Griffithsin (Grft) is an antiviral lectin that has been shown to potently inhibit HIV-1 by binding high-mannose N-linked glycosylation sites on HIV-1 gp120. A key factor for Grft potency is glycosylation at N295 of gp120, which is directly adjacent to N332, a target glycan for an entire class of broadly neutralizing antibodies (bNAbs).
KEYWORDS: HIV inhibition, gp120 glycosylation, griffithsin, high mannose, microbicide
ABSTRACT
Griffithsin (Grft) is an antiviral lectin that has been shown to potently inhibit HIV-1 by binding high-mannose N-linked glycosylation sites on HIV-1 gp120. A key factor for Grft potency is glycosylation at N295 of gp120, which is directly adjacent to N332, a target glycan for an entire class of broadly neutralizing antibodies (bNAbs). Here, we unify previous work on the importance of other glycans to Grft potency against HIV-1 and Grft’s role in mediating the conformational change of gp120 by mutating nearly every glycosylation site in gp120. In addition to a significant loss of Grft activity by the removal of glycosylation at N295, glycan absence at N332 or N448 was found to have moderate effects on Grft potency. Interestingly, in the absence of N295, Grft effectiveness could be improved by a mutation that results in the glycan at N448 shifting to N446, indicating that the importance of individual glycans may be related to their effect on glycosylation density. Grft’s ability to alter the structure of gp120, exposing the CD4 binding site, correlated with the presence of glycosylation at N295 only in clade B strains, not clade C strains. We further demonstrate that Grft can rescue the activity of the bNAbs PGT121 and PGT126 in the event of a loss or a shift of glycosylation at N332, where the bNAbs suffer a drastic loss of potency. Despite targeting the same region, Grft in combination with PGT121 and PGT126 produced additive effects. This indicates that Grft could be an important combinational therapeutic.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) infects about 2 million people per year predominantly in developing areas, such as sub-Saharan Africa (1). No vaccine is yet available, and existing prevention methods, such as daily oral preexposure prophylaxis, are problematic in these regions in terms of availability and user acceptability (2). Microbicides that inhibit HIV-1 infection represent a promising strategy for the prevention of HIV-1 infection, and formulations that are released by insertable items that stop the sexual transmission of the virus are envisioned. For instance, intravaginal rings that release small molecules or antibodies are being developed (3–6), and several proteins that could potentially be used to prevent infection have recently been encapsulated into silk fibroin films (7, 8).
Several major types of proteins have been shown to be highly potent HIV-1 inhibitors, including broadly neutralizing antibodies (bNAbs) (9–12), CCR5 binding proteins (13–16), and lectins (17–20). Each of these types of proteins has moved toward preclinical or clinical trials with some success (21–27).
Griffithsin (Grft) is among the most promising and potent potential microbicidal proteins. This lectin can be produced inexpensively in large quantities from tobacco leaves (28–31), Escherichia coli (32), and rice endosperm (33) and has been shown to have low or no toxicity, an unfolding temperature of 78.8°C, stability at pH 4 to 8, resistance to proteolytic degradation, conservation of potency after incubation at temperatures of up to 50°C, and safety in mice and macaques when applied topically, injected, or ingested (7, 25, 28, 34–43). Multiple groups have begun in-human trials (44, 45). Grft is arguably the most potent lectin HIV-1 inhibitor, showing nanomolar to subnanomolar efficacy against a wide range of HIV-1 strains (18, 28, 39), and has synergistic activity with currently used HIV-1 antiretroviral drugs, including tenofovir, maraviroc, enfuvirtide, and the broadly neutralizing antibody (bNAb) VRC01 (46, 47). Further, due to its ability to bind glycosylated viral surfaces, Grft has been shown to inhibit other viruses, such as severe acute respiratory syndrome coronavirus (SARS-CoV), hepatitis C virus (HCV), herpes simplex virus 2 (HSV-2), Japanese encephalitis virus (JEV), human papillomavirus (HPV), Middle East respiratory syndrome coronavirus (MERS-CoV), as well as HIV-1 and HIV-2 (48–53).
Grft is a dimer (with 121 amino acids per monomer) that has three saccharide binding sites per monomer and binds N-linked high-mannose glycans, such as Man-9, on viral surfaces with a very high affinity (18, 37, 54–56). It has been shown that both subunits of the Grft dimer are required for potent inhibition of HIV-1, despite the tight binding by the individual monomeric subunits to glycosylated gp120 (37, 57). This seeming disconnect between affinity and inhibitory potency has led to the suggestion that while Grft may inhibit HIV-1 infection in a general way by simply binding to any high-mannose site(s) on gp120, Grft may be most effective when it binds to specific regions or when it can cross-link between particular high-mannose sites on gp120, possibly causing (or preventing) a conformational change in gp120 (57–59). Further insight into Grft’s mechanism has come from cryo-electron microscopy studies, where the Bewley group has suggested that Grft can cross-link two separate viruses as part of its inhibition (59, 60).
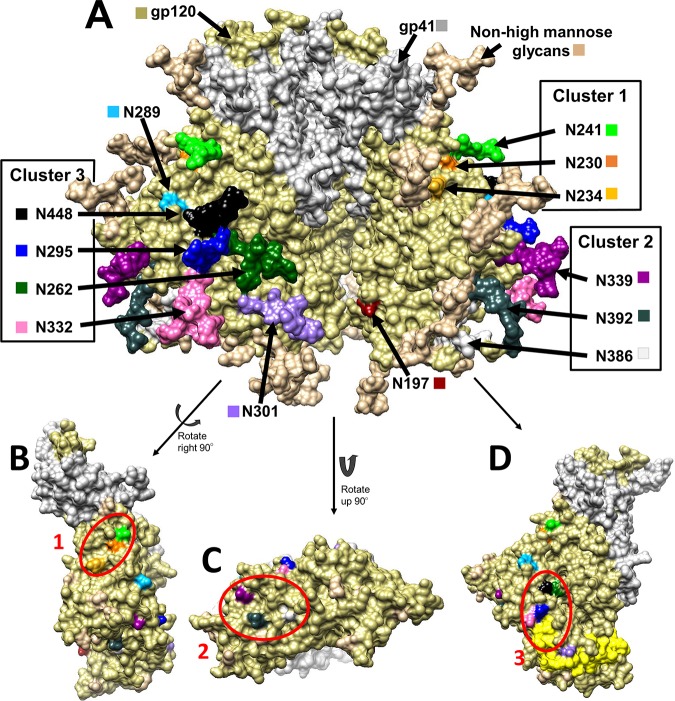
gp120 is found on the HIV-1 surface as a trimer (61, 62), with each monomeric unit having about a dozen relatively conserved high-mannose glycans that can potentially be bound by Grft (63–65) (Fig. 1A). The high-mannose glycans group together to form 3 main clusters, defined by Balzarini et al. (66), as shown in Fig. 1B to D; cluster 1 is composed of glycosylation sites (glycosites) N230, N234, and N241, cluster 2 is comprised of glycosites N339, N386, and N392, and cluster 3 contains glycosites N295, N262, N332, and N448. The exact glycosylation pattern varies by strain; relevant strains are shown in Fig. 2. The gp120 glycosylation site(s) utilized by Grft has been studied by several groups; the consensus is that glycosylation at N295 of gp120 (in cluster 3) is key to Grft potency against HIV-1. Several groups showed a correlation between the presence of the glycosylation sites at N234 and N295 with the very high potency of Grft against several HIV-1 strains (67, 68). Huang and colleagues, working with different viral strains, confirmed the importance of glycosylation at N295 of gp120 but showed little effect of glycosylation at position N234 on the potency of Grft (69), instead pointing to glycosylation at N448 as being important for Grft potency (69, 70). Cluster 3, sitting at the base of the V3 loop, has structural significance, as the V3 loop interacts with the chemokine receptor on the host cell during HIV-1 infection. When Grft binds to cluster 3, it may inhibit the interaction between the V3 loop of gp120 and the chemokine receptor by forming a physical blockade or by impeding the conformational change necessary to expose the V3 loop in the open conformation of the HIV-1 trimer (71–74). Further, cluster 3 includes glycosite N262, which cannot be removed without a complete loss of viral infectivity (75).
FIG 1.
(A) Structure of the HIV gp160 trimer in the unbound, closed state, i.e., in a state not bound to CD4 or CCR5. This structure of clade B JR-FL includes glycosylation (PDB accession number 5FYK) (118) and was prepared using the UCSF Chimera program (119, 120). gp120 is shown in khaki, and gp41 is shown in white. (B to D) gp120 monomers with glycosylation hidden to show the underlying amino acids that fall into clusters. Glycosylation sites (Asn) are shown in a variety of colors, as labeled in the diagram. (B) Cluster 1 encompasses the glycans at N230 (orange), N234 (yellow), and N241 (green). (C) Cluster 2 encompasses the glycans at N339 (dark magenta), N386 (dim gray), and N392 (dark slate gray). (D) Cluster 3 encompasses the glycans at N295 (blue), N332 (hot pink), N262 (dark green), and N448 (black). The V3 loop is highlighted in yellow to demonstrate the proximity of the glycans at N295 and N332.
FIG 2.
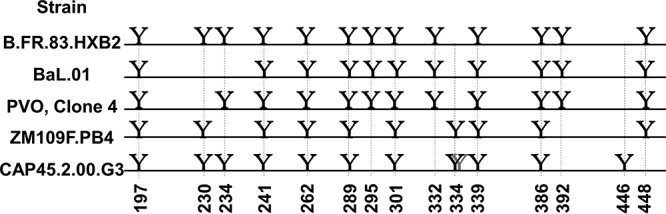
High-mannose glycosylation patterns of HIV strains BaL.01, PVO.4, ZM109, and CAP45. Y, the presence of a likely high-mannose site. The glycan on CAP45 shown in gray is at position 335 and is most likely another high-mannose glycan. Cluster 3 is composed of glycans at N262, N295, N332, and N448. For a full alignment of the sequences, see Fig. 1 posted at http://faculty.ucmerced.edu/pliwang/?page_id=123. High-mannose glycosylation sites were identified through sequence analysis of each strain. The sequences can be found at www.hiv.lanl.gov.
The glycosylation of HIV-1 gp120 has become particularly noteworthy due to several highly potent broadly neutralizing antibodies (bNAbs) that target glycosylation in gp120 and that are being considered for clinical trials (26, 76–80). One class of these antibodies targets the glycan at N332, which is found in cluster 3 next to N295. A loss or a shift of this glycan to N334 upon repetitive bNAb challenge is detrimental to inhibition by these bNAbs (11, 81–84). bNAbs that bind in this area, such as PGT121, have a range of effectiveness against these strains (81–83, 85–87). It has been reported that Grft is synergistic with the bNAb VRC01, which targets the gp120 CD4 binding site (47). However, it is not known if Grft can be used in conjunction with bNAbs that target glycans on gp120 because Grft may compete with these proteins for binding (Fig. 1).
