Summary
Pristine marine environments are highly oligotrophic ecosystems populated by well‐established specialized microbial communities. Nevertheless, during oil spills, low‐abundant hydrocarbonoclastic bacteria bloom and rapidly prevail over the marine microbiota. The genus Alcanivorax is one of the most abundant and well‐studied organisms for oil degradation. While highly successful under polluted conditions due to its specialized oil‐degrading metabolism, it is unknown how they persist in these environments during pristine conditions. Here, we show that part of the Alcanivorax genus, as well as oils, has an enormous potential for biodegrading aliphatic polyesters thanks to a unique and abundantly secreted alpha/beta hydrolase. The heterologous overexpression of this esterase proved a remarkable ability to hydrolyse both natural and synthetic polyesters. Our findings contribute to (i) better understand the ecology of Alcanivorax in its natural environment, where natural polyesters such as polyhydroxyalkanoates (PHA) are produced by a large fraction of the community and, hence, an accessible source of carbon and energy used by the organism in order to persist, (ii) highlight the potential of Alcanivorax to clear marine environments from polyester materials of anthropogenic origin as well as oils, and (iii) the discovery of a new versatile esterase with a high biotechnological potential.
Introduction
In microbial ecology, the idea that ‘everything is everywhere, but, the environment selects’ (Baas‐Becking, 1934; De Wit and Bouvier, 2006) requires that even the rare biota has to persist through time via the assimilation of a source of carbon and energy, unless they are able to remain dormant such as by sporulation. Marine ecosystems are mostly oligotrophic and hostile to non‐adapted microorganisms due to the lack of nutrients and high salinity. The predominant heterotrophic bacteria that inhabit these environments are thus well adapted to such conditions and have mainly specialized in the use of labile substrates released by marine phototrophs (Sharma et al., 2014; Christie‐Oleza et al., 2017; Bakenhus et al., 2018; Zheng et al., 2019).
Alcanivorax is a ubiquitous marine bacterial genus classed as an obligate hydrocarbonoclastic bacterium (OHCB) due to its preference to metabolize hydrocarbons and crude oil derivatives (Yakimov et al., 2019). This genus rapidly blooms and becomes one of the most abundant organisms during marine pollution events and oil‐spills (Kasai et al., 2002; Hara et al., 2003); however, how does Alcanivorax persist within the rare biome of pristine seawater while awaiting favourable conditions? Although members of the Alcanivorax genus can grow – although inefficiently – with more labile substrates such as pyruvate and succinate (Fernández‐Martínez et al., 2003; Naether et al., 2013; Radwan et al., 2019), they would be outcompeted by other heterotrophic bacteria in the natural environment (McGenity et al., 2012; Yakimov et al., 2019). It has been suggested, although, that Alcanivorax spp. may persist in pristine environments by using alkanes released by marine cyanobacteria (Coates et al., 2014; Lea‐Smith et al., 2015) and other hydrocarbon‐producing eukaryotic algae (Sorigué et al., 2016, 2017). The hydrolysis of aliphatic polyesters such as poly(caprolactone) (PCL), poly(hydroxybutylate) (PHB) and poly(butylene succinate) (PBS) by some Alcanivorax isolates has also been shown (Sekiguchi et al., 2011; Zadjelovic et al., 2019), although the underlying molecular mechanisms behind the ability of these strains to degrade such polyesters remained unknown. Interestingly, during the screening for polylactic acid (PLA) esterases, researchers identified that the enzyme ABO2449 encoded by Alcanivorax borkumensis had a strong hydrolytic activity on PLA as well as on a range of other aliphatic polyesters, that is, poly(hydroxybutylate‐co‐valerate) (PHBV), PCL and poly(ethylene succinate) (PES) (Hajighasemi et al., 2016).
A large number of environmental microbes – including members from the genus Alcanivorax – synthesize intracellular aliphatic polyester granules, that is, polyhydroxy alkanoates (PHA), as a strategy to store carbon and energy when nutrient availability is imbalanced (e.g. under high C:N conditions; Fernández‐Martínez et al., 2003; Sabirova et al., 2006; Jendrossek and Pfeiffer, 2014). As well as aliphatic polyesters of natural origin (e.g. the PHAs: PHB and PHBV), in recent years, there has been an increase of industrially manufactured synthetic polyesters (e.g. PLA, PCL, PES or PBS; Flieger et al., 2003; Tseng et al., 2007). Aliphatic polyesters are tagged as ‘biodegradable plastics,’ and although they still represent a small fraction of the global polymer material market, the consumer demand of these environmentally friendly alternatives to traditional non‐biodegradable materials is exponentially growing (Tokiwa et al., 2009; Aeschelmann and Carus, 2015). Like all plastics, aliphatic polyesters also find their way into the oceans (Jambeck et al., 2015; Lebreton et al., 2017). Although considered biodegradable because their linking ester bonds are susceptible to hydrolysis by esterases, lipases or other enzymes (Nakajima‐Kambe et al., 1999; Müller et al., 2001; Zheng et al., 2005; Shah et al., 2014), other factors such as chemical structure, molecular weight, hydrophobicity and crystallinity may hinder degradability of these materials (Tokiwa et al., 2009). Furthermore, polymer degradation in marine environments is even more challenging due to the low temperatures and oligotrophic conditions that hamper microbial activity as well as the reduced encounter rate in such dilute ecosystems. Hence, although some marine microbial isolates have been reported to degrade a range of polyesters (Mabrouk and Sabry, 2001; Ghanem et al., 2005; Sekiguchi et al., 2010), such a process is not as obvious in marine environments as highlighted by studies that failed to observe degradation of some theoretically biodegradable polymers such as PHB, PES and PBS (Sekiguchi et al., 2010; Bagheri et al., 2017).
In this study, we characterize a novel and versatile esterase secreted by Alcanivorax that hydrolyses a number of natural and synthetic polyesters, that is, PHB, PHBV, PES, PBS and PCL. Altogether, the evidence here suggests that the environmental relevance of the genus Alcanivorax goes beyond the degradation of solely oil‐derived compounds.
Results
Polyester degradation by Alcanivorax sp. 24
Alcanivorax sp. 24, isolated from marine plastic debris (Zadjelovic et al., 2019), produced clear degradation halos on plates containing the polyesters PHB, PHBV, PES, PBS and PCL (Fig. 1A and Table 1). Although the degradation of the natural polymers PHB and PHBV was expected because the strain was originally enriched and isolated on PHB (Zadjelovic et al., 2019), the large degradation halos on the synthetic polyesters PES, PBS and PCL within just 3–6 days was remarkable and deserved further investigation. Growth curves of Alcanivorax sp. 24 were conducted with polyesters PHB, PHBV and PES to assess if the bacterium could assimilate and grow on these polymers as well as degrade them (Fig. 1B). Protein quantification was used as a proxy for growth because standard monitoring techniques (e.g. turbidity or colony forming units) could not be performed due to polymer insolubility. Interestingly, Alcanivorax sp. 24 was able to grow on all polymers faster than with the labile control substrate, that is, succinate, particularly with PHB (i.e. ~150 μg of protein/ml at day four; Fig. 1B).
Figure 1.
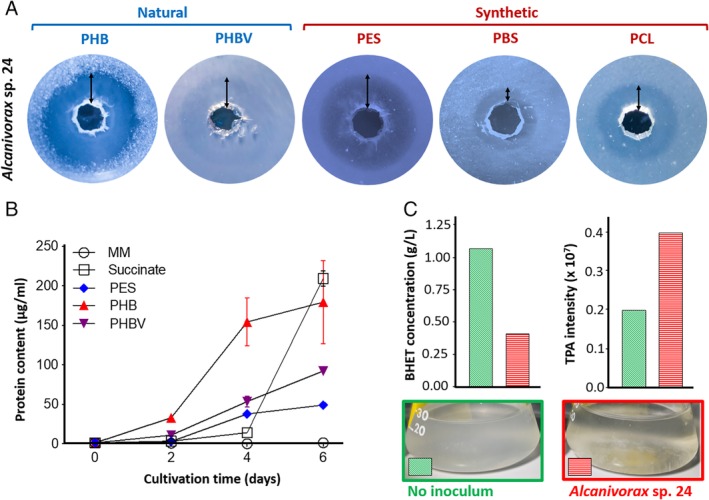
Determination of polyester degradation by Alcanivorax sp. 24.A. Clear zone hydrolysis test of Alcanivorax sp. 24 on the five aliphatic polyesters PHB, PHBV, PES, PBS and PCL. Polyesters were added at 0.3% w/v to BH mineral media containing 1% agarose (w/v). Arrows highlight the hydrolysis halos surrounding the 5 mm‐diameter wells made for Alcanivorax sp. 24 inoculation.B. Growth curves of Alcanivorax sp. 24 when incubated in the presence of three different polyesters, succinate (labile substrate control) and BH mineral media (MM; negative control). Increase in biological biomass was assessed by protein quantification. Error bars indicate the standard deviation of three biological replicates.C. Degradation of the PET intermediate BHET by Alcanivorax sp. 24. The substrate BHET and its hydrolysed product TPA were monitored by LC–MS. ‘No inoculum’ represents the replicate control condition where Alcanivorax sp. 24 was not inoculated. [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 1.
