The rapid emergence of multidrug resistance among bacterial pathogens has become a significant challenge to human health in our century. Therefore, development of next-generation antibacterial compounds is an urgent need. Two-component signal transduction systems (TCS) are stimulus-response coupling devices that allow bacteria to sense and elaborate adaptive responses to changing environmental conditions, including the challenges that pathogenic bacteria face inside the host.
KEYWORDS: PhoP/PhoQ two-component system, Salmonella, antivirulence, drug discovery, quinazolines
ABSTRACT
The rapid emergence of multidrug resistance among bacterial pathogens has become a significant challenge to human health in our century. Therefore, development of next-generation antibacterial compounds is an urgent need. Two-component signal transduction systems (TCS) are stimulus-response coupling devices that allow bacteria to sense and elaborate adaptive responses to changing environmental conditions, including the challenges that pathogenic bacteria face inside the host. The differential presence of TCS, present in bacteria but absent in the animal kingdom, makes them attractive targets in the search for new antibacterial compounds. In Salmonella enterica, the PhoP/PhoQ two-component system controls the expression of crucial phenotypes that define the ability of the pathogen to establish infection in the host. We now report the screening of 686 compounds from a GlaxoSmithKline published kinase inhibitor set in a high-throughput whole-cell assay that targets Salmonella enterica serovar Typhimurium PhoP/PhoQ. We identified a series of quinazoline compounds that showed selective and potent downregulation of PhoP/PhoQ-activated genes and define structural attributes required for their efficacy. We demonstrate that their bioactivity is due to repression of the PhoQ sensor autokinase activity mediated by interaction with its catalytic domain, acting as competitive inhibitors of ATP binding. While noncytotoxic, the hit molecules exhibit antivirulence effect by blockage of S. Typhimurium intramacrophage replication. Together, these features make these quinazoline compounds stand out as exciting leads to develop a therapeutic intervention to fight salmonellosis.
INTRODUCTION
Infection by bacteria of the genus Salmonella constitutes a major global burden of human morbidity and mortality (1). The most recent estimates suggest that nontyphoidal Salmonella serovars are the cause of 155,000 deaths each year (1). One in four diarrheal diseases can trace its origin back to Salmonella serovars (1). Nontyphoidal Salmonella serovars generally cause 2-to-7-day self-limiting illness in immunocompetent individuals. In high-income countries, these infections are characterized by acute fever, abdominal cramps, diarrhea, nausea, and vomiting. However, in low-income, underdeveloped regions, nontyphoidal Salmonella serovars are the most common bacterial bloodstream isolates, and infections by those serovars result in fatality in 20% to 25% cases (2). Successful treatment of these infections has been increasingly hindered by the emergence of strains that are resistant to multiple antibacterial drugs, including those representing the last line of defense, namely, extended-spectrum cephalosporins and fluoroquinolones (3, 4). Salmonella is a growing global health risk with increasing prevalence and continuous emerging resistance. Efforts to identify novel antimicrobial agents and mechanisms to counter Salmonella-borne infections are a key priority to combat infection (5).
There are a number of alternatives to already-exploited bacterial targets. One promising strategy consists of interfering with bacterial pathogenicity traits using pathoblockers, antivirulence compounds that target crucial mechanisms in the development of disease. As these agents would not exert bactericidal or bacteriostatic effects, it is thought that, depending on the importance of the chosen target for bacterial survival throughout the infection path, antivirulence strategies might allow the host to mount an immune response that would favor the clearance of the pathogen (6–8).
In Salmonella enterica, the PhoP/PhoQ two-component system (TCS) orchestrates the expression of phenotypes that allow the pathogen to thrive in the mammalian host. PhoP/PhoQ consists of a canonical form of signal transduction TCS, which couples environmental stimuli to tailored bacterial adaptive responses. This TCS is comprised by PhoQ, a transmembrane sensor with bifunctional (histidine kinase [HK]/phosphatase) activity, and PhoP, a cytoplasmic response regulator. PhoQ and PhoP communicate each other by a phosphorelay mechanism (9). The PhoP/PhoQ regulon comprises more than 100 genes whose expression is directly or indirectly controlled by PhoP activity. These genes include those that are required for Mg2+ homeostasis (10), resistance to acidic pH, and determination of bacterial susceptibility to cationic antimicrobial peptides produced by the host (11, 12). The system is also involved in controlling the mechanism of bacterial entry into intestinal epithelial cells. Once the bacteria are inside either nonphagocytic or phagocytic host cells, expression of PhoP-modulated genes defines the intracellular survival and proliferation capacity of Salmonella (13, 14). Host phagocytes act as vehicles for Salmonella dispersal to vital organs such as liver and spleen (15) and therefore play a major role in the development of systemic infection.
