Abstract
Epigenetic mechanisms are known to regulate gene expression during chondrogenesis. In this study, we have characterized the epigenome during the in vitro differentiation of human mesenchymal stem cells (hMSCs) into chondrocytes. Chromatin immunoprecipitation followed by next‐generation sequencing (ChIP‐seq) was used to assess a range of N‐terminal posttranscriptional modifications (marks) to histone H3 lysines (H3K4me3, H3K4me1, H3K27ac, H3K27me3, and H3K36me3) in both hMSCs and differentiated chondrocytes. Chromatin states were characterized using histone ChIP‐seq and cis‐regulatory elements were identified in chondrocytes. Chondrocyte enhancers were associated with chondrogenesis‐related gene ontology (GO) terms. In silico analysis and integration of DNA methylation data with chondrogenesis chromatin states revealed that enhancers marked by histone marks H3K4me1 and H3K27ac were de‐methylated during in vitro chondrogenesis. Similarity analysis between hMSC and chondrocyte chromatin states defined in this study with epigenomes of cell‐types defined by the Roadmap Epigenomics project revealed that enhancers are more distinct between cell‐types compared to other chromatin states. Motif analysis revealed that the transcription factor SOX9 is enriched in chondrocyte enhancers. Luciferase reporter assays confirmed that chondrocyte enhancers characterized in this study exhibited enhancer activity which may be modulated by DNA methylation and SOX9 overexpression. Altogether, these integrated data illustrate the cross‐talk between different epigenetic mechanisms during chondrocyte differentiation.
Keywords: cartilage, chondrocyte development, chromatin
Abbreviations
- ACAN
aggrecan
- ChIP‐seq
chromatin immunoprecipitation
- COL2A1
collagen type II alpha 1 chain
- DMEM
Dulbecco's modified eagle medium
- ECM
extracellular matrix
- GO
gene ontology
- hMSC
human mesenchymal stem cell
- IGV
integrative genome viewer
- lincRNA
long non coding RNA
- MACS2
model‐based analysis of ChIP‐seq 2
- miRNA
microRNA
- OA
osteoarthritis
- PBS
phosphate buffered saline
- RUNX2
runt‐related transcription factor 2
- SOX9
sex determining region Y box 9
1. INTRODUCTION
Chondrogenesis is the process of differentiation of mesenchymal progenitors into chondrocytes. Articular cartilage, present in synovial joints, comprises an extracellular matrix secreted by chondrocytes and has an important function in aiding the mobility of joints. As the only cell type present in articular cartilage, adult articular chondrocytes are responsible for the homeostasis of cartilage.
During embryogenesis, the skeletal system originates from the mesoderm germ layer. Mesenchymal progenitors differentiate into chondrocytes to form temporary cartilage. During endochondral ossification, these cells generally undergo apoptosis to be replaced by bone. However, cartilage at synovial joints does not ossify and remains throughout the life. Hypertrophic chondrocytes bound for ossification have high expression of COL10A1 and osteoblast markers such as RUNX2, and low expression of cartilage‐specific genes such as COL2A1 and SOX9.1, 2 Chondrogenesis is a multi‐step tightly regulated process mediated by growth and transcription factors, with the SOX9 transcription factor instrumental to the progression of chondrogenic differentiation 3 although not initiation.4 Gene expression during chondrogenesis is in part regulated by dynamic epigenetic mechanisms such as DNA methylation and histone modifications.5, 6 MicroRNAs (miRNAs) and long non‐coding RNAs (lncRNAs) also play a role in chondrogenesis.7, 8, 9 Genome‐wide histone modification changes have been observed during in vitro differentiation of MSCs into chondrocytes.10 As well as development, epigenetic mechanisms are also known to be involved in disease. Cis‐regulatory elements such as gene enhancers have been shown to be disrupted in cartilage pathologies. Deletions in a distal regulatory region of the SOX9 transcription factor gene and within the SOX9 gene itself both lead to campomelic dysplasia in humans.11, 12 Mutations in enhancers of collagen genes are also associated with chondrodysplasias.13, 14 Osteoarthritis (OA), an age‐related cartilage degenerative disease, has a strong genetic component and to date, the vast majority of polymorphisms that confer an increased risk are located in non‐coding regions of the genome, including enhancers.15, 16 There is evidence that the OA phenotype may be linked to the reactivation of developmental pathways.17 Articular cartilage affected by OA shows gene expression changes reminiscent of hypertrophic chondrocytes.1, 18 These studies demonstrate that epigenetic mechanisms regulate gene expression in numerous biological processes. However, how these mechanisms affect gene expression is not fully understood in cartilage development and disease.
Mesenchymal stem cells (MSCs) are able to differentiate into chondrocytes and have been used to study chondrogenesis in vitro. Tissue engineering solutions to cartilage repair include autologous chondrocyte implantation, cartilage autografts, and injection of MSCs into the damaged site.19, 20 However, these methods are not widely used and complications can arise from their application. Further knowledge of the regulatory processes that control gene expression during chondrocyte development is required to develop and improve models for cartilage regeneration. Usage of in vitro models for human chondrogenesis is crucial for understanding the changes that occur during the normal development of human cartilage. Additionally, as in vitro models are used extensively for the study of chondrogenesis, it is important to establish how similar models are to each other and to in vivo chondrogenesis.
In this study, histone ChIP‐seq (H3K4me3, H3K4me1, H3K27ac, H3K27me3, and H3K36me3) was performed in a scaffold‐free in vitro model of human MSC (hMSC) chondrogenesis.21 Analysis of histone ChIP‐seq data revealed that large scale chromatin state changes occur during chondrogenesis and chondrocytes acquire cell‐type‐specific enhancers upon differentiation. Integration of chromatin states with genome‐wide DNA methylation data demonstrated that de‐methylated CpG sites are located within H3K27ac and H3K4me1 marked enhancers during chondrogenesis. Motif analysis revealed that chondrocyte enhancers contain SOX9‐binding motifs. Altogether, our study provides a comprehensive analysis of the global epigenetic changes during MSC chondrogenesis and highlights the role of enhancers in defining cell‐type specificity.
2. MATERIALS AND METHODS
2.1. hMSC culture and chondrogenesis
Bone marrow aspirates (donor n = 2, female, ages 22 & 24) were purchased from LONZA and hMSCs were isolated by adherence to tissue culture flasks for 24 hours. hMSCs were phenotyped by flow cytometry22 and confirmed to have osteoblastogenic and adipogenetic potential as well as chondrogenic. Stem cells were cultured and differentiated into chondrocytes as previously described.23
2.2. Isolation of chondrocytes from cartilage‐like disc
Cartilage discs were digested at Day 14 of chondrogenesis, a time point at which chondrocytes have been determined to be fully differentiated in a pellet model of chondrogenesis.24 Cartilage discs were digested first with 1.5 mL of hyaluronidase (1 mg/mL in sterile PBS) for 15 minutes at 37°C then with 1.5 mL of trypsin (2.5 mg/mL in sterile PBS) at 37°C for 30 minutes. The discs were finally digested with collagenase (2 mg/mL in DMEM media) for 1‐1.5 hours at 37°C until fully digested and the matrix was no longer visible. The digested cartilage containing media was passed through a 100 μm cell strainer to remove any remaining matrix. Each cartilage disc yielded ~250,000‐500,000 cells and multiple discs were pooled together during extraction.
2.3. Chromatin extraction and ChIP‐seq
hMSCs were harvested from monolayer culture using trypsin. Chromatin from hMSCs and differentiated chondrocytes were extracted using the Diagenode iDeal histone ChIP‐seq kit (Diagenode SA, Ougrée, Belgium). Extracted chromatin was sonicated using a Diagenode Bioruptor Standard or Bioruptor Pico to an average size of 200‐500 bp, using 15 sonication cycles (30s on/30 seconds off). ChIP‐seq grade premium antibodies were purchased from Diagenode: H3K4me3 (included in the Diagenode iDeal histone ChIP‐seq kit), H3K4me1 (Cat. no. C15410194), H3K27ac (Cat. no. C15410196), H3K27me3 (Cat. no. C15410195), and H3K36me3 (Cat. no. C15410192). Chromatin immunoprecipitation was performed following the Diagenode iDeal histone ChIP‐seq protocol using chromatin from 1 million cells and 1μg antibody per ChIP. Immunoprecipitated DNA was purified using Agencourt AMPure XP beads (Beckman Coulter (UK) Ltd, High Wycombe, UK). For one hMSC chondrogenesis replicate ChIP‐seq, DNA sequencing libraries were generated using Diagenode MicroPlex v2 kit and single‐ended reads of 50 bp length were generated on an Illumina HiSeq 2500 (Illumina Inc, San Diego, USA). The second experimental replicate was prepared using the NEBNext Ultra II kit (New England Biolabs, Hitchin, UK) and sequenced using an Illumina NextSeq 500 platform, generating 75 bp single‐end reads. For both replicates, 30‐65 million reads were generated per sample (Table S1).
