Abstract
LuxR‐type transcriptional activator proteins frequently regulate the expression of biosynthetic gene clusters (BGCs). With only a fraction of bacterial BGCs being expressed under standard culturing conditions, modulation of LuxRs would provide a powerful approach to activate silent clusters. We show that by exploiting the modular nature of LuxR proteins, it is possible to construct functional chimeric LuxRs, which enables both the rewiring of quorum sensing systems and the activation of silent BGCs. Importantly, our strategy allowed us to identify the novel natural product pseudomonol from a bacterium of the genus Pseudomonas.
Keywords: gene expression, LuxRs, natural products, quorum sensing, transcriptional activation
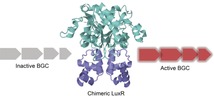
Mixed signals: Modular LuxR‐type transcriptional activator proteins can be engineered to alter the signals to which they respond. This enables the rewiring of quorum‐sensing systems, and the resultant chimeric LuxRs can be applied for the activation of silent biosynthetic gene clusters. In this way, the novel natural product pseudomonol was identified from a bacterium of the genus Pseudomonas.
Ever since the beginning of the golden era of antibiotics, microbial natural products have served as the paradigm to fight infectious diseases in human medicine and agriculture.1, 2 The utility of these natural products goes far beyond antibacterial activity, with many anti‐cancer drugs, immunomodulators, or cholesterol‐lowering drugs being of microbial origin. Thus, the search for novel natural products is more than ever of utmost importance. Genome mining has highlighted the untapped biosynthetic potential of bacterial genomes,3, 4 yet the production of secondary metabolites is a great energy investment for bacteria. As a result, the expression of biosynthetic genes is virtually always tightly regulated. Hence, under laboratory conditions, bacteria typically only express a fraction of the genetically encoded biosynthetic gene clusters.5 Novel approaches to activate these silent gene clusters thus greatly enhance the chances of finding new natural products.5
In this report, we show that regulatory elements that control bacterial biosynthetic gene clusters (BGCs) can be altered in order to modulate and even activate the production of secondary metabolites. We focus on LuxR‐type transcriptional activator proteins, which are often but not exclusively found to flank bacterial BGCs.6 These LuxR‐family proteins serve as key regulatory elements, and a deeper understanding of their effect on the activation of BGCs would allow us to determine the conditions under which they can trigger natural product synthesis. LuxR‐type regulators can be part of quorum sensing (QS) systems that orchestrate BGC expression, leading to the production of multifarious natural products including siderophores, peptide antibiotics, polyketides, phenazines, pigments, and many others.7, 8, 9, 10, 11 These LuxRs are response regulators that act as transcription factors upon binding a signal molecule produced by their cognate LuxI synthase. LuxRs consist of two main domains: the N‐terminal signal‐binding domain (SBD) and C‐terminal DNA‐binding domain (DBD). When binding to a cognate signal molecule (usually derivatives of acyl homoserine lactones, AHLs), the C‐terminal part of the protein undergoes homo‐dimerization and the dimer then acts as a functional transcriptional activator.12 LuxRs specifically interact with discrete palindromic repeat motifs in the promoter region of their target genes, so‐called lux‐boxes.13 The modular nature of LuxRs and response regulators in general makes them an ideal target for synthetic biology experiments.14 Herein, we describe the construction of chimeric LuxR proteins (LuxRCs) and demonstrate their functionality both in a heterologous system and in the native host (Figure S1 in the Supporting Information). We hypothesize that a LuxRC containing a well‐characterized AHL‐binding SBD and DBD of a LuxR associated with an unexpressed BGC could jump‐start the production of the corresponding metabolite upon addition of the respective AHL. This is in particular the case for LuxRs that lack a cognate LuxI synthase, also known as LuxR solos.15 The signals of such LuxR solos are often presumed to be of exogenous nature and thus extremely difficult to identify. Activation of these BGCs without knowledge of the activating signal would provide a potent method for the discovery toolbox of new natural products.