To identify new starting points to target this mechanism, we screened the first and second generations of the GlaxoSmithKline (GSK) published kinase inhibitor set (PKIS) (16) for compounds that could inhibit Salmonella enterica serovar Typhimurium PhoP/PhoQ activity. The PKIS was designed to include a range of chemotypes and eukaryotic kinome inhibition profiles. Compounds in the PKIS were originally prepared in lead optimization efforts to target human serine, threonine, and tyrosine kinases. In fact, scaffolds of the PKIS library are also present in chemotypes that include FDA-approved drugs such as lapatinib (17) and erlotinib (18), tyrosine kinase inhibitors that target the epidermal growth factor receptor.
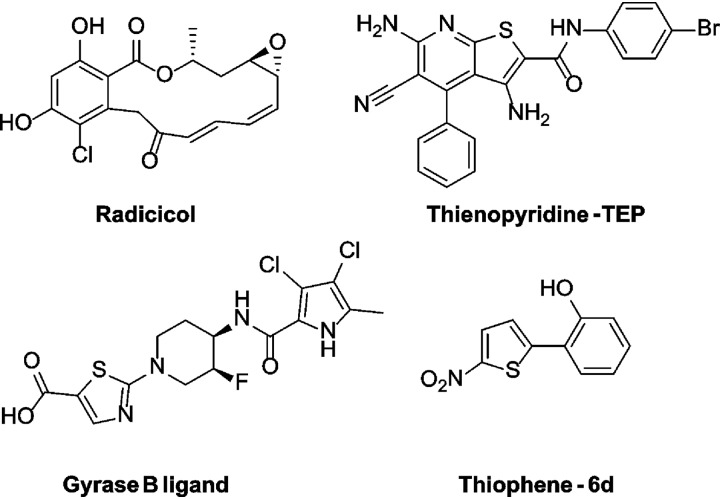
Histidine kinases (HK), including PhoQ, are members of the GHKL family, which also comprises GyrB, Hsp90, and MutL. The HK catalytic domain is characterized by a unique ATP-binding Bergerat fold, which topologically differs from the fold of Ser/Thr/Tyr sensor kinases (19). The Bergerat fold has been explored previously for the generation of HK inhibitors. They include the GHKL inhibitors radicicol and thienopyridone, a modified gyrase B ligand, and a thiophenes-containing scaffold (Fig. 1) (19–21). Each of these scaffolds was shown to inhibit HK activity by competing with ATP in an autophosphorylation reaction. To our knowledge, these series have not been investigated further (22–25). Despite the structural differences between HK and Ser/Thr/Tyr sensor kinases, we expected the PKIS to be a useful screening set to discover lead molecules able to target PhoQ HK activity, resulting in the identification and development of antivirulence agents useful to treat Salmonella infections.
FIG 1.
Chemical structures of previously reported inhibitors of histidine kinases.
RESULTS AND DISCUSSION
Primary screening and selection of lead compounds.
PKIS, a library containing 686 compounds from kinase drug discovery programs, was screened based on our previously established format to identify inhibitors of the Salmonella Typhimurium PhoP/PhoQ signal transduction system (26, 27). In a first round of screening, isogenic Salmonella Typhimurium MS14028 strains carrying transcriptional lacZ fusions to virK, a representative PhoP-activated gene, were grown in lysogeny broth (LB) supplemented with a 50 μM concentration of each compound. The resulting percentage of β-galactosidase activity repression relative to the value obtained without compound was determined (see Data Set S1 in the supplemental material). Simultaneously, we monitored bacterial growth and eliminated from further analysis those compounds that caused alterations in growth curves as described in Materials and Methods (Data Set S1). We set >35% inhibition of virK reporter activity levels as the threshold for compound progression and performed a second round of selection that included a second PhoP-dependent reporter (pagC-lacZ) and a PhoP/PhoQ-unrelated transcriptional reporter (tppB-lacZ), whose expression is regulated by the EnvZ/OmpR TCS (28, 29), to exclude compounds that might cause indiscriminate inhibition of HKs (Data Set S1).
