panama. Subsequent diverging lineages at subgenus level occurred throughout the Pleistocene, with the earliest diverging section Spongocarpus at 2.6 mya and the most recent section Polycystae at 0.4 mya (Fig. 1).
Biogeographic analysis
Richness comparisons based on species included in the phylogenetic analysis revealed that the Central Indo‐Pacific region (A) has the highest species richness with 82 spp., while its neighboring region (B) harbors fewer species (38 spp.). The large area of Central Indo‐Pacific combined with the Indian Ocean (AB) has the highest species richness (17 spp.) in comparison with other area combinations. The Tropical Eastern Pacific (D) has similar species richness (18 spp.) as the Atlantic (E; 17 spp.), while the Eastern Indo‐Pacific (C) has 11 species (Fig. 2). When all 360 extant Sargassum species were considered, we found records of 277 species in the CIP (A) and 111 species in region B. Regions C, D, and E have lower species richness of 12, 35, and 42, respectively.
Figure 2.
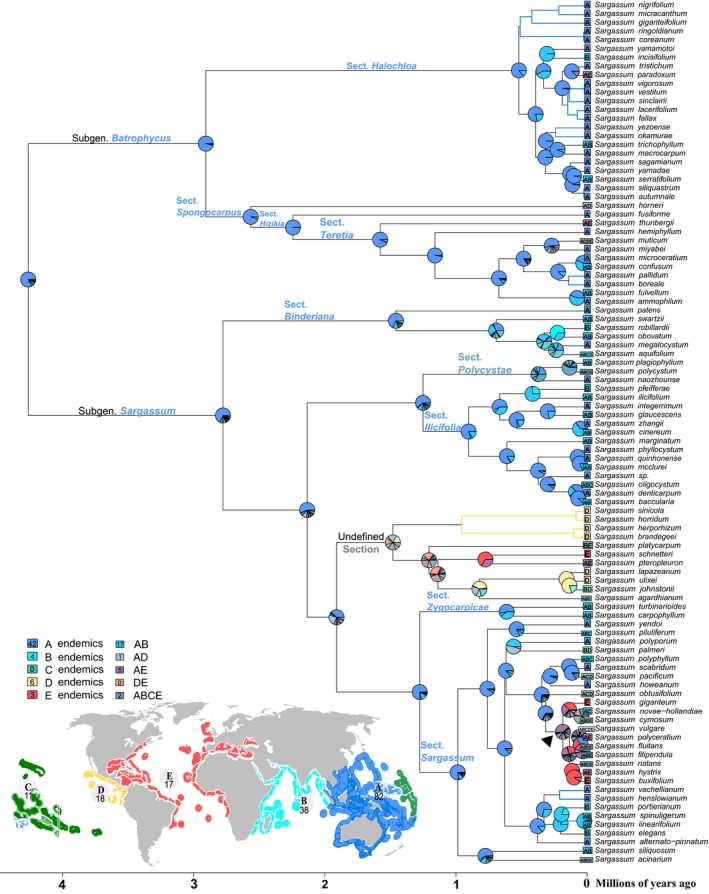
Ancestral range estimation of Sargassum species under the DEC + J model fitted onto a time‐calibrated phylogeny (Fig. 1). Tip symbols represent contemporary geographic ranges of extant taxa, and the nodal pie diagrams reflect ancestral ranges of the common ancestor. Colored taxonomic groups correspond to their ancestral range of the highest probability, and colored branches indicate maximal probability of single area distribution. Marine areas used in the analysis are Central Indo‐Pacific (A), Western Indo‐Pacific (B), Eastern Indo‐Pacific (C), Tropical Eastern Pacific (D), and Atlantic (E).
The DEC model (AIC = 540.3; log‐likelihood = ‐267.5) was identified as being as well fit as the DEC + J model (AIC = 539.1; log‐likelihood = −267.1; p = 0.38) when the five marine areas were analyzed under relaxed constraints for region combinations. The model recovered an unequivocal emphasis placed on stochastic dispersal in the range evolution of species (d = 0.281 > e = 0 > j = 0).
Specifically, the model supported a Central Indo‐Pacific origination of the Sargassum ancestor, as well as for ancestors of subgenera Sargassum and Batrophycus (Fig. 2). Ancestors of the sections within the Sargassum subgenera shared the same ancestral area in the Central Indo‐Pacific, except for section Polycystae – with widespread ancestral range and present globally except in the Tropical Eastern Pacific – and the undefined section whose most recent common ancestor could be from the Central Indo‐Pacific or Tropical Eastern Pacific (Fig. 2). The common ancestor of the subclade (▲ in Fig. 2) within section Sargassum ranged across both the Atlantic and Pacific, with the majority of their descendant taxa occupying both the Pacific and Atlantic Ocean basins.
Overall, results lent support for a Central Indo‐Pacific origination and initial diversification of the Sargassum genus. Lineages began dispersing into the Atlantic less than 1.5 mya in two clades independently (undefined section and subclade ▲ in Fig. 2) and among five phylogenetically disparate species while maintaining their Pacific distribution.