2.4. Luciferase reporter assays
Putative enhancer regions were amplified from human genomic DNA using the primers listed in Table S2 and cloned into the pCpGL‐EF1 plasmid. This plasmid has been modified from the CpG‐free pCpGL‐basic luciferase plasmid by the addition of the EF1 CpG‐free promoter upstream of the luciferase gene25 and can thus be used to analyze DNA methylation effects on non‐promoter regulatory regions. Plasmids were transformed into GT115 E coli (Invitrogen) and DNA isolated using the PureYield Plasmid Midiprep system (Promega). Plasmid DNA was in vitro methylated using CpG Methyltransferase (M SssI, New England Biolabs), with the efficiency of methylation assessed by digestion using HpaII and HhaI methylation‐sensitive restriction enzymes (NEB). The effect of SOX9 on enhancer activity was assessed by transfection with a SOX9 overexpression plasmid (pUT‐FLAG‐SOX9).26 A luciferase reporter (4COL) containing four copies of the Col2a1 48‐bp enhancer was used to confirm SOX9 overexpression.27 SW1353 chondrosarcoma cells were seeded at a cell density of 5 × 103 per well in 96‐well plates. After 24 hours, cells were co‐transfected with 100 ng (DNA methylation) or 50 ng (SOX9 overexpression) of the relevant pCpGL‐EF1 plasmid and 6ng (DNA methylation) or 1.5 ng (SOX9 overexpression) of pRL‐TK Renilla control plasmid and 0.3 μL of FuGENE HD reagent (Promega) per well. Cells were lysed 24hrs post‐transfection and luciferase and the renilla activity measured using the Dual‐Luciferase Reporter Assay kit on a GloMax‐Multi reader. Three experiments with six replicates each were performed for each construct and luciferase/renilla activity normalized to for the empty the pCpGL‐EF1vector.
2.5. ChIP‐seq analysis and chromatin state learning
Quality control of sequencing reads was performed using FastQC (v.0.11.5). All reads passed quality thresholds. Reads were aligned to the reference human genome hg38 using Bowtie2 (v.2.2.4).28 MACS2 (v.2.1.0.2)29 was used to call broad peaks (parameters–broad and –no‐model) using input samples as controls. The ngs.plot program (v.2.61)30 was used to visualize peak enrichment across the genome and at gene expression levels. An Illumina whole‐genome expression array Human HT‐12 V4 was used to determine gene expression levels prior and post chondrogenesis.23 Normalized gene expression signals were categorized into low (signal < 7; 1st quarter), medium (signal between 7 and 9) or high expression (signal > 9; 3rd quarter; Table S3).
ChromHMM (v.1.12)31 was used to train a 16 state model on all histone marks assayed. The number of states was arrived at by running the model with different numbers of states until the separation of chromatin states was seen; as described by the Roadmap Epigenomics Project.32 The Integrative Genomics Viewer (IGV) was used to visualize chromatin state tracks.33 Global chromatin state changes between hMSC and differentiated chondrocytes were visualized using the riverplot package in R. Gene ontology (GO) terms for chromatin states were found using the GREAT tool with default settings.34
Mouse SOX9 ChIP‐seq data (GEO http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE69109) were aligned to mm10 using Bowtie2 (default settings). Aligned reads were converted to hg38 using the UCSC liftOver tool and narrow peaks were called using MACS2 (v.2.1.0.2) using input samples as control.
2.6. Chromatin state comparisons with roadmap epigenomics cell‐types
Chromatin state coordinates from our study were converted to hg19 using UCSC liftOver as Roadmap data were aligned to hg19. Similarity analysis between equivalent chromatin states across hMSC, chondrocyte, and Roadmap cell‐types was performed using the Jaccard index and hierarchical clustering. Roadmap chromatin state data are available to download from the project website (http://www.roadmapepigenomics.org/). For comparisons with human articular chondrocyte enhancers, histone ChIP‐seq data from human fetal and adult articular cartilage were accessed from http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111850.35
2.7. Integration with DNA methylation
An Infinium HumanMethylation450 BeadChip array was used to quantify DNA methylation in the Transwell model of chondrogenesis,36 GEO dataset http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129266. CpG probes from the 450K methylation array were based on human reference genome hg19; therefore, CpG coordinates from the array were first converted to hg38 and intersected with chromatin state coordinates from hMSC and differentiated chondrocytes. A Chi‐square test with 1000 Monte Carlo permutations was used to test the independence of de‐methylated CpG distribution in enhancers. All plots were generated using the ggplot2 package in R.
2.8. Motif analysis
The MEME suite of tools was used for de novo motif searching.37 The analysis of motif enrichment (AME) tool within MEME was used to assess the relative enrichment of SOX9‐binding motifs found in the footprintDB database38 in new chondrocyte enhancers compared to constant enhancers.
2.9. Data availability
ChIP‐seq data have been deposited http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129031. The chondrogenesis 450k DNA methylation array data can be found in http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129266. The chondrogenesis transcriptome analysis using Illumina whole‐genome expression array Human HT‐12 V4 is available upon reasonable request from the authors.
3. RESULTS
3.1. Chromatin state changes during chondrogenesis
Bone marrow‐derived hMSCs were differentiated into chondrocytes over 14 days using an in vitro Transwell model of chondrogenesis. This scaffold‐free model produces a cartilaginous disc which expresses matrix components such as type II collagen and sulphated glycosaminoglycans. The produced matrix assembles cartilage collagens and generates a robust collagen network with the prerequisite covalent cross‐links.39 Chondrogenic genes such as SOX9 have been shown to be induced during the differentiation of hMSCs using this established and reproducible model of chondrogenesis.8, 21, 23 We observed the upregulation of markers of articular chondrocytes such as COL2A1, TNBS4, PRG4, but also markers of hypertrophic chondrocytes such as COL10A1, PTH1R, and ALPL (Figure S1).40, 41, 42
Histone modifications H3K4me3, H3K4me1, H3K27ac, H3K27me3, and H3K36me3 were assayed in hMSCs and differentiated chondrocytes (Day 14) using ChIP‐seq. These histone marks were selected to reflect a wide range of regulatory states. H3K4me3 commonly marks active promoters, H3K4me1 and H3K27ac are found at active enhancers, H3K36me3 are located at actively transcribed regions and H3K27me3 marks transcriptionally repressed regions. The genome‐wide profiles of each histone mark were as expected; the density of each histone mark differs across the genome with the active promoter mark H3K4me3 showing a high density of peaks close to transcriptional start sites (TSS; Figure 1A). Histone modifications are known to influence gene transcription; therefore, histone mark enrichments were correlated with the expression levels of genes in hMSCs and differentiated chondrocytes (Figure 1B; Table S3). Gene expression in hMSCs and differentiated chondrocytes measured by microarray were stratified into groups of low, medium, and highly expressed genes (Table S3). Average read coverages of histone marks across each group were plotted and as expected histone marks and as expected, histone marks typically associated with transcriptional activity were enriched in highly expressed genes (Figure 1C). In contrast, the transcriptionally repressive mark H3K27me3 showed a greater enrichment in genes with low expression levels in differentiated chondrocytes. This demonstrates that the histone ChIP‐seq generated in hMSCs and differentiated chondrocyte exhibit expected genome‐wide profiles and gene expression associations.
Figure 1.
