Summary
Mining operations produce large quantities of wastewater. At a mine site in Northern Finland, two natural peatlands are used for the treatment of mining‐influenced waters with high concentrations of sulphate and potentially toxic arsenic (As). In the present study, As removal and the involved microbial processes in those treatment peatlands (TPs) were assessed. Arsenic‐metabolizing microorganisms were abundant in peat soil from both TPs (up to 108 cells gdw −1), with arsenate respirers being about 100 times more abundant than arsenite oxidizers. In uninhibited microcosm incubations, supplemented arsenite was oxidized under oxic conditions and supplemented arsenate was reduced under anoxic conditions, while little to no oxidation/reduction was observed in NaN3‐inhibited microcosms, indicating high As‐turnover potential of peat microbes. Formation of thioarsenates was observed in anoxic microcosms. Sequencing of the functional genemarkers aioA (arsenite oxidizers), arrA (arsenate respirers) and arsC (detoxifying arsenate reducers) demonstrated high diversity of the As‐metabolizing microbial community. The microbial community composition differed between the two TPs, which may have affected As removal efficiencies. In the present situation, arsenate reduction is likely the dominant net process and contributes substantially to As removal. Changes in TP usage (e.g. mine closure) with lowered water tables and heightened oxygen availability in peat might lead to re‐oxidation and re‐mobilization of bound arsenite.
Introduction
Mining operations produce large amounts of wastewater, which have to be purified prior to their release into the environment (Ledin and Pedersen, 1996). Mining‐affected waters contain a variety of contaminants. The contaminant content depends on the type of ore mined and on the ore beneficiation processes. Typical contaminants in mining‐affected waters include nitrogen compounds (from remnant explosives or the ore beneficiation process), sulphate (from oxidation of sulphidic ores) as well as (potentially toxic) metals and metalloids (Nordstrom, 2011). Arsenic (As) minerals often accompany gold and copper ores, since they all share the same chalcophillic behaviour, and beneficiation of these ores can release As via process waters into the environment (Matschullat, 2000; Bissen and Frimmel, 2003; Nordstrom, 2011). Here, the mobility, bioavailability and toxicity of As strongly depends on As speciation. In most terrestrial environments, the oxyanions arsenate (HxAsVO4 x‐3, x = 1–3) and arsenite (HxAsIIIO3 x‐3, x = 1–3) dominate under oxidizing and reducing conditions respectively (Bissen and Frimmel, 2003).
Wetlands, including natural peatlands, are widely used for the treatment of different kinds of runoffs and wastewaters, as they retard the water flow and provide a large active surface area (Ledin and Pedersen, 1996; Sheoran and Sheoran, 2006; Vymazal, 2011). Contaminants are removed from the water by plant uptake, microbially catalysed redox‐processes, precipitation and adsorption onto plant roots and soil particles (Sheoran and Sheoran, 2006).
Under oxic conditions, mostly in the upper peat layers, As can adsorb to (oxyhydr‐)oxides of iron, manganese, or aluminium (Bissen and Frimmel, 2003). Under suboxic to anoxic conditions and in the presence of sulphur, (hydr‐)oxides may dissolve. The released or newly formed arsenite can either react with zerovalent sulphur and sulphide to form thioarsenates (HxAsVS‐II nO4− nx‐3; n = 1–4; x = 1–3) at neutral to alkaline pH (Planer‐Friedrich et al., 2015; Besold et al., 2018) or bind to natural organic matter (NOM) via sulfhydryl groups at slightly acidic pH (Hoffmann et al., 2012; Langner et al., 2012; Besold et al., 2018). At strongly acidic pH and in extremely reducing microenvironments arsenite eventually precipitate as As‐sulphides. While at acidic conditions, thiol‐binding and As‐sulphide formation are considered rather stable, even if the peat gets intermittently oxic (Langner et al., 2012, 2014), thioarsenate formation may lead to (re‐)mobilization of As at neutral to alkaline pH in presence of reduced inorganic sulphur (Besold et al., 2018).
Microorganisms can use the oxoacids of As in their metabolism (Oremland and Stolz, 2003; Stolz et al., 2006). Arsenite oxidizing microorganisms (e.g. found within the Proteobacteria, Deinococci and Crenarchaeota) conserve energy from the oxidation of arsenite to arsenate and can be autotrophs or heterotrophs (Oremland and Stolz, 2003). Strains capable of arsenite oxidation have been isolated from soils, sediments and As contaminated waters (Salmassi et al., 2006; Garcia‐Dominguez et al., 2008; Osborne et al., 2010), and arsenite oxidizing bacteria can be used for bioremediation purposes (Battaglia‐Brunet et al., 2002). The arsenite oxidase (AioAB) is the key enzyme in arsenite oxidation, and aioA has been frequently used as a genetic marker for arsenite oxidation (e.g. Inskeep et al., 2007; Escudero et al., 2013; Hamamura et al., 2013; Zhang et al., 2015). In wetlands, arsenite oxidation is likely restricted to the uppermost layers, as the water‐saturated conditions prevent diffusion of oxygen into deeper layers.
Under anoxic conditions, microorganisms can conserve energy via dissimilatory reduction of arsenate to arsenite (i.e. arsenate respiration; Oremland and Stolz, 2003; Stolz et al., 2006). Microbial arsenate reduction has been observed in many anoxic soils and sediments (Dowdle et al., 1996; Kulp et al., 2006; Hery et al., 2008), and arsenate respirers have been isolated from these systems (Hery et al., 2008; Kudo et al., 2013; Abin and Hollibaugh, 2017). The dissimilatory arsenate reductase (ArrAB) is the key enzyme in arsenate respiration, and arrA has been frequently used as a genetic marker (e.g. Malasarn et al., 2004; Lear et al., 2007; Song et al., 2009; Escudero et al., 2013). Arsenate respiration is feasible in lower layers of wetlands, where oxygen is scarce and reducing conditions prevail. Furthermore, arsenate can be reduced by the detoxifying arsenate reductase ArsC (Oremland and Stolz, 2003), and arsC has been used as a genetic marker for As detoxification (e.g. Escudero et al., 2013; Costa et al., 2014; Zhang et al., 2015). By catalysing kinetically limited transformations of As species, microorganisms can control the retention of As in treatment peatlands (TPs) and can thus be an important factor in the As biogeochemistry in such systems.
Efficient removal of As in TPs is observed in the field (Palmer et al., 2015; Kujala et al., 2018), but the role of microorganisms in As removal remains elusive. To fill this knowledge gap, As concentrations and As speciation in surface and porewaters of two TPs treating mine process waters (TP A) and drainage waters (TP B) were assessed in the field to elucidate As transformation processes. Then, the potential of peat soil from different depths to oxidize/reduce arsenite/arsenate was investigated in laboratory incubations in order to elaborate potential links of As field speciation to microbial activity. Moreover, microorganisms capable of As turnover and resistance were quantified and the microbial community composition was determined to see how numerous and diverse As‐cycling microorganisms are in TPs and to identify potential key players in As species transformation.
The obtained results about the role of microorganisms in As removal provide valuable information for the continued use of TPs in the treatment of mining‐affected waters.
Results
Total As concentrations and speciation in TPs
As concentrations in surface water and porewater were assessed in two TPs, one receiving mine process water (TP A) and receiving drainage water (TP B; Fig. S1, see Materials and Methods for full description of TPs). Total As concentrations (Astot) in surface water were 21–28 and 33 μg L−1 near the inlet of TP A and TP B respectively (Table S1). In TP A, Astot decreased rapidly with increasing distance from the inlet (TP A1 ➔ TP A7; Fig. S1) to mostly below 1 μg L−1. In TP B, there was only a slight decrease to 28 μg L−1 close to the outlet (TP B3; Fig. S1). Arsenate was the dominant As species in all surface water samples for which As speciation was analysed (TP A1, A2 and TP B1, B2, B3). However, the arsenite contribution increased in TP B with increasing distance from the inlet (TP B1 ➔ TP B3) from below detection limit (TP B1) to 16% of detected species (TP B3; Table S1).
Total As concentrations in the porewater at sampling points/depths used for incubations and microbial community studies ranged from 6.1 to 132 μg L−1 (Table S2). In porewater samples, arsenate was the dominant species at 10 cm depth near the inlet (TP A1), while arsenite was the dominant species at 60 cm depth (Table S2). In addition, inorganic thiolated, methylated and methylthiolated arsenate species were detected in the TP A1 profile, while only methylated As species and arsenite were detected in porewater samples from TP A5 (73%–75% and 6%–7% of detected As species respectively).
Arsenic‐utilization potential of peat microorganisms
The potential of peat microorganisms to oxidize and reduce 50 μM supplemented arsenite and arsenate to peat suspensions respectively, was assessed in microcosm incubations prepared with deionized water. Under oxic conditions, supplemented arsenite was oxidized to arsenate completely within 9 days of incubation in uninhibited microcosms (Fig. 1B). Arsenite oxidation started after 2 days of incubation, indicating a short lag phase in the beginning. In microcosms where sodium azide (NaN3) had been added as an inhibitor, no arsenite oxidation was observed within the 18‐day incubation period (Fig. 1A), indicating that arsenite oxidation was catalysed by microorganisms. NaN3 is a common inhibitor for microbial growth and activity (e.g. Cabrol et al., 2017), which has also been used to inhibit As redox transformations in many As‐NOM studies (Buschmann et al., 2006; Hoffmann et al., 2012; Besold et al., 2018).
Figure 1.
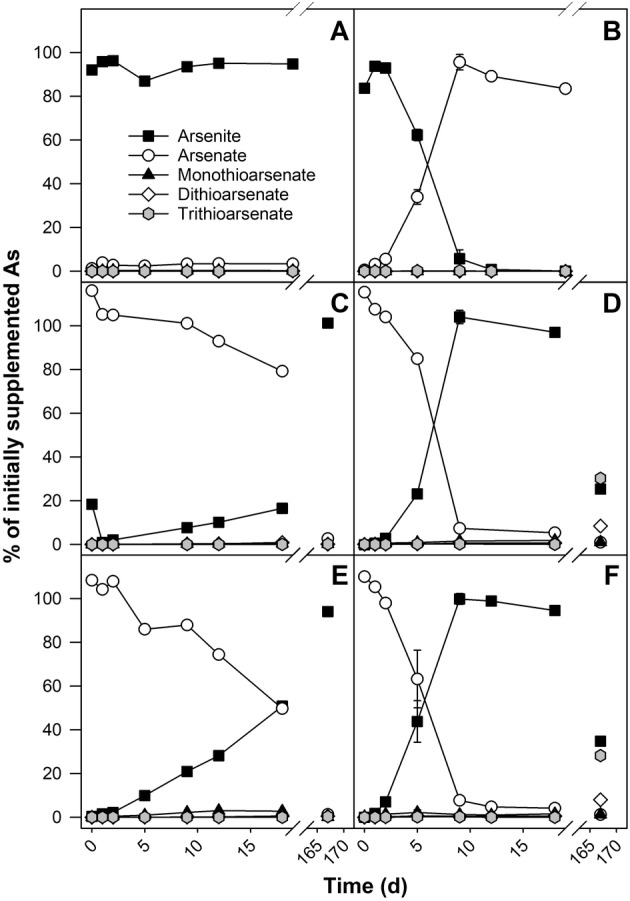
Arsenite‐oxidation (A, B) and arsenate‐reduction (C–F) potential of peat microorganisms. Incubations were conducted under oxic (A, B) or anoxic (C–F) conditions with peat from 0 to 10 cm (A–D) and 60 to 70 cm (E, F) depth. Microcosms were supplemented with NaN3 to inhibit microbial activity (A, C, E) or left without inhibitor (B, D, F) and 50 μM arsenite (A, B) or arsenate (C–F). Incubations were done in triplicate. One replicate was analysed at all sampling timepoints, triplicates were analysed at selected timepoints. Error bars indicate standard errors in cases in which triplicates were analysed.