Discussion
The study of historical biogeographic patterns requires a comprehensive phylogeny that integrates clade age and geographic range data (Moore and Donoghue 2007). With recent advancements in DNA sequencing techniques, molecular analyses have been used to complement morphological analyses for a clearer understanding of the evolutionary relationships among Sargassum sections and species (Mattio and Payri 2011). Thus, the focus of the last decade has been to reconstruct molecular phylogenies that are not fossil‐calibrated and thus not scaled to geological time (Cho et al. 2012). For the first time, we use three commonly sequenced markers and a stem species of Sargassaceae to infer a time‐calibrated phylogeny focusing on the Sargassum genus and, furthermore, produce a set of Bayesian posterior trees for capturing the uncertainties of the tree topology and branch lengths.
The phylogeny recovered here is broadly congruent with the reconstruction in Dixon et al. (2014), which focused on the revision of the subgenera Batrophycus and Sargassum using molecular sequences of ITS‐2, cox3, and rbcLS. Two major clades representing the subgenera Batrophycus and Sargassum diverge into clades corresponding to the taxonomic sections (Fig. 1). Furthermore, in reconstructing the phylogeny of the brown algal crown radiation (BACR) group, Silberfeld et al. (2010) also included two Sargassum species and estimated the age of Sargassum, obtaining a 95% HPD lower bound for the divergence between the two species at 5 mya, which overlaps with the 95% HPD for the age of the diversification inferred here (2.2–6.8 mya). The origination of Sargassum, however, is estimated to be more recent in the present study (95% HPD: 3.4–11.0 mya) compared to the estimate of 22 mya as the lower bound of the 95% HPD by Silberfeld et al. (2010). The difference in time estimation for the origin of Sargassum could be attributed to the inclusion here of a closely related outgroup Turbinaria – not present in the previous study – that causes the descendant nodes (from node 3 in Fig. 1) to be pushed forward in time.
Biogeographic events and fossil records can both be incorporated in time‐calibrated phylogenetic analyses. For example, the formation of the Isthmus of Panama at 3.1 mya (Cowman and Bellwood 2013) can be used to calibrate rates of molecular evolution by applying the time of divergence of populations isolated by the rising land barrier (Knowlton et al. 1993, Knowlton and Weigt 1998, Lessios 2008). Such biogeographic events are essential in driving global marine reorganization and in creating distinct habitats for isolated populations, which then evolve on different trajectories (Lessios 2008). However, we find no reciprocally monophyletic sister clades of Atlantic and Pacific Sargassum species available for dating the origin of Atlantic and Pacific lineages, indicating limited differentiation between ocean realms. Furthermore, this vicariance event appears to be much older than the origin of all the Atlantic Sargassum lineages. Taken together, these phylogenetic results suggest that the dispersal between Pacific and Atlantic Oceans occurred much later after the closure of the Central American Seaway and thus have not been used to calibrate the phylogeny. Fossil records were instead applied to date the origin of Sargassaceae. Uncertainties concerning the age of the fossil were incorporated by specifying a loose maximum bound to avoid overconfidence in the calibration point (Parham et al. 2012). Due to the limited fossil data available for Phaeophyceae (Silberfeld et al. 2010), uncertainties in the origination and diversification dates of Sargassum span several million years (5–7 mya; Table 1).
High diversity regions differ for every marine taxon. However, there are marine taxa with largely concordant diversity hotspots, particularly if they live in the same habitats such as coral reefs (Bellwood and Meyer 2009). A more precise understanding of coral reef biogeography can be achieved by integrating not just coral and fish distributions, which are well studied, but also macroalgal diversity to test the factors driving diversification on reefs (Etti and Schils 2016). Here, ancestral range estimation has revealed the origin of Sargassum to be in the Central Indo‐Pacific (CIP) region with high certainty. From the CIP, Sargassum experienced few and recent dispersal events to the Atlantic in the last 1.5 mya, long after the closure of the Central American Seaway. The macroalgae Lobophora and Portieria show a parallel biogeographic history, originating in the CIP with high diversification within the region, followed by subsequent independent dispersal events into other marine realms (Vieira et al. 2017, Leliaert et al. 2018).
Similarly, for Sargassum, there was a profusion of early speciation events clearly reconstructed to be occurring within the CIP. This region represents the ancestral range of both subgenera Bactrophycus and Sargassum as well as most of their constituent sections (except Polycystae and the undefined section; Fig. 2). The abundance of speciation events in the CIP is likely conservative because it is the most poorly sampled of the five analyzed areas, whereas species from areas C, D, and E (Eastern Indo‐Pacific, Tropical Eastern Pacific, and Atlantic, respectively) are more than 40% sampled on our phylogeny. Clearly, prolific speciation has resulted in high species richness in the CIP, a pattern bearing stark similarity to that of stony corals (Veron et al. 2015). However, maintenance of the richness gradient for corals appears to be driven by range expansion of species into the CIP (Huang et al. 2018), whereas for Sargassum, cladogenesis in the CIP has maintained the high species richness in the region.