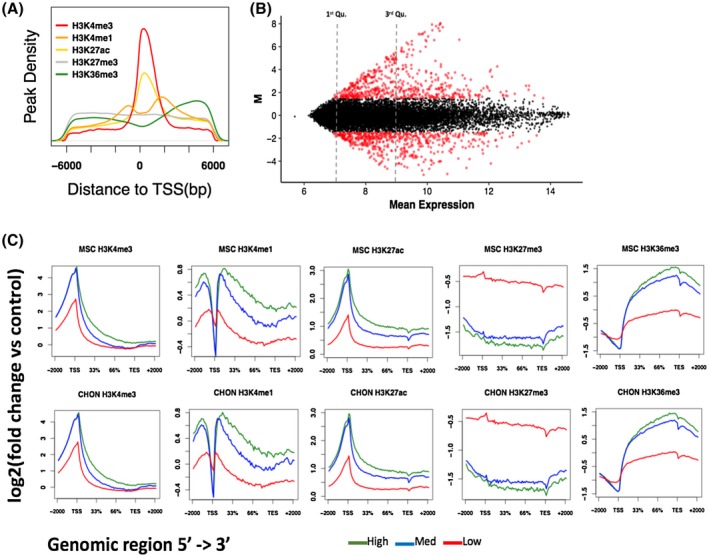
Correlation of histone mark enrichment with gene expression in hMSCs and differentiated chondrocytes. A, Density of histone mark peaks around the TSS (±6 kb). Peaks were called using MACS2 using input samples as controls for background noise. B, MA (log ratio—average expression) plot of differentially expressed genes between hMSCs and differentiated chondrocytes. Gene expression was measured using a cDNA microarray and gene expression is reported as normalized signals. Significantly differentially expressed genes are plotted in red. C, Histone mark enrichments in genes categorized as high expression (expression value > 9; upper quartile), medium expression (expression value > 7 and < 9) and low expression (expression value < 7; lower quartile). Histone marks were associated/overlapped to genes using the ngs.plot tool and plots were generated in R
Combinations of histone modifications can reveal more information about the regulation of gene expression compared to singular histone marks.43 Regulatory elements and chromatin states may be defined by the co‐occurrences of specific histone marks.44 A 16 chromatin state model was trained on the hMSC and differentiated chondrocyte ChIP‐seq data using ChromHMM (Figure 2A). The model yielded a range of chromatin states known to be associated with the histone modifications assayed in this study (model emission probabilities are shown in Table S4). This included promoter states, actively transcribed states and enhancer elements.32 Large scale changes in chromatin states were observed between hMSCs and differentiated chondrocytes, particularly with regards to the quiescent and repressed states becoming transcriptionally active (Figures 2B, S2), demonstrating that genome‐wide histone modification changes occur in the epigenome during chondrogenesis. To elucidate how chromatin states affect gene expression, the GREAT tool34 was used to retrieve gene ontology (GO) terms for each chromatin state. GO terms associated with genes linked to each of the defined chromatin states were non‐specific to cell‐type and mostly encompassed general cell functions, the exception being enhancer states (Figure S3‐S17). In differentiated chondrocytes, the strong active enhancer state (characterized by high enrichment of H3K4me1 and H3K27ac; state 13_EnhS) yielded GO terms related to chondrogenesis and cartilage function (Figure 2C). Previous studies have demonstrated that gene enhancers are cell‐type specific and play an important role in regulating cell‐type specific processes.45 Accordingly, chondrocyte enhancers defined in this study are associated with chondrogenesis related terms, more than promoter or gene transcription chromatin states. Chromatin state changes can clearly be observed around genes that show gene expression changes. For example, we observed the histone modification around the COL2A1 gene switching from repressed/inactive in hMSCs to transcriptionally permissive in chondrocytes (Figure 2D).
Figure 2.
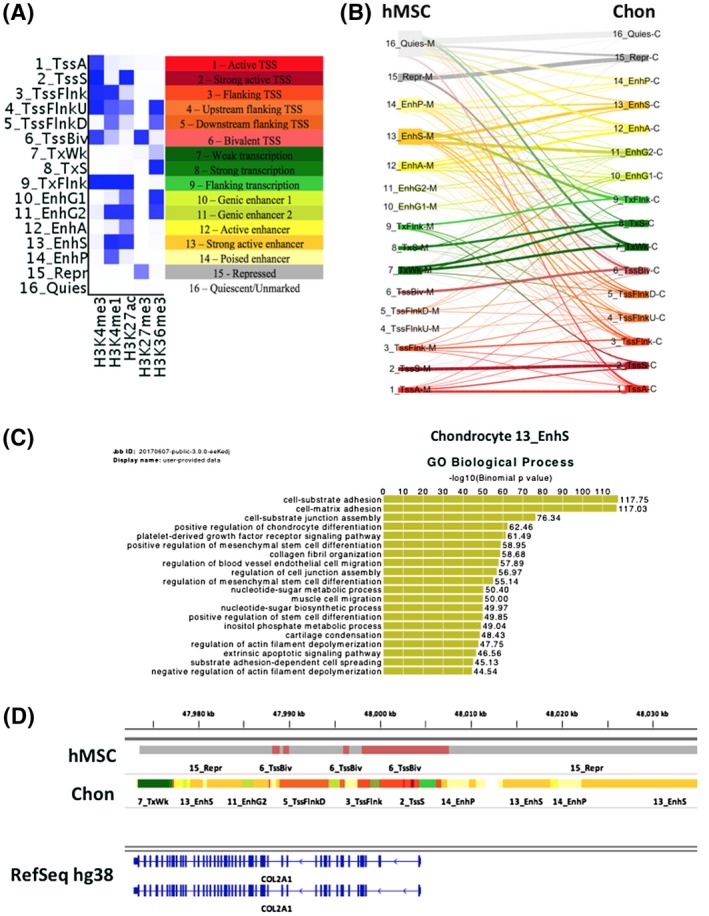
A 16 chromatin state model generated from hMSC and differentiated chondrocyte histone ChIP‐seq. A, Chromatin state model with annotated states. The chromatin state model was generated using the ChromHMM tool with all hMSC and differentiated chondrocyte data using input controls. Intensity of blue within the heatmap represents the model emission probabilities (Table S4). B, Genome‐wide changes in chromatin states during hMSC chondrogenesis. The thickness of the lines represents the frequency of chromatin state changes from hMSC to chondrocytes (Figure S2). C, GREAT biological process GO terms for the chondrocyte strong enhancer (13_EnhS) state. Genome coordinates of the 13_EnhS were used as input into the GREAT tool which associates cis‐regulatory elements with genes. D, IGV genome browser view of hMSC and chondrocyte chromatin states around the COL2A1 gene. Colors for the ChromHMM defined chromatin states are as in panel A
3.2. Comparison to roadmap epigenomics cell types
Several large‐scale consortia have aimed to characterize the epigenomes of various cell‐types including the NIH Roadmap Epigenomics project,32 which defined chromatin states in 127 cell‐types, 98 of which also included the active enhancer mark, H3K27ac. Roadmap cell‐types contained bone marrow‐derived hMSCs and differentiated chondrocytes; therefore, we sought to determine whether the epigenome of our chondrocytes was comparable to those included in the Roadmap project. We compared our 16 chromatin states to the equivalent states of the 18 state model generated by the Roadmap project for their 98 cell‐types that contained the H3K27ac active enhancer mark (Figure S18).The Jaccard similarity coefficient was used to compare equivalent chromatin states across all cell‐types in a pairwise manner. When individual chromatin states except for enhancers were investigated there appeared to be no apparent clustering of cells by type or origin (Figure S19). In contrast, when H3K27ac and H3K4me1 marked enhancers (labeled 13_EnhS in transwell chondrogenesis chromatin state model and 9_EnhA1 in Roadmap 18 state model) were explored, cells clustered with other more closely related cell‐types (Figure 3). Our differentiated chondrocytes (“CHON” in Figure 3) clustered together with the BM‐MSC differentiated chondrocytes from the Roadmap project,10 demonstrating a higher level of similarity to each other than to all other cell types. The Roadmap bone marrow‐derived hMSCs and hMSCs in this study were closely related, contained within a small cluster of primary culture cells consisting of chondrocytes, myocytes, osteoblasts, and fibroblasts (Figure 3). These data corroborate previous studies that report that enhancers are distinct between cell‐types, more than any other regulatory features such as gene promoters.45, 46 Further, enhancers in chondrocytes from different sources showed higher similarity compared to other cell‐type enhancers. Thus, there is a chondrocyte‐specific epigenome based on gene enhancers that can be detected despite differences in chondrogenesis models, laboratory, and MSCs donors.
Figure 3.