Under anoxic conditions, supplemented arsenate was reduced to arsenite completely within 9 days of incubation in uninhibited microcosms with peat from both tested depths (Fig. 1D,F). Arsenate reduction started after a lag phase of 2 days. Only minor arsenate reduction was observed in NaN3 supplemented microcosms with peat soil from 0 to 10 cm depth, while about half of the supplemented arsenate was reduced to arsenite in NaN3 supplemented microcosms with peat soil from 60 to 70 cm depth (Fig. 1C,E). This finding indicates that arsenate reduction was not completely inhibited by NaN3 in peat soil from 60 to 70 cm depth, which might be due to (i) incomplete inhibition caused by rather low NaN3 concentrations, (ii) microorganisms that were not affected by NaN3, or (iii) abiotic arsenate reduction with considerably lower reaction rates. Moreover, the inhibitory effect of NaN3 was not permanent: After 167 days of incubation, arsenate had been reduced to arsenite in the NaN3 supplemented incubations of both depths (Fig. 1C,E).
Sulphide production potential of peat microorganisms
Sulphide production potentials were determined in anoxic microcosms with 0–10 cm peat soil from TP A5 prepared with deionized water and additionally in incubations prepared with mine process water. Initial sulphide concentrations in mine process water were below the detection limit of the assay used for the determination of sulphide concentrations. Sulphide accumulated only in microcosms without supplemented NaN3 (Fig. 2), indicating that NaN3 almost completely inhibited sulphate reduction in peat soil.
Figure 2.
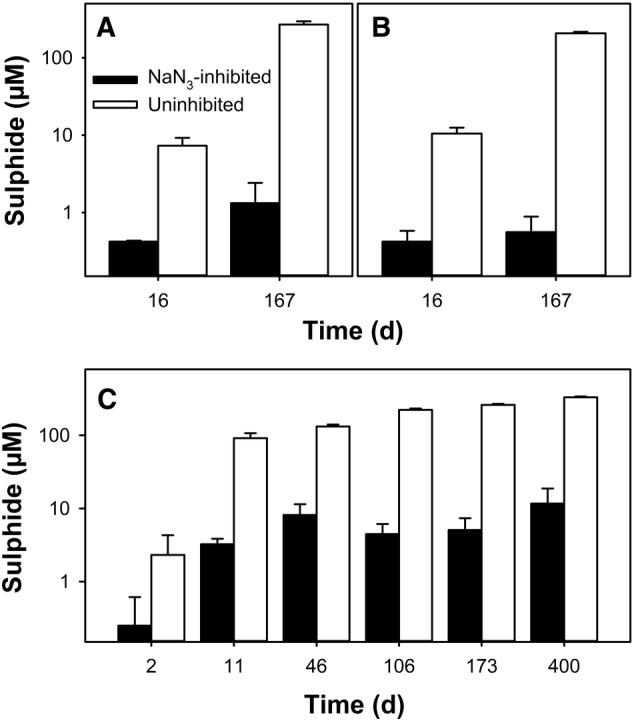
Sulphide production in anoxic microcosms with peat soil from TP A1. Incubations were set up in triplicate with peat from 0 to 10 cm (A, C) and 60 to 70 cm (B). Microcosms were prepared with deionized water (A, B) or mine process water (C). Mean values with standard errors are displayed.
Using deionized water, 270 and 210 μM sulphide accumulated within 167 days of incubation in microcosms with 0–10 and 60–70 cm peat respectively (Fig. 2A and 2B). In microcosms set up with mine process water, 265 μM sulphide had accumulated after 173 days and 330 μM after 433 days of incubation (Fig. 2C). Accumulation of sulphide was faster in microcosms with mine process water than in microcosms with deionized water. This difference occurred possibly due to the more readily available sulphate in incubations with mine process water (as mine process water contains 2100 mg L−1 sulphate). In fact, 91 μM sulphide accumulated within 11 days in microcosms with mine process water, while only 7 μM sulphide accumulated within 16 days in microcosms with deionized water (Fig. 2A,C). However, sulphide accumulation in microcosms with mine process water started to level out at later time points, indicating lower sulphate reduction rates.
Thioarsenate production potential in peat microcosms
Thioarsenate formation was assessed in anoxic microcosms prepared with deionized water and with mine process water. Deionized water was used to avoid possible interference of mine process water contaminants with the formation process and to allow comparison to the arsenite oxidation and arsenate reduction potentials assessed earlier, while mine process water was used to assess the influence of increased sulphate concentrations (2100 mg L−1 in mine process water) on thioarsenate formation. Thioarsenates started to accumulate in uninhibited anoxic microcosms with increasing incubation time. After 167 days of incubation, more than half of the detected As was in the form of thioarsenates in microcosms set up with deionized water, with trithioarsenate being the most abundant species (Fig. 1B,C). In uninhibited anoxic microcosms with mine process water, thioarsenates likewise accumulated with time (Fig. 3B). 68% of the detected As species were thioarsenates after 420 days of incubation, and trithioarsenate was the dominant species as well. No thioarsenates were detected in the NaN3 inhibited incubations with deionized or mine process water (Figs 1C,E and 3A).
Figure 3.
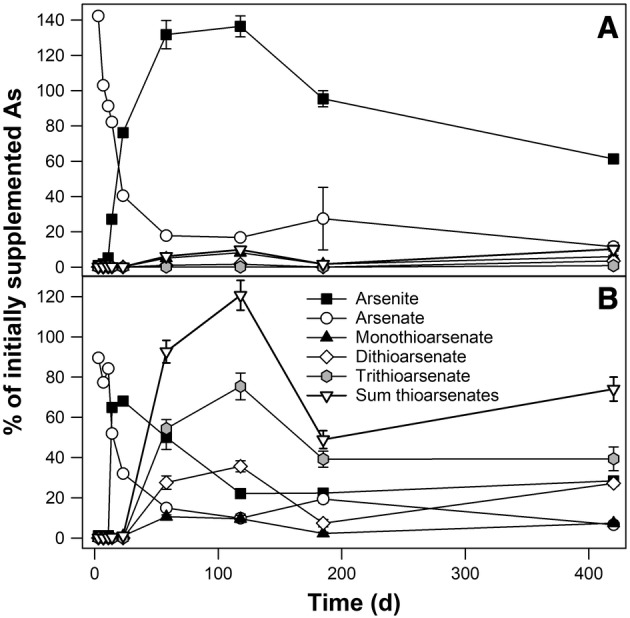
Influence of mine process water on thioarsenate production in anoxic peat microcosms. Triplicate incubations were prepared with 0–10 cm peat soil (TP A1) and mine process water and were supplemented with 50 μM arsenate. Microcosms were supplemented with NaN3 to inhibit microbial activity (A) or left without inhibitor (B). One replicate was analysed at all sampling timepoints, triplicates were analysed at selected timepoints. Error bars indicate standard errors in cases in which triplicates were analysed. At day 185, small amounts of MMA(V) and DMDTA(V) were detected in microcosms without NaN3.
Enumeration of As‐metabolizing microorganisms
As‐tolerating aerobic/aerotolerant microorganisms were abundant in TP A and TP B based on most probable number (MPN) counts (Fig. 4). Up to 3.8 × 107 cells gDW −1 were detected in 0–10 cm peat soil. The cell numbers obtained were slightly (but not significantly) higher with TSB than with the NB growth medium. MPNs of arsenite‐oxidizing microorganisms were about two orders of magnitude lower than for As‐tolerating microorganisms and approximated 1.5 × 105 cells gDW −1 in 0–10 cm peat soil. MPNs of arsenate‐respiring microorganisms ranged from 1.6 × 106 cells gDW −1 to 8.6 × 106 cells gDW −1 (Fig. 4), indicating that arsenate‐respiring microorganisms were more abundant than arsenite‐oxidizing microorganisms in peat soil of both layers. MPNs of As‐tolerating and arsenite‐oxidizing microorganisms were slightly (but not significantly) lower in 60–70 cm peat soil, while this trend was not observed for arsenate‐respiring microorganisms.
Figure 4.
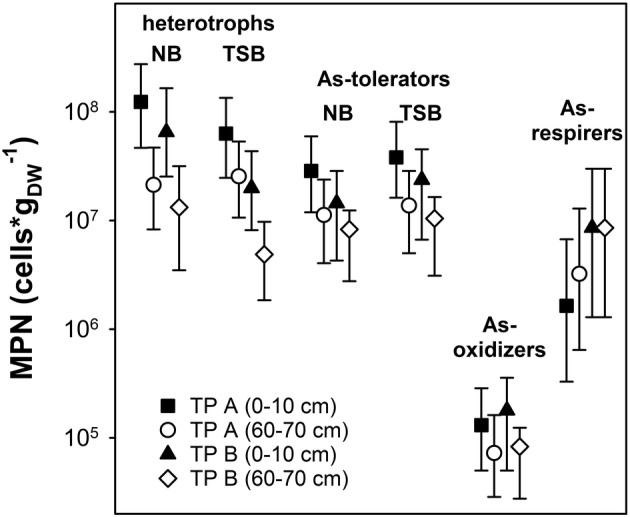
Most probable numbers (MPNs) of aerobic heterotrophic, arsenic‐tolerating, arsenite‐oxidizing and arsenate‐reducing prokaryotes in peat soil from TP A1 and TP B2. MPNs and 95% confidence intervals are shown. Incubations were conducted under oxic (aerobic heterotrophs, arsenic‐tolerating and arsenite‐oxidizing) or anoxic (arsenate‐reducing) conditions. Eight replicates were used for MPNs of aerobic heterotrophs, arsenic‐tolerating microorganisms and arsenite‐oxidizing microorganism; three replicates were used for MPNs of arsenate‐reducers. Aerobic heterotrophs and arsenic‐tolerating MPNs were estimated using two different growth media (nutrient broth = NB and tryptic soy broth = TSB).