The CIP had undergone many climatic fluctuations in the Pleistocene (Pillans et al. 1998). In particular, the drying and subsequent re‐colonization of marine habitats during glacial–interglacial periods of both the Sunda and Sahul shelves created opportunities for the exploitation of new niches (Hofreiter and Stewart 2009, Bowen et al. 2013). Repeated, successive isolation and mixing of populations caused by the fluctuating sea level have long been recognized to promote speciation (Hallam 1985), which in the case of Sargassum coincided with the time of most of its speciation events. Despite the complexity of marine algal biogeography with each taxon exhibiting a distinct diversity pattern (Etti and Schils 2016), the locality of origin for many brown algal (Phaeophyceae) genera appears comparable. Here, the ancestral range of Sargassum is estimated to be in the CIP (Fig. 2). Similarly, the ancestral range of an older brown alga Lobophora, which originated in the Cretaceous period, is in the Indo‐Pacific region (Vieira et al. 2017). Younger brown algae that evolved in the early Cenozoic era, such as Fucus, also have their origination placed in the Pacific (Cánovas et al. 2011).
There have been past speculations on the ancestral ranges of certain taxonomic groups within Sargassum based exclusively on analyzing the species richness, endemism, and oceanic currents among the marine realms. For instance, Phillips (1995) hypothesized that the ancestral area of subgenus Sargassum is possibly in the vicinity of Baja California in the Tropical Eastern Pacific (TEP), evident from the high degree of local endemism and considerable species richness. However, species ranges are dynamic and can change over time (Jablonski et al. 2013). Endemism could also be the result of range contraction, which is associated with paleo‐endemics instead of newly evolved species (Bellwood and Meyer 2009). To account for these dynamics, we use a robust time‐calibrated phylogeny and species range data to infer the CIP origin of subgenus Sargassum with higher certainty than previous studies (Fig. 2).
The subclade within section Sargassum (labelled ▲ in Fig. 2) is inferred to have a joint ancestral range of CIP and Atlantic with relatively high level of certainty. This raises an intriguing hypothesis about its biogeographic history. The relatively recent split of the clade into CIP and Atlantic species (approximately 0.2–0.4 mya) suggests a dispersal into the Atlantic long after the formation of the Isthmus of Panama. A possible route would be across the Pacific Ocean and around the coast of South America via the Antarctic Circumpolar Current (ACC), which has been known to disperse floating macroalgae like Macrocystis and Durvillaea around the coasts of the Subantarctic (Dixon et al. 2014, Hawes et al. 2017). In other words, the ACC may have driven the dispersal of Sargassum – possibly species with temperate distributions (Yamasaki et al. 2014) – into the Atlantic via the Drake Passage (Dixon et al. 2014, Hawes et al. 2017). A more comprehensive phylogeny including all members of the section Sargassum, and incorporating past climate and ocean circulation patterns, is required to test this biogeographic hypothesis more precisely.
Studies utilizing published species records and sequence data are subject to limitations such as inaccurate species identities tagged to the data, which could result in incorrect range estimations and even bias the biogeographic inferences (Costa et al. 2015). However, given that the biogeographic range categories utilized here encompass large marine regions, small uncertainties of individual species records at range boundaries are unlikely to affect the analysis. Furthermore, we have taken steps to curate the species nomenclature and geographic data rigorously, and minimized instances of species misidentification by using only sequence data from algal taxonomists and other carefully vetted sources (datasets and species information available at Zenodo; https://doi.org/10.5281/zenodo.3403402; Tables [Link], [Link], [Link] in the Supporting Information).
Conclusions
This study shows that high Sargassum richness in the Central Indo‐Pacific region is attributable to diversification within the CIP, rather than dispersal from peripheral regions into the CIP. Range evolution into other marine realms appears to have been driven primarily by dispersal. Future studies should focus on more complete taxon sampling and apply more gene markers to place most, if not all, Sargassum species on the phylogeny in order to produce more precise divergence time and ancestral range estimates. Past dispersal routes could also be traced by reconstructing paleoclimatic and paleoceanographic changes. Understanding the biogeographic history of Sargassum will pave the way for projecting range evolution of the macroalgal genus into the future, which is pivotal for developing conservation strategies in the event of climate change‐induced macroalgal encroachment into coral reef habitats.
We thank J.K.Y. Low and J. Fong for providing field photographs and are grateful to F. Leliaert and two anonymous reviewers for constructive reviews. This research is supported by the National Research Foundation, Prime Minister's Office, Singapore under its Marine Science R&D Program (MSRDP‐P03; R‐154‐000‐A25‐281).
Conflict of Interest
The authors declare that they have no conflict of interest, and this article does not contain any studies with animals performed by any of the authors.