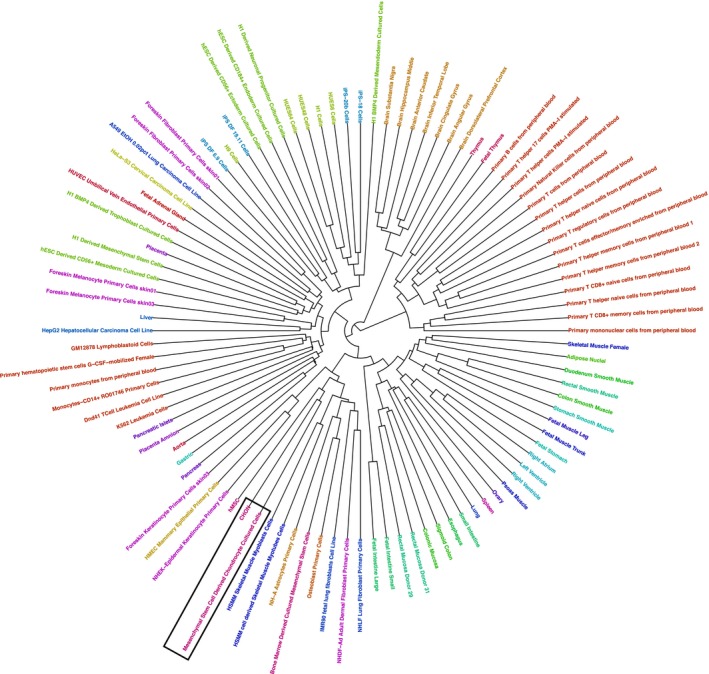
Clustered similarity heatmap of H3K4me1 and H3K27ac enhancers in hMSC and differentiated chondrocytes vs Roadmap cell types. The Jaccard similarity coefficient was calculated between state 13_EnhS in the Transwell chondrogenesis model and 9_EnhA1 in Roadmap 18 state model across cell types. Both states are characterized by high enrichment of H3K4me1 and H3K27ac. Stem cells and differentiated chondrocytes from this study are labeled “hMSC” and “CHON”, respectively. Chondrocytes are indicated by black boxes. A list of Roadmap cell types and their ID codes can be found on the Roadmap project website (http://www.roadmapepigenomics.org)
3.3. Comparison to chondrocyte epigenomes
Roadmap chondrocyte enhancers were intersected with the chondrocyte enhancers identified in this study, resulting in a total of 23 158 enhancer regions common to both types of MSC‐derived chondrocytes (Table S5). We next compared these shared in vitro chondrocyte enhancers with enhancers identified in human fetal and adult articular cartilage35 using the Jaccard similarity coefficient to assess the concordance. The hMSC‐derived chondrocyte enhancer signature was more similar to adult articular chondrocytes compared to either fetal articular chondrocytes or H1 embryonic stem cell‐derived chondrocytes (Figure 4A). A study of differentially accessible chromatin regions in matched intact cartilage (outer region of the lateral tibial plateau) and damaged cartilage (inner region of medial tibial plateau) in OA knee found that enhancers were enriched in significantly differentially accessible regions.47 Of the 77 655 enhancers defined in the study by Liu et al, 14 954 overlapped with enhancers found in the shared enhancers in MSC‐derived chondrocytes. Furthermore, of the 3797 significantly differentially accessible enhancers between intact and damaged cartilage,47 1239 were also found in differentiated chondrocyte enhancers (Figure 4B). This represents a significant overlap of differentially accessible enhancers in knee OA with enhancers in MSC‐derived chondrocytes (hypergeometric test, P < 3.75 × 10−90). This confirms the finding by Lui et al, that dysregulated enhancers in OA are enriched in cell‐type‐specific enhancers.
Figure 4.
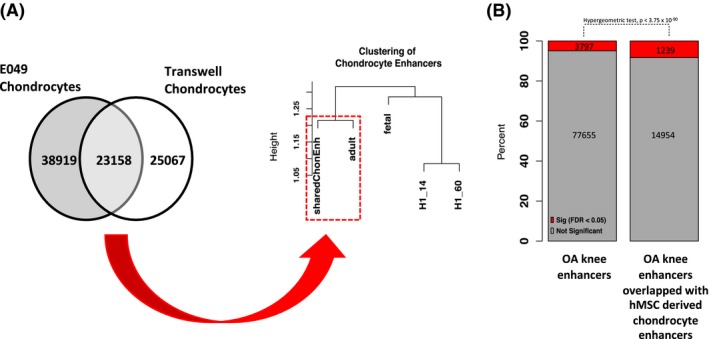
Comparison of shared hMSC‐derived chondrocyte enhancers to human articular chondrocyte and H1‐derived chondrocyte enhancers. A, There were 23 158 shared enhancer regions between differentiated chondrocytes defined in this study and Roadmap E049 enhancers. This shared hMSC‐derived chondrocyte enhancer signature was more similar to adult articular chondrocytes (dashed red box) compared to fetal articular chondrocytes or H1‐derived chondrocytes, as determined by the Jaccard similarity index. Jaccard similarity values were converted to Euclidean distance for hierarchical clustering. The height of the dendrogram represents the distance between samples; nodes joining at lower heights are more similar compared to those joining at greater heights. B, Overlap of shared hMSC‐derived chondrocyte enhancers with enhancers defined in OA knee cartilage. The proportion of enhancers dysregulated in knee OA is shaded in red. A hypergeometric test shows there was a significant overrepresentation of dysregulated enhancers that overlapped with shared hMSC‐derived chondrocyte enhancers (P < 3.75 × 10−90)
3.4. DNA methylation at gene enhancers
Histone modifications are influenced by DNA methylation and vice versa during development.48 DNA methylation occurs at CpG sites in the genome and is typically associated with transcriptional repression. An Illumina Infinium HumanMethylation450K BeadChip array was used to measure DNA methylation. DNA methylation changes during the in vitro Transwell model of chondrogenesis were largely de‐methylation events that were associated with chondrogenesis‐related GO terms.36 We integrated the DNA methylation and ChIP‐seq data in order to investigate the DNA methylation changes in chromatin states during MSC chondrogenesis, focusing on the hypomethylated CpGs (94% of the significantly differentially methylated loci during chondrogenesis) since this is linked to gene transcription activation. Global methylation patterns reflect known trends (Figure 5A,B), for example, gene promoters tend to have low percentage methylation relative to the rest of the genome.49 We observed that enhancers marked by H3K4me1 and H3K27ac (13_EnhS state) were enriched for de‐methylated CpG sites (Figure 5C,D). Fewer than 2% of total CpGs probes present on the Infinium HumanMethylation450K BeadChip were located within chondrocyte chromatin state 13_EnhS (strong enhancers) yet remarkably 41.8% of de‐methylated CpGs were found in this chromatin state (Figure 5E), a highly significant over‐representation (Chi‐square test P < .001). We evaluated the effect of DNA methylation in six selected regions (Table S2) that acquired enhancer status during chondrogenesis in both our model and the chondrogenesis model in Roadmap and also overlapped with a H3K27ac signature during development in of human embryonic limbs.50 The nearest gene to each of these six regions (ASPSCR1, TLE3, WWP2, ZMIZ1, LRP5, and MYEOV) has also been reported to be important in chondrogenesis or cartilage‐related diseases. CpG sites around the ASPSCR1 gene are differentially methylated in human knee OA compared to control.51 TLE3 is a target of the SOX5, SOX6, and SOX9 trio of transcription factors important in chondrogenesis52 and is also involved in osteoblastogenesis.53 Wwp2, an E3 ubiquitin ligase, maintains cartilage homeostasis through the regulation of Adamts‐5.54 The gene is also the host for miR140, which is located toward the 3′ end of the WWP2 gene and is co‐expressed with an isoform of WWP2. MiR140 expression is unique to cartilage and plays an essential role in chondrocyte proliferation.23, 54, 55, 56 ZMIZ1 is differentially expressed in OA chondrocytes following exposure to hyperosmotic conditions.57 LRP5 is involved in MSC differentiation and cartilage degradation.58, 59 MYEOV is associated with multiple myeloma and may also be involved in abnormal bone homeostasis.60 Furthermore, CpG sites within these enhancers show a decrease of DNA methylation during chondrogenesis (Figure S20). The enhancer regions were cloned into a luciferase reporter vector with and without treatment of a CpG methyltransferase. Unmethylated enhancer regions showed increased enhancer activity compared to the empty vector control, confirming that regions classed as enhancers in our model exhibit enhancer activity. With the addition of a CpG methyltransferase, all regions showed a significant decrease in enhancer activity compared to unmethylated regions (Figure 5F).
Figure 5.
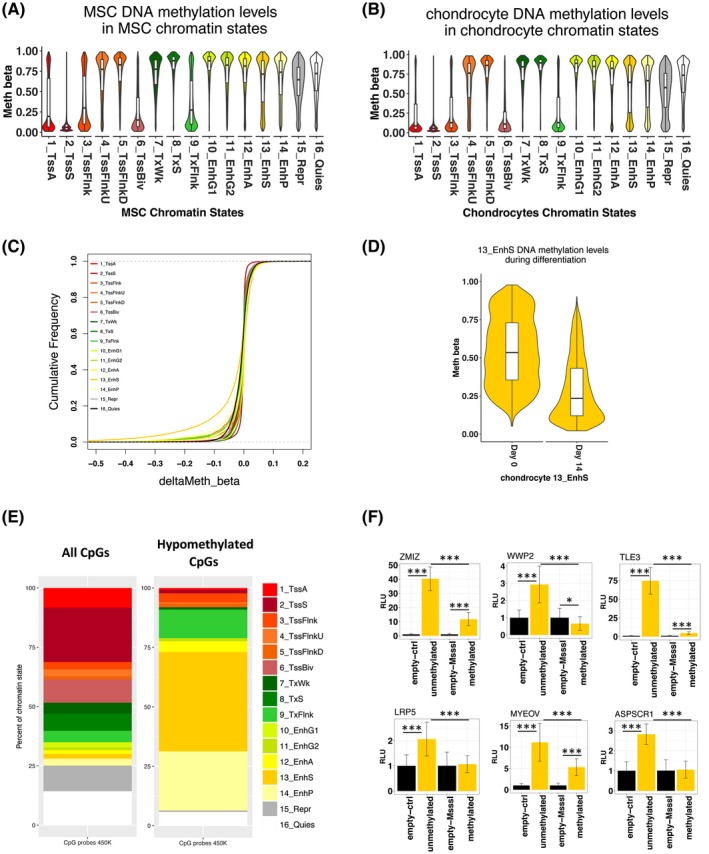
DNA methylation in chondrocyte chromatin states. A,B, Methylation levels of CpGs in the hMSC and chondrocyte chromatin states, respectively. CpG genome coordinates were intersected with chromatin states using BEDTools intersect. C, Empirical cumulative frequency plot of methylation changes (beta values) in chondrocyte chromatin states during hMSC chondrogenesis. D, Significantly methylated CpGs (FDR < 0.05) in between hMSCs and chondrocytes in chondrocyte chromatin states. E, The percentage of all CpGs on the 450k array in each chondrocyte chromatin state and the percentage of de‐methylated CpGs during chondrogenesis in chondrocyte chromatin states. F, Luciferase reporter assay with enhancer regions with and without DNA methylation (n = 6). Enhancers are labeled with their nearest gene. Significance levels: (*) P‐value < .05, (**) P‐value < .01, and (***) P‐value < .001. Error bars are ± standard deviation
3.5. Transcription factor binding at chondrocyte enhancers
Transcription factor binding occurs at gene enhancers to regulate gene expression.61, 62 Therefore, we determined whether chondrocyte enhancers defined in this study contained any transcription factor‐binding motifs. De novo motif searching of the chondrocyte strong enhancer state (13_EnhS) revealed SOX9‐binding motifs (Figure 6A). Motifs found in the strong enhancer state were highly specific to skeletal development with associations to skeletal diseases. For example, there was a positive match to a CREB3L1/OASIS motif, a transcription factor involved in the bone formation.63 Mutations in CREB3L1 have been linked to osteogenesis imperfecta.64, 65, 66 There was a match to ELF3, a transcription factor important during chondrogenesis67 and in cartilage degradation in OA.68, 69 Other matches include the HES and HEY family of transcription factors that are involved in chondrocyte hypertrophy during development 70, 71 (Table S6). The strong promoter state (2_TssS) also contained motifs belonging to transcription factors important in chondrogenesis such as SOX9 and ELF3 but also matched motifs of general transcription factors such as SP1 and the ETS family of transcription factors (Table S7).