We sought to clarify Grft’s usage of glycosylation sites in cluster 3 by extensively mutating gp120 from several strains of HIV-1 having distinct properties: two clade B strains (strain PVO, clone 4 [PVO.4], and strain BaL.01) that are very sensitive to Grft and that have essentially all high-mannose sites present, a clade C strain (strain ZM109F.PB4 [ZM109]) that is less sensitive to Grft and that lacks several glycosylation sites, and a clade C strain (strain CAP45.G3J [CAP45]) that is sensitive to Grft but that does not have a glycosite at 295. We specifically sought to elucidate the effect that the shifting of glycans had on Grft potency. The sum of the PVO.4 mutants is the largest collection of mutants of a single HIV-1 strain to be tested with Grft. We demonstrate Grft’s continued effectiveness against viral strains where the glycosite at N332 and/or N448 has been shifted and that Grft is able to remain effective against strains that are resistant to the bNAbs PGT121 and PGT126. This suggests that Grft could be valuable if used in combination with bNAbs that are more sensitive to such glycosylation changes. We also demonstrate that the Grft-mediated conformational change of gp120 appears to be dependent on the presence of the N295 glycosite in clade B strains but not clade C strains, indicating distinct mechanisms of Grft binding to gp120 and, therefore, potentially distinct mechanisms of HIV-1 inhibition.
RESULTS
Effect of glycosylation at each site in gp120 on Grft sensitivity in a typical HIV-1 strain.
The glycosylation patterns of several strains of HIV-1 are shown in Fig. 2. We chose HIV-1 strain PVO.4 to resolve the effect of glycosylation placement on gp120 in relation to Grft sensitivity. This clade B strain is very sensitive to Grft, having a subnanomolar 50% inhibitory concentration (IC50) of 0.2 ± 0.09 nM, and is also glycosylated at essentially all the sites suggested to be important in Grft inhibition (68–70, 88). In our experiments, each individual high-mannose site in PVO.4 gp120 (except for N262, due to its importance for structural stability [89]) was removed by mutating the saccharide-containing Asn to the closely related Gln, which is not glycosylated by cellular machinery. The single high-mannose site that is not naturally found in PVO.4 (Asn 230) was also mutated to add glycosylation. The sensitivity to Grft inhibition for each PVO.4 variant was measured in single-round inhibition assays and directly compared to that for the wild-type virus. As shown in Table 1, most individual mutations had little effect on the ability of Grft to inhibit PVO.4, except for mutation N295Q in gp120, which led to an 11-fold loss of sensitivity. To a lesser extent (approximately 5-fold), the mutations N448Q and N332Q showed a loss of sensitivity to Grft upon point mutation.
TABLE 1.
Inhibition of HIV gp120 variants by Grft
| Straina | Mutation | Fold differenceb
|
Pc | |
|---|---|---|---|---|
| Mean | SE | |||
| PVO.4 | ||||
| Single mutants | −197 | +2.7 | 0.58 | 1.6E−06 |
| +230 | +2.1 | 0.56 | 0.005 | |
| −234 | +2.2 | 0.58 | 3.7E−06 | |
| −241 | +3.8 | 0.91 | 7.2E−09 | |
| −289 | +2.8 | 0.79 | 0.001 | |
| −295 | +11.1 | 5.83 | < 2.2e−16 | |
| −301 | +1.4 | 0.32 | 6.6E−04 | |
| −332 | +4.6 | 1.22 | 1.4E−08 | |
| −339 | +2.8 | 0.59 | 3.5E−11 | |
| −386 | +1.5 | 0.73 | 0.5 | |
| −392 | +1.7 | 0.35 | 4.5E−05 | |
| −448 | +5.1 | 0.91 | 9.7E−12 | |
| Double and triple mutants | −234, −295 | +9.8 | 3.2 | 1.7E−07 |
| −234, −448 | +6.4 | 2.4 | 9.2E−10 | |
| −295, −332 | +42.9 | 6.9 | 2.2E−16 | |
| −295, −448 | +23.2 | 7.8 | 2.7E−10 | |
| −332, −448 | +35.5 | 7.1 | 9.4E−07 | |
| −234, −295, −448 | +28.2 | 5.0 | 3.15E−13 | |
| −295, −332, −448 | +88.2 | 10.7 | 8.1E−14 | |
| Shift of glycosylation site mutants | 332 > 334 | +1.3 | 0.49 | 0.2 |
| 448 > 446 | +1.7 | 0.79 | 0.04 | |
| −295, 332 > 334 | +8.0 | 2.3 | 8.1E−09 | |
| −295, 448 > 446 | +3.2 | 0.9 | 6.6E−09 | |
| 332 > 334, −448 | +6.6 | 2.7 | 1.1E−07 | |
| −332, 448 > 446 | +10.2 | 3.3 | 1.4E−11 | |
| 332 > 334, 448 > 446 | +5.4 | 3.7 | 8.8E−05 | |
| −295, 332 > 334, −448 | +21.2 | 18.0 | 3.8E−05 | |
| −295, −332, 448 > 446 | +29.4 | 11.5 | 4.3E−09 | |
| −295, 332 >334, 448 >446 | +21.5 | 4.7 | 2.8E−06 | |
| BaL.01 | −295 | +14.1 | 1.9 | 1.0E−10 |
| −332 | +2.1 | 0.6 | 0.002 | |
| −448 | +1.0 | 0.3 | 0.57 | |
| ZM109 | +234 | −10.2 | 2.6 | 1.0E−11 |
| +295 | −52.9 | 12.6 | 8.1E−13 | |
| +332 | −1.0 | 0.3 | 0.8 | |
| −334 | +1.4 | 0.2 | 0.005 | |
| +446 | +1.2 | 0.1 | 0.03 | |
| −448 | −4.3 | 1.2 | 1.4E−11 | |
| +234, +295 | −157 | 66 | 2.6E−16 | |
| +234, +446 | −20.6 | 6.3 | 1.1E−11 | |
| CAP45 | −234 | +1.2 | 0.3 | 0.4 |
| +295 | −3.2 | 1.2 | 7.3E−10 | |
| +332 | −3.2 | 1.2 | 4.7E−08 | |
| −334 | −2.3 | 0.9 | 1.1E−03 | |
| −446 | +2.0 | 0.9 | 1.5E−03 | |
| +448 | −1.3 | 0.6 | 0.1 | |
The Gfrt IC50 for wild-type strains PVO.4, BaL.01, ZM109, and CAP45 are 0.2 ± 0.09, 0.26 ± 0.08, 26.9 ± 6.2, and 0.54 ± 0.24 nM (standard error), respectively.
Mean is the average of triplicate data replicated a minimum of three times. A plus sign indicates that Grft inhibits the wild-type virus better than the mutant virus. A minus sign indicates that Grft inhibits the wild-type virus less well than the mutant virus (the mutant is more sensitive to Grft).
P values were determined by an analysis of variance with the Bonferroni correction.
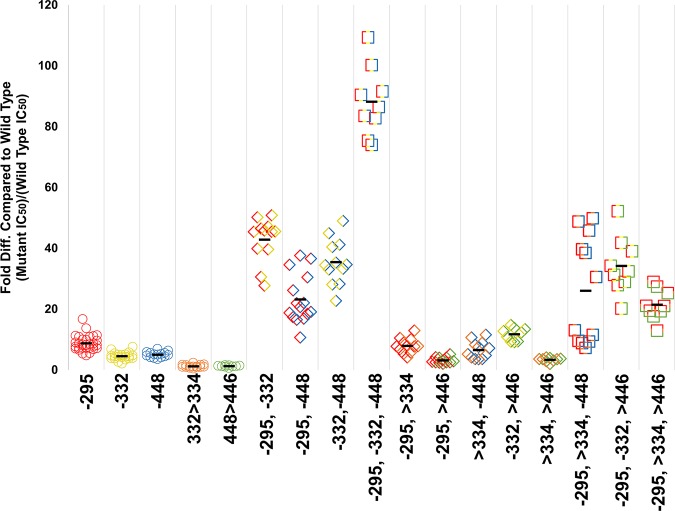
To determine the combined effect of each of these potentially important glycosylation sites, a double mutation was made in every combination of the sites N295, N448, and N332, as well as with the site N234, which had previously been observed to correlate with Grft inhibition (68, 88). Each of these double deletions showed a larger effect on the ability of Grft to inhibit PVO.4 than each of the single mutations, except N234Q/N295Q, which was effectively unchanged. The double deletion with the largest effect, N295Q/N332Q, showed a Grft IC50 43-fold higher than that for the wild-type virus (Table 1 and Fig. 3). In general, mutants with each combination that included a loss of the glycosylation site at position 295 showed a relatively high loss of sensitivity to Grft (Table 1 and Fig. 3), with each lost glycosite at positions 332 and 448 providing some additive effect. Mutations to remove all three of the proximal glycosylation sites (N295Q/N332Q/N448Q) in HIV-1 gp120 were made in PVO.4 (Table 1 and Fig. 3), resulting in an 88-fold loss of sensitivity to Grft. In addition, a triple mutant in which the glycosite at position 234, as well as the glycosites at positions 295 and 448 (N234Q/N295Q/N448Q), was removed showed a 28-fold decrease in sensitivity (Table 1).
FIG 3.
Grft inhibition against PVO.4 gp120 mutants. Each individual mutation is represented by a color, as follows: red, N295; yellow, N332; blue, N448; orange, N332 > N334; green, N448 > N446. A minus sign indicates that glycan has been removed. The greater than sign (>) indicates that glycan has been shifted. These colors are combined to represent combinational mutants. Circles indicate single mutants, diamonds indicate double mutants, and squares indicate triple mutants. Glycan deletion mutants were made by mutating the Asn to Gln. For statistical analysis, see Table 1 posted at http://faculty.ucmerced.edu/pliwang/?page_id=123.
Removal of more than three glycosylation sites from gp120 resulted in increasingly poor inhibition by Grft. Deletion of the gp120 glycosylation site N301, in addition to the existing N295Q/N332Q/N448Q triple mutation, resulted in a 55-fold difference in inhibition from that of the wild type. The removal of the glycan at position 386, in conjunction with the triple mutation, revealed a 129-fold difference in inhibition from that of the wild type. Combining these mutations (N295Q/N301Q/N332Q/N386Q/N448Q) led to a 126-fold difference in inhibition from that of the wild type. Continuing in a serial fashion, glycans at Asn392 and then Asn339 were removed, resulting in IC50s increasing up to 500-fold and 1,000-fold respectively (see Fig. 2 posted at http://faculty.ucmerced.edu/pliwang/?page_id=123). However, mutants with mutations beyond the triple mutation N295Q/N332Q/N448Q had a decrease in infectivity as well as a loss of sensitivity to Grft, so the results must be interpreted with caution (see Fig. 2 posted at the website mentioned above). The low infectivity of the latter mutants made it unfeasible to continue with serial deletions.
Several point mutations in gp120 in the commonly used HIV-1 strain BaL.01 were also carried out to remove the glycosylation at glycosylation cluster 3 (66). As expected, removal of glycosylation at position 295 showed the largest effect. Little effect was seen when glycosylation at either position 332 or position 448 was individually removed (Table 1). This finding differs from the results of Huang et al. (69), who observed a loss of potency of Grft when gp120 was mutated to remove glycosylation at position 448 in this strain. This disparity may be due to differences in the IC50 assay techniques used (see Fig. 5 posted at the website mentioned above).