Supporting information
Table S1. GenBank accession numbers of sequences used in the analyses.
Table S2. Locality data of five marine realms; where 1 represents presence and 0 absence: A) Central Indo‐Pacific, B) Western Indo‐Pacific, C) Eastern Indo‐Pacific, D) Tropical Eastern Pacific, and E) Atlantic.
Table S3. Geographic records for each species, adapted from AlgaeBase.
Table S4. Sources of the sequences used in this study and their respective voucher specimens and publication information.
Table S5. Sources of species taxonomy, adapted from AlgaeBase.
Contributor Information
Zhi Ting Yip, Email: zhiting.yip@gmail.com.
Danwei Huang, Email: huangdanwei@nus.edu.
References
- Ajisaka, T. 2002. Sargassum specimens from Singapore and Malaysia in the Herbarium of the Bishop Museum In Abbott I. A. & McDermid K. J. [Eds.] Taxonomy of Economic Seaweeds With Reference to Some Pacific Species. Volume VIII California Sea Grant College Program, University of California, La Jolla, pp. 77–88. [Google Scholar]
- Ajisaka, T. , Noro, T. , Trono, Jr, G. C. , Chiang, Y. M. & Yoshida, T. 1994. Several Sargassum species (Subgenus Sargassum) in east Asia with furcately branching leaves In Abbott I. A. [Ed.] Taxonomy of Economic Seaweeds With Reference to Some Pacific Species. Volume IV California Sea Grant College Program, University of California, La Jolla, pp. 9–22. [Google Scholar]
- Ajisaka, T. , Noro, T. & Yoshida, T. 1995. Zygocarpic Sargassum species (subgenus Sargassum) from Japan In Abbott I. A. [Ed.] Taxonomy of Economic Seaweeds With Reference to Some Pacific Species. Volume V California Sea Grant College Program, University of California, La Jolla, pp. 11–44. [Google Scholar]
- Ajisaka, T. , Phang, S. M. & Yoshida, T. 1999. Preliminary report of Sargassum species collected from Malaysian coasts In Abbott I. A. [Ed.] Taxonomy of Economic Seaweeds With Reference to Some Pacific Species. Volume VII California Sea Grant College Program, University of California, La Jolla, pp. 23–41. [Google Scholar]
- Altekar, G. , Dwarkadas, S. , Huelsenbeck, J. P. & Ronquist, F. 2004. Parallel Metropolis‐coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20:407–15. [DOI] [PubMed] [Google Scholar]
- Azevedo, C. A. A. , Carneiro, M. A. A. , Oliveira, S. R. & Marinho‐Soriano, E. 2011. Macrolgae as an indicator of the environmental health of the Pirangi reefs, Rio Grande do Norte, Brazil. Revista Brasileira de Farmacognosia. 21:323–8. [Google Scholar]
- Bellwood, D. R. , Hughes, T. P. , Connolly, S. R. , Tanner, J. & Worm, B. 2005. Environmental and geometric constraints on Indo‐Pacific coral reef biodiversity. Ecol. Lett. 8:643–51. [Google Scholar]
- Bellwood, D. R. & Meyer, C. P. 2009. Searching for heat in a marine biodiversity hotspot. J. Biogeogr. 36:569–76. [Google Scholar]
- Bellwood, D. R. , Renema, W. & Rosen, B. R. 2012. Biodiversity hotspots, evolution and coral reef biogeography: a review In Gower D., Johnson K., Richardson J., Rosen B., Rüber L. & Williams S. [Eds.] Biotic Evolution and Environmental Change in Southeast Asia. Cambridge University Press, Cambridge, pp. 216–45. [Google Scholar]
- Bouckaert, R. , Heled, J. , Kühnert, D. , Vaughan, T. , Wu, C. H. , Xie, D. , Suchard, M. A. , Rambaut, A. & Drummond, A. J. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10:e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen, B. W. , Rocha, L. A. , Toonen, R. J. & Karl, S. A. 2013. The origins of tropical marine biodiversity. Trends Ecol. Evol. 28:359–66. [DOI] [PubMed] [Google Scholar]
- Cánovas, F. G. , Mota, C. F. , Serrão, E. A. & Pearson, G. A. 2011. Driving south: a multi‐gene phylogeny of the brown algal family Fucaceae reveals relationships and recent drivers of a marine radiation. BMC Evol. Biol. 11:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho, O. , Mattio, L. , Draisma, S. , Fredericq, S. & Diaz‐Pulido, G. 2015. Morphological and molecular assessment of Sargassum (Fucales, Phaeophyceae) from Caribbean Colombia, including the proposal of Sargassum giganteum sp. nov., Sargassum schnetteri comb. nov. and Sargassum section Cladophyllum sect. nov. Syst. Biodiv. 13:105–30. [Google Scholar]