Figure 6.
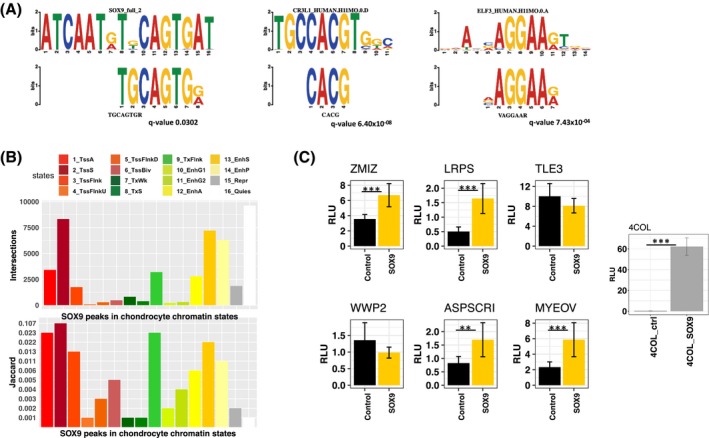
Transcription factor binding in chondrocyte chromatin states. A, Transcription factor motifs found in the chondrocyte strong enhancer state. TF motifs found include SOX9, CREB3L1 and ELF3 (Table S6). De novo motif analysis was performed using MEME. B, Numbers of SOX9 peaks derived from mouse rib chondrocyte ChIP‐seq data in chondrocyte chromatin states. Jaccard index of similarity between SOX9 peaks and chondrocyte chromatin states. C, Luciferase reporter assay with enhancer regions ± SOX9 overexpression (n = 6). Enhancers are labeled with their nearest gene. 4COL is a luciferase reporter containing four copies of the Col2a1 48‐bp enhancer and was used to confirm SOX9 overexpression27 Significance levels: (**) P‐value < .01 and (***) P‐value < .001. Error bars are ± standard deviation
SOX9 is a pivotal transcription factor driving chondrogenesis and interacts with promoters and enhancers to promote chondrogenesis.72 To further characterize chondrocyte enhancers, they were classified into two groups: new enhancers, defined by a change in chromatin state from quiescent or repressed to active enhancers during chondrogenesis, and constant enhancers; regions which were active enhancers both prior and post chondrogenesis. The analysis of motif enrichment (AME) algorithm implemented in the MEME suite of motif searching tools was used to contrast relative SOX9 motif enrichment in these two classes of enhancers found in chondrocytes. We found that both SOX9 motifs were significantly more enriched in the new enhancer class compared to the constant enhancer class (Figure S21). This suggests that enhancers have different properties depending on whether they acquired enhancer status upon differentiation or if they were enhancers beforehand.
To investigate whether SOX9 binds to motifs found in chondrocyte enhancers we used a publicly available mouse rib chondrocyte SOX9 ChIP‐seq dataset72 and converted the data to human genome coordinates. SOX9 is an evolutionary conserved transcription factor with conserved binding sites.73, 74, 75 De novo motif searching using lifted over SOX9 peaks recovered human SOX9 motifs (Table S7), as was the case with the original mouse analysis.72 This is evidence that the SOX9 binding site is conserved and the lifted over sequences contain SOX9 motifs, rather than being lifted over due to regional homology of the sequences around the motif. The majority of SOX9 peaks derived from mouse ChIP‐seq data were found in the chondrocyte strong promoter (2_TssS) state, strong active enhancer state (13_EnhS) state and quiescent state (16_Quies); the latter simply being due to the high percentage of the genome marked quiescent. Accounting for the size of chromatin states, there was more SOX9 enrichment in promoter and enhancer states (Figure 6B). This confirms that the chondrocyte promoters and enhancers identified in our study contain real and conserved SOX9‐binding sites. The impact of SOX9 overexpression was assessed on the previously cloned enhancer regions and a SOX9‐responsive Col2a1 enhancer reporter27 (Figure 6F). Four out of six enhancers exhibited increased enhancer activity with SOX9 overexpression (Figure 6C). All regions except one (nearest gene TLE3) have a SOX9‐binding site in the lifted over SOX9 ChIP‐seq data; as predicted, the TLE3 region did not show increased enhancer activity upon SOX9 overexpression.
Previously, analysis of mouse Sox9 ChIP‐seq found that AP‐1 factors, Jun, and Fos were found to co‐localize with Sox9.72, 76 The authors found that whilst Sox9 and AP‐1 factors can form protein‐protein complexes, co‐localization primarily occurred through the binding of factors to the same binding sites. Positive matches to JUN and FOS motifs were also found in the de novo motif search of lifted over SOX9 peaks (Table S8), demonstrating that this mechanism is conserved between the two species.
4. DISCUSSION
There are numerous in vitro models of chondrogenesis and although some models utilize scaffolds for cells to grow, scaffold‐free models are reported to better reflect the conditions during in vivo chondrogenesis during development.77 Chondrocytes from this study were derived from a scaffold‐free chondrogenesis model using bone marrow‐derived hMSCs. Other scaffold‐free models include the micromass and pellet culture system. In contrast, chondrocytes from the Roadmap project were derived from human BM‐MSCs in a 3D alginate chondrogenesis model.10 Whilst there has been some gene expression comparisons between models,78, 79 no comparison has been made about changes in their epigenetic landscape. Here, we show that chondrocyte gene enhancers across two different models are highly concordant relative to other cell‐types. This is indicative of a unique chondrocyte epigenetic signature, independent of model and laboratory‐specific effects. Although hMSC‐derived chondrocyte enhancer concordance is evidence that chondrogenic models are reliable and comparable, further work is required to establish their likeness to in vivo chondrocytes. We observed both articular and growth plate chondrocyte gene expression markers in our differentiated chondrocytes. However, although the classical gene for hypertrophy, COL10A1, is upregulated, protein production or matrix deposition appears to be limited.21, 39 An upregulation of markers of hypertrophy is also commonly observed in pellet models of hMSC chondrogenesis.80, 81, 82 More work is needed to determine whether in vitro systems reflect chondrocytes which undergo endochondral ossification or articular chondrocytes found in adult synovial joints. However, we have identified in vitro chondrocyte enhancers that overlap with enhancers found in knee cartilage and corroborated that enhancers dysregulated in OA are more likely to be cell‐type‐specific enhancers.47 The concordance between enhancers identified between hMSC chondrogenesis models and OA suggests that in vitro models have an important role in studies into cartilage development and disease.