Characteristics and chemical structure of the polymers used in this study.
| Polymer | Form | Formula | Chemical structure | Degradation products | Origin |
|---|---|---|---|---|---|
| Poly[3‐hydroxybutyrate] (PHB)a | Powder | [C4H6O2]n |
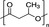
|
3‐Hydroxybutyrate | Natural |
| Poly[3‐hydroxybutyrate‐co‐3‐hydroxyvalerate] (PHBV)a | Pellets | [C4H6O2]x[C5H8O2]y |
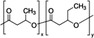
|
3‐Hydroxybutyrate 3‐Hydroxyvalerate |
Natural |
| Polyethylene succinate (PES)a | Chunks | [C6H8O4]n |
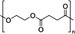
|
Ethylene glycol Succinate |
Synthetic |
| Polybutylene succinate (PBS)a | Pellets | [C8H12O4]n |
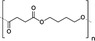
|
Butanodiol Succinate |
Synthetic |
| Polycaprolactone (PCL)b | Chunks | [C6H10O2]n |
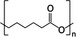
|
6‐Hydroxyhexanoate | Synthetic |
| Bis[2‐Hydroxyethyl] terephthalate (BHET)b | Flakes | C12H14O6 |
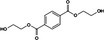
|
Ethylene glycol Terephthalate (TPA) |
Synthetic |
Product supplied by Sigma‐Aldrich®.
Product supplied by Goodfellow©.
We also assayed if Alcanivorax sp. 24 could degrade bis[2‐hydroxyethyl] terephthalate (BHET; Table 1), an intermediate of the recalcitrant polyester polyethylene terephthalate (PET). When grown on BHET, Alcanivorax sp. 24 produced apparent clumpy growth in liquid cultures and it decreased the white turbidity of this insoluble compound (Fig. 1C). Unfortunately, BHET interfered with the protein quantification method producing a large background signal, and hence, growth quantification was not possible. To overcome the difficulties in measuring bacterial growth, a metabolic analysis was carried out to determine if the strain was able to degrade the PET intermediate. The hydrolysis of BHET generates ethylene glycol, which could be assimilated by Alcanivorax, and terephthalic acid (TPA; Table 1), a sub‐product that should accumulate in the media as the bacterium does not encode for the necessary catabolic pathway for TPA degradation (Zadjelovic et al., 2019). BHET and TPA were measured by LC‐MS confirming that Alcanivorax sp. 24 was active in hydrolysing the BHET sidechains as ~60% of this compound disappeared from the culture medium and, proportionally, TPA concentration increased (Fig. 1C and Supplementary Fig. S1). The incomplete degradation of BHET and clumping of Alcanivorax sp. 24 in the presence of this substrate could be attributed to a possible toxic effect of TPA as it built up in the medium.
Proteomic analysis to identify the secreted esterase(s) responsible for polymer hydrolysis
The exoproteomic analysis of Alcanivorax grown in the presence of succinate, PHB, PHBV, PES and BHEt allowed the identification of 1250 proteins of which the 13 most abundant ones already represented over 50% of the total exoproteomic fraction, all with a predicted secretion signal (Supplementary Table S1). A PCA of the exoproteomes showed how the conditions PHB, PHBV and PES strongly differed from the labile control (i.e. succinate) and BHET condition (PC1, which explained 53% of the variability; Fig. 2A). This difference was mainly driven by the abundantly detected PHB depolymerase ALC24_4107 (Fig. 2B). This secreted esterase represented 10%–13% of Alcanivorax's exoproteome when grown in the presence of PHB, PHBV and PES as opposed to the other conditions (<0.05% in succinate and BHET), flagging this enzyme as the main candidate in driving the aliphatic polyesters depolymerisation observed in Fig. 1A and, thus, was selected for further investigating (see below). Three other hydrolase/esterase proteins were detected in the exoproteome of Alcanivorax sp. 24, although in much lower abundance (i.e. ALC24_3279, ALC24_3988 and ALC24_4209; Table 2).
Figure 2.
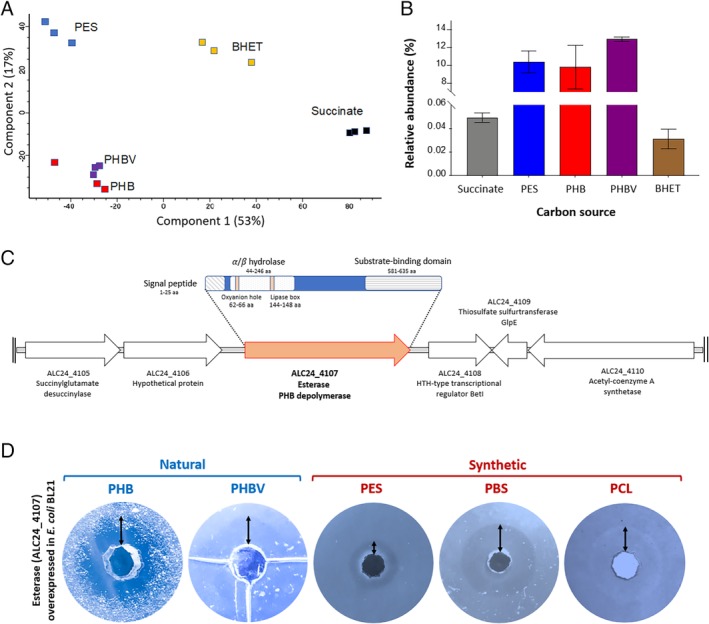
Proteomic analysis and identification of the esterase (i.e. ALC24_4107) involved in the aliphatic polyester degradation of Alcanivorax sp. 24.A. PCA of the exoproteomes produced by Alcanivorax sp. 24 when grown in the presence of different substrates including the three aliphatic polyesters PES, PHB and PHBV as well as BHET and succinate.B. Relative abundance of the esterase ALC24_4107 in each one of the exoproteomes of Alcanivorax sp. 24 when grown in the presence of different substrates. Error bars indicate the standard deviation of three biological replicates.C. Protein domains and genomic context of ALC24_4107.D. Hydrolytic activity of the heterologously overexpressed ALC24_4107 in E. coli BL21 assessed by a clear zone hydrolysis test on five different aliphatic polyesters. Halos surround 5 mm‐diameter wells. [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 2.
Subset of relevant proteins detected in the exoproteome of Alcanivorax sp. 24 when exposed to different polymers.