- Cho, S. M. , Lee, S. M. , Ko, Y. D. , Mattio, L. & Boo, S. M. 2012. Molecular systematic reassessment of Sargassum (Fucales, Phaeophyceae) in Korea using four gene regions. Bot. Mar. 55:473–84. [Google Scholar]
- Connolly, S. R. , Bellwood, D. R. & Hughes, T. P. 2003. Indo‐Pacific biodiversity of coral reefs: deviations from a mid‐domain model. Ecology 84:2178–90. [Google Scholar]
- Costa, H. , Foody, G. , Jiménez, S. & Silva, L. 2015. Impacts of species misidentification on species distribution modeling with presence‐only data. ISPRS Int. J. Geoinf. 4:2496–518. [Google Scholar]
- Cowman, P. & Bellwood, D. 2011. Coral reefs as drivers of cladogenesis: expanding coral reefs, cryptic extinction events, and the development of biodiversity hotspots. J. Evol. Biol. 24:2543–62. [DOI] [PubMed] [Google Scholar]
- Cowman, P. F. & Bellwood, D. R. 2013. Vicariance across major marine biogeographic barriers: temporal concordance and the relative intensity of hard versus soft barriers. Proc. R. Soc. B Biol. Sci. 280:20131541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba, D. , Taboada, G. L. , Doallo, R. & Posada, D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R. R. M. , Mattio, L. , Huisman, J. M. , Payri, C. E. , Bolton, J. J. & Gurgel, C. F. D. 2014. North meets south – taxonomic and biogeographic implications of a phylogenetic assessment of Sargassum subgenera Arthrophycus and Bactrophycus (Fucales, Phaeophyceae). Phycologia 53:15–22. [Google Scholar]
- Drummond, A. J. & Rambaut, A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etti, R. T. & Schils, T. 2016. Global biogeography of marine algae with applications for coral reef connectivity. Proceedings of the 13th International Coral Reef Symposium, pp. 28–47.
- Guindon, S. & Gascuel, O. 2003. A simple, fast and accurate method to estimate large phylogenies by maximum‐likelihood. Syst. Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Guiry, M. D. & Guiry, G. M. 2018. AlgaeBase. World‐wide electronic publication, National University of Ireland, Galway: http://www.algaebase.org; searched on August 24, 2018. [Google Scholar]
- Hallam, A. 1985. Jurassic molluscan migration and evolution in relation to sea level changes In Friedman G. M. [Ed.] Sedimentary and Evolutionary Cycles. Springer, Berlin, pp. 4–5. [Google Scholar]
- Hawes, N. A. , Taylor, D. I. & Schiel, D. R. 2017. Transport of drifting fucoid algae: Nearshore transport and potential for long distance dispersal. J. Exp. Mar. Biol. Ecol. 490:34–41. [Google Scholar]
- Hay, M. E. 1997. The ecology and evolution of seaweed‐herbivore interactions on coral reefs. Coral Reefs 16:S67–76. [Google Scholar]
- Hofreiter, M. & Stewart, J. 2009. Ecological change, range fluctuations and population dynamics during the Pleistocene. Curr. Biol. 19:R584–94. [DOI] [PubMed] [Google Scholar]
- Huang, D. , Goldberg, E. E. , Chou, L. M. & Roy, K. 2018. The origin and evolution of coral species richness in a marine biodiversity hotspot. Evolution 72:288–302. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J. P. , Ronquist, F. , Nielsen, R. & Bollback, J. P. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310–4. [DOI] [PubMed] [Google Scholar]
- Hughes, T. P. , Bellwood, D. R. & Connolly, S. R. 2002. Biodiversity hotspots, centres of endemicity, and the conservation of reefs. Ecol. Lett. 5:775–84. [Google Scholar]
- Jablonski, D. 1993. The tropics as a source of evolutionary novelty through geological time. Nature 364:142–4. [Google Scholar]
- Jablonski, D. , Belanger, C. L. , Berke, S. K. , Huang, S. , Krug, A. Z. , Roy, K. , Tomasovych, A. & Valentine, J. W. 2013. Out of the tropics, but how? Fossils, bridge species, and thermal ranges in the dynamics of the marine latitudinal diversity gradient Proc. Natl. Acad. Sci. USA 110:10487–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski, D. , Roy, K. & Valentine, J. W. 2006. Out of the tropics: evolutionary dynamics of the latitudinal diversity gradient. Science 314:102–6. [DOI] [PubMed] [Google Scholar]
- Katoh, K. & Standley, D. M. 2013. MAFFT: multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, S. A. , Baird, A. H. , Hughes, T. P. , Madin, J. S. & Connolly, S. R. 2013. Faunal breaks and species composition of Indo‐Pacific corals: the role of plate tectonics, environment and habitat distribution. Proc. R. Soc. B Biol. Sci. 280:20130818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerswell, A. 2006. Global biodiversity patterns of benthic marine algae. Ecology 87:2479–88. [DOI] [PubMed] [Google Scholar]