Shifting gp120 glycosylation sites around N295 has little effect on sensitivity to Grft.
The N332 glycosylation site was further explored due to the importance of the glycan at this site as a target for broadly neutralizing antibodies (81–83, 85–87). As shown in Table 1, Grft maintained potency against PVO.4 when glycosite N332 was shifted to position 334. An analysis of thousands of published HIV-1 strains (245 clade A, 2,231 clade B, and 1,449 clade C strains) in the HIV-1 sequence database was carried out, looking at the conservation of each high-mannose glycan and the frequency of shifts occurring at each site. The glycans in cluster 3 showed the most movement in their location. Approximately 12% of all HIV-1 strains have a glycan located at N334 (referred to here as the N332 > N334 shift; see Fig. 3 posted at the website mentioned above). The other most common relocation is found primarily in clade C strains: 26% of clade C strains do not have glycosylation at position 448, and 72% of those exhibit a glycan at N446 instead (referred to here as the N448 > N446 shift; see Fig. 3C posted at the website mentioned above). Since removal of glycosite N448 in strain PVO.4 showed a clear decrease in Grft potency, we evaluated the N448 > N446 shift as well. Table 1 shows that shifting the glycosylation site in PVO.4 (N448 > N446) did not have a significant effect on Grft sensitivity.
We then investigated individual shift mutations (N332 > N334, N448 > N446) in combination with glycan deletions at other sites in cluster 3, N295Q, N332Q, and N448Q (see Table 1 for the IC50s and Fig. 3 for a graphical representation of the results; see Table 1 at http://faculty.ucmerced.edu/pliwang/?page_id=123 for the results of statistical analysis). In addition, two mutants that had both shifts, one in the presence and the other in the absence of glycosite N295, were generated (Table 1 and Fig. 3). Overall, the N332 > N334 shift had little effect on Grft potency. In general, the N448 > N446 shift also had little effect, with the notable exception that this shift increased the sensitivity of the virus to Grft (lowered the IC50) when the virus also lacked the glycan at position 295.
Effect of glycosylation on HIV-1 strain ZM109F.PB4.
While Grft has been shown to be effective against strains from all clades tested, it has shown variable effectiveness against strains from clade C. This has been ascribed to differences in glycosylation patterns across clades, particularly the lack of glycosylation site at position 295 for 79% of clade C strains (see Fig. 3C posted at the website mentioned above). Among the strains tested, ZM109 had the lowest sensitivity to Grft with an IC50 of 27 ± 6 nM. This strain lacks the glycosylation site at positions 234 and 295 and also exhibits the N332 > N334 shift in glycosylation (90) (Fig. 2). To investigate the role of glycosylation in sensitivity to Grft, several mutations were carried out, focusing on the critical glycosylation cluster comprised of N295/N33(2)4/N448. As shown in Table 1, placing a glycan at position 332 or removing one at position 334 has little effect on sensitivity to Grft, while adding a glycosylation site at position 295 dramatically improved sensitivity to Grft, with a 53-fold reduction in the IC50.
Interestingly, when the glycosylation site at position N448 in ZM109 gp120 is shifted to position 446 in the virus (which naturally lacks glycosylation at position 295), a 4-fold increase in sensitivity to Grft was observed (Table 1). Similar to the results with strain PVO.4, this indicates that a shift of glycosylation to the 446 position may be able to partially rescue the loss of the glycosylation site at position 295 in terms of Grft action (Fig. 4).
FIG 4.
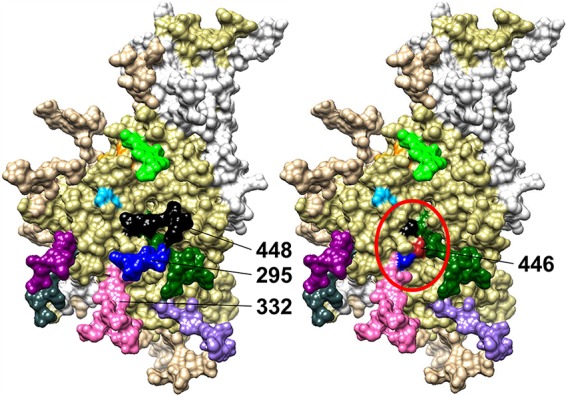
A visual representation of how the shift of a glycan from N448 to N446 in the absence of a glycan at N295 may lead to a higher glycan density and, therefore, a better binding environment for Grft. (Left) gp120 monomer, with the relevant glycan sites in cluster 3 (N295, N332, N448) being indicated. (Right) gp120 with the amino acid at position 446 (red) to show its proximity to cluster 3 (circled in red) if it is mutated to Asn to become a glycosite. The structure has PDB accession number 5FYK (118) and was modeled using the UCSF Chimera program (119, 120).
The glycosylation site at position N234 in gp120 has also been described to be important in some strains, despite this site being in cluster 1 and distal to the critical cluster 3 (Fig. 1) (66, 68). Although ZM109 was not mentioned in that study, when a glycosylation site is added at position 234, a 10-fold increase in sensitivity to Grft was observed. Most striking was the finding that when glycosylation was enabled at both position 234 and position 295, a 157-fold increase in Grft sensitivity was observed (Table 1).
Effect of glycosylation on gp120 in CAP45.G3J, a Grft-sensitive strain that naturally lacks glycosylation at 295.
Grft is highly potent across many strains of HIV-1, including some that naturally lack the glycosylation site at position 295. One such strain is CAP45.G3J (which is referred to here as CAP45), a clade C virus that lacks glycosylation sites at positions 295, 332, and 392 and yet is inhibited by Grft with an IC50 of 0.54 ± 0.23 nM. To investigate the role of glycosylation in such a strain, we made mutations in the important glycan cluster 3. When the glycosylation site at N295 was added back to this gp120, sensitivity to Grft was moderately improved by a factor of 3.2-fold (Table 1). The other two glycans in cluster 3 are naturally in the shifted positions, N334 and N446 (Fig. 2). Returning the glycan at N334 back to its canonical site at N332 or deleting it entirely also moderately decreased the IC50. Movement of a glycosite to N448 produced no difference in sensitivity from that of the wild type, whereas deletion of the glycan at that location was one of the few mutations that caused a slight increase in the Grft IC50.
Grft is compatible with broadly neutralizing antibodies targeting glycan at gp120 N332.
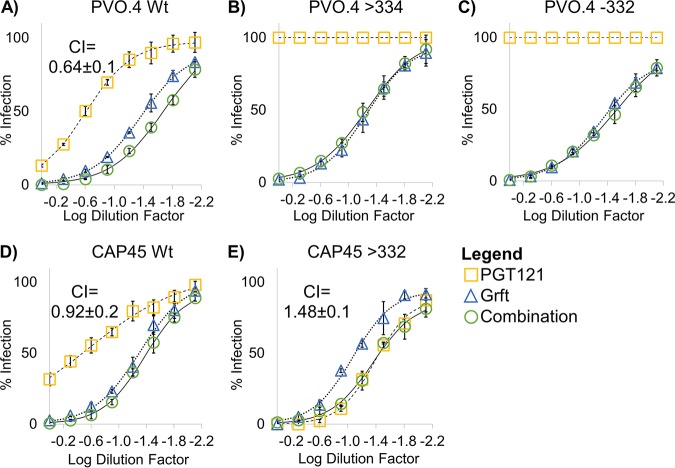
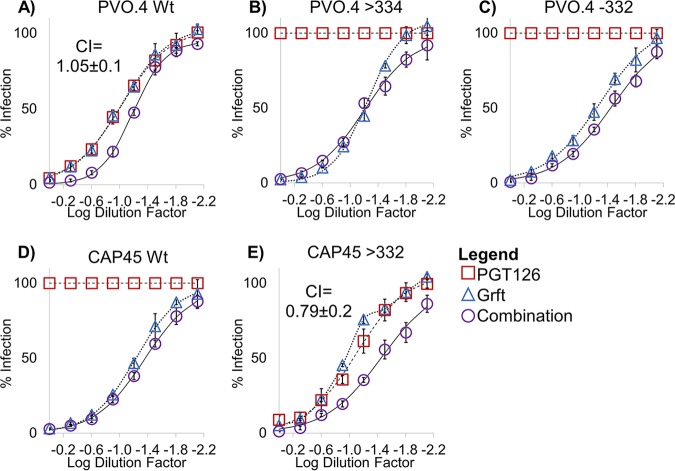
We next investigated the potential of Grft as a combinational therapy with bNAbs. It has previously been shown that Grft is synergistic with the bNAb VRC01, which targets a separate area of gp120 (the CD4 binding site), so we sought to determine if Grft is compatible with another major class of bNAbs which target the glycan at N332 and which may therefore be antagonistic with Grft. As shown in Fig. 5, in strains in which the activity of Grft is relatively weak, the activities of PGT121 and PGT126 are potent, and vice versa. We then analyzed the HIV-1 neutralization activity of Grft when it was used in combination with the two bNAbs in a constant 1:1 IC50 ratio. Grft was able to rescue the activity when bNAbs became ineffective after the loss or the shift of the glycan at N332 (Fig. 6B and C and 7B and D). The effects of the combinations ranged from synergistic to mildly antagonistic, with combination index (CI) values ranging from 0.64 to 1.48 (Fig. 6 and 7).
FIG 5.
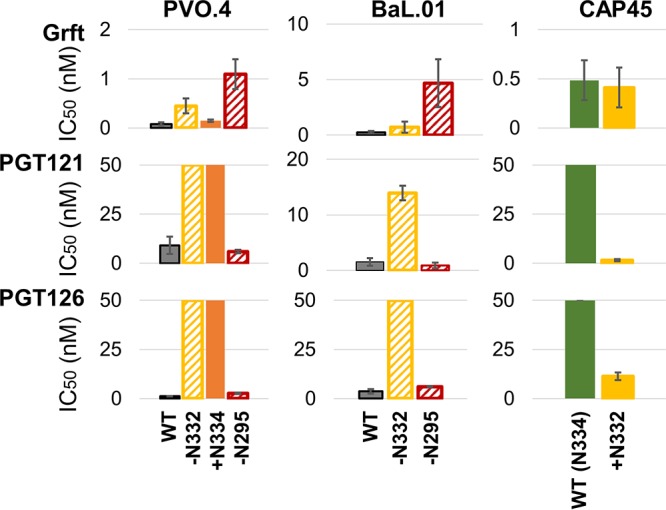
Grft and bNAbs PGT121 and PGT126 have complementary activity against a range of viral variants. IC50 values for Grft, PGT121, and PGT126 in single-round infection assays against various strains and mutants involving glycosylation at N332 and N295 of gp120 are shown. Wild-type strain CAP45 naturally has N334; therefore, +N332 indicates a shift of the glycan to N332. Solid bars indicate the presence of the glycan, and slashed bars indicate the absence of the glycan (also labeled on the x axis, where a plus sign indicates the presence of a glycan and a minus sign indicates the absence of a glycan at that site).
FIG 6.