Functional genes involved in As metabolism in TPs
Amplicon sequence libraries were obtained from two points (one in TP A and one in TP B) and two depths per TP in four replicates (i.e. total number of 16 amplicon libraries per primer set) for bacterial 16S rRNA genes and genes involved in As turnover (aioA, arrA and arsC) with eight different primer sets (Table 1). Since two nested approaches for arrA (As1f/r + As2f/r and As1f/2r + As 2f/r) used the same primers in the second PCR step, the sequences from those approaches could not be distinguished after sequencing and are thus combined in the same library. The percentages of correct sequences (i.e. sequences that match the targeted gene) were on average 50%, 93% and 97% for aioA, arrA and arsC respectively. The dada2 algorithm used for quality filtering created high‐quality amplicon sequence variants (ASVs). For the following analyses, these ASVs were used unclustered. Amplicon sequencing of genes involved in As turnover was most successful with the two primer sets targeting arsC (arsC1 and arsC2), which yielded a total of 178 470 sequences and 895 ASVs for the non‐rarefied dataset (arsC1 + arsC2) after quality filtering and removal of non‐target sequences. For aioA (aioA1 and aioA2) and arrA (arrA1 and arrA2), less sequences were obtained, with 39 691 sequences in 582 ASVs for aioA (aioA1 + aioA2) and 65 839 sequences in 418 ASVs for arrA (arrA1 + arrA2). PERMANOVA analysis indicated differences in community composition obtained with different primer sets for each gene (p < 0.001), indicating that the use of two different primer systems per gene likely captured a higher diversity than the use of a single primer system.
Table 1.
Primers, PCR reactions and PCR conditions used to PCR amplify fragments of aioA, arsC, arrA and 16S rRNA genes for IonTorrent sequencing.
| Gene | PCR‐type | Primer set | Name for primer set in this study | Forward primer | Reverse primer | PCR conditions | PCR product length (bp) | References |
|---|---|---|---|---|---|---|---|---|
| aioA | Initial amplification | M13‐aroA #1F/aroA #1R | aioA1 | TGTAAAACGACGGCCAGTGTSGGBTGYGGMTAYCABGYCTA | TTGTASGCBGGNCGRTTRTGRAT | 95°C 10 min, 35 cycles (95°C/45 s, 50°C/45 s, 72°C/90 s) 72°C/5 min | 500 | Inskeep et al. (2007) |
| Initial amplification | M13‐aroA #2F/aroA #2R | aioA2 | TGTAAAACGACGGCCAGTGTCGGYYGYGGMTAYCAYGYYTA | YTCDGARTTGTAGGCYGGBCG | 95°C 10 min, 35 cycles (95°C/45 s, 50°C/45 s, 72°C/90 s) 72°C/5 min | 500 | Inskeep et al. (2007) | |
| arsC | Initial amplification | M13‐arsC‐4f/arsC‐4r | arsC1 | TGTAAAACGACGGCCAGTTCHTGYCGHAGYCAAATGGCHGAAG | GCCATGCACCWCCTCT | 95°C/5 min, 35 cycles (95°C/90 s, 46°C/90 s, 72°C/3 min), 72°C/5 min | 300–400 | Escudero et al. (2013) |
| Initial amplification | M13‐arsC‐5f/arsC‐5r | arsC2 | TGTAAAACGACGGCCAGTGGHAAYTCHTGYCGNAGYCAAATGGC | GCNGGATCVTCRAAWCCCCARNWC | 95°C/5 min, 35 cycles (95°C/90 s, 58°C/90 s, 72°C/3 min), 72°C/5 min | 300–400 | Escudero et al. (2013) | |
| arrA | Initial amplification | AS1f/AS1r | arrA1 | CGAAGTTCGTCCCGATHACNTGG | GGGGTGCGGTCYTTNARYTC | 95°C 10 min, 35 cycles (95°C/45 s, 50°C/45 s, 72°C/90 s) 72°C/5 min | Lear et al. (2007) | |
| Nested amplification | M13‐AS2f/AS1r | arrA1 | TGTAAAACGACGGCCAGTGTCCCNATBASNTGGGANRARGCNMT | GGGGTGCGGTCYTTNARYTC | 95°C 10 min, 30 cycles (95°C/45 s, 55°C/45 s, 72°C/90 s) 72°C/5 min | 625 | Lear et al. (2007) | |
| Initial amplification | AS1f/AS2r | arrA1 | CGAAGTTCGTCCCGATHACNTGG | ATANGCCCARTGNCCYTGNG | 95°C 10 min, 35 cycles (95°C/45 s, 55°C/45 s, 72°C/2 min) 72°C/5 min | Song et al. (2009) | ||
| Nested amplification | M13‐AS2f/AS2r | arrA1 | TGTAAAACGACGGCCAGTGTCCCNATBASNTGGGANRARGCNMT | ATANGCCCARTGNCCYTGNG | 95°C 10 min, 30 cycles (95°C/45 s, 55°C/45 s, 72°C/90 s) 72°C/5 min | 625 | Song et al. (2009) | |
| Initial amplification | M13‐ArrPSRfwd/ArrPSRrev | arrA2 | TGTAAAACGACGGCCAGTAGTTCGTSCCSATCWSSTGGGAC | ACTCSGGSGTSYKGTCCTTSAG | 95°C 5 min, 35 cycles (95°C/60 s, 59°C/60 s, 72°C/90 s) 72°C/5 min | 550–600 | Kudo et al. (2013) | |
| All functional genes | Addition of sequencing adapters | IonA_IonXpressBarcode_M13/P1_reverse primer | CCATCTCATCCCTGCGTGTCTCCGAC‐barcode‐TAAAACGACGGCCAGT | CCTCTCTATGGGCAGTCGGTGAT‐ reverse primer | Same temperatures as for initial PCR, 10 cycles | Kujala et al. (2018); Mäki et al. (2016) | ||
| Bacterial 16S rRNA gene | Initial amplification | 27f/338r | 16S | AGAGTTTGATCMTGGCTCAG | TGCTGCCTCCCGTAGGAGT | 95°C 10 min, 30 cycles (95°C/30 s, 52°C/30 s, 72°C/60 s) 72°C/10 min | 300 | Universal primers |
| Addition of barcodes and sequencing adapters | IonA_IonXpressBarcode_27f/P1_338r | CCATCTCATCCCTGCGTGTCTCCGAC‐barcode‐AGAGTTTGATCMTGGCTCAG | CCTCTCTATGGGCAGTCGGTGAT TGCTGCCTCCCGTAGGAGT | Same temperatures as for initial PCR, 10 cycles | Kujala et al. (2018); Mäki et al. (2016) |
All amplifications were run in a CFX96 touch qPCR cycler (Bio‐Rad).
For the estimation of diversity indicators that are comparable between different samples, genes and primer sets, ASV was tables rarified to a depth of 600 sequences. In the rarified ASV tables, the number of observed ASVs was highest for arsC (Table 2). While for aioA the number of detected ASV was only slightly lower, significantly less ASV were detected for arrA (p = 0.04; Table 2). Similarly, Faith's PD was significantly higher for aioA and arsC than for arrA (p < 0.01), indicating that the phylogenetic diversity (PD) recovered was higher for aioA and arsC. Shannon diversity was only slightly higher for arrA and arsC than for aioA (not significant), while Evenness was significantly higher for arrA and arsC than for aioA (p < 0.05).
Table 2.
Diversity of different microbial groups along the gradients in TP A and TP B.
| No. of sequences | ASVs | Faith PD | Shannon | Evenness | |||
|---|---|---|---|---|---|---|---|
| (observed) | |||||||
| Arsenite oxidase (aioA) | aioA1 (220)a | TP A (0–10 cm) | 3102 (1901–3696) | 38 ± 15 | 7.33 ± 2.25 | 3.29 ± 1.03 | 0.63 ± 0.13 |
| TP A (60–70 cm) | 2298 (398–3497)b | 29 ± 13 | 7.93 ± 3.08 | 2.43 ± 0.92 | 0.50 ± 0.12 | ||
| TP B (0–10 cm) | 1746 (951–3570) | 34 ± 5 | 6.58 ± 1.06 | 4.12 ± 0.75 | 0.81 ± 0.16 | ||
| TP B (60–70 cm) | 1025 (197–2964)b | 25 | 8.23 | 2.13 | 0.46 | ||
| aioA2 (220) | TP A (0–10 cm) | 729 (263–1145)b | 24 ± 1 | 7.05 ± 1.38 | 3.99 ± 0.11 | 0.87 ± 0.01 | |
| TP A (60–70 cm) | 887 (167–2897)c | 20 | 3.58 | 2.27 | 0.52 | ||
| TP B (0–10 cm) | 1853 (346–3341)b | 21 ± 5 | 7.12 ± 2.04 | 2.3 ± 0.82 | 0.54 ± 0.25 | ||
| TP B (60–70 cm) | 767 (106–1847)c | 9 ± 6 | 6.32 ± 1.99 | 0.51 ± 0.31 | 0.16 ± 0.05 | ||
| Dissimilatory arsenate reductase (arrA) | arrA1 (220) | TP A (0–10 cm) | 1944 (768–4362) | 12 ± 3 | 4.36 ± 2.32 | 2.45 ± 0.94 | 0.69 ± 0.21 |
| TP A (60–70 cm) | 1626 (764–2245) | 11 ± 3 | 4.29 ± 0.78 | 2.32 ± 0.49 | 0.67 ± 0.08 | ||
| TP B (0–10 cm) | 5476 (4769–6050) | 29 ± 5 | 4.68 ± 2.07 | 3.96 ± 0.33 | 0.82 ± 0.03 | ||
| TP B (60–70 cm) | 3408 (2057–4509) | 19 ± 4 | 3.67 ± 0.98 | 3.4 ± 0.36 | 0.8 ± 0.04 | ||
| arrA2 (220) | TP A (0–10 cm) | 488 (231–856)d | 13 | 4.34 | 2.96 | 0.80 | |
| TP A (60–70 cm) | 635 (444–977)c | 21 | 4.36 | 3.74 | 0.85 | ||
| TP B (0–10 cm) | 1821 (1493–2153) | 17 ± 5 | 2.51 ± 0.43 | 3.23 ± 0.34 | 0.8 ± 0.03 | ||
| TP B (60–70 cm) | 1353 (1016–2042) | 21 ± 7 | 4.71 ± 2.25 | 3.54 ± 0.57 | 0.81 ± 0.04 | ||
| Detoxifying arsenate reductase (arsC) | arsC1 (270) | TP A (0–10 cm) | 4857 (4455–5279) | 23 ± 5 | 6.41 ± 0.92 | 3.15 ± 0.13 | 0.70 ± 0.02 |
| TP A (60–70 cm) | 5672 (4349–6855) | 26 ± 2 | 8.24 ± 0.85 | 3.36 ± 0.28 | 0.71 ± 0.04 | ||
| TP B (0–10 cm) | 3587 (2544–4149) | 21 ± 7 | 6.79 ± 1.72 | 2.77 ± 0.51 | 0.64 ± 0.04 | ||
| TP B (60–70 cm) | 3647 (2320–5191) | 17 ± 5 | 6.33 ± 1.04 | 2.80 ± 0.20 | 0.69 ± 0.05 | ||
| arsC2 (270) | TP A (0–10 cm) | 6470 (2092–8350) | 25 ± 12 | 7.71 ± 5.15 | 3.36 ± 1.22 | 0.74 ± 0.16 | |
| TP A (60–70 cm) | 6022 (4454–7721) | 30 ± 6 | 13.75 ± 3.29 | 3.90 ± 0.30 | 0.80 ± 0.04 | ||
| TP B (0–10 cm) | 10,446 (8961–12,068) | 48 ± 11 | 13.11 ± 4.90 | 4.34 ± 0.55 | 0.78 ± 0.05 | ||
| TP B (60–70 cm) | 6385 (3445–8165) | 27 ± 8 | 7.93 ± 2.23 | 3.79 ± 0.37 | 0.81 ± 0.03 |
Number of sequences is obtained from the original ASV tables, while all other diversity indicators are based on rarified ASV tables (rarefied at a depth of 600 sequences).