- Knowlton, N. & Weigt, L. A. 1998. New dates and new rates for divergence across the Isthmus of Panama. Proc. R. Soc. B Biol. Sci. 265:2257–63. [Google Scholar]
- Knowlton, N. , Weigt, L. A. , Solorzano, L. A. , Mills, D. K. & Bermingham, E. 1993. Divergence in proteins, mitochondrial DNA, and reproductive compatibility across the Isthmus of Panama. Science 260:1629–32. [DOI] [PubMed] [Google Scholar]
- Lee, K. & Yoo, S. A. 1992. Korean species of Sargassum subgenus Bactrophycus J. Agardh (Sargassaceae, Fucales) In Abbott I. A. [Ed.] Taxonomy of Economic Seaweeds With Reference to Some Pacific and Western Atlantic Species. Volume III California Sea Grant College Program, University of California, La Jolla, pp. 139–47. [Google Scholar]
- Leliaert, F. , Payo, D. A. , Gurgel, C. F. D. , Schils, T. , Draisma, S. G. , Saunders, G. W. & Le Gall, L. 2018. Patterns and drivers of species diversity in the Indo‐Pacific red seaweed Portieria . J. Biogeogr. 45:2299–313. [Google Scholar]
- Leprieur, F. , Descombes, P. , Gaboriau, T. , Cowman, P. F. , Parravicini, V. , Kulbicki, M. , Melián, C. J. , de Santana, C. N. , Heine, C. , Mouillot, D. , et al. 2016. Plate tectonics drive tropical reef biodiversity dynamics. Nat. Commun. 7:11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessios, H. A. 2008. The great American schism: divergence of marine organisms after the rise of the Central American Isthmus. Annu. Rev. Ecol. Evol. Syst. 39:63–91. [Google Scholar]
- Maddison, W. P. & Maddison, D. R. 2017. Mesquite: a modular system for evolutionary analysis. Version 3.2. http://mesquiteproject.org
- Mattio, L. , Anderson, R. J. & Bolton, J. J. 2015. A revision of the genus Sargassum (Fucales, Phaeophyceae) in South Africa. S. Afr. J. Bot. 98:95–107. [Google Scholar]
- Mattio, L. & Payri, C. E. 2009. Taxonomic revision of Sargassum species (Fucales, Phaeophyceae) from New Caledonia based on morphological and molecular analyses. J. Phycol. 45:1374–88. [DOI] [PubMed] [Google Scholar]
- Mattio, L. , Payri, C. , Verlaque, M. & Reviers, B. 2010. Taxonomic revision of Sargassum sect. Acanthocarpicae (Fucales, Phaeophyceae). Taxon 59:896–904. [Google Scholar]
- Mattio, L. & Payri, C. E. 2011. 190 years of Sargassum taxonomy, facing the advent of DNA phylogenies. Bot. Rev. 77:31–70. [Google Scholar]
- Mattio, L. , Payri, C. E. & Stiger‐Pouvreau, V. 2008. Taxonomic revision of Sargassum (Fucales, Phaeophyceae) from French Polynesia based on morphological and molecular analyses. J. Phycol. 44:1541–55. [DOI] [PubMed] [Google Scholar]
- Mattio, L. , Payri, C. E. & Verlaque, M. 2009. Taxonomic revision and geographic distribution of the subgenus Sargassum (Fucales, Phaeophyceae) in the Western and Central Pacific islands based on morphological and molecular analyses. J. Phycol. 45:1213–27. [DOI] [PubMed] [Google Scholar]
- Mattio, L. , Zubia, M. , Loveday, B. , Crochelet, E. , Duong, N. , Payri, C. E. , Bhagooli, R. & Bolton, J. J. 2013. Sargassum (Fucales, Phaeophyceae) in Mauritius and Réunion, western Indian Ocean: taxonomic revision and biogeography using hydrodynamic dispersal models. Phycologia 52:578–94. [Google Scholar]
- Matzke, N. J. 2013. BioGeoBEARS: biogeography with Bayesian (and Likelihood) Evolutionary Analysis in R Scripts. R Package, Version 0.2 1. [Google Scholar]
- Matzke, N. J. 2014. Model selection in historical biogeography reveals that founder‐event speciation is a crucial process in island clades. Syst. Biol. 63:951–70. [DOI] [PubMed] [Google Scholar]
- McCook, L. J. , Jompa, J. & Diaz‐Pulido, G. 2001. Competition between corals and algae on coral reefs: a review of evidence and mechanisms. Coral Reefs 19:400–17. [Google Scholar]
- McManus, J. W. & Polsenberg, J. F. 2004. Coral–algal phase shifts on coral reefs: ecological and environmental aspects. Oceanography 60:263–79. [Google Scholar]
- Miller, M. A. , Pfeiffer, W. & Schwartz, T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop, 14 Nov. 2010, New Orleans, pp. 1–8.