Activity of the combination of Grft and PGT121 against select wild-type and mutant strains of HIV. (A to E) Dose-response curves of Grft and PGT121 singly and in combination against wild-type HIV strains or strains with the indicated mutations to demonstrate Grft’s ability to rescue in the absence or a shift of a glycan at N332 without antagonistic effects. The combinations were tested by dilution of Grft and PGT121 at a fixed 1:1 ratio proportional to their IC50s. Relatively high starting concentrations were serially diluted to get a dose-response curve. Starting concentrations of Grft were as follows: 2 nM for the PVO.4 wild type (Wt) and PVO.4 with a glycan shift to position 334 (>334), 12 nM for PVO.4 with a deletion of the glycan at 332 (−332), 5.4 nM for the CAP45 wild type, and 3.4 nM for CAP45 with a glycan shifted back to 332 (>332). The starting concentrations of PGT121 were as follows: 60 nM for all PVO.4 variants, 107 nM for the CAP45 wild type, and 20 nM for CAP45 with a glycan shifted back to 332.
FIG 7.
Activity of the combination of Grft and PGT126 against select strains and mutations of HIV. (A to E) Dose-response curves of Grft and PGT126 singly and in combination against wild-type HIV strains or strains with the indicated mutations to demonstrate Grft’s ability to rescue in the absence or shift of a glycan at N332 without antagonistic effects. The combinations were tested by dilution of Grft and PGT126 at a fixed 1:1 ratio proportional to their IC50s. Relatively high starting concentrations were serially diluted to get a dose-response curve. The starting concentrations of Grft were as follows: 1 nM for the PVO.4 wild type (Wt), 2 nM for PVO.4 with a glycan shift to 334 (>334), 12 nM for PVO.4 with a deletion of the glycan at 332 (−332), 5.4 nM for the CAP45 wild type. and 3.4 nM for CAP45 with a glycan shifted back to 332 (>332). Starting concentrations of PGT126 were as follows: 11 nM for the PVO.4 wild type, 60 nM for PVO.4 with the glycan shifted to position 334 or removed from position 332, 60 nM for the CAP45 wild type, and 120 nM for CAP45 with a glycan shifted back to position 332.
Conformational changes induced by griffithsin.
It was previously demonstrated that the naturally occurring presence of glycosite 295 in some strains of gp120 seemed to be a strong determinant of the ability of Grft to induce a conformational change in gp120, exposing the CD4 binding site, and that this correlated well with inhibition potency (57, 58). These experiments used virion capture by a conformationally sensitive antibody (the b12 antibody) after incubation of the virion with Grft. We sought to confirm this finding using the same virion capture assay against wild-type and mutated viruses (Fig. 8). Clade B strains PVO.4 and BaL.01 showed a significant decrease in CD4 site exposure upon incubation with Grft when glycosite N295 was removed (Fig. 8). However, clade C viruses did not show a correlation between the presence of glycosylation at position 295 in gp120 and exposure of the CD4 binding site upon incubation with Grft (Fig. 8), despite the previous results showing that the addition of glycosite N295 to ZM109 did increase its sensitivity to Grft (Table 1).
FIG 8.
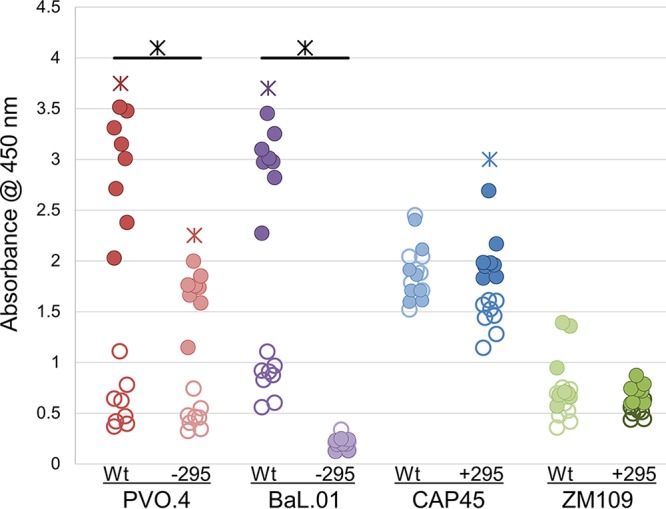
Grft mediates CD4 binding site exposure in clade B viruses. A virion capture assay using MAb b12 to capture HIV virions with an exposed CD4 binding site was performed. Filled circles indicate samples with 4 nM Grft. Open circles indicate samples with 0 nM Grft. PVO.4 and BaL.01 naturally have N295, while CAP45 and ZM109 do not. Colored asterisks (found over PVO.4 WT, PVO.4 -295, BaL.01 Wt, and Cap45 + 295) indicate a comparison of filled versus open circles and that the mean for the samples with 4 nM Grft was significantly (P < 0.0001) different from that for the samples with no Grft. Black asterisks indicate a comparison between the wild-type and mutant of a single strain, showing where the results for the wild-type and mutant samples at 4 nM Grft for a single strain were significantly different from each other (P < 0.0001). Significance was calculated using a one-way analysis of variance with a post hoc pairwise t test and the Bonferroni correction.
DISCUSSION
In the fight against HIV-1, the use of the same drug(s) in both HIV-1 prevention and treatment leads to concerns about the development of viral resistance. Therefore, it is critical to develop other means of HIV-1 prevention that can be cheaply produced (28–30, 32, 33) in a sustained-release, heat-stable formulation (3–7, 91, 92) so that they can be easily distributed in sub-Saharan Africa. The lectin Grft is a strong candidate for use in such a prevention technique. A successful microbicide must be effective against a wide range of HIV-1 strains, so it is imperative to understand the components of Grft inhibition. Grft potency is independent of coreceptor usage by the virus; however, Grft’s direct binding to high-mannose glycans on gp120 necessitates the determination of which glycosylation sites are involved in Grft inhibition, with the goal of optimizing Grft usage either alone or in combination with other inhibitors, such as bNAbs, that have separate modes of action (46, 93).
It has previously been reported that glycosylation at N295 on gp120 is critical for the potency of Grft and other lectins against HIV-1 (35, 67–69, 88, 94, 95). However, there is disagreement about the importance of glycosylation at other sites, with some reports demonstrating that glycosylation at the sites N448, N339, and N234 is also important for Grft inhibition (67–70). Our work provides detailed and comprehensive data on how glycosylation at various sites on gp120 affects Grft potency. In so doing, we also gain insight into the mechanism of Grft action against HIV-1 entry. In the typical strain PVO.4, each high-mannose site was individually eliminated through mutation. It was found that the most important individual site was, as expected, N295; moderate effects were also seen upon individual elimination of glycosites N332 and N448 of gp120. Grft sensitivity to mutations at other high-mannose sites diminished with the distance from the center of the high-mannose patch at cluster 3 (Table 1). Combinations of mutations at this glycan cluster 3 site impaired Grft potency, with the mutant with the triple mutations N295Q/N332Q/N448Q showing an 88-fold reduction in sensitivity. The effectiveness of Grft was also tested against two strains that naturally lack glycosylation at gp120 position 295. One of these strains is less sensitive to Grft (ZM109), and the other is quite sensitive to Grft (CAP45). It was found that adding a glycosylation site at position 295 did increase the sensitivity of both strains to Grft, although the effect was much larger for ZM109 (Table 1).
These results suggest that Grft potency and the importance of glycosylation at N295 are at least partially due to the density of the glycan cluster. The location of N295 places it near the center of the high-mannose patch of gp120 (96, 97). Previous research has shown that the N295 site in gp120 stabilizes the high-mannose patch and that the loss of glycosylation at this site leads to increased enzymatic processing of the glycans in the area (98, 99) (Fig. 4). This would explain the disparate results obtained by other researchers, since the glycan locations in each strain would have slightly different impacts on glycan processing based on the gp120 conformation. Thermodynamic studies have shown that Grft binds more tightly to a saccharide that is closely related to Man-9 than to one that more resembles a processed site, such as Man-5 (37). Also, glycosite N295 is placed at the base of the V3 loop, which means that binding to this site would likely allow Grft to inhibit the gp120 interaction with CCR5, possibly through hindrance of a gp120 conformational change or a CCR5-V3 loop interaction. This reasoning is corroborated by the rescue of Grft potency by the glycan shift from N448 to N446, in the absence of N295, in strains PVO.4 and ZM109 (Table 1 and Fig. 3). The presence of a glycan at N446 may fill the hole in the high-mannose patch created by the loss of N295 and so facilitate Grft binding in an orientation and location similar to those when a glycan is present at N295 (Fig. 4). CAP45, which is sensitive to Grft and which is not glycosylated at position 295, may retain a high-density glycan patch in cluster 3 by virtue of having an additional, nonconserved glycan in the region at N335, which is located near the center of the glycan cluster 3 (Fig. 2). This additional glycan may help to maintain the density of the glycan patch, thereby minimizing glycan processing. Further mutational studies in conjunction with analysis of the glycan composition by mass spectrometry would substantiate these hypotheses.
The glycosylation at site N332 in HIV-1 gp120 is particularly important in the prevention of HIV-1 infection because it provides the binding site for several highly potent bNAbs which have been proposed to be possible microbicides or injectable therapies (85, 86). In some viral strains, the natural glycosite is at N332, while others show a glycan at N334 instead. A range of effectiveness is observed for bNAbs that bind this area, with the antibodies retaining potency against some strains while losing efficacy against others (81, 84, 85). We show that Grft is largely insensitive to shifts in the glycosite at either position 332 or position 448, indicating that Grft could be used to complement bNAbs as part of a plan for the prevention of HIV-1 infection (Table 1 and Fig. 2). Indeed, we found that bNAbs complement the activity of Grft against a variety of strains: the combination of PGT121 or PGT126 with Grft resulted in potent inhibition of HIV-1, regardless of the presence of glycosylation at site N295 or N332, making such a combination valuable for inhibiting a wide range of viral strains. Grft was able to rescue inhibition when the bNAbs lost potency after the shift or removal of the glycan at N332, and the bNAbs had minimal sensitivity to the loss of N295. Despite their preference for neighboring glycans (and, therefore, the possibility of antagonism between Grft and the bNAbs), Grft displayed primarily additive effects when combined with these bNAbs. There was only one instance (the return of a glycan at N332 in CAP45) where Grft and PGT121 showed mildly antagonistic effects when combined. Since only the full removal (of primarily N295) and not the shifting of glycans inhibits Grft potency, Grft’s addition to any combinational therapy appears to be consistently beneficial, as Grft can rescue in instances where shifting glycans negate antibody potency. It has previously been shown that viral escape from Grft primarily requires the complete removal of glycans (88), which would increase viral susceptibility to the immune system.