| ID (ALC24_) | Annotated function | Prediction for secretion a | Succinate | PHB | PHBV | PES | BHET | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Abundance (%; n = 3) | Abundance (%; n = 3) | FCb (log2) | Abundance (%; n = 3) | FCb (log2) | Abundance (%; n = 3) | FCb (log2) | Abundance (%; n = 3) | FCb (log2) | |||||||||
| 4107 | Esterase PHB depolymerase | SP | NCS | Extracellular | 0.05 | 9.83 | 6.8 | + | 12.94 | 7.4 | + | 10.40 | 7.1 | + | 0.03 | −2.0 | + |
| 3279 | putative hydrolase YxeP | SP | NCS | Unknown | 0.01 | 0.07 | 0.2 | 0.07 | 0.4 | 0.41 | 2.8 | + | 0.15 | 0.8 | |||
| 3988 | Alpha/beta hydrolase | NCS | Unknown | 0.00 | 0.02 | 2.7 | + | 0.04 | 3.3 | + | 0.19 | 5.6 | + | 0.13 | 4.0 | + | |
| 4209 | Ferri‐bacillibactin esterase BesA | SP | Unknown | 0.00 | 0.04 | 3.9 | + | 0.04 | 4.5 | + | 0.01 | 3.2 | + | 0.00 | −0.7 | ||
| 2998 | Periplasmic binding protein | SP | Unknown | 0.32 | 7.33 | 4.4 | + | 8.74 | 4.7 | + | 1.42 | 2.2 | + | 0.78 | 0.3 | ||
| 2082 | Alpha‐keto acid periplasmic SBP | Unknown | 0.12 | 1.60 | 7.6 | + | 1.60 | 7.7 | + | 0.06 | 0.7 | 0.00 | −3.0 | + | |||
| 2735 | C4‐dicarboxylate periplas. SBP | SP | Periplasmic | 0.12 | 0.06 | −0.7 | 0.07 | −0.4 | 3.55 | 4.7 | + | 0.10 | −1.2 | ||||
| 3132 | Alcohol dehydrogenase | NCS | Periplasmic | 0.00 | 0.15 | 2.7 | 0.25 | 3.5 | + | 13.72 | 9.3 | + | 0.94 | 4.3 | + | ||
| 1432 | Alcohol dehydrogenase | SP | NCS | Periplasmic | 0.05 | 0.10 | 0.0 | 0.16 | 0.9 | 1.38 | 3.8 | + | 0.32 | 0.8 | |||
| 1781 | Flagellin 2 | NCS | Extracellular | 1.70 | 10.38 | 1.7 | 12.25 | 2.1 | + | 10.69 | 2.0 | + | 20.46 | 1.6 | |||
| 2995 | NHL repeat protein | SP | NCS | Outer Memb | 0.28 | 15.79 | 5.4 | + | 18.09 | 5.7 | + | 2.19 | 2.3 | + | 0.41 | −0.7 | |
| 1102 | Type VI secretion system | NCS | Extracellular | 0.12 | 4.84 | 4.1 | + | 1.53 | 3.0 | + | 0.09 | −0.1 | 1.64 | 2.7 | + | ||
| 3348 | Neisseria PilC protein | SP | NCS | Unknown | 0.03 | 0.55 | 2.8 | + | 1.39 | 4.2 | + | 0.55 | 3.1 | + | 0.10 | −0.1 | |
| 2260 | Fimbria adhesin protein | SP | NCS | Extracellular | 0.08 | 1.04 | 2.8 | + | 0.60 | 2.2 | + | 0.16 | 0.6 | 0.30 | 0.3 | ||
| 0797 | Unknown, adherence | SP | NCS | Extracellular | 0.00 | 0.16 | 4.9 | + | 0.19 | 5.2 | + | 0.21 | 5.4 | + | 0.16 | 4.3 | + |
| 2342 | hypothetical protein | Unknown | 0.00 | 0.71 | 11.3 | + | 3.26 | 13.6 | + | 0.39 | 10.4 | + | 0.00 | 1.0 | |||
Protein secretion systems and localization. SP, signal peptide. NCS, non‐classical secretion.
Fold change (FC) of each protein in each one of the conditions vs. the succinate control. Significant values are indicated (+; q‐value < 0.05).
The exoproteomic analysis also highlighted the substrate‐binding component of three membrane transporters that could be involved in importing the specific subproducts from polymer hydrolysis (Table 2). Although transporters ALC24_2998 and, more specifically, ALC24_2082 could be involved in transporting 3‐hydroxybutyrate and 3‐hydroxyvalerate from PHB and PHBV depolymerisation, ALC24_2735 may import derivatives generated from PES hydrolysis. The strong increase of alcohol dehydrogenases in the periplasm of the bacterium in the presence of PES (ALC24_3132 and ALC24_1432 representing 13.7% and 1.4% protein abundance respectively; Table 2) suggests that its degradation product ethylene glycol is transformed into glyoxylate before being imported and catabolized within the cell (Salvador et al., 2019). Interestingly, the alcohol dehydrogenase ALC24_3132 was also significantly upregulated when Alcanivorax was grown in the presence of BHET (log2 fold change of 4.3), suggesting a possible generation of ethylene glycol from the hydrolysis of BHET as hinted by the detection of TPA during the metabolomics analysis (Fig. 1C). An increased secretion of proteins involved in adhesion when Alcanivorax sp. 24 was grown in the presence of the different aliphatic polyesters was also detected (e.g. ALC24_3348, ALC24_2260 and ALC24_0797; Table 2). Although the proteomic data has helped us identify those enzymes and transporters that are possibly involved in assimilating polyester intermediates, further biochemical experimentation is required to confirm their function and substrate specificity.
A previously characterized esterase from Alcanivorax borkumensis that showed a strong hydrolytic activity on aliphatic polyesters (ABO2449; Hajighasemi et al., 2016) and that had a conserved homologue in our Alcanivorax sp. 24, i.e. ALC_2069 (E‐value 10−118), was not detected in the exoproteome under any condition (Supplementary Table S1). We analysed the cellular proteome of Alcanivorax sp. 24 (Supplementary Table S2) to investigate if the enzyme ALC_2069, which we initially believed was responsible for the observed polyester hydrolysis, was contained within the cell. The cellular proteome produced 2590 detected proteins amongst which ALC_2069, although detected, only represented <0.004% of the proteome.
The proteomic data highlighted ALC24_4107 as the main candidate responsible for the phenotype observed in Fig. 1A, and hence, this enzyme was selected for further confirmation and characterization.
Protein structure and genomic context of the abundantly secreted esterase ALC24_4107
Apart from a clear signal peptide for secretion on the protein's N‐terminal (25 amino acids‐long signal peptide involved in conventional secretion systems), ALC24_4107 contained an α/β‐hydrolase domain between amino acids leucine and threonine (position 44 to 246; Fig. 2C). Although α/β‐hydrolase domains are a diverse superfamily which includes esterases, proteases, lipases, dehalogenases and epoxide‐hydrolases, ALC24_4107 contained the characteristic catalytic triad of amino acids serine, aspartic acid and histidine found in the active site of hydrolases involved in polyester degradation (Jendrossek, 1998). The enzyme's structural analysis also revealed the presence of a substrate‐binding domain in position 581 to 635 (Fig. 2C) which is essential for the hydrolysis of non‐soluble substrates – such as when the polyesters are found in the extracellular milieu (Shinomiya et al., 1997) – and may provide the enzyme with its substrate specificity.
ALC24_4107 was analysed using the Depolymerase Engineering Database, DED (Knoll et al., 2009), revealing it belonged to the extracellular depolymerase group e‐dPHAscl (type 1) homologous family 8. Nevertheless, ALC24_4107 and its closest homologues (i.e. AAB40611.1 from Alcaligenes faecalis and Alcanivorax dieselolei) formed a distinctive branch within the family of depolymerases (Supplementary Fig. S2).
The genomic context of the esterase ALC24_4107 in Alcanivorax sp. 24 showed a transcriptional regulator immediately downstream from the gene (Fig. 2C) which could be involved in regulating the strong induction of the enzyme observed by proteomics. Enzymes involved in short‐chain fatty acid metabolism, and which may have a role in processing the hydrolysed intermediates of the polyesters, were also encoded further downstream from ALC24_4107.
ALC24_4107 has a promiscuous hydrolytic activity on aliphatic polyesters
To confirm the hydrolytic activity of esterase ALC24_4107, the enzyme was overexpressed in E. coli BL21. Although the enzyme was successfully produced and secreted when it was cloned with its original signal peptide for secretion, best results were obtained when this peptide was replaced by the host's phoA signal peptide (Supplementary Fig. S3; Ahn et al., 2006). The highest overexpression, assessed by clear zone halos produced by E. coli supernatants on PHB agarose plates, were obtained after 72 h of incubation after inducing with 1 mM of IPTG at 27°C (Supplementary Fig. S3).
The overexpressed esterase degraded all polymers i.e. PHB, PHBV, PES, PBS and PCL (Fig. 2D) confirming the broad range activity of this enzyme on aliphatic polyesters.
Distribution and activity of homologous esterases of ALC24_4107 in other bacteria
A BLASTp search of the esterase ALC24_4107 showed the presence of homologous copies of the enzyme in other Alcanivorax strains (i.e. with 71% to 93% identity), as well as in other genus, that is, Pseudoalteromonas, Aestuariibacter, Microbulbifer and Alteromonas (55%, 58%, 59% and 60% identity respectively; Fig. 3A). However, the closest related protein of ALC24_4107 found in the NCBI databases was a PHB depolymerase encoded by Alcaligenes faecalis (99% identity). All proteins included in the phylogenetic tree were annotated as PHB depolymerases or had esterase‐like domains (Fig. 3A). Interestingly, no homologue of ALC24_4107 was observed in the model strain Alcanivorax borkumensis.
Figure 3.