- Moore, B. R. & Donoghue, M. J. 2007. Correlates of diversification in the plant clade Dipsacales: geographic movement and evolutionary innovations. Am. Nat. 170:S28–55. [DOI] [PubMed] [Google Scholar]
- Mora, C. , Chittaro, P. M. , Sale, P. F. , Kritzer, J. P. & Ludsin, S. A. 2003. Patterns and processes in reef fish diversity. Nature 421:933–6. [DOI] [PubMed] [Google Scholar]
- Moura, A. E. , Nielsen, S. C. A. , Vilstrup, J. T. , Moreno‐Mayar, J. V. , Gilbert, M. T. P. , Gray, H. , Natoli, A. , Möller, L. & Hoelzel, A. R. 2013. Recent diversification of a marine genus (Tursiops spp.) tracks habitat preference and environmental change. Syst. Biol. 62:865–77. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. T. 2014. Seaweed diversity in Vietnam, with an emphasis on the brown algal genus Sargassum. PhD Thesis. Ghent University Faculty of Sciences, Department of Biology, Phycology Research Group: Gent, 191 pp.
- Noiraksar, T. & Ajisaka, T. 2008. Taxonomy and distribution of Sargassum (Phaeophyceae) in the Gulf of Thailand. J. Appl. Phycol. 20:963–77. [Google Scholar]
- Okamura, K. 1932. The distribution of marine algae in Pacific waters. Kokusai Shuppan Insatsusha. 4:30–150. [Google Scholar]
- Parham, J. F. , Donoghue, P. C. J. , Bell, C. J. , Calway, T. D. , Head, J. J. , Holroyd, P. A. , Inoue, J. G. et al. 2012. Best practices for justifying fossil calibrations. Syst. Biol. 61:346–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, B. C. & Dawson, E. Y. 1965. Non‐calcareous marine algae from California Miocene deposits. Nova Hedwigia 10:273–295. [Google Scholar]
- Phillips, N. 1995. Biogeography of Sargassum (Phaeophyta) in the Pacific basin In Abbott I. A. [Ed.] Taxonomy of Economic Seaweeds With Reference to Some Pacific Species. Volume V California Sea Grant College Program, University of California, La Jolla, pp. 107–44. [Google Scholar]
- Pillans, B. , Chappell, J. & Naish, T. R. 1998. A review of the Milankovitch climatic beat: template for Plio‐Pleistocene sea‐level changes and sequence stratigraphy. Sediment. Geol. 122:5–21. [Google Scholar]
- R Core Team 2013. R: A Language and Environment for Statistical Computing. Available from: http://www.R-project.org. [Google Scholar]
- Rambaut, A. , Suchard, M. A. , Xie, D. & Drummond, A. J. 2014. Tracer: MCMC trace analysis tool. Tracer v.1.6.
- Ree, R. & Smith, S. 2008. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57:4–14. [DOI] [PubMed] [Google Scholar]
- Rothman, M. D. , Mattio, L. , Anderson, R. J. & Bolton, J. J. 2017. A phylogeographic investigation of the kelp genus Laminaria (Laminariales, Phaeophyceae), with emphasis on the South Atlantic Ocean. J. Phycol. 53:778–89. [DOI] [PubMed] [Google Scholar]
- Sanciangco, J. C. , Carpenter, K. E. , Etnoyer, P. J. & Moretzsohn, F. 2013. Habitat availability and heterogeneity and the Indo‐Pacific warm pool as predictors of marine species richness in the Tropical Indo‐Pacific. PLoS ONE 8:e56245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberfeld, T. , Leigh, J. W. , Verbruggen, H. , Cruaud, C. , de Reviers, B. & Rousseau, F. 2010. A multi‐locus time‐calibrated phylogeny of the brown algae (Heterokonta, Ochrophyta, Phaeophyceae): investigating the evolutionary nature of the “brown algal crown radiation”. Mol. Phylogenet. Evol. 56:659–74. [DOI] [PubMed] [Google Scholar]
- Silva, P. C. , Basson, P. W. & Moe, R. L. 1996. Catalogue of the benthic marine algae of the Indian Ocean. Univ of California Press. 79:1–1259. [Google Scholar]
- Smith, J. E. , Brainard, R. , Carter, A. , Grillo, S. , Edwards, C. , Harris, J. , Lewis, L. et al. 2016. Re‐evaluating the health of coral reef communities: baselines and evidence for human impacts across the central Pacific. Proc. R. Soc. B 283:20151985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding, M. , Fox, H. , Allen, G. , Davidson, N. , Ferdaña, Z. , Finlayson, M. , Halpern, B. , Jorge, M. , Lombana, A. , Lourie, S. , et al. 2007. Marine ecoregions of the world: A bioregionalization of coastal and shelf areas. BioScience 57:573–83. [Google Scholar]
- Stebbins, G. L. 1974. Flowering Plants: Evolution Above The Species Level. Belknap Press, Cambridge, MA, 480 pp. [Google Scholar]