As a proof of concept, we aimed at constructing a rewired, QS system that regulates a known metabolite. As a test system, we used the mupirocin (Figure 1) BGC of Pseudomonas sp. QS1027, which is regulated by a QS machinery that responds to 3‐oxo‐C10 HSL (Figure 1).16 To test the functionality of the LuxRCs with a SBD and a DBD from two different LuxRs (Figure 2), we created a chimera that contained the DBD of LasR, a well‐studied LuxR of Pseudomonas aeruginosa,17 and the SBD of the mupirocin LuxR, MupR. The vector encoding this LuxRC MupR‐SBD::LasR‐DBD was then co‐transformed into Escherichia coli DH5α along with a specific promoter probe vector (PPV) encoding the lux‐box‐containing promoter region from lasI (plasI). The gene lasI encodes the AHL synthase of the LasR/LasI QS circuit of P. aeruginosa. The expression of this system is upregulated by the transcriptional activator LasR. Thus, this LasR‐responsive promoter, when cloned upstream to the reporter gene lacZ or gene cluster luxABCDE, controls the expression of the enzyme β‐galactosidase or the luciferase gene cluster, respectively. If functional, the resulting system (E. coli co‐transformed with LuxRC MupR‐SBD::LasR‐DBD + plasI in PPV) should be activated by 3‐oxo‐C10 HSL. We subjected different LuxRC constructs to a concentration of 10 μm of 3‐oxo‐C10 HSL, and the resulting reporter fusion read‐outs were quantified. A number of variants of LuxRCs, differing in the length of SBD or DBD, were tested and the results provided an insight into the modular nature of LuxRs. It was evident that the overall size of the luxR C genes played an important role in generating a functional LuxRC. The size of LuxRC should match that of the native LuxR, which it aims to emulate; otherwise no transcriptional activity was observed (Figure 2 c,d). LuxRCs that were able to dimerize and showed good transcriptional activity, comparable to the corresponding native LuxR, (Figure 2) were then subjected to homology modeling (Figure S2a–d in the Supporting Information)
Figure 1.
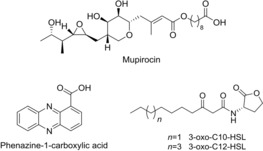
Structures of natural products from Pseudomonas spp. and AHL signal molecules.
Figure 2.
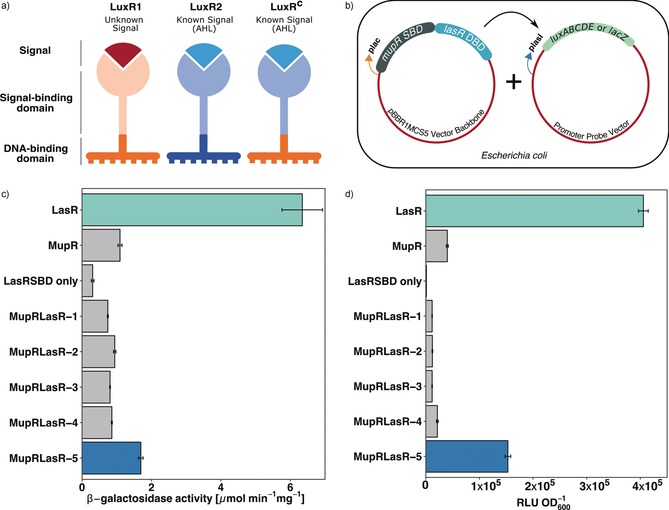
a) Schematic representation of a LuxR chimera (LuxRCs) obtained by synthetically fusing the signal‐binding domain (SBD) of one LuxR with the DNA‐binding domain (DBD) of another. b) Experimental set‐up in the heterologous host E. coli DH5α. The coding sequence of LuxRCs is inserted downstream to a constitutive promoter plac in the plasmid pBBR1MCS5, which is then cloned into a promoter probe vector (PPV) upstream to a reporter gene. 10 μm of the AHL 3‐oxo‐C10 HSL was added to the media and the culture was grown at 37 °C for 3 hours starting from OD600=0.1. LasR SBD alone served as a negative control. c) The beta‐galactosidase activity of various MupR SBD+LasR DBD LuxRCs was calculated and compared to the complete LuxR counterparts (LasR or MupR). PPV pFU62 encoding a lacZ gene downstream to plasI promoter was used. d) Bioluminescence emitted by interaction of different LuxRCs with plasI promoter introduced upstream to luxABCDE cassette in pFU36 was quantified. RLU=relative luminescence units. Differences in the biological functionality of the LuxRC variants is more evident here than in (c) because this assay is more sensitive.
These preliminary studies regarding the generation of functional LuxRCs allowed us to design LuxRCs for in‐host use. With the aim of rewiring the regulatory machinery of the mupirocin‐producing mup BGC in Pseudomonas sp. QS1027, we used a LuxRC with the LasR SBD from P. aeruginosa, which binds to 3‐oxo‐C12 HSL (Figure 1), and the MupR DBD. In principle, this would allow us to change the signal that leads to mupirocin production from 3‐oxo‐C10 HSL to 3‐oxo‐C12 HSL. Thus, we introduced luxR C variants episomally into Pseudomonas sp. QS1027 ΔmupR. Addition of the LasR signal, 3‐oxo‐C12 HSL, should then lead to the expression of the mup BGC, which in turn leads to the production of mupirocin. Indeed, addition of 3‐oxo‐C12 HSL to Pseudomonas sp. QS1027ΔmupR, which produces LuxRC LasR‐SBD::MupR‐DBD, induced the biogenesis of mupirocin (Figure 3 a), whereas addition of the original signal 3‐oxo‐C10 HSL, resulted in only marginal production levels of mupirocin (Figure S10).