Combinations of histone modifications can define regulatory elements and regulate genes through modulating chromatin remodeling to allow or block access to transcription factors. However, histone modifications also rely on other epigenetic mechanisms such as DNA methylation and vice versa.48 Crosstalk between the two epigenetic mechanisms allows for greater control of gene transcription and it is important to consider histone modifications in the wider context of the whole epigenome. Traditionally, studies into DNA methylation focused on gene promoters where CpG islands are more likely to be found and array probe design is biased toward promoters. Although our data are extensive, we only compared ~ 450,000 (1.6%) of the ~28 million CpG sites in the human genome.83 Reduced representation bisulfite sequencing (RRBS) in chondrogenesis only identified limited CpG methylation changes in gene promoters.10 However, RRBS is heavily biased toward promoters and whole‐genome bisulfite sequencing remains the only method that can universally capture almost the entire DNA methylome. We show in this study that significant changes occur at distal gene enhancers during chondrogenesis. DNA demethylation at enhancer regions has also been observed during other stem cell differentiation processes, including differentiation of intestinal epithelium progenitors,84 hematopoietic stem cells 85 and embryonic stem cells 86 but also due to MSC age and culture conditions.87 DNA demethylation at enhancers is associated with the development of most human organs.88 Aberrant DNA methylation in enhancers has been implicated in diseases such as cancer89, 90, 91 and osteoarthritis (OA).51, 92
Motif discovery at chondrocyte enhancers recovered motifs of transcription factors known to be involved in cartilage development and diseases such as CREB3L1, ELF3, and SOX9. We utilized a mouse Sox9 ChIP‐seq dataset to assess whether enhancers defined in our study contained SOX9‐binding sites. SOX9 has a highly conserved DNA‐binding motif and function.93, 94, 95, 96 Therefore, we considered the liftover of mouse reads to human genome coordinates to be appropriate for our analysis. Indeed, we recovered a human SOX9 motif from lifted over peaks, illustrating that the DNA‐binding sites and motif of SOX9 are highly conserved between human and mouse. Using liftover, species‐specific SOX9‐binding information is lost but conserved sites are retained, these sites arguably being the most important, as evolutionary conservation is a marker of essentiality. We found SOX9 motifs in our chondrocyte enhancers via de novo motif searching as well as conserved SOX9 binding using mouse SOX9 ChIP‐seq. SOX9 acts in conjunction with transcription factors SOX5 and SOX6 in chondrogenesis,97 to bind to super enhancers promoting chondrogenesis.98 Super enhancers are loosely defined as multiple enhancers in close proximity exhibiting high levels of active enhancer markers such as H3K27ac or transcription factors (Pott and Lieb, 2015). SOX9 bound enhancers have previously been proposed to be important for defining the chondrocyte phenotype. Furthermore, mutations of Sox9‐binding motifs within distal Acan enhancers in transgenic mice resulted in a loss of chondrocyte‐specific expression.99
Enhancers are thought to regulate their target genes by forming a loop to physically contact the gene promoter within topologically associating domains100, 101; an interaction mediated by transcription factors.102 Gene enhancers can be located distal from their target promoters and therefore, target gene prediction can be challenging without chromatin conformation data.103 Although we have validated that enhancers identified in this study do, indeed, possess enhancer activity that may be modulated by DNA methylation and SOX9 binding, further functional work is required to elucidate their gene target(s) and importance in cartilage development. In this study, we show that enhancers are dynamic during chondrogenesis and may serve as potential targets for modulating hMSC differentiation.
To conclude, the integration of ChIP‐seq with methylation data revealed that gene enhancers are de‐methylated during an in vitro Transwell model of chondrogenesis. Comparison of chromatin states across hMSCs and chondrocytes generated in this study along with those from the Roadmap Epigenomics project revealed that enhancers marked by H3K4me1 and H3K27ac are more cell‐type specific compared to other chromatin states. Chondrocytes from the Epigenomics Roadmap project and this study showed a more similarity of enhancers with each other than other cell‐types despite being from different models. We have established that chondrocyte enhancers contain motifs to which SOX9 binds in vivo. Additional investigations are needed to elucidate further the epigenetic landscape of chondrocytes originating from other in vitro models and to determine whether these are comparable to the epigenome of human articular chondrocytes. A link between reactivation of developmental pathways and OA has been suggested17; more research is needed to fully explore the association between development and disease.
5. CONCLUSION
Human mesenchymal stem cells are able to differentiate into chondrocytes, the cell type found in cartilage, making them an accessible system to study gene regulation during this process. Epigenetic mechanisms such as histone modifications and DNA methylation together with transcription factor binding play a role in activating and repressing gene expression. In this study, we investigated the genome‐wide histone modification changes during chondrocyte differentiation. Integration of this data with DNA methylation and SOX9 transcription factor ChIP‐seq revealed epigenetic changes at gene enhancer elements. Regions of the genome that transition from non‐enhancers to enhancers in chondrocytes are enriched for SOX9 transcription factor‐binding sites. Luciferase reporter assays revealed that enhancer activity may be modulated by manipulating DNA methylation and SOX9 expression. This study has defined important regulatory elements in chondrocytes which could serve as targets for future mechanistic studies.
CONFLICT OF INTEREST
None.
AUTHOR CONTRIBUTIONS
Kathleen Cheung: Collection and/or assembly of data, data analysis and interpretation, manuscript writing. Matthew J. Barter: Collection and/or assembly of data, provision of study material or patients. Julia Falk: Collection and/or assembly of data. Carole Proctor: Conception and design. Louise N. Reynard: Conception and design, provision of study material or patients. David. A. Young: Conception and design, final approval of the manuscript.
Supporting information
ACKNOWLEDGMENTS
This work was supported by the Medical Research Council and Arthritis Research UK as part of the MRC‐Arthritis Research UK Centre for Integrated Research into Musculoskeletal Ageing (CIMA, grant references JXR 10641 and MR/P020941/1); Arthritis Research UK [grant numbers 18746 and 19424]; the JGW Patterson Foundation; The Dunhill Medical Trust; and the NIHR Newcastle Biomedical Research.
Cheung K, Barter MJ, Falk J, Proctor CJ, Reynard LN, Young DA. Histone ChIP‐Seq identifies differential enhancer usage during chondrogenesis as critical for defining cell‐type specificity. The FASEB Journal. 2020;34:5317–5331. 10.1096/fj.201902061RR
Contributor Information
Kathleen Cheung, Email: kat.cheung@ncl.ac.uk.
David A. Young, Email: david.young@ncl.ac.uk.
DATA AVAILABILITY STATEMENT
ChIP‐seq data have been submitted into the NCBI GEO data repository with accession http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129031.
REFERENCES
- 1. van der Kraan PM, van den Berg WB. Chondrocyte hypertrophy and osteoarthritis: role in initiation and progression of cartilage degeneration? Osteoarthr Cartil. 2012;20:223‐232. [DOI] [PubMed] [Google Scholar]
- 2. Mackie EJ, Ahmed YA, Tatarczuch L, Chen K‐S, Mirams M. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol. 2008;40:46‐62. [DOI] [PubMed] [Google Scholar]
- 3. Akiyama H. Control of chondrogenesis by the transcription factor Sox9. Mod Rheumatol. 2008;18:213‐219. [DOI] [PubMed] [Google Scholar]
- 4. Liu C‐F, Angelozzi M, Haseeb A, Lefebvre V. SOX9 is dispensable for the initiation of epigenetic remodeling and the activation of marker genes at the onset of chondrogenesis. Development. 2018;145:dev164459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Furumatsu T, Ozaki T. Epigenetic regulation in chondrogenesis. Acta Med Okayama. 2010;64:155‐161. [DOI] [PubMed] [Google Scholar]
- 6. Hata K. Epigenetic regulation of chondrocyte differentiation. Jpn Dent Sci Rev. 2015;51:105‐113. [Google Scholar]
- 7. Yang BO, Guo H, Zhang Y, Chen L, Ying D, Dong S. MicroRNA‐145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS ONE. 2011;6:e21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barter MJ, Gomez R, Hyatt S, et al. The long non‐coding RNA ROCR contributes to SOX9 expression and chondrogenic differentiation of human mesenchymal stem cells. Development. 2017;144:4510‐4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tian YE, Guo R, Shi B, Chen L, Yang L, Fu Q. MicroRNA‐30a promotes chondrogenic differentiation of mesenchymal stem cells through inhibiting Delta‐like 4 expression. Life Sci. 2016;148:220‐228. [DOI] [PubMed] [Google Scholar]
- 10. Herlofsen SR, Bryne JC, Høiby T, et al. Genome‐wide map of quantified epigenetic changes during in vitro chondrogenic differentiation of primary human mesenchymal stem cells. BMC Genom. 2013;14:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meyer J, Südbeck P, Held M, et al. Mutational analysis of the SOX9 gene in campomelic dysplasia and autosomal sex reversal: lack of genotype/phenotype correlations. Hum Mol Genet. 1997;6:91‐98. [DOI] [PubMed] [Google Scholar]