Average values of 1–4 replicates per sampling point are given.
Number in parenthesis indicates sequence length after dada2.
Three replicates used for diversity calculations.
Two replicates used for diversity calculations.
One replicate used for diversity calculations.
Phylogenetic analysis indicated five, seven and 11 groups for aioA, arrA and arsC respectively, with individual groups containing 2–44 ASVs or 0.5%–34% of all sequences per gene (Fig. 5). The aioA ASVs were related to Burkholderia multivorans and Sinorhizobium sp. (group 1; 34%), Thiobacillus sp. and Bradyrhizobium sp. (group 3; 8.2%), Pseudomonas arsenicoxydans, Thiomonas sp. and Ralstonia syzygii (group 4; 3.7%) as well as to Limnobacter sp. and Hydrogenophaga sp. (group 5; 1.1%; Fig. 5A). The arrA ASVs were related to arrA of Sulfuritalea hydrogenivorans and Exiguobacterium sp. (groups 1 and 4; Fig. 5B), Geobacter spp. and Desulfuromonas sp. (group 3), Geobacter uraniireducens, Citrobacter sp. and Aeromonas sp. (group 6), or were not closely related to arrA of cultured organisms (groups 2, 5 and 7). The arsC ASVs were related to Syntrophobacter sp., Syntrophus spp. and Smithella sp. (groups 1 and 5; Fig. 5C), Sedimentisphaera spp. and Chitinispirillum alkaliphilum (group 2), fungal arsC (group 3), Bosea thiooxidans (group 6), Bacillus spp. and Lactobacillus spp. (group 7) and Geobacillus pelophilus (group 8). Sequences of group 4 were not closely related to arsC of cultured organisms. 16S rRNA gene amplicons were dominated by Chloroflexi, Proteobacteria and Acidobacteria (Fig. 6).
Figure 5.
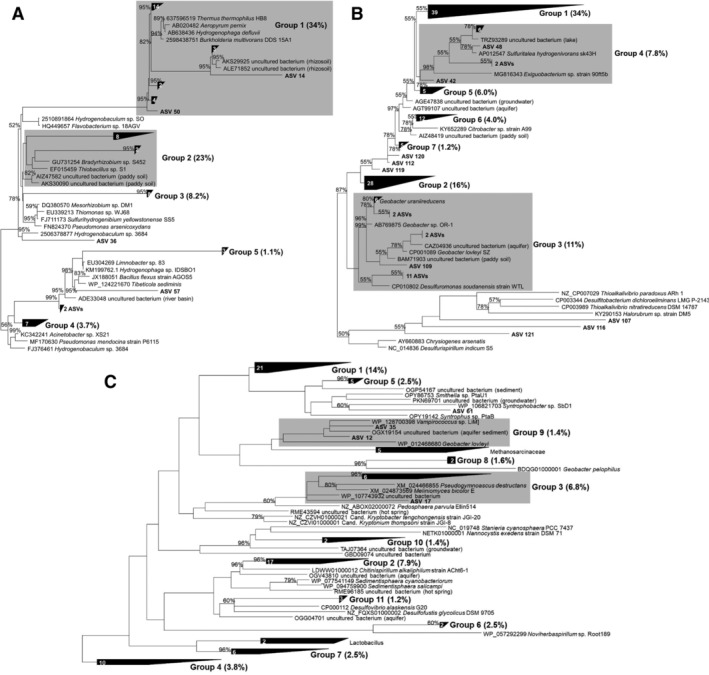
Phylogenetic trees of aioA (A), arrA (B) and arsC (C) representative sequences detected in TPs (TP A1 and TP B2). Reference sequences from cultured species and uncharacterized microorganisms were obtained from public databases. Neighbour Joining trees were constructed in ARB from translated amino acid sequences of full‐length references sequences using frequency‐based position filters (390, 410 and 115 alignment positions used for aioA, arrA and arsC respectively), and own sequences were added to the reference trees using ARB parsimony and frequency‐based position filters tailored to the length of the added sequences. ASVs generated with the two primer sets per gene were combined after normalization to relative abundances. Bootstrap values (1000 replications) are indicated, bootstrap values <50% have been omitted. Reference sequences not closely related to TP ASVs were omitted from the final tree representation to improve readability.
Figure 6.
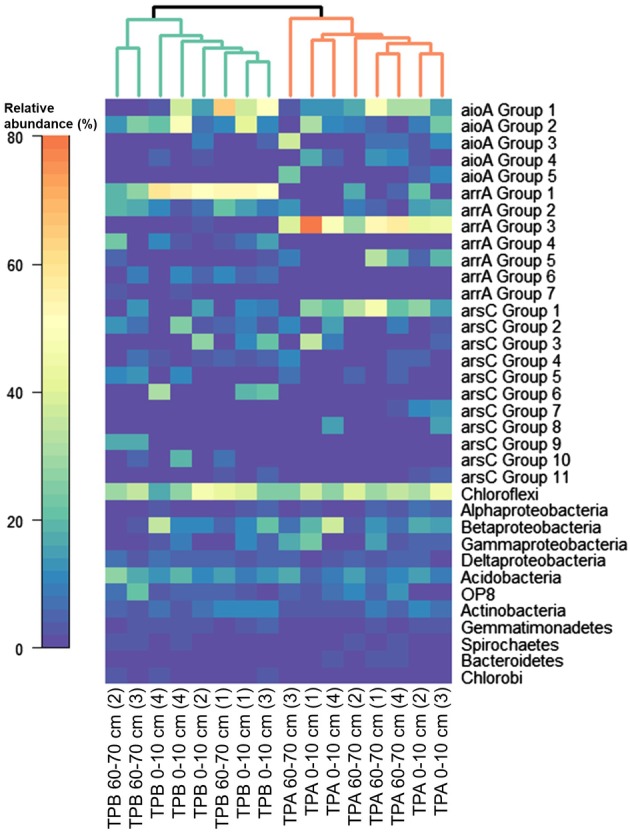
Heatmap showing the relative abundance of aioA, arrA and arsC groups as well as 16S rRNA gene phyla in TPs receiving mining‐affected waters (TP A1 and TP B2). The heatmap was generated in R (‘annHeatmap2’) from relative abundance data of the observed groups. Average relative abundances per group were calculated from relative abundances obtained for the two primer sets per gene. Columns were clustered using average linkage hierarchical clustering based on the Bray–Curtis dissimilarity matrix of the dataset (‘vegdist’).
For all As genes and primer sets, PERMANOVA analysis indicated significant differences in community composition between the two TPs (p < 0.05; Fig. 6). For aioA and arrA, depth did not affect community composition (p ≥ 0.1), while for arsC communities differed between depths (p < 0.01). ANCOM did not indicate any specific ASVs characteristic for either one of the TPs or depth, indicating that the differences in community composition were not caused by single ASVs but rather by groups of ASVs. The aioA groups 1 and 2 were on average more abundant in TP B, while aioA group 4 was on average more abundant in TP A (Fig. 6). The arrA groups 3 and 5 were more prominent in TP A, while arrA groups 1, 4 and 6 were more prominent in TP B. The arsC groups 1, 7 and 8 were on average more abundant in TP A, while arsC groups 2 and 6 were on average more abundant in TP B (Fig. 6). Based on 16S rRNA gene analysis, bacterial phyla were of similar relative abundance in both TPs (Fig. 6).
Discussion
Wetlands including peatlands are widely used for the treatment of metal‐/metalloid‐contaminated waters (Sheoran and Sheoran, 2006; Vymazal, 2011). The studied peatlands have been rather effectively removing As from mining‐affected waters for up to 10 years and have accumulated As in the peat (Palmer et al., 2015). High retention of As was confirmed in the present study since most of the As was retained near the inlet in mine process water receiving TP A (less in TP B; Table S1), and most of the As accumulated in 0–10 cm peat. Natural peatlands have been reported to accumulate very high concentrations of As of more than 3000 mg kgdw −1 (González et al., 2006; Bauer et al., 2008), indicating the potential of the TPs to accumulate even more As in future years and thus allowing for their continued use as a purification system for mining‐affected waters.
Microorganisms are capable of using different As species in their energy metabolism or when detoxifying As (Stolz et al., 2006) and may affect As speciation in peat. Changes in As speciation within the TPs and/or along depth profiles can thus indicate microbial As turnover in peat. This was indeed observed in the field: As enters the TPs mainly as arsenate due to the strongly oxidizing nature of the gold extraction process (cyanide leaching), and mainly arsenate was found in surface waters as arsenate reduction was likely prevented in most parts by the rather oxic conditions in those waters. However, a higher percentage of arsenite was found near the outlet of TP B, indicating ongoing arsenate reduction even in rather oxic surface waters. Moreover, arsenite was detected in all profiles and was the dominant As species in the TP A1 profile in 50 and 60 cm depth (Table S2), which shows that most of the arsenate reduction in TPs is likely found in deeper, anoxic peat layers.
The high potential of the TP peat for microbially catalysed arsenate reduction under anoxic conditions was demonstrated also in microcosm incubations (Fig. 1), and arsenate respirers as well as As‐tolerating microorganisms were abundant in TPs (106–107 and ~107 cells gdw −1 respectively) and accounted for about 36% and 50% of the general aerobic heterotrophs (Fig. 4). The high abundance of arsenate respirers and As‐tolerators is likely responsible for fast arsenate reduction in deeper peat layers and might thus contribute to efficient As removal in the TPs (see below for discussion of removal processes).
Arsenate reduction has been reported from a variety of anoxic soils and sediments (Dowdle et al., 1996; Oremland and Stolz, 2003; Kulp et al., 2006), and arsenate respirers have been isolated from many anoxic As‐contaminated environments (Oremland and Stolz, 2003). However, limited information is available on arsenate reduction and arsenate reducers in peatlands (natural or disturbed). Thus, the present study contributes to a better understanding of arsenate reducers in peatlands. Arsenate can be reduced by arsenate respiring as well as arsenate detoxifying microorganisms. The ability to detoxify As is important for the survival and functioning of microorganisms in environments with elevated As concentrations such as the studied TPs, since As can be toxic at rather low concentrations (Paez‐Espino et al., 2009). Moreover, soil processes such as respiration, microbial biomass as well as functioning of enzymes like alkaline phosphatase are inhibited by As concentrations as low as 40–100 mg As kgDW −1 (Ghosh et al., 2004; Bhattacharyya et al., 2008; Liu et al., 2019), which is much lower than the concentrations that were detected in TP peat (around 200 mg kgDW −1). As‐tolerating bacteria have been detected and isolated from a variety of As‐contaminated soils and can tolerate up to 15 g L−1 As (Villegas‐Torres et al., 2011; Escudero et al., 2013; Xiao et al., 2017). On the other hand, As‐sensitive microorganisms such as Aliivibrio fischeri are inhibited already by As concentrations <2 mg L−1 (Fulladosa et al., 2005). Thus, there is a high need to detoxify As in TPs, which would explain the high observed diversity and abundance of As‐tolerating microorganisms. Sequences of arsC from TPs were related to arsC from bacteria, archaea and fungi (Fig. 5C), highlighting the importance of an arsenic detoxification mechanism for microorganisms in TPs. Arsenic detoxification moreover contributes to arsenate reduction and might be in part responsible for arsenate reduction observed in microcosm studies or in the field.