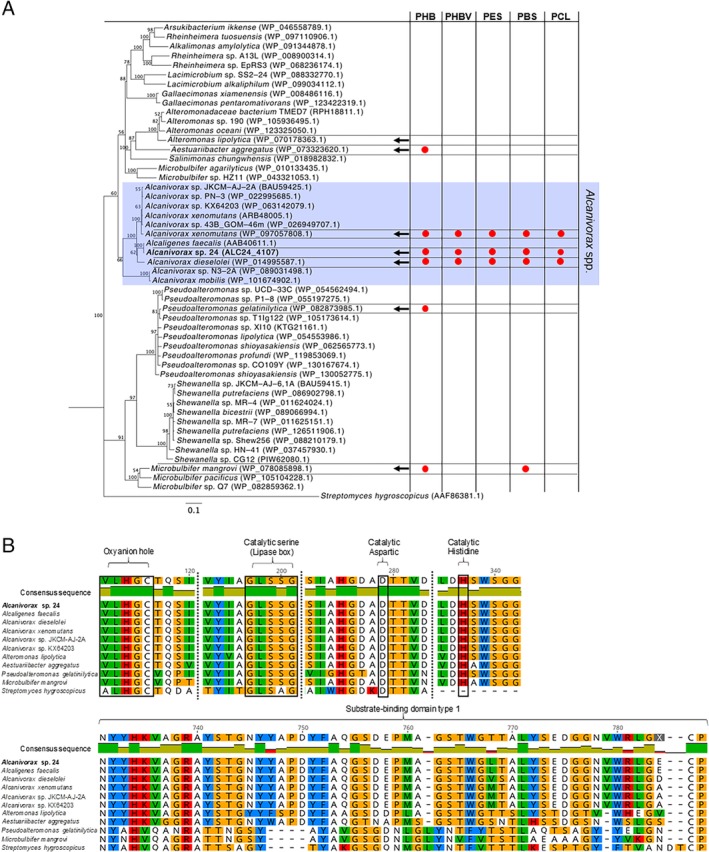
Similarities and hydrolytic activity of ALC24_4107 homologous esterases.A. Phylogenetic tree of ALC24_4107 and its closest homologues in other bacteria retrieved from an NCBI BLASTp search. The tree was generated using Neighbour‐Joining and Jukes‐Cantor as the generic distance, with bootstrap set to 1000 replicates represented at the base of the nodes, and using the PHB‐depolymerase from Streptomyces hygrogroscopicus as the outgroup. Arrows indicate the strains that were purchased and for which hydrolytic activity was assessed on each one of the five aliphatic polyesters. The tests that gave a positive clearing halo (information available in Supplementary Fig. S4) are indicated by red circles.B. Multiple alignment of ALC24_4107 with 10 relevant homologues, including the esterases encoded by the six strains tested for their hydrolytic activity. Only the catalytic and substrate‐binding domains are shown. [Color figure can be viewed at http://wileyonlinelibrary.com]
Six strains encoding esterases from different branches of the phylogenetic tree (Fig. 3A) were purchased in an effort to determine their ability to degrade the different aliphatic polyesters. Out of the six strains assessed, only the Alcanivorax isolates (Alcanivorax xenomutans JC109 and Alcanivorax dieselolei B‐5) were able to produce clear zone halos in all polymers, that is, PHB, PHBV, PES, PBS and PCL (Fig. 3A and Supplementary Fig. S4). Those strains encoding esterases from different branches (i.e. Microbulbifer mangrovi DD13, Pseudoalteromonas gelatinilytica NH153 and Aestuariibacter aggregatus WH169) only produced clearing halos on PHB plates, although M. mangrovi also produced halos on PBS (Supplementary Fig. S4). Despite the absence of halos could be due to a lack of enzyme induction, it is most likely that the homologous PHB depolymerases from different branches are not as promiscuous as those encoded by Alcanivorax. The sequence analysis of each one of the tested esterases revealed that, although they all conserved the catalytic triad of amino acids, the substrate binding domain diverged between the copies encoded by Alcanivorax and the rest of the strains (Fig. 3B).
Discussion
Over the past few decades, the identification of PHA depolymerases has received large attention (Knoll et al., 2009; García‐Hidalgo et al., 2013; Martínez‐Tobón et al., 2018; Sayyed et al., 2019). Despite the extant sequence variability amongst these enzymes, all intra‐ and extracellular PHA depolymerases share a common α/β‐hydrolase fold and a catalytic triad (Knoll et al., 2009). The key finding of this study is the vast promiscuity of substrates shown by the abundantly secreted PHA depolymerase ALC24_4107, which is conserved in a number of Alcanivorax strains (Fig. 3). This enzyme, highly induced in the presence of polymers (representing over >10% of the exoproteome; Fig. 2B), was able to hydrolyse a large range of aliphatic polyesters of natural and synthetic origin, that is, PHB, PHBV, PES, PBS and PCL (as proven by heterologous overexpression; Fig. 2D). It had already been reported that Alcanivorax strains could degrade a variety of these polyesters (i.e. PHB, PBS and PCL) (Sekiguchi et al., 2011), although the mechanisms involved were not previously identified. In another study that performed an in vitro screening for PLA esterases, researchers identified that the enzyme ABO2449 encoded by Alcanivorax borkumensis had a strong hydrolytic activity on PLA as well as on a range of other aliphatic polyesters, that is, PHBV, PCL and PES (Hajighasemi et al., 2016). A homologue of the ABO2449 α/β‐hydrolase was encoded by Alcanivorax sp. 24 (ALC_2069) although this enzyme was not detected in the exoproteome (Supplementary Table S1), and its abundance was extremely low in the cellular proteome (< 0.004%; Supplementary Table S2). Hence, we prove here that ALC24_4107, and not ALC_2069, is responsible for aliphatic polyester biodegradation by Alcanivorax sp. 24.
Biosynthesis of natural polyesters, that is, PHAs, is a widely distributed mechanism of carbon storage amongst environmental microorganisms (Jendrossek and Pfeiffer, 2014), and therefore, it may be an important source of carbon and energy to those organisms able to biodegrade them when PHA producers are lysed, for example, after phage infection or inefficient grazing. For example, 95% of members of the Roseobacter clade, an abundant and versatile group of marine heterotrophs (Buchan et al., 2005; Christie‐Oleza et al., 2012) that pioneer the colonization of marine surfaces including marine plastic debris (Elifantz et al., 2013; Erni‐Cassola et al., 2019), encode phaC, the polymerase that catalyses PHA biosynthesis (i.e. 711 of 750 Roseobacter genomes encoded a phaC homologue, E‐value <10−100; Supplementary Table S3). Alcanivorax sp. 24 also encodes phaC (ALC24_0403 and ALC24_1241) as well as for the other genes involved in PHA biosynthesis (i.e. phaA, phaB and phasin) and, hence, is likely to produce and store PHAs as proven in other Alcanivorax strains (Fernández‐Martínez et al., 2003; Sabirova et al., 2006). Alcanivorax sp. 24, isolated from marine plastic debris, may be able to persist and thrive in marine biofilms by assimilating natural PHAs produced by its surrounding microorganisms as well as biodegrade the material it colonizes if made of polyester chains.
Because its isolation and characterization, the genus Alcanivorax has become a reference of marine oil‐degradation (Yakimov et al., 1998; de Lorenzo, 2006; Schneiker et al., 2006; Gregson et al., 2019). Although Alcanivorax spp. are found in low abundance under normal environmental conditions, these microbes rapidly bloom in oil‐contaminated marine ecosystems (Kasai et al., 2002; Hara et al., 2003). Although Alcanivorax shows a clear preference for oils and has been classified as an ‘obligate’ hydrocarbonoclastic organism (Yakimov et al., 2007), members of this genus can also grow on some more‐labile substrates (Radwan et al., 2019), although they are likely to be outcompeted by other members of the marine microbial community. It was suggested that Alcanivorax may be able to persist in pristine environments by using alkanes produced by marine cyanobacteria (Lea‐Smith et al., 2015; Valentine and Reddy, 2015). Nevertheless, in this study, we show that, as well as an oil biodegrader, Alcanivorax is a specialist in biodegrading aliphatic polyesters via the secretion of an abundant and substrate‐promiscuous α/β‐hydrolase (Fig. 4). Here, we suggest that this esterase would allow this organism to persist using naturally occurring polyesters and, currently, it may also confer the potential to biodegrade polyester plastics of anthropogenic origin.
Figure 4.