- 12. Wunderle VM, Critcher R, Hastie N, Goodfellow PN, Schedl A. Deletion of long‐range regulatory elements upstream of SOX9 causes campomelic dysplasia. Proc Natl Acad Sci USA. 1998;95:10649‐10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bell DM, Leung KKH, Wheatley SC, et al. SOX9 directly regulates the type‐ll collagen gene. Nat Genet. 1997;16:174‐178. [DOI] [PubMed] [Google Scholar]
- 14. Li F, Lu Y, Ding M, et al. Runx2 contributes to murine Col10a1 gene regulation through direct interaction with its cis‐enhancer. J Bone Miner Res. 2011;26:2899‐2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Capellini TD, Chen H, Cao J, et al. Ancient selection for derived alleles at a GDF5 enhancer influencing human growth and osteoarthritis risk. Nat Genet. 2017;49:1202‐1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reynard LN, Ratnayake M, Santibanez‐Koref M, Loughlin J. Functional characterization of the osteoarthritis susceptibility mapping to CHST11‐A bioinformatics and molecular study. PLoS ONE. 2016;11:e0159024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012;64:1697‐1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tchetina EV, Squires G, Poole AR. Increased type II collagen degradation and very early focal cartilage degeneration is associated with upregulation of chondrocyte differentiation related genes in early human articular cartilage lesions. J Rheumatol. 2005;32:876‐886. [PubMed] [Google Scholar]
- 19. Yamasaki S, Mera H, Itokazu M, Hashimoto Y, Wakitani S. Cartilage repair with autologous bone marrow mesenchymal stem cell transplantation. Cartilage. 2014;5:196‐202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y, Zhou G, Cao Y. Recent progress in cartilage tissue engineering—our experience and future directions. Engineering. 2017;3:28‐35. [Google Scholar]
- 21. Murdoch AD, Grady LM, Ablett MP, Katopodi T, Meadows RS, Hardingham TE. Chondrogenic differentiation of human bone marrow stem cells in transwell cultures: generation of scaffold‐free cartilage. Stem Cells. 2007;25:2786‐2796. [DOI] [PubMed] [Google Scholar]
- 22. Zeybel M, Hardy T, Wong YK, et al. Multigenerational epigenetic adaptation of the hepatic wound‐healing response. Nat Med. 2012;18:1369‐1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barter MJ, Tselepi M, Gómez R, et al. Genome‐wide MicroRNA and gene analysis of mesenchymal stem cell chondrogenesis identifies an essential role and multiple targets for miR‐140‐5p. Stem Cells. 2015;33:3266‐3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. In vitro chondrogenesis of bone marrow‐derived mesenchymal progenitor cells. Exp Cell Res. 1998;238:265‐272. [DOI] [PubMed] [Google Scholar]
- 25. Klug M, Rehli M. Functional analysis of promoter CPG‐methylation using a CpG‐free luciferase reporter vector. Epigenetics. 2006;1:127‐130. [DOI] [PubMed] [Google Scholar]
- 26. Lefebvre VR, Huang W, Harley VR, et al. SOX9 is a potent activator of the chondrocyte‐specific enhancer of the pro1(II) collagen. Gene. 1997;17:2336‐2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lefebvre V, Zhou G, Mukhopadhyay K, et al. An 18‐base‐pair sequence in the mouse proalpha1(II) collagen gene is sufficient for expression in cartilage and binds nuclear proteins that are selectively expressed in chondrocytes. Mol Cell Biol. 1996;16:4512‐4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langmead B, Salzberg SL. Fast gapped‐read alignment with Bowtie 2. Nat Methods. 2012;9:357‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feng J, Liu T, Qin BO, Zhang Y, Liu XS. Identifying ChIP‐seq enrichment using MACS. Nat Protoc. 2012;7:10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen LI, Shao N, Liu X, Nestler E. ngs.plot: quick mining and visualization of next‐generation sequencing data by integrating genomic databases. BMC Genom. 2014;15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ernst J, Kellis M. ChromHMM: automating chromatin‐state discovery and characterization. Nat Meth. 2012;9:215‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kundaje A, Meuleman W, Ernst J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high‐performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McLean CY, Bristor D, Hiller M, et al. GREAT improves functional interpretation of cis‐regulatory regions. Nat Biotechnol. 2010;28:495‐501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ferguson GB, Van Handel B, Bay M, et al. Mapping molecular landmarks of human skeletal ontogeny and pluripotent stem cell‐derived articular chondrocytes. Nat Commun. 2018;9:3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barter MJ, Bui C, Cheung K, et al. Hypomethylation during MSC chondrogenesis occurs predominantly at enhancer regions. Sci Rep. 2020;625954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ma W, Noble WS, Bailey TL. Motif-based analysis of large nucleotide data sets using MEMEChIP. Nat Protoc. 2014;9:1428–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sebastian A, Contreras‐Moreira B. footprintDB: a database of transcription factors with annotated cis elements and binding interfaces. Bioinformatics. 2014;30:258‐265. [DOI] [PubMed] [Google Scholar]
- 39. Murdoch AD, Hardingham TE, Eyre DR, Fernandes RJ. The development of a mature collagen network in cartilage from human bone marrow stem cells in Transwell culture. Matrix Biol. 2016;50:16‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hissnauer TN, Baranowsky A, Pestka JM, et al. Identification of molecular markers for articular cartilage. Osteoarthr Cartil. 2010;18:1630‐1638. [DOI] [PubMed] [Google Scholar]
- 41. Lian C, Wang X, Qiu X, et al. Collagen type II suppresses articular chondrocyte hypertrophy and osteoarthritis progression by promoting integrin β1−SMAD1 interaction. Bone Res. 2019;7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kozhemyakina E, Zhang M, Ionescu A, et al. Identification of a Prg4 ‐expressing articular cartilage progenitor cell population in mice. Arthritis Rheumatol. 2015;67:1261‐1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol. 2005;21:133‐153. [DOI] [PubMed] [Google Scholar]
- 44. Baker M. Making sense of chromatin states. Nat Methods. 2011;2011:89. [DOI] [PubMed] [Google Scholar]
- 45. Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type‐specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vandel J, Cassan O, Lèbre S, Lecellier C‐H, Bréhélin L. Probing transcription factor combinatorics in different promoter classes and in enhancers. BMC Genom. 2019;20:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu YE, Chang J‐C, Hon C‐C, et al. Chromatin accessibility landscape of articular knee cartilage reveals aberrant enhancer regulation in osteoarthritis. Sci Rep. 2018;8:15499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295‐304. [DOI] [PubMed] [Google Scholar]
- 49. Sharifi‐Zarchi A, Gerovska D, Adachi K, et al. DNA methylation regulates discrimination of enhancers from promoters through a H3K4me1‐H3K4me3 seesaw mechanism. BMC Genom. 2017;18:964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cotney J, Leng J, Yin J, et al. The evolution of lineage‐specific regulatory activities in the human embryonic limb. Cell. 2013;154:185‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alvarez‐Garcia O, Fisch KM, Wineinger NE, et al. Increased DNA methylation and reduced expression of transcription factors in human osteoarthritis cartilage. Arthritis Rheumatol (Hoboken, NJ). 2016;68:1876‐1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee WJ, Chatterjee S, Yap SP, et al. An integrative developmental genomics and systems biology approach to identify an in vivo sox trio‐mediated gene regulatory network in murine embryos. Biomed Res Int. 2017;2017:8932583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kokabu S, Sato T, Ohte S, et al. Expression of TLE3 by bone marrow stromal cells is regulated by canonical Wnt signaling. FEBS Lett. 2014;588:614‐619. [DOI] [PubMed] [Google Scholar]
- 54. Mokuda S, Nakamichi R, Matsuzaki T, et al. Wwp2 maintains cartilage homeostasis through regulation of Adamts5. Nat Commun. 2019;10:2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang J, Qin S, Yi C, et al. MiR‐140 is co‐expressed with Wwp2‐C transcript and activated by Sox9 to target Sp1 in maintaining the chondrocyte proliferation. FEBS Lett. 2011;585:2992‐2997. [DOI] [PubMed] [Google Scholar]
- 56. Miyaki S, Sato T, Inoue A, et al. MicroRNA‐140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173‐1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tew SR, Vasieva O, Peffers MJ, Clegg PD. Post‐transcriptional gene regulation following exposure of osteoarthritic human articular chondrocytes to hyperosmotic conditions. Osteoarthr Cartil. 2011;19:1036‐1046. [DOI] [PubMed] [Google Scholar]
- 58. Shin Y, Huh Y, Kim K, et al. Low‐density lipoprotein receptor‐related protein 5 governs Wnt‐mediated osteoarthritic cartilage destruction. Arthritis Res Ther. 2014;16:R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhou S, Eid K, Glowacki J. Cooperation between TGF‐β and Wnt pathways during chondrocyte and adipocyte differentiation of human marrow stromal cells. J Bone Miner Res. 2003;19:463‐470. [DOI] [PubMed] [Google Scholar]
- 60. Ealy M, Chen W, Ryu G‐Y, et al. Gene expression analysis of human otosclerotic stapedial footplates. Hear Res. 2008;240:80‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Grossman SR, Engreitz J, Ray JP, Nguyen TH, Hacohen N, Lander ES. Positional specificity of different transcription factor classes within enhancers. Proc Natl Acad Sci U S A. 2018;115:E7222‐E7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Palstra R‐J, Grosveld F. Transcription factor binding at enhancers: shaping a genomic regulatory landscape in flux. Front Genet. 2012;3:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Murakami T, Saito A, Hino S‐I, et al. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol. 2009;11:1205‐1211. [DOI] [PubMed] [Google Scholar]