Arsenate respirers from TPs as assessed by arrA sequencing were mostly not related to arrA of cultured microorganisms (Fig. 5B), indicating a high potential of yet unknown arsenate respirers in TPs. These hitherto uncharacterized arsenate respirers are likely well adapted to the conditions in the TPs, including cold climate conditions and high contaminant loads. Some detected groups of arrA were related to Geobacter spp. and Sulfuritalea hydrogenivorans, species which are known to reduce arsenate under anoxic conditions and have been detected in many As‐contaminated environments (Lear et al., 2007; Hery et al., 2008; Ohtsuka et al., 2013; Watanabe et al., 2017). However, based on 16S rRNA gene sequencing, the overall abundance of Geobacter and Sulfuritalea is rather low in the studied TPs (0.5%–0.9% relative abundance; Kujala et al., 2018, this study), highlighting the importance of the yet unknown arsenate respirers in the system and their possible contribution to As removal. Even though arsenate respirers were almost equally abundant in TP A and TP B (Fig. 4), the arsenate respirer community differed strongly between the two TPs (Fig. 6). Different kinds of arsenate respirers likely differ in their habitat preferences and arsenate turnover rates. Differences in TP usage (e.g. different flow regimes, different inflow water composition different TP ages), as well as initial differences between the TPs (e.g. peat type, peat depth), might have selected for different microbial communities which in turn might contribute to the observed differences in As removal efficiencies. Arsenate respirers of arrA group 3 (related to Geobacter spp. and Desulfuromonas sp.) and arsenate tolerators of group 1 (related to Syntrophobacter sp., Syntrophus spp. and Smithella sp.) were more abundant in TP A (receiving mine process water), indicating that those groups might be in part responsible for the higher As removal in TP A. However, microbial arsenic reduction is likely not the only factor determining removal efficiencies of As in TPs, since other factors like hydraulic load and water residence time also contribute. Since hydraulic load is much higher for TP B than for TP A (38 vs. 6.1 mm day−1; Palmer et al., 2015), this also negatively affects As removal in TP B.
Oxic microcosm incubations with supplemented arsenite demonstrated the potential for arsenite oxidation in peat soil (Fig. 1) and the arsenite oxidizer community was rather diverse (Figs 5A and 6). The potential arsenite oxidation rate was comparable to the potential arsenate reduction rate assessed in anoxic microcosm incubations with supplemented arsenate (all arsenite/arsenate oxidized/reduced within 9 days, Fig. 1), indicating a similar process potential for both arsenite oxidation and arsenate reduction. However, arsenite oxidizers were less abundant in the studied TPs (105 cells gdw −1, ~0.5% of general aerobic heterotrophs; Fig. 4). As the TPs are permanently water‐saturated, oxygen availability in the peat is likely low, which would restrict the growth of arsenite oxidizers. Abundant aioA were related to Hydrogenophaga sp., Thiocapsa marina and Bradyrhizobium sp. (Fig. 5A). These organisms are known arsenite oxidizers and have been detected and/or isolated from a variety of As‐contaminated environments including soils, sediments and Hot Creeks (Salmassi et al., 2006; Garcia‐Dominguez et al., 2008). However, many ASVs were not closely related to known aioA, indicating phylogenetic novelty of arsenite‐oxidizers in the studied TPs. These novel arsenite‐oxidizers are likely well‐adapted to the conditions in the TPs.
Both arsenite oxidation and arsenate reduction are feasible processes in the studied TPs. Moreover, it is conceivable that As cycling occurs within the peatlands, leading to a reduction of arsenate in the lower, anoxic peat layers or anoxic microenvironments in the upper layers followed by oxidation of arsenite in the upper, oxic layers. Such internal cycling might even occur repeatedly. Arsenite oxidizers, arsenate respirers and arsenic‐tolerating microorganisms were detected in both layers based on MPN counts and functional gene analysis (Figs 4 and 6), indicating that those functional groups are present in the same layers, which might allow for switching between oxidizing and reducing processes even within the same layer with possible changes of environmental parameters. However, it was not possible to determine if and how often oxidation–reduction cycling occurred in situ, and arsenate reduction to arsenite was the observed net process. Thus, for net As cycling, arsenate reduction is likely the dominating process as (i) arsenate is the dominating As species in the inflow, (ii) anoxic conditions prevail through most of the peat which would support arsenate reduction, (iii) arsenite is found in the porewater likely as a result of arsenate reduction, and (iv) arsenite oxidizers are less abundant in TPs than arsenate reducers (arsenate respirers and arsenate tolerators). However, the contribution arsenite oxidation to net As cycling might increase when changes in the water table become more prominent, as might be the case when the use of the TPs for water treatment is discontinued (e.g. due to mine closure or changes in the water treatment process). Lowered water tables would lead to enhanced diffusion of oxygen into the peat and might thus allow for the re‐oxidation and re‐mobilization of bound arsenite (Langner et al., 2014), which might, in turn, lead to leaching of bound As from the TPs.
In systems with high concentrations of arsenite and reduced sulphur species, thioarsenate formation may occur. Thioarsenates have been studied extensively in extreme environments like hot springs (e.g. Planer‐Friedrich et al., 2007; Ullrich et al., 2013) or in groundwaters (Wallschläger and Stadey, 2007; Planer‐Friedrich et al., 2018), while peatlands have received less attention. However, thioarsenates have been detected in a natural As‐enriched peatland just recently (Besold et al., 2018), indicating that they may be indeed widespread in anoxic, sulphidic environments. In the studied TPs, sulphate concentrations are very high, especially in TP A, which receives mine process waters (Palmer et al., 2015). Anoxic conditions in deeper peat layers allow for microbial sulphate reduction to elemental sulphur and further on to sulphide. Indeed, sulphide accumulation was observed in anoxic microcosms (Fig. 2). After microbially catalysed arsenate reduction to arsenite (Figs 1 and 3), high sulphide concentrations can then lead to an abiotic formation of thioarsenates at neutral to alkaline pH (Planer‐Friedrich et al., 2010) and a subsequent mobilization of As in soils (Sun et al., 2016; ThomasArrigo et al., 2016). Abiotic reduction of arsenate with sulphide can be neglected in the microcosms (pH 6–7) as it becomes kinetically relevant only at much lower pH (Rochette et al., 2000).
Thioarsenate formation was observed in anoxic, uninhibited microcosms, and thioarsenates contributed to up to 80% of the detected As species (Figs 1 and 3). Di‐ and tri‐thioarsenate were the dominating thioarsenates formed in the microcosms. Earlier studies have shown that high S:As ratios favour the formation of di‐ and tri‐thioarsenate rather than mono‐thioarsenate (Planer‐Friedrich et al., 2010). S:As ratios in the microcosm incubations were >10 in all cases, thus the observed dominance of di‐ and tri‐thioarsenate is in line with the earlier studies.
Even though the results obtained from the microcosm incubations cannot be transferred directly to the situation in situ, (re‐)mobilization of bound arsenite due to thioarsenate formation is feasible in the TPs. This study found first indications that in peat layers where (microbially formed) arsenite coincides with high microbial sulphate reduction rates, thioarsenate formation may occur.
Conclusions and outlook
The present study characterized the microbial community composition within two TPs receiving mining‐affected waters using functional gene analysis and their As oxidation and reduction potential. The most important findings are summarized in Fig. 7. Both TPs efficiently removed As, even though removal efficiencies were lower in TP B. In TP A, total As concentrations were high and arsenate was the major As species in the inflow water, while total As concentrations were low in the outflow. In TP B, differences in the total As concentration between inflow and outflow were low, reflecting the smaller removal efficiencies of TP B. Like in TP A, arsenate was the dominant As species, however, the contribution of arsenite increased in TP B with increasing distance from the inflow.
Figure 7.
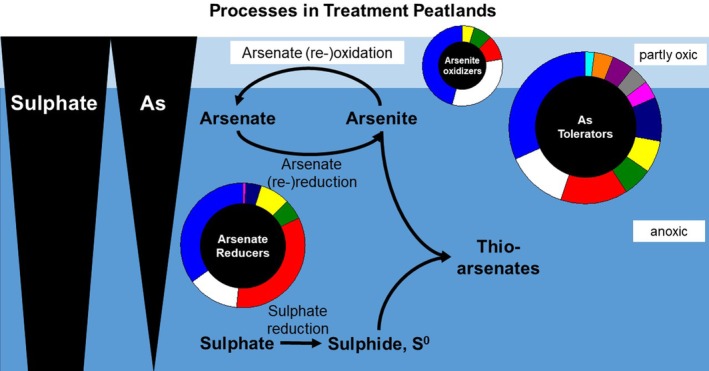
Conceptual model of microbial As turnover in TPs. Processes are indicated by arrows. Microbial groups contributing to As turnover are represented as spheres. The relative abundance of an Asmetabolizing group is indicated by the size of the sphere, while the diversity of the group is indicated by the number of pie sectors. The peat is water‐saturated and thus mainly anoxic. Arsenic and sulphate pore water concentrations decrease with increasing peat depth. Under anoxic conditions, arsenate is reduced to arsenite. Repeated As cycling, i.e. (re‐)reduction and (re‐)oxidation might occur. Sulphate is reduced in anoxic peat layers, leading to the production of reduced sulphur species and the subsequent formation of thioarsenates.
Peat microorganisms showed the potential to oxidize and reduce arsenite and arsenate respectively, and especially arsenate reducers were abundant in peat. Based on the analysis of functional genes, As‐metabolizing microbes in TPs were diverse and many detected ASVs were not closely related to cultured microorganisms. Sulphide and thioarsenate production were observed in anoxic microcosms, and the formation of thioarsenates might lead to lowered As removal or even As remobilization. Thus, the collective data indicate that (i) net arsenate reduction occurs in situ and is likely catalysed by As‐metabolizing microorganisms, (ii) hitherto uncultivated arsenic‐metabolizing microorganisms contribute to As removal in TPs, (iii) thioarsenate formation might lead to remobilization of bound As, and (iv) changes in water table (e.g. after mine closure) might lead to oxidation of bound arsenite by hitherto uncultured arsenite oxidizers and subsequent remobilization of bound As.
Even though the present study gives a good insight into the As‐metabolizing potential of peat microorganisms, open questions remain that should be addressed in future research. Future approaches might include (i) the isolation and characterization of As‐metabolizing microorganisms from peat soil, (ii) metagenomic analyses to overcome the issue of primer bias encountered in amplicon‐based approaches, (iii) in situ detection and quantification of thioarsenates in TPs, and (iv) investigation of in situ binding mechanisms and As speciation in TP peat.