Grft is able to induce a conformational change in gp120 that is generally correlated to inhibitory potency, increasing gp120 binding to the conformationally sensitive antibody b12 (57, 58). In the current work, we studied this conformational change by making mutations to a set of viruses, allowing a direct comparison in the presence or the absence of particular glycosites in each virus rather than a comparison of separate strains and inference of a correlation. We show that for clade B strains, the conformational change mediated by Grft is dependent on the presence of the glycosylation at position 295 and is correlated with inhibitory potency. However, the clade C strains CAP45 and ZM109 did not show a significant difference in conformational change regardless of the presence or absence of glycosylation at position 295, although they have different properties. CAP45 may be in a b12-binding conformation regardless of the presence of Grft (Fig. 8), and ZM109 shows little conformational change regardless of the presence of glycosylation at the N295 site, but the presence of glycosylation at N295 dramatically enhances its sensitivity to Grft. Taken together, these data suggest that Grft is capable of binding clade B viruses differently than clade C viruses. However, the link between conformational change and inhibition of clade B viruses may be an instance of correlation and not causation.
Overall, this work delineates several modes of interaction between Grft and gp120 and shows that vulnerabilities in Grft inhibition are complementary to vulnerabilities in some bNAbs that bind to glycosylation sites on gp120, such as PGT121 and PGT126. A combination of Grft with either bNAb has additive effects, and Grft can fully rescue inhibition in the presence of glycan shifts in gp120 that inactivate these bNAbs. Therefore, Grft is a suitable candidate for combinational therapies with bNAbs.
MATERIALS AND METHODS
Protein production and purification.
Plasmids containing the Grft gene with an associated N-terminal histidine tag were transformed into Escherichia coli BL21(DE3) (Novagen) competent cells and expressed in minimal medium with 15NH4Cl as the sole nitrogen source. After induction with isopropyl β-d-1-thiogalactopyranoside (IPTG), the cells were incubated at 22°C for 16 h. Once harvested, the pelleted cells were resuspended in 6 M guanidine hydrochloride, 200 mM NaCl, and 50 mM Tris (pH 8.0), which allowed complete solubilization of the proteins from both the inclusion and the supernatant upon cell disruption. The resulting solution was passed through a French press three times at 16,000 lb/in2 and then centrifuged at 27,000 × g for 1 h. The supernatant was loaded onto a nickel chelating column (Qiagen, Hilden, Germany) that had been equilibrated with the same resuspension buffer. The flowthrough was collected and rerun over the column. The column was washed with the resuspension buffer and then a wash buffer with 6 M guanidine hydrochloride, 200 mM NaCl, and 20 mM NaPi (pH 7.2). Grft was eluted using 6 M guanidine hydrochloride, 200 mM NaCl, and 20 mM sodium acetate (pH 4.0) and then added dropwise to a refolding buffer (200 mM NaCl, 1 mM EDTA, 550 mM arginine hydrochloride, 60 mM Tris, pH 8.0) and allowed to stir overnight at 4°C. The solution was then dialyzed in 4 liters 200 mM NaCl and Tris (pH 8.0) for 5 h at 4°C and then transferred to new dialysis buffer (4 liters), and the mixture was stirred at 4°C overnight. The protein solution was then transferred to 4 liters of a low-salt dialysis buffer (80 mM NaCl, 20 mM Tris, pH 8.0). The protein was finally purified on a C4 reversed-phase chromatography column (Vydac, Hesperia, CA) and lyophilized in a Labconco freeze-dry system for long-term storage. All samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) at each step of purification to confirm the proper size of the Grft construct. Nuclear magnetic resonance (NMR) spectroscopy was used to determine that griffithsin was properly folded, as described previously (57).
Viruses and reagents.
Viral plasmids containing the env gene from HIV-1 were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, as follows: PVO, clone 4, strain SVPB11 (referred to as PVO.4), from David Montefiori and Feng Gao (100); ZM109F.PB4, strain SVPC13 (referred to as ZM109), from C. A. Derdeyn and E. Hunter (101); CAP45.2.00.G3, strain SVPC16 (referred to as CAP45), from L. Morris, K. Mlisana, and D. Montefiori (102); and HIV-1 clone BaL.01, catalog number 11445 (referred to as BaL.01), from J. R. Mascola (103). The viral genome pSG3ΔEnv, used to make single-round virus, was from John C. Kappes and Xiaoyun Wu (104, 105). Sequencing primers for all viruses were generated using the Integrated DNA Technologies OligoAnalyzer (https://www.idtdna.com/calc/analyzer) and ordered from ELIM Biopharm (https://www.elimbio.com/services/oligo-synthesis/). TZM-bl cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc. (104, 106–109). 239-FT cells were obtained from Invitrogen (110–112). Both cell lines were cultured in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum, 25 mM HEPES with penicillin-streptomycin or Geneticin, respectively. The anti-HIV-1 gp120 monoclonal antibody (MAb; IgG1 b12) was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Dennis Burton and Carlos Barbas (113–116).
Generation of mutant env-pseudotyped virus DNA.
All mutants were generated by a 2-step PCR using reagents from New England Biolabs Inc. (MA, USA). Step 1 of the PCR used Phusion polymerase to generate fragments of the gene overlapping at the site of the mutational primer, using the protocol suggested by New England Biolabs Inc. Step 2 of the PCR combined the two fragments without primers for 10 cycles, and then an additional 30 cycles were run under normal conditions (the end primers of the gene were added). The products of both steps of PCR were purified using a GeneJET PCR purification kit (Thermo Scientific, MA, USA). DNA was digested with restriction enzymes from New England Biolabs (MA, USA) and then ligated into pCDNA3.1 (for strains PVO.4, BaL.01, and CAP45) or pCR3.1 (for strain ZM109). The presence of mutations was confirmed by DNA sequencing.
Generation of env-pseudotyped single-round virus.
293-FT cells were grown in DMEM with 500 μg/ml Geneticin and cotransfected with 10 ng of the desired envelope plasmid and 10 ng of the pSG3ΔEnv plasmid using the X-tremeGENE HP DNA transfection reagent (Roche/Sigma-Aldrich, St. Louis, MO, USA). Transfected cells grew for 24 to 48 h, depending on the strain. The supernatant was collected, centrifuged, and then filtered through 0.45-μm-pore-size membranes. Viral stocks were stored in low-binding tubes at −80°C.
Single-cycle neutralization assay (TZM-bl cell assay).
On day 1, 104 TZM-bl cells (grown in DMEM with 1% penicillin-streptomycin) were seeded into a 96-well plate and allowed to incubate at 37°C for 18 to 20 h. On day 2, the medium was changed at 1 h 40 min before the time of the assay and added to a final volume of 50 μl per well. A dilution series of Grft, tailored to the strain and mutation of virus to be added, was made in phosphate-buffered saline (PBS), and 20 μl was added to the wells (or 20 μl of PBS for controls). Thirty microliters of single-round virus (at a dilution that would result in a read at an absorbance of 570 nm at approximately 40 min) was added 20 min later, with three rows of the plate being used for the mutated virus and three rows being used for the wild type of the virus being tested. The wells on the edge of the plate were not used due to evaporation effects. On day 3, after 18 to 20 h, the medium was changed and the cells were allowed to incubate for 48 h. On day 5, the cells were lysed using 0.5% NP-40 in PBS, and a substrate solution was added (20 mM KCl, 100 mM 2-mercaptoethanol, 8.3 mM chlorophenol red-β-d-galactopyranoside [Calbiochem] in PBS). The absorbance signal was measured at 570 nm and 630 nm when the reading at 570 nm was approximately 2. The ratio of the signal at 570 nm over that at 630 nm was calculated and compared to the ratio for the controls to determine the percent infection for each well. The 50% inhibitory concentration was determined using a linear equation fitted between the two data points surrounding 50% inhibition for each row, and the three values were averaged for a final IC50. All mutants were tested in triplicate three times or more. After approximately 2 years of data collection, the average IC50 for all strains dropped by approximately 2-fold. To our best determination, this was caused by a change in the composition of the medium from the commercial source. However, the ratios between the wild type and the mutants remained consistent (see Fig. 4 posted at http://faculty.ucmerced.edu/pliwang/?page_id=123); therefore, the data are presented as the fold difference from the wild type. The data were plotted using Microsoft Excel software. For synergy studies, combinations of Grft with bNAbs PGT121 and PGT126 were tested by use of a dilution series at a fixed 1:1 ratio proportional to their IC50s. The combination index (CI) was calculated using CompuSyn software, based on the equations of Chou and Talalay (117). The data were plotted using Microsoft Excel software, with curve fit equation generated in Matlab software.
p24 ELISA and relative infectivity.
Virus samples were diluted to a final volume of 225 μl, and then 25 μl of 5% Triton X-100 was added to the samples. The samples were then vortexed for 20 s, before being incubated at 37°C for 30 min. The amount of p24 was measured using a human immunodeficiency virus type 1 (HIV-1) p24/capsid protein p24 enzyme-linked immunosorbent assay (ELISA) kit from Sinobiological (Beijing, China) according to the manufacturer’s instructions. Relative infectivity was determined by a single-cycle neutralization assay, with all viral dilutions being normalized by the p24 concentrations. The results were calculated using a four-parameter logistic curve on the website for MyAssays analysis software.
Virion capture ELISA.
Exposure of the gp120 CD4 binding site was measured as described previously by the Morris and colleagues (68) and LiWang and colleagues (57). Briefly, high-binding 96-well flat-bottomed plates (Thermo Scientific, MA, USA) were coated overnight at 4°C with 1 μg/well of MAb b12 in PBS. On day 2, the plate was washed and blocked with bovine serum albumin. Pseudovirus samples were preincubated with Grft at 4 nM and 20 nM and a medium-only control for 1 h at 37°C and then added to the b12-coated plate and incubated for another 2 h at 37°C. Then, the plate was washed, and bound virus was lysed with 150 μl 0.5% Triton X-100. One hundred microliters of virus lysate was transferred to a human immunodeficiency virus type 1 (HIV-1) p24/capsid protein p24 ELISA kit from Sinobiological (Beijing, China) and tested according to the manufacturer’s instructions.
ACKNOWLEDGMENTS
The authors gratefully acknowledge James McIntyre and Sage Sorenson for technical assistance.
The work was funded by the National Institutes of Health (grant 1R01AI112011).