- 64. Keller RB, Tran TT, Pyott SM, et al. Monoallelic and biallelic CREB3L1 variant causes mild and severe osteogenesis imperfecta, respectively. Genet Med. 2018;20:411‐419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cayami FK, Maugeri A, Treurniet S, et al. The first family with adult osteogenesis imperfecta caused by a novel homozygous mutation in CREB3L1 . Mol Genet Genomic Med. 2019;7:e823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Symoens S, Malfait F, D’hondt S, et al. Deficiency for the ER‐stress transducer OASIS causes severe recessive osteogenesis imperfecta in humans. Orphanet J Rare Dis. 2013;8:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Otero M, Peng H, El HK, et al. ELF3 modulates type II collagen gene (COL2A1) transcription in chondrocytes by inhibiting SOX9‐CBP/p300‐driven histone acetyltransferase activity. Connect Tissue Res. 2017;58:15‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wondimu EB, Culley KL, Quinn J, et al. Elf3 contributes to cartilage degradation in vivo in a surgical model of post‐traumatic osteoarthritis. Sci Rep. 2018;8:6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Otero M, Plumb DA, Tsuchimochi K, et al. E74‐like factor 3 (ELF3) impacts on matrix metalloproteinase 13 (MMP13) transcriptional control in articular chondrocytes under proinflammatory stress. J Biol Chem. 2012;287:3559‐3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rutkowski TP, Kohn A, Sharma D, Ren Y, Mirando AJ, Hilton MJ. HES factors regulate specific aspects of chondrogenesis and chondrocyte hypertrophy during cartilage development. J Cell Sci. 2016;129:2145‐2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Haller R, Schwanbeck R, Martini S, et al. Notch1 signaling regulates chondrogenic lineage determination through Sox9 activation. Cell Death Differ. 2012;19:461‐469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ohba S, He X, Hojo H, McMahon A. Distinct transcriptional programs underlie Sox9 regulation of the mammalian chondrocyte. Cell Rep. 2015;12:229‐243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pask AJ, Harry JL, Graves JAM, et al. SOX9 has both conserved and novel roles in marsupial sexual differentiation. Genesis. 2002;33:131‐139. [DOI] [PubMed] [Google Scholar]
- 74. Kanai Y, Koopman P. Structural and functional characterization of the mouse Sox9 promoter: implications for campomelic dysplasia. Hum Mol Genet. 1999;8:691‐696. [DOI] [PubMed] [Google Scholar]
- 75. Tew SR, Clegg PD, Brew CJ, Redmond CM, Hardingham TE. SOX9 transduction of a human chondrocytic cell line identifies novel genes regulated in primary human chondrocytes and in osteoarthritis. Arthritis Res Ther. 2007;9:R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. He X, Ohba S, Hojo H, McMahon AP. AP‐1 family members act with Sox9 to promote chondrocyte hypertrophy. Development. 2016;143:3012‐3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Whitney GA, Mera H, Weidenbecher M, Awadallah A, Mansour JM, Dennis JE. Methods for producing scaffold‐free engineered cartilage sheets from auricular and articular chondrocyte cell sources and attachment to porous tantalum. Biores Open Access. 2012;1:157‐165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang L, Su P, Xu C, Yang J, Yu W, Huang D. Chondrogenic differentiation of human mesenchymal stem cells: a comparison between micromass and pellet culture systems. Biotechnol Lett. 2010;32:1339‐1346. [DOI] [PubMed] [Google Scholar]
- 79. Watts AE, Ackerman‐Yost JC, Nixon AJ. A comparison of three‐dimensional culture systems to evaluate in vitro chondrogenesis of equine bone marrow‐derived mesenchymal stem cells. Tissue Eng Part A. 2013;19:2275‐2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Huynh NPT, Zhang B, Guilak F. High‐depth transcriptomic profiling reveals the temporal gene signature of human mesenchymal stem cells during chondrogenesis. FASEB J. 2019;33:358‐372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mueller MB, Tuan RS. Functional characterization of hypertrophy in chondrogenesis of human mesenchymal stem cells. Arthritis Rheum. 2008;58:1377‐1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mueller MB, Fischer M, Zellner J, et al. Hypertrophy in mesenchymal stem cell chondrogenesis: Effect of TGF‐β isoforms and chondrogenic conditioning. Cells Tissues Organs. 2010;192:158‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lövkvist C, Dodd IB, Sneppen K, Haerter JO. DNA methylation in human epigenomes depends on local topology of CpG sites. Nucleic Acids Res. 2016;44:5123‐5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sheaffer KL, Kim R, Aoki R, et al. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 2014;28:652‐664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bock C, Beerman I, Lien W‐H, et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell. 2012;47:633‐647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Petell CJ, Alabdi L, He M, et al. An epigenetic switch regulates de novo DNA methylation at a subset of pluripotency gene enhancers during embryonic stem cell differentiation. Nucleic Acids Res. 2016;44:7605‐7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pasumarthy KK, Doni Jayavelu N, Kilpinen L, et al. Methylome analysis of human bone marrow MSCs reveals extensive age‐ and culture‐induced changes at distal regulatory elements. Stem Cell Reports. 2017;9:999‐1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Li C, Fan Y, Li G, et al. DNA methylation reprogramming of functional elements during mammalian embryonic development. Cell Discov. 2018;4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bell RE, Golan T, Sheinboim D, et al. Enhancer methylation dynamics contribute to cancer plasticity and patient mortality. Genome Res. 2016;26:601‐611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Heyn H, Vidal E, Ferreira HJ, et al. Epigenomic analysis detects aberrant super‐enhancer DNA methylation in human cancer. Genome Biol. 2016;17:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Qu Y, Siggens L, Cordeddu L, et al. Cancer‐specific changes in DNA methylation reveal aberrant silencing and activation of enhancers in leukemia. Blood. 2017;129:e13‐e25. [DOI] [PubMed] [Google Scholar]
- 92. Zhang Y, Fukui N, Yahata M, et al. Genome‐wide DNA methylation profile implicates potential cartilage regeneration at the late stage of knee osteoarthritis. Osteoarthr Cartil. 2016;24:835‐843. [DOI] [PubMed] [Google Scholar]
- 93. Cremazy F, Soullier S, Berta P, et al. Further complexity of the human SOX gene family revealed by the combined use of highly degenerate primers and nested PCR. FEBS Lett. 1998;438:311‐314. [DOI] [PubMed] [Google Scholar]
- 94. Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409‐1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Oh C‐D, Maity SN, Lu J‐F, et al. Identification of SOX9 interaction sites in the genome of chondrocytes. PLoS ONE. 2010;5:e10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yasuda H, Oh C‐D, Chen DI, de Crombrugghe B, Kim J‐H. A novel regulatory mechanism of Type II collagen expression via a SOX9‐dependent enhancer in intron 6. J Biol Chem. 2017;292:528‐538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Leung VYL, Gao BO, Leung KKH, et al. SOX9 governs differentiation stage‐specific gene expression in growth plate chondrocytes via direct concomitant transactivation and repression. PLoS Genet. 2011;7:e1002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Liu C‐F, Lefebvre V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome‐wide through super‐enhancers to drive chondrogenesis. Nucleic Acids Res. 2015;43:8183‐8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Li IMH, Liu KE, Neal A, Clegg PD, De Val S, Bou‐Gharios G. Differential tissue specific, temporal and spatial expression patterns of the Aggrecan gene is modulated by independent enhancer elements. Sci Rep. 2018;8:950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Smith E, Lajoie B, Jain G, Dekker J. Invariant TAD boundaries constrain cell‐type‐specific looping interactions between promoters and distal elements around the CFTR locus. Am J Hum Genet. 2016;98:185‐201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Dixon JR, Gorkin DU, Ren B. Chromatin domains: The unit of chromosome organization. Mol Cell. 2016;62:668‐680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M, Thanos D. Transcription factors mediate long‐range enhancer‐promoter interactions. Proc Natl Acad Sci. 2009;106:20222‐20227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Javierre BM, Burren OS, Wilder SP, et al. Lineage‐specific genome architecture links enhancers and non‐coding disease variants to target gene promoters. Cell. 2016;167(1369–1384):e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP‐seq data have been deposited http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129031. The chondrogenesis 450k DNA methylation array data can be found in http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129266. The chondrogenesis transcriptome analysis using Illumina whole‐genome expression array Human HT‐12 V4 is available upon reasonable request from the authors.
ChIP‐seq data have been submitted into the NCBI GEO data repository with accession http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129031.