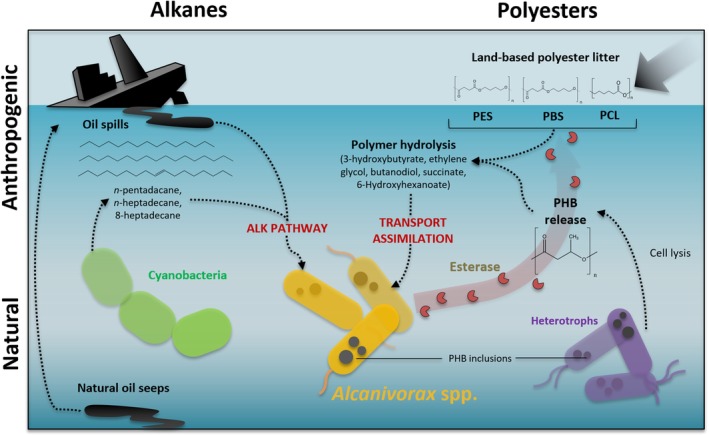
Ecological strategy of Alcanivorax spp. in the environment.The biodegradation and assimilation of hydrocarbons (left) and polyesters (right) of both natural (bottom) and anthropogenic origin (top) is depicted. [Color figure can be viewed at http://wileyonlinelibrary.com]
Experimental procedures
Bacterial strains and culture conditions
Alcanivorax sp. 24 (accession number SNUA0000000; Zadjelovic et al., 2019) as well as the six strains obtained from the Korean and German culture collections KCTC and DSMZ [i.e. Microbulbifer mangrovi DD13 (KCTC 23483), Alcanivorax xenomutans JC109 (KCTC 23751), Alteromonas lipolytica JW12 (KCTC 52408), Alcanivorax dieselolei B‐5 (DSM 16502), Aestuariibacter aggregatus WH169 (DSM 23094), Pseudoalteromonas gelatinilytica NH153 (DSM 100951)] were routinely grown in Marine Broth or on Marine Agar plates (BD Difco™). E. coli BL21 was grown using LB broth (Sigma‐Aldrich®).
The analysis of polymer degradation was performed with Bushnell‐Hass basal mineral media (BH; Bushnell and Haas, 1941), adjusted to pH 7.0 and supplemented with NaCl 30 g L−1 and 1 ml L−1 of an ASW trace metal solution (Wyman et al., 1985). Agarose 1% (w/v) was added as the solidifying agent. BH mineral media was amended with sodium succinate 0.5% w/v or each one of the polymers listed in Table 1 at 0.3% w/v as the source of carbon and energy. BHET was added at a final concentration of 0.1% w/v.
Polymer preparation and biodegradation test (clear zone test)
Aliphatic polyesters were purchased from Sigma‐Aldrich® and Goodfellow©. Polymers were available in the forms detailed in Table 1 and emulsified in the mineral media as previously described (Nishida et al., 1998; Tansengco and Tokiwa, 1998). Briefly, 0.3% w/v of PHB was directly added as a powder suspension to BH media containing 1% agarose (Nishida and Tokiwa, 1993), autoclaved and mechanically homogenized by blending before pouring into Petri dishes. PBS, PES and PCL were pre‐dissolved in acetone and PHBV in dichloromethane before adding at 0.3% final polymer concentration to autoclaved media, after which it was blended and poured into Petri dishes. Dissolvent was evaporated for 20 min at 80°C.
Clear zone degradation tests were conducted as described previously (Augusta et al., 1993). Briefly, wells (approximately 5 mm diameter) were made in the solid media in which resuspended cells in minimal BH medium were used as the inoculum. Inoculated plates were left to dry and then incubated at 30°C for 7 days.
Growth curves and monitoring
The liquid cultures were prepared in 50 ml glass Erlenmeyer flasks filled with 30 ml of BH mineral media containing the different sources of carbon and inoculated with washed bacterial cells, all performed in independent biological triplicates. Cultures were incubated for 7 days at 30°C and shaking at 200 rpm. Due to the clumps formed by the insoluble polymers, growth was monitored by protein quantification using the QuantiPro™ BCA Assay kit (Sigma‐Aldrich®) following the supplier's recommendations. LIVE/DEAD Cell Viability Assay (Invitrogen™) was used to visualize polymer colonization and check the cell viability (Supplementary Fig. S5). A T‐test analysis was performed using Prism 7 to determine the significance of the growth curve results (p‐value set at p ≤ 0.05).
Degradation of BHET was performed as described for other polymers although the monitoring of growth was not possible by protein quantification due to the large background signal given by the substrate. BHET degradation was measured via metabolite consumption and TPA formation using LC‐MS as described below.
Metabolite detection of BHET and TPA using LC‐MS
Samples were prepared and processed as previously described (Yoshida et al., 2016) with minor modifications. BHET and TPA were extracted from 5 ml of culture samples using 10 ml of ethyl acetate as a solvent. The organic phase was recovered and evaporated (speedVac, Genevac™ EZ‐2). The residual product was resuspended in a mix of 150 μl of dimethyl sulfoxide (DMSO), 100 μl of 16 mM phosphate buffer adjusted at pH 2.5 and 50 μl of acetonitrile (total resuspension volume 300 μl). The sample was acidified using HCl (pH 2.0) and pre‐filtered (0.22 μm Spin Cups‐Cellulose Acetate Filters) before analysis via reversed‐phase liquid chromatography on a Dionex UltiMate 3000 HPLC (ThermoScientific) equipped with a Zorbax Eclipse Plus C18 column (dimensions 4.6 mm × 150 mm, 5 μm particle size; Agilent Technologies) coupled to an amaZon SL Ion Trap MS (Bruker), operated in positive mode with a scanning range for molecular ions of 100–1000 m/z. A gradient elution of mobile phases A (water, 0.1% formic acid) and B (acetonitrile, 0.1% formic acid) was used at a flow rate of 1 ml min−1 as follows: the ratio of solvent A:B was lineally decreased from 95:5 to 30:70 over 5 min, followed by a second decrease from 30:70 to 20:80 over 10 min and finally from 20:80 to 5:95 over 12 min. The injection volume was 10 μl at a temperature of 25°C. MS data was processed with the Bruker Compass DataAnalysis software version 4.2 (Bruker). Calibration curves were constructed using different concentrations of BHET and TPA in DMSO (Supplementary Fig. S1).
Exoproteome and cellular proteome preparation for proteomics
At day 7, 20 ml of the culture from each culture was centrifuged at 4000 rpm, 15 min at 4°C. Cell pellets were immediately frozen on dry ice and stored at −20°C. Supernatants (exoproteomes) were further filtered through 0.22 μm pore size hydrophilic filters (Minisart® Syringe Filters). Proteins in the supernatants were precipitated using a trichloroacetic acid (TCA) and sodium deoxycholate (DOC) protocol as previously described (Christie‐Oleza and Armengaud, 2010). Both exoproteome and cell pellets were dissolved in 70 μl and 300 μl of lithium dodecyl sulphate (1 × LDS with 1% β‐mercaptoethanol, Invitrogen™) respectively. Samples were incubated at 95°C for 5 min and vortexed for three cycles before loading 30 μl of each sample onto 10% NuPAGE™ Bis‐Tris precast gels (1.0 mm, 10‐well, Invitrogen™). Gels were allowed a short migration to enter the polyacrylamide gel (1 cm) before staining with SimplyBlue™ SafeStain (Invitrogen™). Band sections containing the entire proteome were cut and placed into 1.5 ml microcentrifuge tubes to be stored at −20°C until use (Christie‐Oleza and Armengaud, 2010).
Tryptic digestion and shotgun proteomics
Proteomes contained in the gel bands were reduced (dithiothreitol) and alkylated (iodoacetamide) before digestion using trypsin (Christie‐Oleza and Armengaud, 2010). Tryptic peptides were recovered from the gels using an extraction buffer (formic acid/acetonitrile; (Shevchenko et al., 2007) and analysed by nanoLC‐ESI‐MS/MS using an Ultimate 2000 LC system (Dionex‐LC Packings) coupled to an Orbitrap Fusion mass spectrometer (Thermo‐Scientific). An LC separation of 60 min for exoproteomes or 120 min for cellular proteomes were performed on a 25 cm column before MS/MS analysis using settings as described previously (Christie‐Oleza et al., 2015).
Comparative proteomic analysis
Peptide spectrum profiles were identified and quantified using MaxQuant (v1.5.5.1) within the framework of the Label Free Quantification (LFQ) method (Cox and Mann, 2008). Parameters were set by default although including the match between run function. Spectra were searched against the protein database of Alcanivorax sp. 24 (Zadjelovic et al., 2019). Perseus (v1.5.6.0) was used to perform the comparative proteomic analysis (Tyanova et al., 2016), where a two‐sample T‐test was used to determine protein variations between each condition when compared to the succinate control condition. Statistical analysis was set using a false discovery rate (FDR) of 0.05 and minimal log2 fold change of 2. A protein was considered valid when present in every replicate of at least one condition. The list of polypeptides, LFQ intensities, relative abundance and differential detection is listed in Supplementary Tables S1 and S2.