REFERENCES
- 1.WHO. 2019. Progress report on HIV, viral hepatitis and sexually transmitted infections, 2016–2021. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Marrazzo JM, Ramjee G, Richardson BA, Gomez K, Mgodi N, Nair G, Palanee T, Nakabiito C, van der Straten A, Noguchi L, Hendrix CW, Dai JY, Ganesh S, Mkhize B, Taljaard M, Parikh UM, Piper J, Mâsse B, Grossman C, Rooney J, Schwartz JL, Watts H, Marzinke MA, Hillier SL, McGowan IM, Chirenje ZM. 2015. Tenofovir-based preexposure prophylaxis for HIV infection among African women. N Engl J Med 372:509–518. doi: 10.1056/NEJMoa1402269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith JM, Rastogi R, Teller RS, Srinivasan P, Mesquita PMM, Nagaraja U, McNicholl JM, Hendry RM, Dinh CT, Martin A, Herold BC, Kiser PF. 2013. Intravaginal ring eluting tenofovir disoproxil fumarate completely protects macaques from multiple vaginal simian-HIV challenges. Proc Natl Acad Sci U S A 110:16145–16150. doi: 10.1073/pnas.1311355110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nel A, van Niekerk N, Kapiga S, Bekker L-G, Gama C, Gill K, Kamali A, Kotze P, Louw C, Mabude Z, Miti N, Kusemererwa S, Tempelman H, Carstens H, Devlin B, Isaacs M, Malherbe M, Mans W, Nuttall J, Russell M, Ntshele S, Smit M, Solai L, Spence P, Steytler J, Windle K, Borremans M, Resseler S, Van Roey J, Parys W, Vangeneugden T, Van Baelen B, Rosenberg Z. 2016. Safety and efficacy of a dapivirine vaginal ring for HIV prevention in women. N Engl J Med 375:2133–2143. doi: 10.1056/NEJMoa1602046. [DOI] [PubMed] [Google Scholar]
- 5.Baeten JM, Palanee-Phillips T, Brown ER, Schwartz K, Soto-Torres LE, Govender V, Mgodi NM, Matovu Kiweewa F, Nair G, Mhlanga F, Siva S, Bekker L-G, Jeenarain N, Gaffoor Z, Martinson F, Makanani B, Pather A, Naidoo L, Husnik M, Richardson BA, Parikh UM, Mellors JW, Marzinke MA, Hendrix CW, van der Straten A, Ramjee G, Chirenje ZM, Nakabiito C, Taha TE, Jones J, Mayo A, Scheckter R, Berthiaume J, Livant E, Jacobson C, Ndase P, White R, Patterson K, Germuga D, Galaska B, Bunge K, Singh D, Szydlo DW, Montgomery ET, Mensch BS, Torjesen K, Grossman CI, Chakhtoura N, Nel A, Rosenberg Z, McGowan I, Hillier S. 2016. Use of a vaginal ring containing dapivirine for HIV-1 prevention in women. N Engl J Med 375:2121–2132. doi: 10.1056/NEJMoa1506110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunawardana M, Baum MM, Smith TJ, Moss JA. 2014. An intravaginal ring for the sustained delivery of antibodies. J Pharm Sci 103:3611–3620. doi: 10.1002/jps.24154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, Herrera C, Coburn J, Olejniczak N, Ziprin P, Kaplan DL, LiWang PJ. 2017. Stabilization and sustained release of HIV inhibitors by encapsulation in silk fibroin disks. ACS Biomater Sci Eng 3:1654–1665. doi: 10.1021/acsbiomaterials.7b00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yavuz B, Morgan JL, Showalter L, Horng KR, Dandekar S, Herrera C, LiWang P, Kaplan DL. 2018. Pharmaceutical approaches to HIV treatment and prevention. Adv Ther 1:1800054. doi: 10.1002/adtp.201800054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhiman JN, Lynch RM. 2017. Broadly neutralizing antibodies as treatment: effects on virus and immune system. Curr HIV/AIDS Rep 14:54–62. doi: 10.1007/s11904-017-0352-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton DR, Mascola JR. 2015. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol 16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wibmer CK, Moore PL, Morris L. 2015. HIV broadly neutralizing antibody targets. Curr Opin HIV AIDS 10:135–143. doi: 10.1097/COH.0000000000000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Kang BH, Ishida E, Zhou T, Griesman T, Sheng Z, Wu F, Doria-Rose NA, Zhang B, McKee K, O'Dell S, Chuang G-Y, Druz A, Georgiev IS, Schramm CA, Zheng A, Joyce MG, Asokan M, Ransier A, Darko S, Migueles SA, Bailer RT, Louder MK, Alam SM, Parks R, Kelsoe G, Von Holle T, Haynes BF, Douek DC, Hirsch V, Seaman MS, Shapiro L, Mascola JR, Kwong PD, Connors M. 2016. Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity 45:1108–1121. doi: 10.1016/j.immuni.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocchi F, Devico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 14.Haqqani AA, Tilton JC. 2013. Entry inhibitors and their use in the treatment of HIV-1 infection. Antiviral Res 98:158–170. doi: 10.1016/j.antiviral.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Jin H, Kagiampakis I, Li P, Liwang PJ. 2010. Structural and functional studies of the potent anti-HIV chemokine variant P2-RANTES. Proteins 78:295–308. doi: 10.1002/prot.22542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao B, Mankowski MK, Snyder BA, Ptak RG, Liwang PJ. 2011. Highly potent chimeric inhibitors targeting two steps of HIV cell entry. J Biol Chem 286:28370–28381. doi: 10.1074/jbc.M111.234799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyd MR, Gustafson KR, McMahon JB, Shoemaker RH, O'Keefe BR, Mori T, Gulakowski RJ, Wu L, Rivera MI, Laurencot CM, Currens MJ, Cardellina JH, Buckheit RW, Nara PL, Pannell LK, Sowder RC, Henderson LE. 1997. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob Agents Chemother 41:1521–1530. doi: 10.1128/AAC.41.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori T, O'Keefe BR, Sowder RC, Bringans S, Gardella R, Berg S, Cochran P, Turpin JA, Buckheit RW, McMahon JB, Boyd MR. 2005. Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J Biol Chem 280:9345–9353. doi: 10.1074/jbc.M411122200. [DOI] [PubMed] [Google Scholar]
- 19.Balzarini J. 2006. Inhibition of HIV entry by carbohydrate-binding proteins. Antiviral Res 71:237–247. doi: 10.1016/j.antiviral.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Akkouh O, Ng TB, Singh SS, Yin C, Dan X, Chan YS, Pan W, Cheung R. 2015. Lectins with anti-HIV activity: a review. Molecules 20:648–668. doi: 10.3390/molecules20010648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar R, Qureshi H, Deshpande S, Bhattacharya J. 2018. Broadly neutralizing antibodies in HIV-1 treatment and prevention. Ther Adv Vaccines Immunother 6:61–68. doi: 10.1177/2515135518800689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lasnik A. 2013. Preclinical safety assessment of griffithsin-based vaginal microbicides. Master’s thesis. University of Louisville, Louisville, KY. [Google Scholar]
- 23.Tsai C-C, Emau P, Jiang Y, Agy MB, Shattock RJ, Schmidt A, Morton WR, Gustafson KR, Boyd MR. 2004. Cyanovirin-N inhibits AIDS virus infections in vaginal transmission models. AIDS Res Hum Retroviruses 20:11–18. doi: 10.1089/088922204322749459. [DOI] [PubMed] [Google Scholar]
- 24.Riddler SA, Zheng L, Durand CM, Ritz J, Koup RA, Ledgerwood J, Bailer RT, Koletar SL, Eron JJ, Keefer MC, Macatangay BJC, Cyktor JC, Mellors JW, Hite M, Clark J, Currin D, Tipton M, Weinman R, Onesi S, Hurley C, Bunce CA, Storey S, Dunaway S, Lambert N, Berzins B, Gottesman J, Leonard M, Ray G, Kittelson P, Benson C, Muttera L, Flynn T, Sbrolla A, Wiggins I, Howard J, Arduino RC, Villamil AE, Kessels L, Spitz T, Campbell D, Kudumu M, Sise T, Nair A, Baer J, Epperson K, Perelson A, Jennings C, Tressler R, Acosta E, Casazza J, Benns A. 2018. Randomized clinical trial to assess the impact of the broadly neutralizing HIV-1 monoclonal antibody VRC01 on HIV-1 persistence in individuals on effective ART. Open Forum Infect Dis 5:ofy242. doi: 10.1093/ofid/ofy242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girard L, Birse K, Holm JB, Gajer P, Humphrys MS, Garber D, Guenthner P, Noël-Romas L, Abou M, McCorrister S, Westmacott G, Wang L, Rohan LC, Matoba N, McNicholl J, Palmer KE, Ravel J, Burgener AD. 2018. Impact of the griffithsin anti-HIV microbicide and placebo gels on the rectal mucosal proteome and microbiome in non-human primates. Sci Rep 8:8059. doi: 10.1038/s41598-018-26313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caskey M, Klein F, Nussenzweig MC. 2019. Broadly neutralizing anti-HIV-1 monoclonal antibodies in the clinic. Nat Med 25:547–553. doi: 10.1038/s41591-019-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kish-Catalone TM, Lu W, Gallo RC, DeVico AL. 2006. Preclinical evaluation of synthetic −2 RANTES as a candidate vaginal microbicide to target CCR5. Antimicrob Agents Chemother 50:1497–1509. doi: 10.1128/AAC.50.4.1497-1509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Keefe BR, Vojdani F, Buffa V, Shattock RJ, Montefiori DC, Bakke J, Mirsalis J, d'Andrea A-L, Hume SD, Bratcher B, Saucedo CJ, McMahon JB, Pogue GP, Palmer KE. 2009. Scaleable manufacture of HIV-1 entry inhibitor griffithsin and validation of its safety and efficacy as a topical microbicide component. Proc Natl Acad Sci U S A 106:6099–6104. doi: 10.1073/pnas.0901506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuqua JL, Hamorsky K, Khalsa G, Matoba N, Palmer KE. 2015. Bulk production of the antiviral lectin griffithsin. Plant Biotechnol J 13:1160–1168. doi: 10.1111/pbi.12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuqua JL, Wanga V, Palmer KE. 2015. Improving the large scale purification of the HIV microbicide, griffithsin. BMC Biotechnol 15:12. doi: 10.1186/s12896-015-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vafaee Y, Staniek A, Mancheno-Solano M, Warzecha H. 2014. A modular cloning toolbox for the generation of chloroplast transformation vectors. PLoS One 9:e110222. doi: 10.1371/journal.pone.0110222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giomarelli B, Schumacher KM, Taylor TE, Sowder RC, Hartley JL, McMahon JB, Mori T. 2006. Recombinant production of anti-HIV protein, griffithsin, by auto-induction in a fermentor culture. Protein Expr Purif 47:194–202. doi: 10.1016/j.pep.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 33.Vamvaka E, Arcalis E, Ramessar K, Evans A, O'Keefe BR, Shattock RJ, Medina V, Stöger E, Christou P, Capell T. 2016. Rice endosperm is cost-effective for the production of recombinant griffithsin with potent activity against HIV. Plant Biotechnol J 14:1427–1437. doi: 10.1111/pbi.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derby N, Lal M, Aravantinou M, Kizima L, Barnable P, Rodriguez A, Lai M, Wesenberg A, Ugaonkar S, Levendosky K, Mizenina O, Kleinbeck K, Lifson JD, Peet MM, Lloyd Z, Benson M, Heneine W, O'Keefe BR, Robbiani M, Martinelli E, Grasperge B, Blanchard J, Gettie A, Teleshova N, Fernández-Romero JA, Zydowsky TM. 2018. Griffithsin carrageenan fast dissolving inserts prevent SHIV HSV-2 and HPV infections in vivo. Nat Commun 9:3881. doi: 10.1038/s41467-018-06349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balzarini J, Tochikura T, Yoshida O, Oki T, Yamamoto N, Takeuchi T. 2005. Targeting the glycans of gp120: a novel approach aimed at the Achilles heel of HIV. Lancet Infect Dis 5:726–731. doi: 10.1016/S1473-3099(05)70271-1. [DOI] [PubMed] [Google Scholar]
- 36.Moncla BJ, Pryke K, Rohan LC, Graebing PW. 2011. Degradation of naturally occurring and engineered antimicrobial peptides by proteases. Adv Biosci Biotechnol 2:404–408. doi: 10.4236/abb.2011.26059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moulaei T, Shenoy SR, Giomarelli B, Thomas C, McMahon JB, Dauter Z, O'Keefe BR, Wlodawer A. 2010. Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity. Structure 18:1104–1115. doi: 10.1016/j.str.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lal M, Lai M, Ugaonkar S, Wesenberg A, Kizima L, Rodriguez A, Levendosky K, Mizenina O, Fernández-Romero J, Zydowsky T. 2018. Development of a vaginal fast-dissolving insert combining griffithsin and carrageenan for potential use against sexually transmitted infections. J Pharm Sci 107:2601–2610. doi: 10.1016/j.xphs.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Emau P, Tian B, O'Keefe BR, Mori T, McMahon JB, Palmer KE, Jiang Y, Bekele G, Tsai CC. 2007. Griffithsin, a potent HIV entry inhibitor, is an excellent candidate for anti-HIV microbicide. J Med Primatol 36:244–253. doi: 10.1111/j.1600-0684.2007.00242.x. [DOI] [PubMed] [Google Scholar]
- 40.Kouokam JC, Huskens D, Schols D, Johannemann A, Riedell SK, Walter W, Walker JM, Matoba N, O'Keefe BR, Palmer KE. 2011. Investigation of griffithsin’s interactions with human cells confirms its outstanding safety and efficacy profile as a microbicide candidate. PLoS One 6:e22635. doi: 10.1371/journal.pone.0022635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kouokam JC, Lasnik AB, Palmer KE. 2016. Studies in a murine model confirm the safety of griffithsin and advocate its further development as a microbicide targeting HIV-1 and other enveloped viruses. Viruses 8:311–315. doi: 10.3390/v8110311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barton C, Kouokam JC, Lasnik AB, Foreman O, Cambon A, Brock G, Montefiori DC, Vojdani F, McCormick AA, O'Keefe BR, Palmer KE. 2014. Activity of and effect of subcutaneous treatment with the broad-spectrum antiviral lectin griffithsin in two laboratory rodent models. Antimicrob Agents Chemother 58:120–127. doi: 10.1128/AAC.01407-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barton C, Kouokam JC, Hurst H, Palmer KE. 2016. Pharmacokinetics of the antiviral lectin griffithsin administered by different routes indicates multiple potential uses. Viruses 8:E331. doi: 10.3390/v8120331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lal M, Lai M, Ugaonkar S, Wesenberg A, Kizima L, Rodriguez A, Levendosky K, Mizenina O, Fernández-Romero J, Zydowsky T. 2018. Self-administered griffithsin and carrageenan containing microbicide fast-dissolving insert as pre-exposure prophylaxis against HIV and HPV infections, p 71, abstr OA20.04. Abstr HIV Res Prev Meet, Madrid, Spain. Mary Ann Liebert, Inc, New Rochelle, NY. [Google Scholar]
- 45.Palmer KE. 2014. Griffithsin-based rectal microbicides for prevention of viral entry (PREVENT). http://grantome.com/grant/NIH/U19-AI113182-04.