- Stiger, V. , Horiguchi, T. , Yoshida, T. , Coleman, A. W. & Masuda, M. 2003. Phylogenetic relationships inferred from ITS‐2 nrDNA comparisons within the genus Sargassum (Fucales, Phaeophyceae) from the Pacific basin, with an emphasis on the taxonomic subdivision of the genus. Phycol. Res. 51:1–10. [Google Scholar]
- Tornabene, L. , Valdez, S. , Erdmann, M. & Pezold, F. 2015. Support for a “Center of Origin” in the Coral Triangle: cryptic diversity, recent speciation, and local endemism in a diverse lineage of reef fishes (Gobiidae: Eviota). Mol. Phylogenet. Evol. 82:200–10. [DOI] [PubMed] [Google Scholar]
- Trono, Jr, G. . 1992. The genus Sargassum in the Philippines In Abbott I. A. [Ed.] Taxonomy of Economic Seaweeds With Reference to Some Pacific and Caribbean Species. Volume II California Sea Grant College Program, University of California, La Jolla, pp. 43–94. [Google Scholar]
- Tseng, C. K. & Lu, B. 1988. Studies on Chinese species of zygocarpic Sargassum In Abbott I. A. [Ed.] Taxonomy of Economic Seaweeds With Reference to Some Pacific and Caribbean Species. Volume II California Sea Grant College Program, University of California, La Jolla, pp. 23–54. [Google Scholar]
- Tseng, C. K. , Yoshida, T. & Chiang, Y. M. 1985. East Asiatic species of Sargassum subgenus Bactrophycus J.Agardh (Sargassaceae, Fucales), with keys to the sections and species In Abbott I. A. & Norris J. N. [Eds.] Taxonomy of Economic Seaweeds With Reference to Some Pacific and Caribbean Species. Volume I California Sea Grant College Program, University of California, La Jolla, pp. 1–14. [Google Scholar]
- Veron, J. , Stafford‐Smith, M. , DeVantier, L. & Turak, E. 2015. Overview of distribution patterns of zooxanthellate Scleractinia. Front. Mar. Sci. 1:435. [Google Scholar]
- Vieira, C. , Camacho, O. , Sun, Z. , Fredericq, S. , Leliaert, F. , Payri, C. & De Clerck, O. 2017. Historical biogeography of the highly diverse brown seaweed Lobophora (Dictyotales, Phaeophyceae). Mol. Phylogenet. Evol. 110:81–92. [DOI] [PubMed] [Google Scholar]
- Wood, H. M. , Matzke, N. J. , Gillespie, R. G. & Griswold, C. E. 2013. Treating fossils as terminal taxa in divergence time estimation reveals ancient vicariance patterns in the palpimanoid spiders. Syst. Biol. 62:264–84. [DOI] [PubMed] [Google Scholar]
- Yamasaki, M. , Aono, M. , Ogawa, N. , Tanaka, K. , Imoto, Z. & Nakamura, Y. 2014. Drifting algae and fish: implications of tropical Sargassum invasion due to ocean warming in western Japan. Estuar. Coast. Shelf Sci. 147:32–41. [Google Scholar]
- Yip, Z. T. , Quek, Z. B. R. , Low, J. K. Y. , Wilson, B. , Bauman, A. G. , Chou, L. M. , Todd, P. A. & Huang, D. 2018. Diversity and phylogeny of Sargassum (Fucales, Phaeophyceae) in Singapore. Phytotaxa 369:200–10. [Google Scholar]
- Yoshida, T. 1988. Japanese and Taiwanese Species of Sargassum subgenus Sargassum In Abbott I. A. [Ed.] Taxonomy of Economic Seaweeds With Reference to Some Pacific and Caribbean Species. Volume II California Sea Grant College Program, University of California, La Jolla, pp. 5–21. [Google Scholar]
- Yoshida, T. 1989. Systematics of Sargassum (Fucales, Phaeophyceae. Korean J. Phycol. 4:107–10. [Google Scholar]
- Yoshida, T. , Stiger, V. , Ajisaka, T. & Noro, T. 2002. A molecular study of section‐level classification of Sargassum subgenus Bactrophycus (Sargassaceae, Phaeophyta) In Abbott I. A. & McDermid K. J. [Eds.] Taxonomy of Economic Seaweeds With Reference to Some Pacific Species. Volume VIII California Sea Grant College Program, University of California, La Jolla, pp. 89–94. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. GenBank accession numbers of sequences used in the analyses.
Table S2. Locality data of five marine realms; where 1 represents presence and 0 absence: A) Central Indo‐Pacific, B) Western Indo‐Pacific, C) Eastern Indo‐Pacific, D) Tropical Eastern Pacific, and E) Atlantic.
Table S3. Geographic records for each species, adapted from AlgaeBase.
Table S4. Sources of the sequences used in this study and their respective voucher specimens and publication information.
Table S5. Sources of species taxonomy, adapted from AlgaeBase.