Heterologous overexpression of ALC24_4107 in E. coli BL21
The esterase gene ALC24_4107 was codon optimized, and three variants were synthesized by GenScript: (1) using the wild type signal‐peptide for secretion, (2) using no signal‐peptide, and (3) using a signal‐peptide for secretion from the host's phoA (MKQSTIALALLPLLFTPVTKA; Ahn et al., 2006). Synthesized genes were ligated into the NdeI/XhoI restriction sites of the overexpression vector pET‐24a(+). Overexpression of ALC24_4107 in E. coli BL21 was optimized by testing different temperatures (27 and 37°C) and IPTG concentrations (0.2, 0.5 and 1 mM). IPTG induction was carried out when cultures reached an OD600 of 0.6. The culture cell pellet and supernatant of each one of the conditions were recovered after 24 h by centrifugation (8000 rpm, 5 min). Cell pellets were disrupted using 1× BugBuster® Protein Extraction Reagent in combination with three sonication steps of 5 min. Supernatants and cell lysates were screened for their hydrolytic activity using the clear zone test on PHB plates (Supplementary Fig. S3). After determining the optimal conditions for ALC24_4107 overexpression (i.e. supernatants from cultures containing the phoA signal peptide, incubated at 37°C and induced with 1 mM of IPTG), its activity was tested against polyesters PHB, PHBV, PES, PBS and PCL using the clear zone test (Fig. 2D).
Proteomic and genomic in silico analysis
The motifs and domains in ALC24_4107 were searched using the Conserved Domain Analysis tool from NCBI. Protein secretion was predicted using the servers: SignalP 4.1. (Petersen et al., 2011), SecretomeP 2.0a (Bendtsen et al., 2005), LipoP 1.0. (Juncker et al., 2003) and PSORTb v3.0.2. (Yu et al., 2010). The phylogenetic tree of ALC24_4107 and its closest homologues in the NCBI database (as determined by BLASTp) was built using Geneious Prime applying Jukes‐Cantor and Neighbour‐Joining.
Supporting information
Fig S1 BHET degradation by Alcanivorax sp. 24 assessed by LC–MS. (A) BHET standard curve. (B) Extracted ion chromatogram of BHET and TPA obtained from cultures where Alcanivorax sp. 24 was absent (panels 1 and 2) and present (panels 3 and 4).
Fig. S2. Phylogenetic context of the PHB‐depolymerase ALC24_4101 from Alcanivorax sp. 24 and Alcanivorax dieselolei (highlighted in blue) with the closest PHA depolymerases identified by the Depolymerase Engineering Database (DED; Knoll et al., 2009). The sequence AAB40611.1 from Alcaligenes faecalis and homologues from Shewanella sp. MR4 and MR7 (ABI40356.1 and ABI41661.1, respectively) –all three present in the DED database– are also highlighted in blue. The tree was generated using Neighbour‐Joining and Jukes‐Cantor as the generic distance, with bootstrap set to 1000 replicates represented at the base of the nodes. The scale bar shows nucleotide/amino acids substitutions per 100 residues.
Fig. S3. PHB clear zone test to screen for the activity of the heterologously overexpressed esterase from Alcanivorax sp. 24 (ALC24_4107) in E. coli BL21. Plastic square petri dishes 13 × 13 cm.
Fig. S4. Aliphatic polyester clear zone test using microorganisms encoding close homologue esterases to ALC24_4107. Polymers PHB (A), PESu (B), PBSu (C), PHBV (D) and PCL (E) were tested. Alcanivorax sp. 24 on PHBV and PCL (F and G, respectively) were performed separately.
Fig. S5. Determination of cells viability using LIVE/DEAD™ BacLight™ Bacterial Viability Kit. Staining procedure applied to Alcanivorax sp. 24 biofilms grown on PHB, PES and BHET. Green staining represents viable cells whereas red staining represents dead cell or those with compromised membrane integrity.
Supplementary Table S1 Exoproteome of Alcanivorax sp. 24 exposed to different polyesters.
Supplementary Table S2 Cellular proteome of Alcanivorax sp. 24 exposed to different polyesters.
Supplementary Table S3 Presence of phaC as a proxy of PHA biosynthesis in 750 Roseobacter genomes.
Acknowledgements
VZ was supported by CONICYT‐BECAS CHILE/Doctorado Becas Chile en el Extranjero, Folio 72160583. JAC‐O was supported by the NERC Independent Research Fellowship NE/K009044/1, NERC research project NE/S005501/1, and Ramón y Cajal contract RYC‐2017‐22452 (funded by the Ministry of Science, Innovation and Universities, the National Agency of Research, and the European Social Fund). RB was supported by the MINECO project CTM2015‐70180‐R (FEDER co‐funding). We would like to acknowledge Robyn Wright and Gabriel Erni‐Cassola for their contribution during this research.
Contributor Information
Vinko Zadjelovic, Email: v.zadjelovic@warwick.ac.uk.
Joseph A. Christie‐Oleza, Email: joseph.christie@uib.eu.
References
- Aeschelmann, F. , and Carus, M. (2015) Biobased building blocks and polymers in the world: capacities, production, and applications–status quo and trends towards 2020. Ind Biotechnol 11: 154–159. [Google Scholar]
- Ahn, J.‐H. , Hwang, M.‐Y. , Lee, K.‐H. , Choi, C.‐Y. , and Kim, D.‐M. (2006) Use of signal sequences as an in situ removable sequence element to stimulate protein synthesis in cell‐free extracts. Nucleic Acids Res 35: 21–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas‐Becking, L.G.M. (1934) Geobiologie of inleiding tot de milieukunde. The Hague, the Netherlands: Van Stockkum & Zoon, Den Haag: W.P. Van Stockum & Zoon. [Google Scholar]
- Bagheri, A.R. , Laforsch, C. , Greiner, A. , and Agarwal, S. (2017) Fate of so‐called biodegradable polymers in seawater and freshwater. Glob Challenges 1: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakenhus, I. , Dlugosch, L. , Giebel, H.A. , Beardsley, C. , Simon, M. , and Wietz, M. (2018) Distinct biogeographic patterns of bacterioplankton composition and single‐cell activity between the subtropics and Antarctica. Environ Microbiol 20: 3100–3108. [DOI] [PubMed] [Google Scholar]
- Bendtsen, J.D. , Kiemer, L. , Fausbøll, A. , and Brunak, S. (2005) Non‐classical protein secretion in bacteria. BMC Microbiol 5: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan, A. , González, J.M. , and Moran, M.A. (2005) Overview of the marine Roseobacter lineage. Appl Environ Microbiol 71: 5665–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell, L.D. , and Haas, H.F. (1941) The utilization of certain hydrocarbons by microorganisms. J Bacteriol 41: 653–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , and Armengaud, J. (2010) In‐depth analysis of exoproteomes from marine bacteria by shotgun liquid chromatography‐tandem mass spectrometry: the Ruegeria pomeroyi DSS‐3 case‐study. Mar Drugs 8: 2223–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , Miotello, G. , and Armengaud, J. (2012) High‐throughput proteogenomics of Ruegeria pomeroyi: seeding a better genomic annotation for the whole marine Roseobacter clade. BMC Genomics 13: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , Scanlan, D.J. , and Armengaud, J. (2015) “You produce while I clean up”, a strategy revealed by exoproteomics during Synechococcus‐Roseobacter interactions. Proteomics 15: 3454–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie‐Oleza, J.A. , Sousoni, D. , Lloyd, M. , Armengaud, J. , and Scanlan, D.J. (2017) Nutrient recycling facilitates long‐term stability of marine microbial phototroph‐heterotroph interactions. Nat Microbiol 2: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates, R.C. , Podell, S. , Korobeynikov, A. , Lapidus, A. , Pevzner, P. , Sherman, D.H. , et al (2014) Characterization of cyanobacterial hydrocarbon composition and distribution of biosynthetic pathways. PLoS One 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J. , and Mann, M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 26: 1367–1372. [DOI] [PubMed] [Google Scholar]