- 46.Férir G, Palmer KE, Schols D. 2011. Synergistic activity profile of griffithsin in combination with tenofovir, maraviroc and enfuvirtide against HIV-1 clade C. Virology 417:253–258. doi: 10.1016/j.virol.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 47.Hamorsky KT, Grooms-Williams TW, Husk AS, Bennett LJ, Palmer KE, Matoba N. 2013. Efficient single tobamoviral vector-based bioproduction of broadly neutralizing anti-HIV-1 monoclonal antibody VRC01 in Nicotiana benthamiana plants and utility of VRC01 in combination microbicides. Antimicrob Agents Chemother 57:2076–2086. doi: 10.1128/AAC.02588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Keefe BR, Giomarelli B, Barnard DL, Shenoy SR, Chan PKS, McMahon JB, Palmer KE, Barnett BW, Meyerholz DK, Wohlford-Lenane CL, McCray PB. 2010. Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J Virol 84:2511–2521. doi: 10.1128/JVI.02322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meuleman P, Albecka A, Belouzard S, Vercauteren K, Verhoye L, Wychowski C, Leroux-Roels G, Palmer KE, Dubuisson J. 2011. Griffithsin has antiviral activity against hepatitis C virus. Antimicrob Agents Chemother 55:5159–5167. doi: 10.1128/AAC.00633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nixon B, Stefanidou M, Mesquita PMM, Fakioglu E, Segarra T, Rohan L, Halford W, Palmer KE, Herold BC. 2013. Griffithsin protects mice from genital herpes by preventing cell-to-cell spread. J Virol 87:6257–6269. doi: 10.1128/JVI.00012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishag HZA, Li C, Huang L, Sun M, Wang F, Ni B, Malik T, Chen P, Mao X. 2013. Griffithsin inhibits Japanese encephalitis virus infection in vitro and in vivo. Arch Virol 158:349–358. doi: 10.1007/s00705-012-1489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levendosky K, Mizenina O, Martinelli E, Jean-Pierre N, Kizima L, Rodriguez A, Kleinbeck K, Bonnaire T, Robbiani M, Zydowsky TM, O'Keefe BR, Fernández-Romero JA. 2015. Griffithsin and carrageenan combination to target HSV-2 and HPV. Antimicrob Agents Chemother 59:7290–7298. doi: 10.1128/AAC.01816-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Millet JK, Séron K, Labitt RN, Danneels A, Palmer KE, Whittaker GR, Dubuisson J, Belouzard S. 2016. Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin. Antiviral Res 133:1–8. doi: 10.1016/j.antiviral.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ziółkowska NE, O'Keefe BR, Mori T, Zhu C, Giomarelli B, Vojdani F, Palmer KE, McMahon JB, Wlodawer A. 2006. Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding. Structure 14:1127–1135. doi: 10.1016/j.str.2006.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziółkowska NE, Shenoy SR, O'Keefe BR, Wlodawer A. 2007. Crystallographic studies of the complexes of antiviral protein griffithsin with glucose and N-acetylglucosamine. Protein Sci 16:1485–1489. doi: 10.1110/ps.072889407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xue J, Gao Y, Hoorelbeke B, Kagiampakis I, Zhao B, Demeler B, Balzarini J, LiWang PJ. 2012. The role of individual carbohydrate-binding sites in the function of the potent anti-HIV lectin griffithsin. Mol Pharm 9:2613–2625. doi: 10.1021/mp300194b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xue J, Hoorelbeke B, Kagiampakis I, Demeler B, Balzarini J, LiWang PJ. 2013. The griffithsin dimer is required for high-potency inhibition of HIV-1: evidence for manipulation of the structure of gp120 as part of the griffithsin dimer mechanism. Antimicrob Agents Chemother 57:3976–3989. doi: 10.1128/AAC.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alexandre KB, Gray ES, Pantophlet R, Moore PL, McMahon JB, Chakauya E, O'Keefe BR, Chikwamba R, Morris L. 2011. Binding of the mannose-specific lectin, griffithsin, to HIV-1 gp120 exposes the CD4-binding site. J Virol 85:9039–9050. doi: 10.1128/JVI.02675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lusvarghi S, Lohith K, Morin-Leisk J, Ghirlando R, Hinshaw JE, Bewley CA. 2016. Binding site geometry and subdomain valency control effects of neutralizing lectins on HIV-1 viral particles. ACS Infect Dis 2:882–891. doi: 10.1021/acsinfecdis.6b00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lusvarghi S, Bewley C. 2016. Griffithsin: an antiviral lectin with outstanding therapeutic potential. Viruses 8:296. doi: 10.3390/v8100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Center RJ, Schuck P, Leapman RD, Arthur LO, Earl PL, Moss B, Lebowitz J. 2001. Oligomeric structure of virion-associated and soluble forms of the simian immunodeficiency virus envelope protein in the prefusion activated conformation. Proc Natl Acad Sci U S A 98:14877–14882. doi: 10.1073/pnas.261573898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu P, Chertova E, Bess J, Lifson JD, Arthur LO, Liu J, Taylor KA, Roux KH. 2003. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci U S A 100:15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Go EP, Irungu J, Zhang Y, Dalpathado DS, Liao HX, Sutherland LL, Munir Alam S, Haynes BF, Desaire H. 2008. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes’ accessibility. J Proteome Res 7:1660–1674. doi: 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, Burton DR, Crispin M, Scanlan C. 2011. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One 6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem 265:10373–10382. [PubMed] [Google Scholar]
- 66.Balzarini J, Van Laethem K, Peumans WJ, Van Damme EJM, Bolmstedt A, Gago F, Schols D. 2006. Mutational pathways, resistance profile, and side effects of cyanovirin relative to human immunodeficiency virus type 1 strains with N-glycan deletions in their gp120 envelopes. J Virol 80:8411–8421. doi: 10.1128/JVI.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moulaei T, Alexandre KB, Shenoy SR, Meyerson JR, Krumpe LR, Constantine B, Wilson J, Buckheit RW, McMahon JB, Subramaniam S, Wlodawer A, O'Keefe BR. 2015. Griffithsin tandemers: flexible and potent lectin inhibitors of the human immunodeficiency virus. Retrovirology 12:6. doi: 10.1186/s12977-014-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alexandre KB, Gray ES, Lambson BE, Moore PL, Choge IA, Mlisana K, Karim SSA, McMahon J, O'Keefe B, Chikwamba R, Morris L. 2010. Mannose-rich glycosylation patterns on HIV-1 subtype C gp120 and sensitivity to the lectins, griffithsin, cyanovirin-N and scytovirin. Virology 402:187–196. doi: 10.1016/j.virol.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang X, Jin W, Griffin GE, Shattock RJ, Hu Q. 2011. Removal of two high-mannose N-linked glycans on gp120 renders human immunodeficiency virus 1 largely resistant to the carbohydrate-binding agent griffithsin. J Gen Virol 92:2367–2373. doi: 10.1099/vir.0.033092-0. [DOI] [PubMed] [Google Scholar]
- 70.Hu B, Du T, Li C, Luo S, Liu Y, Huang X, Hu Q. 2015. Sensitivity of transmitted and founder HIV-1 envelopes to carbohydrate binding agents griffithsin, cyanovirin-N and Galanthus nivalis agglutinin. J Gen Virol 96:3660–3666. doi: 10.1099/jgv.0.000299. [DOI] [PubMed] [Google Scholar]
- 71.Stanfield R, Cabezas E, Satterthwait A, Stura E, Profy A, Wilson I. 1999. Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing Fabs. Structure 7:131–142. doi: 10.1016/S0969-2126(99)80020-3. [DOI] [PubMed] [Google Scholar]
- 72.Wilen CB, Tilton JC, Doms RW. 2012. Molecular mechanisms of HIV entry. Adv Exp Med Biol 726:223–242. doi: 10.1007/978-1-4614-0980-9_10. [DOI] [PubMed] [Google Scholar]
- 73.Ma X, Lu M, Gorman J, Terry DS, Hong X, Zhou Z, Zhao H, Altman RB, Arthos J, Blanchard SC, Kwong PD, Munro JB, Mothes W. 2018. HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. Elife 7:e34271. doi: 10.7554/eLife.34271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kong L, Wilson IA, Kwong PD. 2015. Crystal structure of a fully glycosylated HIV-1 gp 120 core reveals a stabilizing role for the glycan at Asn262. Proteins 83:590–596. doi: 10.1002/prot.24747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Julg B, Sok D, Schmidt SD, Abbink P, Newman RM, Broge T, Linde C, Nkolola J, Le K, Su D, Torabi J, Pack M, Pegu A, Allen TM, Mascola JR, Burton DR, Barouch DH. 2017. Protective efficacy of broadly neutralizing antibodies with incomplete neutralization activity against SHIV in rhesus monkeys. J Virol 91:e01187-17. doi: 10.1128/JVI.01187-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Badamchi-Zadeh A, Tartaglia LJ, Abbink P, Bricault CA, Liu P-T, Boyd M, Kirilova M, Mercado NB, Nanayakkara OS, Vrbanac VD, Tager AM, Larocca RA, Seaman MS, Barouch DH. 2018. Therapeutic efficacy of vectored PGT121 gene delivery in HIV-1-infected humanized mice. J Virol 92:e01925-17. doi: 10.1128/JVI.01925-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang H-W, Shekhar K, Gupta S, Nkolola JP, Seaman MS, Smith KM, Borducchi EN, Cabral C, Smith JY, Blackmore S, Sanisetty S, Perry JR, Beck M, Lewis MG, Rinaldi W, Chakraborty AK, Poignard P, Nussenzweig MC, Burton DR. 2013. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stephenson KE, Julg B, Ansel J, Walsh SR, Tan CS, Maxfield L, Abbink P, Gelderblom HC, Priddy F, DeCamp AC, Arduino R, DeJesus E, Tomaras G, Seaman MS, Barouch D. 2019. Therapeutic activity of PGT121 monoclonal antibody in HIV-infected adults, abstr 145. Abstr Conf Retroviruses Opportunist Infect. CROI Foundation/IAS-USA, Seattle, WA. [Google Scholar]