Experimental procedures
Sampling site and peat sampling
Samples were taken from two peatlands treating mining‐affected waters near a gold mine in Finnish Lapland (about 68° northern latitude; Fig. S1). Treatment peatland A (TP A) is used for purification of pretreated process waters, while treatment peatland B (TP B) is used for the purification of drainage water. Average inflow concentrations of As were 1.3 and 0.4 μM to TP A and TP B respectively and average removal efficiencies were 98% and 57% in TP A and TP B respectively (average 2013–2017, obtained from monitoring data provided by the mining company). The pH in surface water and porewater of both TPs varied from 5.0 to 7.5 and the redox potential varied from −150 to 0 mV in TP A and −100 to 100 mV in TP B. In TP A, porewater sulphide concentrations were 2.0 and 11 μM in 10 and 60 cm depth respectively, while porewater Fe(II) concentrations were 2.9 and 113 μM in 10 and 60 cm depth respectively. In TP B, porewater sulphide concentrations were 0.2 and 1.0 μM in 10 and 60 cm depth respectively, while porewater Fe(II) concentrations were 226 and 1190 μM in 10 and 60 cm depth respectively. A more detailed site description is provided in Palmer and colleagues (2015). Peat samples were taken on several occasions (MPN counts and molecular work September 2015, microcosm incubations March 2016) with a soil corer from 0 to 10 cm peat and 60 to 70 cm peat. Peat As concentrations were 196 and 5.1 mg kg−1 in 0–10 and 60–70 cm peat respectively, at sampling point TP A1, and 65.7 and 31.0 mg kg−1 in 0–10 and 60–70 cm peat respectively, at point TP B2 (Table S2). Samples intended for DNA extraction and subsequent analysis of the As metabolizing microbial community were flash‐frozen on dry ice in the field and stored at −20°C. Samples intended for determination of cell numbers and for incubation experiments were stored at 4°C in the dark until sample processing. Incubation experiments were started within 2–3 weeks after sampling to prevent the impact of storage on process potentials.
Determination of aqueous total As concentrations and speciation in TPs
Surface water was sampled in summer 2017 at seven sampling points along TP A and three sampling points along TP B (Fig. S1). For total As determination, samples were filtered through 0.2 μm filters (cellulose acetate, CA; Macherey‐Nagel), stabilized in 0.5% H2O2 and 0.8% HNO3, and analysed by inductively coupled mass spectroscopy (ICP‐MS). Surface water samples for As speciation were filtered through 0.2 μm filters and acidified with 0.2% (v/v) HCl to avoid precipitation of Fe mineral phases and As loss due to co‐precipitation or sorption. All samples with total As concentrations >0.5 μg L−1 were analysed for their As speciation using anion exchange chromatography with a 20–100 mM gradient NaOH eluent and suppressor coupled to ICP‐MS (AEC‐ICP‐MS) as described earlier (Planer‐Friedrich et al., 2007). Additionally, pore water samples were obtained at sampling points TP A1 and TP A5 at the depths where samples were collected for incubation experiments and microbial community studies (see below). Since the potential occurrence of thioarsenates was expected in these pore water samples and acidification leads to their precipitation as AsS mineral phases (Smieja and Wilkin, 2003), filtration and on‐site flash‐freezing on dry ice for species preservation was used instead of HCl acidification as described earlier (Planer‐Friedrich et al., 2007). Total As for these samples was analysed as described above. All samples with total As concentrations >0.1 μg L−1 were analysed for their As speciation using AEC‐ICP‐MS as well but with a 2.5–100 mM gradient NaOH eluent leaving out the suppressor, to enable detection of methylated species, as described earlier (Planer‐Friedrich et al., 2007).
Incubation experiments
Soil slurry incubations were set up with field‐fresh peat soil from sampling point TP A5 (0–10 cm depth) and TP A1 (60–70 cm depth) to test for the potential of peat microorganisms to oxidize and reduce arsenite or arsenate respectively. 0–10 cm peat from TP A5 was chosen to prevent masking of target processes by potential leaching of the high initial As content of surface peat from TP A1. One gram wet peat soil was diluted 1:60 with deionized water in 125 mL incubation bottles. Incubations were set up in triplicate. Oxic and anoxic incubations supplemented with 50 μM arsenite and arsenate respectively, were set up with and without inhibitor. Incubations with inhibitor were supplemented with 0.75 mmol molcarbon −1 sodium azide (NaN3) to inhibit biological arsenite oxidation/arsenate reduction (controls). The concentration of NaN3 was chosen based on previous studies investigating As binding and transformation in organic‐rich environments (Buschmann et al., 2006; Hoffmann et al., 2012; Besold et al., 2018). For oxic incubations, slurries were prepared with ambient air in the headspace. Bottles were closed with rubber stoppers, and stoppers were removed regularly for aeration (1–2 times day−1) as well as for sampling. For anoxic incubations, slurries were prepared in a glovebag (Coy Laboratory Products), which was operated with a gas mix of 95% N2 and 5% H2. Bottles were closed with butyl‐rubber stoppers and crimp‐sealed to preserve anoxic conditions. Anoxic incubations were sampled inside the glovebag using a syringe. Peat soil slurries were incubated at room temperature on a shaker in the dark and sampled after 0, 1, 2, 5, 9, 12, 19 and 167 days. Concentrations of produced sulphide were determined after 16 and 167 days using the methylene blue method (Cline, 1969). Additional incubations were set up with peat soil and mine process water (sulphate = 2100 mg L−1) to assess the effect of increased sulphate concentrations on thioarsenate formation. These incubations were set up with 0–10 cm peat soil under anoxic conditions with and without NaN3 and were supplemented with 50 μM arsenate. Samples were taken at least six times from 2 to 433 days for determination of As speciation and sulphide concentration. Samples were filtered (0.2 μm, Chromafil CA; Macherey‐Nagel, Germany) and flash‐frozen immediately to prevent changes in As speciation. After thawing the flash‐frozen samples in a glovebag, aqueous As speciation was analysed by AEC‐ICP‐MS as described in Planer‐Friedrich and colleagues (2007).
Enumeration of As‐utilizing prokaryotes by MPN counts
MPN counts were set up targeting arsenite‐oxidizers, aerobic As‐tolerating microorganisms and arsenate respirers. Incubations were set up with soil from TP A1 and TP B2 (0–10 and 60–70 cm depth). An artificial porewater solution containing minerals, trace elements and vitamins (modified from Balch et al., 1979; Kuhner et al., 1996) was prepared with the following concentrations (in mg L−1): (NH4)2 SO4, 12.6; Na2SO4, 13.5; CaCl2 × 2 H2O, 10.0; MgCl2 × 2 H2O, 10.0; FeCl2 × 4 H2O, 10.0; KH2PO4, 0.4; MnSO4 × 1 H2O, 5.0; FeSO4 × 7 H2O, 1.0; CoCl2 × 6 H2O, 1.0; CaCl2 × 2 H2O, 1.0; ZnSO4 × 7 H2O, 1.0; CuSO4 × 5 H2O, 0.1; AlK(SO4)2 × 12 H2O, 0.2; H3BO3, 0.1; Na2MoO4 × 2 H2O, 0.1; and nitrilotriacetic acid, 15; biotin, 0.004; folic acid, 0.004; pyridoxine‐HCl 0.02; thiamine‐HCl, 0.01; riboflavin, 0.01; niacin, 0.01; DL‐Ca‐pantothenic acid, 0.01; vitamin B12, 0.0002; p aminobenzoic acid, 0.01; lipoic acid, 0.01. Growth medium was prepared using artificial porewater solution supplemented with 2 mM arsenite and 2 mM HCO3 − for arsenite‐oxidizers, with 2 mM arsenite, 2 mM arsenate and nutrient broth (NB, Sigma‐Aldrich; to a final concentration of 2.5 g L−1) or tryptic soil broth (TSB, Sigma‐Aldrich; to a final concentration of 3.0 g L−1) for aerobic As‐tolerating microorganisms, with 2 mM arsenate, 2 mM lactate and 2 mM acetate for arsenate respirers, and with NB (at a final concentration of 2.5 g L−1) and TSB (at a final concentration of 3.0 g L−1) for general aerobic heterotrophs. The pH of all growth media was adjusted to 7.0. Incubations for arsenite‐oxidizers and As‐tolerating microorganisms were conducted in 96‐well plates with eight replicates per dilution step. Incubations for arsenate respirers were conducted in triplicates in 50 ml serum bottles with sterile N2 in the headspace. Plates and bottles were incubated at 20°C for 3 months in the dark. Production of arsenate or arsenite was qualitatively determined in the arsenite oxidizer and arsenate respirers MPN respectively, using a KMnO4 screening technique (Salmassi et al., 2002, 2006). Forty microliters or 60 μl of 0.01 M KMnO4 was added to 300 or 500 μl of an arsenite oxidizer or arsenate respirer incubation respectively. A purple colour indicated the presence of arsenate, while an orange colour indicated the presence of arsenite. Arsenite‐oxidizer incubations were scored positive, when the KMnO4 test indicated mainly arsenate in the growth medium, arsenate respirer incubations were scored positive, when the KMnO4 test indicated mainly arsenite in the growth medium, and As‐tolerating and general aerobic heterotrophic incubations were scored positive when growth was observed in the wells by visual inspection.
DNA extraction and barcoded amplicon sequencing
DNA was extracted from four replicate samples per site (TP A1 and TP B2) and depth (0–10 and 60–70 cm) using the PowerLyzer PowerSoil DNA isolation kit (MoBio Laboratories). Samples were freeze‐dried, homogenized, and rewetted (0.1 g of dried soil +150 μl water) prior to extraction. In total, 17 previously published primer sets for genes involved in microbial As metabolism were tested, and functional genes were successfully amplified and sequenced with two (aioA), two (arsC), and three (arrA) primer sets (Table 1). Moreover, bacterial 16S rRNA genes were amplified from all samples as described earlier (Kujala et al., 2018). PCRs were set up in 25 μl reactions with the following composition: Maxima SYBR Green/Fluorescein qPCR 2× Master Mix (Thermo Scientific), 0.4 μM of each primer (Biomers), and 0.02% Bovine Serum Albumin (Thermo Scientific), 1–5 ng template DNA. Primers and PCR conditions are given in Table 1. Additional primer sets were tested (Table S3), but no PCR product was obtained with those despite thorough testing, or no correct sequences were obtained from the PCR products. Following the initial PCR, barcodes and sequencing primers were added in a second PCR (Table 1). PCR products from the second PCR were purified and pooled in equimolar concentrations prior to sequencing on the Ion Torrent PGM. Sequencing templates were prepared using the Ion PGM Hi‐Q OT2 Kit. For sequencing, the Ion PGM Hi‐Q OT2 Kit and an Ion 316 Chips v2 (Thermo Fisher) were used as described earlier (Mäki et al., 2016; Kujala et al., 2018).