- De Wit, R. , and Bouvier, T. (2006) “Everything is everywhere, but, the environment selects”; what did Baas Becking and Beijerinck really say? Environ Microbiol 8: 755–758. [DOI] [PubMed] [Google Scholar]
- Elifantz, H. , Horn, G. , Ayon, M. , Cohen, Y. , and Minz, D. (2013) Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in eastern Mediterranean coastal seawater. FEMS Microbiol Ecol 85: 348–357. [DOI] [PubMed] [Google Scholar]
- Erni‐Cassola, G. , Wright, R.J. , Gibson, M.I. , and Christie‐Oleza, J.A. (2019) Early colonization of weathered polyethylene by distinct bacteria in marine coastal seawater. Microb Ecol: 1–10. 10.1007/s00248-019-01424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Martínez, J. , Pujalte, M.J. , García‐Martínez, J. , Mata, M. , Garay, E. , and Rodríguez‐Valera, F. (2003) Description of Alcanivorax venustensis sp. nov. and reclassification of Fundibacter jadensis DSM 12178T (Bruns and Berthe‐Corti 1999) as Alcanivorax jadensis comb. nov., members of the emended genus Alcanivorax . Int J Syst Evol Microbiol 53: 331–338. [DOI] [PubMed] [Google Scholar]
- Flieger, M. , Kantorová, M. , Prell, A. , Řezanka, T. , and Votruba, J. (2003) Biodegradable plastics from renewable sources. Rev Folia Microbiol 48: 27–44. [DOI] [PubMed] [Google Scholar]
- García‐Hidalgo, J. , Hormigo, D. , Arroyo, M. , and de la Mata, I. (2013) Novel extracellular PHB depolymerase from Streptomyces ascomycinicus: PHB copolymers degradation in acidic conditions. PLoS One 8: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem, N.B. , Mabrouk, M.E.S. , Sabry, S.A. , and El‐Badan, D.E.S. (2005) Degradation of polyesters by a novel marine Nocardiopsis aegyptia sp. nov.: application of Plackett‐Burman experimental design for the improvement of PHB depolymerase activity. J Gen Appl Microbiol 51: 151–158. [DOI] [PubMed] [Google Scholar]
- Gregson, B.H. , Metodieva, G. , Metodiev, M.V. , and McKew, B.A. (2019) Differential protein expression during growth on linear versus branched alkanes in the obligate marine hydrocarbon‐degrading bacterium Alcanivorax borkumensis SK2T. Environ Microbiol 21: 2347–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajighasemi, M. , Nocek, B.P. , Tchigvintsev, A. , Brown, G. , Flick, R. , Xu, X. , et al (2016) Biochemical and structural insights into enzymatic depolymerization of polylactic acid and other polyesters by microbial carboxylesterases. Biomacromolecules 17: 2027–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, A. , Syutsubo, K. , and Harayama, S. (2003) Alcanivorax which prevails in oil‐contaminated seawater exhibits broad substrate specificity for alkane degradation. Environ Microbiol 5: 746–753. [DOI] [PubMed] [Google Scholar]
- Jambeck, J.R. , Geyer, R. , Wilcox, C. , Siegler, T.R. , Perryman, M. , Andrady, A. , et al (2015) Plastic waste inputs from land into the ocean. Mar Pollut 347: 768–771. [DOI] [PubMed] [Google Scholar]
- Jendrossek, D. (1998) Microbial degradation of polyesters: a review on extracellular poly(hydroxyalkanoic acid) depolymerases. Polym Degrad Stab 59: 317–325. [Google Scholar]
- Jendrossek, D. , and Pfeiffer, D. (2014) New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3‐hydroxybutyrate). Environ Microbiol 16: 2357–2373. [DOI] [PubMed] [Google Scholar]
- Juncker, A.S. , Willenbrock, H. , von Heijne, G. , Brunak, S. , Nielsen, H. , and Krogh, A. (2003) Prediction of lipoprotein signal peptides in gram‐negative bacteria. Protein Sci 101: 1652–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai, Y. , Kishira, H. , Sasaki, T. , Syutsubo, K. , Watanabe, K. , and Harayama, S. (2002) Predominant growth of Alcanivorax strains in oil‐contaminated and nutrient‐supplemented sea water. Environ Microbiol 4: 141–147. [DOI] [PubMed] [Google Scholar]
- Knoll, M. , Hamm, T.M. , Wagner, F. , Martinez, V. , and Pleiss, J. (2009) The PHA depolymerase engineering database: a systematic analysis tool for the diverse family of polyhydroxyalkanoate (PHA) depolymerases. BMC Bioinform 10: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea‐Smith, D.J. , Biller, S.J. , Davey, M.P. , Cotton, C.A.R. , Perez Sepulveda, B.M. , Turchyn, A.V. , et al (2015) Contribution of cyanobacterial alkane production to the ocean hydrocarbon cycle. Proc Natl Acad Sci 112: 13591–13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton, L.C.M. , Van Der Zwet, J. , Damsteeg, J.W. , Slat, B. , Andrady, A. , and Reisser, J. (2017) River plastic emissions to the world's oceans. Nat Commun 8: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo, V. (2006) Blueprint of an oil‐eating bacterium. Nat Biotechnol 24: 952–953. [DOI] [PubMed] [Google Scholar]
- Mabrouk, M.M. , and Sabry, S.A. (2001) Degradation of poly (3‐hydroxybutyrate) and its copolymer poly (3‐hydroxybutyrate‐co‐3‐hydroxyvalerate) by a marine Streptomyces sp. SNG9. Microbiol Res 156: 323–335. [DOI] [PubMed] [Google Scholar]
- Martínez‐Tobón, D.I. , Gul, M. , Elias, A.L. , and Sauvageau, D. (2018) Polyhydroxybutyrate (PHB) biodegradation using bacterial strains with demonstrated and predicted PHB depolymerase activity. Appl Microbiol Biotechnol 102: 8049–8067. [DOI] [PubMed] [Google Scholar]
- McGenity, T.J. , Folwell, B.D. , McKew, B.A. , and Sanni, G.O. (2012) Marine crude‐oil biodegradation: a central role for interspecies interactions. Aquat Biosyst 8: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, R.J. , Kleeberg, I. , and Deckwer, W.D. (2001) Biodegradation of polyesters containing aromatic constituents. J Biotechnol 86: 87–95. [DOI] [PubMed] [Google Scholar]
- Naether, D.J. , Slawtschew, S. , Stasik, S. , Engel, M. , Olzog, M. , Wick, L.Y. , et al (2013) Adaptation of the hydrocarbonoclastic bacterium Alcanivorax borkumensis SK2 to alkanes and toxic organic compounds: a physiological and transcriptomic approach. Appl Environ Microbiol 79: 4282–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima‐Kambe, T. , Shigeno‐Akutsu, Y. , Nomura, N. , Onuma, F. , and Nakahara, T. (1999) Microbial degradation of polyurethane, polyester polyurethanes and polyether polyurethanes. Appl Microbiol Biotechnol 51: 134–140. [DOI] [PubMed] [Google Scholar]
- Nishida, H. , Suzuki, S. , and Tokiwa, Y. (1998) Distribution of poly(β‐propiolactone) aerobic degrading microorganisms in different environments. J Environ Polym Degrad 6: 43–58. [Google Scholar]
- Nishida, H. , and Tokiwa, Y. (1993) Distribution of poly(β‐hydroxybutyrate) and poly(ɛ‐caprolactone) aerobic degrading microorganisms in different environments. J Polym Environ 1: 227–233. [Google Scholar]