- 80.Moldt B, Le KM, Carnathan DG, Whitney JB, Schultz N, Lewis MG, Borducchi EN, Smith KM, Mackel JJ, Sweat SL, Hodges AP, Godzik A, Parren P, Silvestri G, Barouch DH, Burton DR. 2016. Neutralizing antibody affords comparable protection against vaginal and rectal simian/human immunodeficiency virus challenge in macaques. AIDS 30:1543–1551. doi: 10.1097/QAD.0000000000001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams M-R, Lambson BE, Ranchobe N, Ping L, Ngandu N, Abdool Karim Q, Abdool Karim SS, Swanstrom RI, Seaman MS, Williamson C, Morris L. 2012. Evolution of an HIV glycan—dependent broadly neutralizing antibody epitope through immune escape. Nat Med 18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sok D, Doores KJ, Briney B, Le KM, Saye-Francisco KL, Ramos A, Kulp DW, Julien J-P, Menis S, Wickramasinghe L, Seaman MS, Schief WR, Wilson IA, Poignard P, Burton DR. 2014. Promiscuous glycan site recognition by antibodies to the high-mannose patch of gp120 broadens neutralization of HIV. Sci Transl Med 6:236ra63. doi: 10.1126/scitranslmed.3008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rademeyer C, Korber B, Seaman MS, Giorgi EE, Thebus R, Robles A, Sheward DJ, Wagh K, Garrity J, Carey BR, Gao H, Greene KM, Tang H, Bandawe GP, Marais JC, Diphoko TE, Hraber P, Tumba N, Moore PL, Gray GE, Kublin J, McElrath MJ, Vermeulen M, Middelkoop K, Bekker L-G, Hoelscher M, Maboko L, Makhema J, Robb ML, Abdool Karim S, Abdool Karim Q, Kim JH, Hahn BH, Gao F, Swanstrom R, Morris L, Montefiori DC, Williamson C. 2016. Features of recently transmitted HIV-1 clade C viruses that impact antibody recognition: implications for active and passive immunization. PLoS Pathog 12:e1005742. doi: 10.1371/journal.ppat.1005742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bouvin-Pley M, Morgand M, Moreau A, Jestin P, Simonnet C, Tran L, Goujard C, Meyer L, Barin F, Braibant M. 2013. Evidence for a continuous drift of the HIV-1 species towards higher resistance to neutralizing antibodies over the course of the epidemic. PLoS Pathog 9:e1003477. doi: 10.1371/journal.ppat.1003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garces F, Sok D, Kong L, McBride R, Kim HJ, Saye-Francisco KF, Julien J-P, Hua Y, Cupo A, Moore JP, Paulson JC, Ward AB, Burton DR, Wilson IA. 2014. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell 159:69–79. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Garces F, Lee JH, de Val N, de la Pena AT, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, Sanders RW, Ward AB, Wilson IA. 2015. Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity 43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kong L, Lee JH, Doores KJ, Murin CD, Julien J-P, McBride R, Liu Y, Marozsan A, Cupo A, Klasse P-J, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. 2013. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alexandre KB, Moore PL, Nonyane M, Gray ES, Ranchobe N, Chakauya E, McMahon JB, O'Keefe BR, Chikwamba R, Morris L. 2013. Mechanisms of HIV-1 subtype C resistance to GRFT, CV-N and SVN. Virology 446:66–76. doi: 10.1016/j.virol.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang X, Jin W, Hu K, Luo S, Du T, Griffin GE, Shattock RJ, Hu Q. 2012. Highly conserved HIV-1 gp120 glycans proximal to CD4-binding region affect viral infectivity and neutralizing antibody induction. Virology 423:97–106. doi: 10.1016/j.virol.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 90.Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 91.Morrow RJ, Woolfson AD, Donnelly L, Curran R, Andrews G, Katinger D, Malcolm RK. 2011. Sustained release of proteins from a modified vaginal ring device. Eur J Pharm Biopharm 77:3–10. doi: 10.1016/j.ejpb.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Friend DR. 2011. Intravaginal rings: controlled release systems for contraception and prevention of transmission of sexually transmitted infections. Drug Deliv Transl Res 1:185–193. doi: 10.1007/s13346-011-0024-4. [DOI] [PubMed] [Google Scholar]
- 93.Férir G, Huskens D, Palmer KE, Boudreaux DM, Swanson MD, Markovitz DM, Balzarini J, Schols D. 2012. Combinations of griffithsin with other carbohydrate-binding agents demonstrate superior activity against HIV type 1, HIV type 2, and selected carbohydrate-binding agent-resistant HIV type 1 strains. AIDS Res Hum Retroviruses 28:1513–1523. doi: 10.1089/AID.2012.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Balzarini J, Van Laethem K, Hatse S, Froeyen M, Van Damme E, Bolmstedt A, Peumans W, De Clercq E, Schols D. 2005. Marked depletion of glycosylation sites in HIV-1 gp120 under selection pressure by the mannose-specific plant lectins of Hippeastrum hybrid and Galanthus nivalis. Mol Pharmacol 67:1556–1565. doi: 10.1124/mol.104.005082. [DOI] [PubMed] [Google Scholar]
- 95.Balzarini J, Van Laethem K, Hatse S, Froeyen M, Peumans W, Van Damme E, Schols D. 2005. Carbohydrate-binding agents cause deletions of highly conserved glycosylation sites in HIV GP120: a new therapeutic concept to hit the Achilles heel of HIV. J Biol Chem 280:41005–41014. doi: 10.1074/jbc.M508801200. [DOI] [PubMed] [Google Scholar]
- 96.Lemmin T, Soto C, Stuckey J, Kwong PD. 2017. Microsecond dynamics and network analysis of the HIV-1 SOSIP Env trimer reveal collective behavior and conserved microdomains of the glycan shield. Structure 25:1631–1639. doi: 10.1016/j.str.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 97.Behrens A-J, Crispin M. 2017. Structural principles controlling HIV envelope glycosylation. Curr Opin Struct Biol 44:125–133. doi: 10.1016/j.sbi.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coss KP, Vasiljevic S, Pritchard LK, Krumm SA, Glaze M, Madzorera S, Moore PL, Crispin M, Doores KJ. 2016. HIV-1 glycan density drives the persistence of the mannose patch within an infected individual. J Virol 90:11132–11144. doi: 10.1128/JVI.01542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pritchard LK, Vasiljevic S, Ozorowski G, Seabright GE, Cupo A, Ringe R, Kim HJ, Sanders RW, Doores KJ, Burton DR, Wilson IA, Ward AB, Moore JP, Crispin M. 2015. Structural constraints determine the glycosylation of HIV-1 envelope trimers. Cell Rep 11:1604–1613. doi: 10.1016/j.celrep.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human Immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 102.Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BTM, Hunter E, Hahn BH, Montefiori DC. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in southern Africa. J Virol 80:11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li Y, Svehla K, Mathy NL, Voss G, Mascola JR, Wyatt R. 2006. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J Virol 80:1414–1426. doi: 10.1128/JVI.80.3.1414-1426.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC, Wu X. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/aac.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 106.Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. 2009. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol 83:8289–8292. doi: 10.1128/JVI.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takeuchi Y, McClure MO, Pizzato M. 2008. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J Virol 82:12585–12588. doi: 10.1128/JVI.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol 74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Graham FL, Smiley J, Russell WC, Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 111.Harrison T, Graham F, Williams J. 1977. Host-range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology 77:319–329. doi: 10.1016/0042-6822(77)90428-7. [DOI] [PubMed] [Google Scholar]
- 112.Naldini L, Blömer U, Gage FH, Trono D, Verma IM. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci U S A 93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Burton DR, Barbas CF, Persson MA, Koenig S, Chanock RM, Lerner RA, Lerner RA. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci U S A 88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Barbas CF, Björling E, Chiodi F, Dunlop N, Cababa D, Jones TM, Zebedee SL, Persson MA, Nara PL, Norrby E. 1992. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc Natl Acad Sci U S A 89:9339–9343. doi: 10.1073/pnas.89.19.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roben P, Moore JP, Thali M, Sodroski J, Barbas CF III, Burton DR. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol 68:4821–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Burton D, Pyati J, Koduri R, Sharp S, Thornton G, Parren P, Sawyer L, Hendry R, Dunlop N, Nara P. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 117.Chou T-C, Talalay P. 1984. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 118.Stewart-Jones GBE, Soto C, Lemmin T, Chuang G-Y, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens A-J, Bylund T, Choi CWW, Davison JRR, Georgiev ISS, Joyce MGG, Do Kwon YD, Pancera M, Taft J, Yang Y, Zhang B, Shivatare SSS, Shivatare VSS, Lee C-C, Wu C-Y, Bewley CAA, Burton DRR, Koff WCC, Connors M, Crispin M, Baxa U, Korber BTT, Wong C-H, Mascola JRR, Kwong P. 2016. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 120.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. 2018. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci 27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]