Figure 3.

UHPLC‐MS‐based analysis of natural product formation by candidate luxR deletion mutants carrying luxR C variants. a) Comparison of mupirocin formation in different strain of Pseudomonas sp. QS1027 using SIM at m/z 501. WT mupirocin synthesis was set as 100 %. LuxRC LasR‐SBD::MupR‐DBD variant 2 showed the best ability to induce mupirocin production. b) Phenazine‐1‐carboxylic acid formation by P. chlororaphis WT and ΔphzR producing different LuxRC was monitored using SIM at m/z 225. LuxRC LasR‐SBD::PhzR‐DBD variant 2 showed the best ability to induce phenazine production. T‐tests were used to compare the test strains with the best functional LuxRC to the no AHL control, 3‐oxo C10 HSL control (for QS1027), and C6 HSL control (for DSM19603). ***=p‐value <0.001.
The production titers for mupirocin were surprisingly high, almost matching wild‐type (WT) production levels (Figure 3 a). This clearly showed that well‐designed LuxRC are functional and effectively enable the rewiring of natural product regulatory pathways. To further show the scope of our approach, we rewired the regulatory system linked to phenazine production in Pseudomonas chlororaphis subsp. aurantiaca (DSM19603). Production of orange‐colored phenazines, specifically phenazine‐1‐carboxylic acid, requires the LuxR PhzR. Creation of ΔphzR mutant completely abolished phenazine production resulting in a strain that makes cream‐colored colonies (Figure S4). Expression of a luxR C LasR‐SBD::PhzR‐DBD in Pseudomonas chlororaphis ΔphzR along with exogenous addition of 3‐oxo‐C12 HSL re‐established the production of phenazine‐1‐carboxylic acid (4, Figure 3 b). Here also, addition of the original signal led only to marginal production titers of phenazine (Figure 3 b).
Quorum‐sensing‐controlled natural product production is widespread amongst Gram‐negative bacteria, in particular in Pseudomonas species.18 Variants of these QS systems are known where LuxRs have no cognate signal synthase, so‐called orphan LuxRs or LuxR solos.6, 15 LuxR‐solo‐regulated BGCs are widespread yet often silent since the corresponding LuxR signal is unknown.15 The strategy described in this paper, clearly shows that it is possible to generate a functional chimeric LuxR using parts of a LuxR solo, consequently making a receptor that responds to known signals. Therefore, the next step was to demonstrate that chimeric LuxR solos can activate silent BGCs in bacterial strains. We chose a strain that we had isolated from a soil sample, Pseudomonas sp. SZ57. This strain has a large number of genomically encoded biosynthetic gene clusters, yet cultivation of this strain under different conditions led to the isolation of only one family of natural products: syringafactins A and C (Figure 4). Hidden Markov Model (HMM)‐based search for all LuxR‐like proteins present in the genome of this bacterium led us to a potential candidate, which was structurally akin to a LuxR solo (Figure 4). This putative LuxR, later named PseR, was thus chosen to be activated by the generation of a chimera. To this end, LasR SBD was fused to the DBD of this new candidate LuxR, and the LuxRC was then introduced into a wild‐type strain of Pseudomonas sp. SZ57. Production of the plasmid‐encoded LuxRC was under the control of an arabinose‐inducible promoter and it was responsive to 3‐oxo‐C12 HSL as its signal molecule. As anticipated, the introduction of this LuxRC (LuxRC LasR‐SBD::PseR‐DBD) into Pseudomonas sp. SZ57 led to the production of a compound that was not produced in the WT strain. Importantly, neither the addition of 3‐oxo‐C12 HSL alone, nor the introduction of the chimera without the respective signal led to the production of this metabolite (Figure S9).
Figure 4.
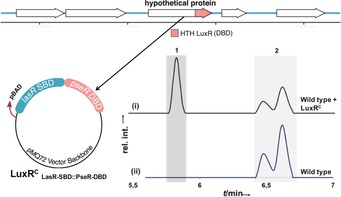
Creation of a luxRC from PseR for activation of a silent BGC. The LasR SBD was fused with the PseR DBD in the pMQ72 vector backbone and introduced into WT SZ57. The luxRC was induced with 100 mm arabinose and activated by adding 10 μm 3‐oxo‐C12 HSL. When cultured in in LB medium, SZ57 produces syringafactins A and C [peaks 2 in the HPLC spectra], and no other significant lipophilic secondary metabolites are detected. With LuxRC, an additional metabolite was produced [peak 1 in spectrum (i)]. UV detection was at λ=210 nm.