- Petersen, T.N. , Brunak, S. , von Heijne, G. , and Nielsen, H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Radwan, S.S. , Khanafer, M.M. , and Al‐awadhi, H.A. (2019) Ability of the so‐called obligate hydrocarbonoclastic bacteria to utilize nonhydrocarbon substrates thus enhancing their activities despite their misleading name. BMC Microb 41: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirova, J.S. , Ferrer, M. , Lünsdorf, H. , Wray, V. , Kalscheuer, R. , Steinbüchel, A. , et al (2006) Mutation in a “tesB‐like” hydroxyacyl‐coenzyme A‐specific thioesterase gene causes hyperproduction of extracellular polyhydroxyalkanoates by Alcanivorax borkumensis SK2. J Bacteriol 188: 8452–8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador, M. , Abdulmutalib, U. , Gonzalez, J. , Kim, J. , Smith, A.A. , Faulon, J.‐L. , et al (2019) Microbial genes for a circular and sustainable bio‐PET economy. Genes (Basel) 10: 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayyed, R.Z. , Wani, S.J. , Alyousef, A.A. , Alqasim, A. , Syed, A. , and El‐Enshasy, H.A. (2019) Purification and kinetics of the PHB depolymerase of Microbacterium paraoxydans RZS6 isolated from a dumping yard. PLoS One 14: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiker, S. , Bartels, D. , Bekel, T. , Brecht, M. , Buhrmester, J. , Chernikova, T.N. , et al (2006) Genome sequence of the ubiquitous hydrocarbon‐ degrading marine bacterium Alcanivorax borkumensis . Nat Biotechnol 24: 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi, T. , Saika, A. , Nomura, K. , Watanabe, T. , Watanabe, T. , Fujimoto, Y. , et al (2011) Biodegradation of aliphatic polyesters soaked in deep seawaters and isolation of poly(ε‐caprolactone)‐degrading bacteria. Polym Degrad Stab 96: 1397–1403. [Google Scholar]
- Sekiguchi, T. , Sato, T. , Enoki, M. , Kanehiro, H. , Uematsu, K. , and Kato, C. (2010) Isolation and characterization of biodegradable plastic degrading bacteria from deep‐sea environments. JAMSTEC Rep Res Dev 11: 33–41. [Google Scholar]
- Shah, A.A. , Kato, S. , Shintani, N. , Kamini, N.R. , and Nakajima‐Kambe, T. (2014) Microbial degradation of aliphatic and aliphatic‐aromatic co‐polyesters. Appl Microbiol Biotechnol 98: 3437–3447. [DOI] [PubMed] [Google Scholar]
- Sharma, A.K. , Becker, J.W. , Ottesen, E.A. , Bryant, J.A. , Duhamel, S. , Karl, D.M. , et al (2014) Distinct dissolved organic matter sources induce rapid transcriptional responses in coexisting populations of Prochlorococcus, Pelagibacter and the OM60 clade. Environ Microbiol 16: 2815–2830. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A. , Tomas, H. , Havliš, J. , Olsen, J.V. , and Mann, M. (2007) In‐gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860. [DOI] [PubMed] [Google Scholar]
- Shinomiya, M. , Iwata, T. , Kasuya, K.I. , and Doi, Y. (1997) Cloning of the gene for poly(3‐hydroxybutyric acid) depolymerase of Comamonas testosteroni and functional analysis of its substrate‐binding domain. FEMS Microbiol Lett 154: 89–94. [DOI] [PubMed] [Google Scholar]
- Sorigué, D. , Légeret, B. , Cuiné, S. , Blangy, S. , Moulin, S. , Billon, E. , et al (2017) Acids to hydrocarbons. Science 357: 903–907. [DOI] [PubMed] [Google Scholar]
- Sorigué, D. , Légeret, B. , Cuiné, S. , Morales, P. , Mirabella, B. , Guédeney, G. , et al (2016) Microalgae synthesize hydrocarbons from long‐chain fatty acids via a light‐dependent pathway. Plant Physiol 171: 2393–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansengco, M.L. , and Tokiwa, Y. (1998) Thermophilic microbial degradation of polyethylene succinate. World J Microbiol Biotechnol 14: 133–138. [Google Scholar]
- Tokiwa, Y. , Calabia, B.P. , Ugwu, C.U. , and Aiba, S. (2009) Biodegradability of plastics. Int J Mol Sci 10: 3722–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, M. , Hoang, K.C. , Yang, M.K. , Yang, S.F. , and Chu, W.S. (2007) Polyester‐degrading thermophilic actinomycetes isolated from different environment in Taiwan. Biodegradation 18: 579–583. [DOI] [PubMed] [Google Scholar]
- Tyanova, S. , Temu, T. , Sinitcyn, P. , Carlson, A. , Hein, M.Y. , Geiger, T. , et al (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740. [DOI] [PubMed] [Google Scholar]
- Valentine, D.L. , and Reddy, C.M. (2015) Latent hydrocarbons from cyanobacteria. Proc Natl Acad Sci 112: 13434–13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman, M. , Gregory, R.P.F. , and Carr, N.G. (1985) Novel role for phycoerythrin in marine cyanobacterium Synechococcus strain DC2. Science 230: 4–6. [DOI] [PubMed] [Google Scholar]
- Yakimov, M.M. , Golyshin, P.N. , Crisafi, F. , Denaro, R. , and Giuliano, L. (2019) Marine, aerobic hydrocarbon‐degrading Gammaproteobacteria: the family Alcanivoracaceae In: McGenity T. (eds) Taxonomy, Genomics and Ecophysiology of Hydrocarbon‐Degrading Microbes. Handbook of Hydrocarbon and Lipid Microbiology, Cham: Springer. [Google Scholar]
- Yakimov, M.M. , Golyshin, P.N. , Lang, S. , Moore, E.R. , Abraham, W.R. , Lünsdorf, H. , and Timmis, K.N. (1998) Alcanivorax borkumensis gen nov, sp nov, a new, hydrocarbon‐degrading and surfactant producing marine bacterium. Int J Syst Bacteriol 48: 339–348. [DOI] [PubMed] [Google Scholar]
- Yakimov, M.M. , Timmis, K.N. , and Golyshin, P.N. (2007) Obligate oil‐degrading marine bacteria. Curr Opin Biotech 18: 257–266. [DOI] [PubMed] [Google Scholar]
- Yoshida, S. , Hiraga, K. , Takanaha, T. , Taniguchi, I. , Yamaji, H. , Maeda, Y. , et al (2016) A bacterium that degrades and assimilates poly(ethyleneterephthalate). Science 351: 1196–1199. [DOI] [PubMed] [Google Scholar]
- Yu, N.Y. , Wagner, J.R. , Laird, M.R. , Melli, G. , Rey, S. , Lo, R. , et al (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26: 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadjelovic, V. , Gibson, M.I. , Dorador, C. , and Christie‐Oleza, J.A. (2019) Genome of Alcanivorax sp. 24: a hydrocarbon degrading bacterium isolated from marine plastic debris. Mar Genomics 49: 1–4. [Google Scholar]
- Zheng, Q. , Chen, Q. , Cai, R. , He, C. , Guo, W. , Wang, Y. , et al (2019) Molecular characteristics of microbially mediated transformations of Synechococcus‐derived dissolved organic matter as revealed by incubation experiments. Environ Microbiol 21: 2533–2543. [DOI] [PubMed] [Google Scholar]
- Zheng, Y. , Yanful, E.K. , and Bassi, A.S. (2005) A review of plastic waste biodegradation. Crit Rev Biotechnol 25: 243–250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1 BHET degradation by Alcanivorax sp. 24 assessed by LC–MS. (A) BHET standard curve. (B) Extracted ion chromatogram of BHET and TPA obtained from cultures where Alcanivorax sp. 24 was absent (panels 1 and 2) and present (panels 3 and 4).
Fig. S2. Phylogenetic context of the PHB‐depolymerase ALC24_4101 from Alcanivorax sp. 24 and Alcanivorax dieselolei (highlighted in blue) with the closest PHA depolymerases identified by the Depolymerase Engineering Database (DED; Knoll et al., 2009). The sequence AAB40611.1 from Alcaligenes faecalis and homologues from Shewanella sp. MR4 and MR7 (ABI40356.1 and ABI41661.1, respectively) –all three present in the DED database– are also highlighted in blue. The tree was generated using Neighbour‐Joining and Jukes‐Cantor as the generic distance, with bootstrap set to 1000 replicates represented at the base of the nodes. The scale bar shows nucleotide/amino acids substitutions per 100 residues.
Fig. S3. PHB clear zone test to screen for the activity of the heterologously overexpressed esterase from Alcanivorax sp. 24 (ALC24_4107) in E. coli BL21. Plastic square petri dishes 13 × 13 cm.
Fig. S4. Aliphatic polyester clear zone test using microorganisms encoding close homologue esterases to ALC24_4107. Polymers PHB (A), PESu (B), PBSu (C), PHBV (D) and PCL (E) were tested. Alcanivorax sp. 24 on PHBV and PCL (F and G, respectively) were performed separately.
Fig. S5. Determination of cells viability using LIVE/DEAD™ BacLight™ Bacterial Viability Kit. Staining procedure applied to Alcanivorax sp. 24 biofilms grown on PHB, PES and BHET. Green staining represents viable cells whereas red staining represents dead cell or those with compromised membrane integrity.
Supplementary Table S1 Exoproteome of Alcanivorax sp. 24 exposed to different polyesters.
Supplementary Table S2 Cellular proteome of Alcanivorax sp. 24 exposed to different polyesters.
Supplementary Table S3 Presence of phaC as a proxy of PHA biosynthesis in 750 Roseobacter genomes.