Sequence analysis
Sequence analysis was done in Qiime2 (Boylen et al., 2019). After demultiplexing, during which sequences with incorrect primer (>1 mismatch) or barcode (>1 mismatch) were removed, sequences were quality‐filtered using the dada2 plugin for IonTorrent sequences (‘denoise‐pyro’) with the following modifications to the default parameters: At the 5′ end of the sequences, primers were removed (‘‐p‐trim‐left’; 41–44 bp, depending on analysed gene) and sequences were trimmed at the 3′ end to achieve equal lengths for all sequences of a gene (‘‐p‐trun‐len’; 261–314 bp depending on gene). The length for trimming was selected to retain a reasonable number of sequences with the fragments being as long as possible. This resulted in final sequence lengths after using dada2 of 220, 220 and 270 bp for aioA, arrA and arsC respectively. ASVs obtained after dada2 quality filtering were used without further clustering unless indicated otherwise. Feature classifiers were trained using aioA, arrA and arsC sequences of pure cultures (114, 35 and 600 sequences respectively) with the Qiime2 plugin ‘feature‐classifier’. For classifier training, only the part of the sequence corresponding to the amplified genes was used. Classifiers were used mainly to identify and remove non‐target sequences, which accounted for 50%, 7% and 3% of all aioA, arrA and arsC amplicons. While trained classifiers allowed the identification of amplicon sequences on the phylum level, they did not allow for classification at lower taxonomic levels. Taxonomic affiliations of amplicon sequences on lower taxonomic levels were obtained by BLAST search of sequences and by the construction of phylogenetic trees. Closely related reference sequences to ASV representative sequences were identified by BLAST (Altschul et al., 1990; Table S4). For the construction of phylogenetic trees, the same collection of pure culture sequences was used as for the classifier training, albeit with full‐length sequences. Alignment of in silico translated reference and own sequences, as well as the construction of phylogenetic trees, was done in ARB (Ludwig et al., 2004). ARB allows for the construction of trees based on full‐length reference sequences, thus creating more robust phylogenetic trees. Frequency‐based position filters were applied for the construction of reference phylogenies, i.e. only alignment positions in which the majority of the sequences had an amino acid were considered. This resulted in the use of 390, 410 and 115 alignment positions for aioA, arrA and arsC respectively. Individual shorter sequences (i.e. own sequences from the present study and related environmental sequences) were added to the existing tree using ARB parsimony using a frequency‐based position filter for those shorter sequences (resulting in the use of 73, 72 and 86 alignment positions for aioA, arrA and arsC respectively). The addition of shorter sequences does not affect the overall tree topology. This approach of adding shorter sequencing to a preexisting tree has been frequently used in studies of ribosomal RNA as well as functional genes (see e.g. Glöckner and Meyerdierks, 2006; Pester et al., 2012; Gutierrez et al., 2013; Pjevac et al., 2017). Own sequences were grouped based on their positioning in the phylogenetic trees. A heatmap showing the occurrence of the aforementioned clusters of aioA, arrA and arsC as well as the major phyla (>1% relative abundance) based on 16S rRNA gene sequencing were constructed in R using ‘annHeatmap2’ of the Heatplus package (Ploner, 2015).
ASV tables were rarefied, i.e. subsampled at a depth of 600 sequences for all genes to allow for comparison of diversity in the two TPs and depths and between the three genes. Alpha diversity measures (number of observed ASVs, Faith's PD, Shannon diversity index H′, Pielou's Evenness) were calculated based on these rarified ASV tables. Library coverage and richness estimators were not calculated since the dada2 algorithm removes singletons during its chimera detection step. The non‐rarefied libraries were tested for community differences between peatlands, depths and the two primer sets per gene using adonis PERMANOVA on weighted unifrac distance matrices (Anderson, 2001; Oksanen et al., 2018). ANCOM (Mandal et al., 2015) was used to identify ASVs that differed in abundance between the two TPs or depths. Sequences were deposited in the NCBI sequence read archive under accession number SRP158809.
Supporting information
Appendix S1: Supporting Information
Acknowledgements
Funding for this work was provided by the Academy of Finland (Project 287397 ‘Microbial transformations of arsenic and antimony in Northern natural peatlands treating mine waste waters’). Additional support was provided by the European Research Council (ERC grant awarded to MT, Grant Agreement No. 615146, FP/2007‐2013), the Bavarian Funding Program for the Initiation of International Projects and the German Research Foundation (grants awarded to BPF, BayIntAn_UBT_2017_23 and DFG PL 302/20‐1). Furthermore, the authors would like to thank Anne Eberle for providing some aqueous As data from her master thesis.
References
- Abin, C.A. , and Hollibaugh, J.T. (2017) Desulfuribacillus stibiiarsenatis sp. nov., an obligately anaerobic, dissimilatory antimonate‐ and arsenate‐reducing bacterium isolated from anoxic sediments, and emended description of the genus Desulfuribacillus . Int J Syst Evol Microbiol 67: 1011–1017. [DOI] [PubMed] [Google Scholar]
- Altschul, S. , Gish, W. , Miller, W. , Myers, E.W. , and Lipman, D.J. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Anderson, M.J. (2001) A new method for non‐parametric multivariate analysis of variance. Austral Ecol 26: 32–46. [Google Scholar]
- Balch, W.E. , Fox, G.E. , Magrum, L.J. , Woese, C.R. , and Wolfe, R.S. (1979) Methanogens: reevaluation of a unique biological group. Microbiol Rev 43: 260–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia‐Brunet, F. , Dictor, M.C. , Garrido, F. , Crouzet, C. , Morin, D. , Dekeyser, K. , et al (2002) An arsenic(III)‐oxidizing bacterial population: selection, characterization, and performance in reactors. J Appl Microbiol 93: 656–667. [DOI] [PubMed] [Google Scholar]
- Bauer, M. , Fulda, B. , and Blodau, C. (2008) Groundwater derived arsenic in high carbonate wetland soils: sources, sinks, and mobility. Sci Tot Environ 401: 109–120. [DOI] [PubMed] [Google Scholar]
- Besold, J. , Biswas, A. , Suess, E. , Scheinost, A.C. , Rossberg, A. , Mikutta, C. , et al (2018) Monothioarsenate transformation kinetics determines arsenic sequestration by sulfhydryl groups of peat. Environ Sci Technol 52: 7317–7326. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, P. , Tripathy, S. , Kim, K. , and Kim, S.H. (2008) Arsenic fractions and enzyme activities in arsenic‐contaminated soils by groundwater irrigation in West Bengal. Ecotoxicol Environ Saf 71: 149–156. [DOI] [PubMed] [Google Scholar]
- Bissen, M. , and Frimmel, F.H. (2003) Arsenic – a review. Part I: occurrence, toxicity, speciation, mobility. Acta Hydrochim Hydrobiol 31: 9–18. [Google Scholar]
- Boylen, E. , Rideout, J.R. , Dillon, M.R. , Bokulich, N.A. , Abnet, C.C. , Al‐Ghalith, G.A. , et al (2019) Reproducible, interactive, scalable and extensive micobiome data science using QIIME 2. Nat Biotechnol 37: 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann, J. , Kappeler, A. , Lindauer, U. , Kistler, D. , Berg, M. , and Sigg, L. (2006) Arsenite and arsenate binding to dissolved humic acids: influence of pH, type of humic acid, and aluminum. Environ Sci Technol 40: 6015–6020. [DOI] [PubMed] [Google Scholar]
- Cabrol, L. , Quéméneur, M. , and Misson, B. (2017) Inhibitory effects of sodium azide on microbial growth in experimental resuspension of marine sediment. J Microbiol Methods 133: 62–65. [DOI] [PubMed] [Google Scholar]
- Cline, J.D. (1969) Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr 14: 454–458. [Google Scholar]
- Costa, P.S. , Scholte, L.L.S. , Reis, M.P. , Chaves, A.V. , Oliveira, P.L. , Itabayana, L.B. , et al (2014) Bacteria and genes involved in arsenic speciation in sediment impacted by long‐term gold mining. PLoS One 9: e95655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle, P.R. , Laverman, A.M. , and Oremland, R.S. (1996) Bacterial dissimilatory reduction of arsenic(V) to arsenic(III) in anoxic sediments. Appl Environ Microbiol 62: 1664–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero, L.V. , Casamayor, E.O. , Chong, G. , Pedros‐Alio, C. , and Demergasso, C. (2013) Distribution of microbial arsenic reduction, oxidation and extrusion genes along a wide range of environmental arsenic concentrations. PLoS One 8: e78890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulladosa, E. , Murrat, J.C. , Martínez, M. , and Villaescusa, I. (2005) Patterns of metals and arsenic poisoning in Vibrio fischeri bacteria. Chemosphere 60: 43–48. [DOI] [PubMed] [Google Scholar]
- Garcia‐Dominguez, E. , Mumford, A. , Rhine, E.D. , Paschal, A. , and Young, L.Y. (2008) Novel autotrophic arsenite‐oxidizing bacteria isolated from soil and sediments. FEMS Microbiol Ecol 66: 401–410. [DOI] [PubMed] [Google Scholar]
- Ghosh, A.K. , Bhattacharyya, P. , and Pal, R. (2004) Effect of arsenic contamination on microbial biomass and its activities in arsenic contaminated soils of Gangetic West Bengal, India. Environ Int 30: 491–499. [DOI] [PubMed] [Google Scholar]
- Glöckner, F.O. , and Meyerdierks, A. (2006) Metagenome analyses In Molecular Identification, Systematics and Population Structure of Prokaryotes, Stackebrandt E. (ed). Heidelberg: Springer, pp. 261–281. [Google Scholar]
- González, Z.I. , Krachler, M. , Cheburkin, A.K. , and Shotyk, W. (2006) Spatial distribution of natural enrichments of arsenic, selenium, and uranium in a minerotrophic peatland, Gola di Lago, Canton Ticino, Switzerland. Environ Sci Technol 40: 6568–6574. [DOI] [PubMed] [Google Scholar]
- Gutierrez, T. , Berry, D. , Yang, T. , Mishamandani, S. , McKay, L. , Teske, A. , and Aitken, M.D. (2013) Role of bacterial exopolysaccharides (EPS) in the fate of the oil released during the Deepwater horizon oil spill. PLos One 8: e67717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamura, N. , Fukushima, K. , and Itai, T. (2013) Identification of antimony‐ and arsenic‐oxidizing bacteria associated with antimony mine tailing. Microbes Environ 28: 257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hery, M. , Gault, A.G. , Rowland, H.A.L. , Lear, G. , Polya, D.A. , and Lloyd, J.R. (2008) Molecular and cultivation‐dependent analysis of metal‐reducing bacteria implicated in arsenic mobilization in south‐east Asian aquifers. Appl Geochem 23: 3215–3223. [Google Scholar]
- Hoffmann, M. , Mikutta, C. , and Kretzschmar, R. (2012) Bisulfide reaction with natural organic matter enhances arsenite sorption: insights from X‐ray adsorption spectroscopy. Environ Sci Technol 46: 11788–11797. [DOI] [PubMed] [Google Scholar]
- Inskeep, W.P. , Macur, R.E. , Hamamura, N. , Warelow, T.P. , Ward, S.A. , and Santini, J.M. (2007) Detection, diversity and expression of aerobic bacterial arsenite oxidase genes. Environ Microbiol 9: 934–943. [DOI] [PubMed] [Google Scholar]
- Kudo, K. , Yamaguchi, N. , Makino, T. , Ohtsuka, T. , Kimura, K. , Dong, D.T. , and Amachi, S. (2013) Release of arsenic from soil by a novel dissimilatory arsenate‐reducing bacterium, Anaeromyxobacter sp. strain PSR‐1. Appl Environ Microbiol 79: 4635–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner, C.H. , Drake, H.L. , Alm, E. , and Raskin, L. (1996) Methane production and oxidation by soils from acidic forest wetlands of east‐central Germany. 96th General Meeting of the American Society for Microbiology, p. 304.