We isolated a compound with a pseudo‐molecular ion peak at m/z=876.6916, which corresponds to the molecular formula C52H94NO9. LC–MSn analysis suggested that this compound contains seven repeated hydrated isoprenoid units. Extensive spectroscopic analysis was employed to propose the structure shown in Figure 5. This previously undescribed compound is a meroterpenoid, which we named pseudomonol. Interestingly, this structural class has not been described in pseudomonads so far (Figure 5 and Figures S5, S6).
Figure 5.
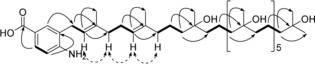
Structure of pseudomonol and selected correlations observed by 2D NMR spectroscopy. Bold lines=1H‐1H COSY correlations; solid arrows=HMBC correlations; dashed arrow=nuclear Overhauser effect (NOE) correlations.
In summary, we show that it is possible to generate functional LuxR chimera that respond to a predefined signal molecule. We have demonstrated that it is possible to rewire QS systems by changing the SBD of a LuxR. In a second step we applied this approach to generate functionally active chimeras of LuxR solos. This allowed us to activate the production of a new natural product: pseudomonol. We thus present a powerful strategy to access new secondary metabolites that would not be produced under standard laboratory conditions. Furthermore, our approach is particularly useful for synthetic biologists who want to rewire the QS‐mediated regulatory machineries and create artificial systems with precise control of signals that active specific BGCs.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We are grateful for financial support from the Leibniz Association. This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy—EXC 2051—Project‐ID 390713860 and the collaborative research cluster ChemBioSys SFB1127 as well as an Exploration Grant of the Boehringer Ingelheim Foundation (BIS). We are grateful to the Europäischer Fonds für regionale Entwicklung (EFRE) for an instrument grant and to the DECHEMA for a Max‐Buchner‐Fellowship. We also thank A. Perner and M. Flak for LC‐HRMS and LC‐MS/MS analysis, and H. Heineke for NMR measurements.
R. Mukherji, S. Zhang, S. Chowdhury, P. Stallforth, Angew. Chem. Int. Ed. 2020, 59, 6192.
References
- 1. Newman D. J., Cragg G. M., Snader K. M., Nat. Prod. Rep. 2000, 17, 215–234. [DOI] [PubMed] [Google Scholar]
- 2. Newman D. J., Cragg G. M., J. Nat. Prod. 2012, 75, 311–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown E. D., Wright G. D., Nature 2016, 529, 336–343. [DOI] [PubMed] [Google Scholar]
- 4. Winter J. M., Behnken S., Hertweck C., Curr. Opin. Chem. Biol. 2011, 15, 22–31. [DOI] [PubMed] [Google Scholar]
- 5. Rutledge P. J., Challis G. L., Nat. Rev. Microbiol. 2015, 13, 509–523. [DOI] [PubMed] [Google Scholar]
- 6. Brotherton C. A., Medema M. H., Greenberg E. P., MSystems 2018, 3, e00208-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McRose D. L., Baars O., Seyedsayamdost M. R., Morel F. M. M., Proc. Natl. Acad. Sci. USA 2018, 115, 7581–7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Popat R., Harrison F., da Silva A. C., Easton S. A. S., McNally L., Williams P., Diggle S. P., Philos. Trans. R. Soc. London Ser. B 2017, 284, 20170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Williams P., Microbiology 2007, 153, 3923–3938. [DOI] [PubMed] [Google Scholar]
- 10. Klaus J. R., Deay J., Neuenswander B., Hursh W., Gao Z., Bouddhara T., Williams T. D., Douglas J., Monize K., Martins P., et al., J. Bacteriol. 2018, 200, e00008-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Higgins S., Heeb S., Rampioni G., Fletcher M. P., Williams P., Cámara M., Front. Cell. Infect. Microbiol. 2018, 8, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Churchill M. E. A., Chen L., Chem. Rev. 2011, 111, 68–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Whiteley M., Greenberg E. P., J. Bacteriol. 2001, 183, 5529–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmidl S. R., Ekness F., Sofjan K., Daeffler K. N.-M., Brink K. R., Landry B. P., Gerhardt K. P., Dyulgyarov N., Sheth R. U., Tabor J. J., Nat. Chem. Biol. 2019, 15, 690–698. [DOI] [PubMed] [Google Scholar]
- 15. Subramoni S., Venturi V., Microbiology 2009, 155, 1377–1385. [DOI] [PubMed] [Google Scholar]
- 16. Arp J., Götze S., Mukherji R., Mattern D. J., García-Altares M., Klapper M., Brock D. A., Brakhage A. A., Strassmann J. E., Queller D. C., Bardl B., Willing K., Peschel G., Stallforth P., Proc. Natl. Acad. Sci. USA 2018, 115, 3758–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kiratisin P., Tucker K. D., Passador L., J. Bacteriol. 2002, 184, 4912–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gross H., Loper J. E., Nat. Prod. Rep. 2009, 26, 1408–1446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