- Kujala, K. , Mikkonen, A. , Saravesi, K. , Ronkanen, A.K. , and Tiirola, M. (2018) Microbial diversity along a gradient in peatlands treating mining‐affected waters. FEMS Microbiol Ecol 94:fiy145. [DOI] [PubMed] [Google Scholar]
- Kulp, T.R. , Hoeft, S.E. , Miller, L.G. , Saltikov, C. , Murphy, J.N. , Han, S. , et al (2006) Dissimilatory arsenate and sulfate reduction in sediments of two hypersaline, arsenic‐rich soda lakes: mono and Searles lakes, California. Appl Environ Microbiol 72: 6514–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner, P. , Mikutta, C. , and Kretzschmar, R. (2012) Arsenic sequestration by organic sulphur in peat. Nat Geosci 5: 66–73. [Google Scholar]
- Langner, P. , Mikutta, C. , and Kretzschmar, R. (2014) Oxidation of organosulfur‐coordinated arsenic and realgar in peat: implications for the fate of arsenic. Environ Sci Technol 48: 2281–2289. [DOI] [PubMed] [Google Scholar]
- Lear, G. , Song, B. , Gault, A.G. , Polya, D.A. , and Lloyd, J.R. (2007) Molecular analysis of arsenate‐reducing bacteria within Cambodian sediments following amendments with acetate. Appl Environ Microbiol 73: 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledin, M. , and Pedersen, K. (1996) The environmental impact of mine wastes – roles of microorganisms and their significance in treatment of mine wastes. Earth Sci Rev 41: 67–108. [Google Scholar]
- Liu, C. , Tian, H. , Li, H. , Xie, W. , Wang, Z. , Megharaj, M. , and He, W. (2019) The accuracy in the assessment of arsenic toxicity using soil alkaline phosphatase depends on soil water contents. Ecol Indic 102: 457–465. [Google Scholar]
- Ludwig, W. , Strunk, O. , Westram, R. , Richter, L. , Meier, H. , Yadhukumar, et al (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäki, A. , Rissanen, A.J. , and Tiirola, M. (2016) A practical method for barcoding and size‐trimming PCR templates for amplicon sequencing. Biotechniques 60: 88–90. [DOI] [PubMed] [Google Scholar]
- Malasarn, D. , Saltikov, W. , Campbell, K.M. , Santini, J.M. , Hering, J.G. , and Newman, D.K. (2004) arrA is a reliable marker for As(V) respiration. Science 306: 455–465. [DOI] [PubMed] [Google Scholar]
- Mandal, S. , Van Treuren, W. , White, R.A. , Eggesbø, M. , Knight, R. , and Peddada, S.D. (2015) Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis 26: 27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschullat, J. (2000) Arsenic in the geosphere – a review. Sci Tot Environ 249: 297–312. [DOI] [PubMed] [Google Scholar]
- Nordstrom, D.K. (2011) Hydrogeochemical processes governing the origin, transport and fate of major and trace elements from mine wastes and mineralized rock to surface waters. Appl Geochem 26: 1777–1791. [Google Scholar]
- Ohtsuka, T. , Yamaguchi, N. , Makino, T. , Sakurai, K. , Kimura, K. , Kudo, K. , et al (2013) Arsenic dissolution from Japanese paddy soil by a dissimilatory arsenate‐reducing bacterium Geobacter sp. OR‐1. Environ Sci Technol 47: 6263–6271. [DOI] [PubMed] [Google Scholar]
- Oksanen, J. , Blanchet, F.G. , Kindt, R. , Legendre, P. , Minchin, P.R. , and O'Hara, R.B. (2018) vegan: community ecology package. R package version 2.3‐5, 2016.
- Oremland, R.S. , and Stolz, J.F. (2003) The ecology of arsenic. Science 300: 939–944. [DOI] [PubMed] [Google Scholar]
- Osborne, T.H. , Jamieson, H.E. , Hudson‐Edwards, K.A. , Nordstrom, D.K. , Walker, R.S. , Ward, S.A. , and Santini, J.M. (2010) Microbial oxidation of arsenite in a subarctic environment: diversity of arsenite oxidase genes and identification of a psychrotolerant arsenite oxidizer. BMC Microbiol 10: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Páez‐Espino, D. , Tamames, J. , de Lorenzo, V. , & Cánovas, D. (2009) Microbial responses to environmental arsenic. BioMetals 22:117–130. [DOI] [PubMed] [Google Scholar]
- Palmer, K. , Ronkanen, A.K. , and Kløve, B. (2015) Efficient removal of arsenic, antimony and nickel from mine wastewaters in Northern treatment peatlands and potential risks in their long‐term use. Ecol Eng 75: 350–364. [Google Scholar]
- Pester, M. , Knorr, K.‐H. , Friedrich, M.W. , Wagner, M. , and Loy, A. (2012) Sulfate‐reducing microorganisms in wetlands – fameless actors in carbon cycling and climate change. Front Microbiol 3: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pjevac, P. , Schauberger, C. , Poghosyan, L. , Herbold, C.W. , van Kessel, M.A.H.J. , Daebeler, A. , et al (2017) AmoA‐targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment. Front Microbiol 8: 1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planer‐Friedrich, B. , Härtig, C. , Lohmayer, R. , Suess, E. , McCann, S.H. , and Oremland, R. (2015) Anaerobic chemolithotrophic growth of the haloalkaliphilic bacterium strain MLMS‐1 by disproportionation of monothioarsenate. Environ Sci Technol 49: 6554–6563. [DOI] [PubMed] [Google Scholar]
- Planer‐Friedrich, B. , London, J. , McCleskey, R.B. , Nordstrom, D.K. , and Wallschläger, D. (2007) Thioarsenates in geothermal waters of Yellowstone National Park: determination, preservation, and geochemical importance. Environ Sci Technol 41: 5245–5251. [DOI] [PubMed] [Google Scholar]
- Planer‐Friedrich, B. , Schaller, J. , Wismeth, F. , Mehlhorn, J. , and Hug, S. (2018) Monothioarsenate occurrence in Bangladesh groundwater and its removal by ferrous and zero‐valent iron technologies. Environ Sci Technol 52: 5931–5939. [DOI] [PubMed] [Google Scholar]
- Planer‐Friedrich, B. , Suess, E. , Scheinost, A.C. , and Wallschläger, D. (2010) Arsenic speciation in sulfidic waters: reconciling contradictory spectroscopic and chromatographic evidence. Anal Chem 82: 10228–10235. [DOI] [PubMed] [Google Scholar]
- Ploner, A (2015) Heatplus: Heatmaps with row and/or column covariates and colored clusters. R package version 2.26.0. [Google Scholar]
- Rochette, E.A. , Bostick, B.C. , Li, G. , and Fendorf, S. (2000) Kinetics of arsenate reduction by dissolved sulfide. Environ Sci Technol 34: 4714–4720. [Google Scholar]
- Salmassi, T.M. , Venkateswaren, K. , Satomi, M. , Newman, D.K. , and Hering, J.G. (2002) Oxidation of arsenite by Agrobacterium albertimagni, AOL15, sp. nov., isolated from Hot Creek, California. Geomicrobiol J 19: 53–66. [Google Scholar]
- Salmassi, T.M. , Walker, J.J. , Newman, D.K. , Leadbetter, J.R. , Pace, N.R. , and Hering, J.G. (2006) Community and cultivation analysis of arsenite oxidizing biofilms at Hot Creek. Environ Microbiol 8: 50–59. [DOI] [PubMed] [Google Scholar]
- Sheoran, A.S. , and Sheoran, V. (2006) Heavy metal removal mechanism of acid mine drainage in wetlands: a critical review. Miner Eng 19: 105–116. [Google Scholar]
- Smieja, J.A. , and Wilkin, R.T. (2003) Preservation of sulfidic waters containing dissolved As(III). J Environ Monitor 5: 913–916. [DOI] [PubMed] [Google Scholar]
- Song, B. , Chyun, E. , Jaffe, P.R. , and Ward, B.B. (2009) Molecular methods to detect and monitor dissimilatory arsenate‐respiring bacteria (DARB) in sediments. FEMS Microbiol Ecol 68: 108–117. [DOI] [PubMed] [Google Scholar]
- Stolz, J.E. , Basu, P. , Santini, J.M. , and Oremland, R.S. (2006) Arsenic and selenium in microbial metabolism. Annu Rev Microbiol 60: 107–130. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Quicksall, A.N. , Chillrud, S.N. , Mailloux, B.J. , and Bostick, B.C. (2016) Arsenic mobilization from sediments in microcosms under sulfate reduction. Chemosphere 153: 254–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ThomasArrigo, L.K. , Mikutta, C. , Lohmayer, R. , Planer‐Friedrich, B. , and Kretzschmar, R. (2016) Sulfidization of organic freshwater flocs from a minerotrophic peatland: speciation changes of iron, sulfur, and arsenic. Environ Sci Technol 50: 3607–3616. [DOI] [PubMed] [Google Scholar]
- Ullrich, M. , Pope, J. , Seward, T. , Wilson, N. , and Planer‐Friedrich, B. (2013) Sulfur redox chemistry governs diurnal antimony and arsenic cycles at champagne Pool, Waiotapu, New Zealand. J Volcanol Geoth Res 262: 164–177. [Google Scholar]
- Villegas‐Torres, M.F. , Bedoya‐Reina, O.C. , Salazar, C. , Vives‐Florez, M.J. , and Dussan, J. (2011) Horizontal arsC gene transfer among microorganisms isolated from arsenic polluted soil. Int Biodeterior Biodegrad 65: 147–152. [Google Scholar]
- Vymazal, J. (2011) Constructed wetlands for wastewater treatment: five decades of experience. Environ Sci Technol 45: 61–69. [DOI] [PubMed] [Google Scholar]
- Wallschläger, D. , and Stadey, C.J. (2007) Determination of (oxy)thioarsenates in sulfidic waters. Anal Chem 79: 3873–3880. [DOI] [PubMed] [Google Scholar]
- Watanabe, T. , Miura, A. , Iwata, T. , Kojima, H. , and Fukui, M. (2017) Dominance of Sulfuritalea species in nitrate‐depleted water of a stratified freshwater lake and arsenate respiration ability within the genus. Env Microbiol Rep 9: 522–527. [DOI] [PubMed] [Google Scholar]
- Xiao, E.Z. , Krumins, V. , Xiao, T.F. , Dong, Y.R. , Tang, S. , Ning, Z.P. , et al (2017) Depth‐resolved microbial community analyses in two contrasting soil cores contaminated with antimony and arsenic. Environ Pollut 221: 244–255. [DOI] [PubMed] [Google Scholar]
- Zhang, S.Y. , Zhao, F.J. , Sun, G.X. , Su, J.Q. , Yang, X.R. , Li, H. , and Zhu, Y.G. (2015) Diversity and abundance of arsenic biotransformation genes in paddy soils from southern China. Environ Sci Technol 49: 4138–4146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information