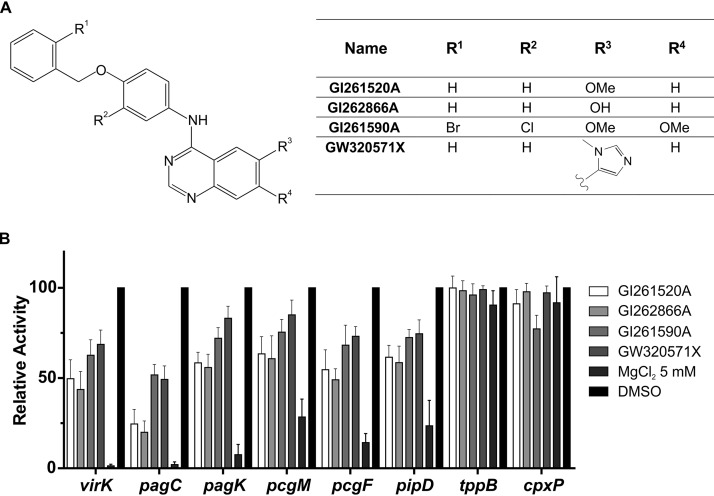
Next, we selected molecules that showed ≥50% downregulation of the activity levels of the virK and pagC reporters and no significant alteration of tppB reporter activity levels. This step resulted in the identification of two hit compounds, GI261520A and GI262866A, differing in the substitution of the 6 position of the quinazoline scaffold, having a methoxy or an alcohol substituent, respectively (Fig. 2A). To verify the significance of this selection for the PhoP/PhoQ activity, we added four additional reporters from the PhoP/PhoQ regulon (pagK, pcgM, pcgF, and pipD) and reinforced the selectivity criterion by addition of another unrelated transcriptional reporter from the CpxAR TCS regulon, cpxP-lacZ, to our β-galactosidase assays. We also added stringency to the test by lowering the concentration of the compounds to 25 μM (Fig. 2B). To obtain preliminary hints regarding identification of the chemical moieties responsible for the inhibitory effect of the selected leads, we searched the PKIS library for additional, related quinazoline analogs and found GI261590A and GW320571X. These two compounds were included in the comparisons performed in subsequent assays (Fig. 2B). These results further substantiated the selection of compounds, showing that the two hit compounds exerted a 36% to 80% repressive effect on the β-galactosidase activity levels measured from the six PhoP-controlled reporters tested. This result indicated that substituents in positions R1, R2, R3, and R4 were relevant for the core quinazoline moiety to confer the capacity to downregulate PhoP/PhoQ.
FIG 2.
Inhibitory action of selected compounds from the primary screen of the PKIS molecules. (A) Chemical structures of four molecules selected in the screening of the published kinase inhibitor set (PKIS) from GlaxoSmithKline (GSK). Numbers correspond to the reference code for each molecule assigned by GSK. (B) Inhibition action was calculated on the basis of the β-galactosidase activity of lacZ transcriptional fusions to six different PhoP-activated reporter genes (virK, pagC, pagK, pcgM, pcgF, and pipD) and two PhoP-unrelated control reporter genes (tppB and cpxP). Cells were grown overnight either in LB supplemented with 0.25% DMSO (vehicle for the compounds) or in LB plus the indicated compound at a concentration of 25 μM. Relative activity levels were calculated by taking the values obtained with the sole addition of 0.25% DMSO as 100%. Results represent averages from three independent assays performed in duplicate, and error bars correspond to standard deviations (SD).
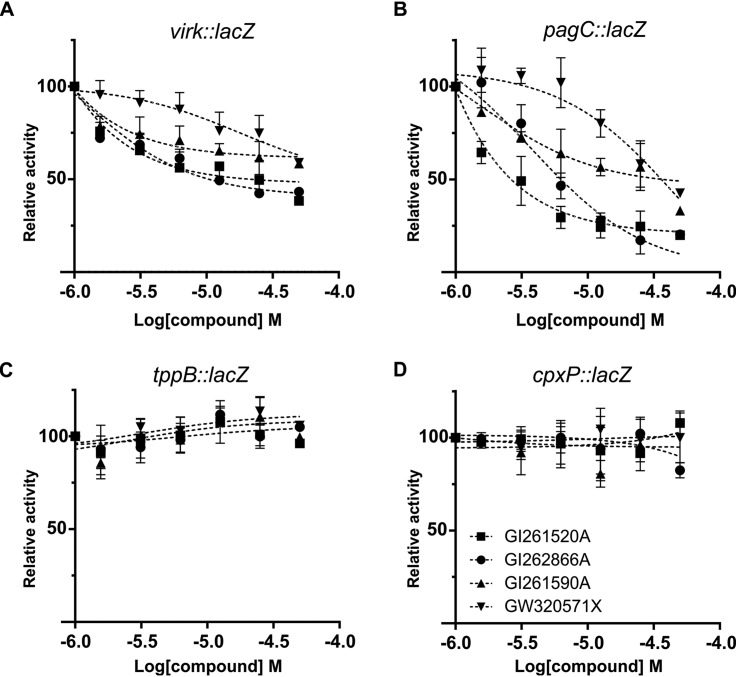
Dose-dependent inhibition was verified by performing β-galactosidase assays from virK and pagC reporters using a final concentration of the compounds in the 0 to 50 μM range (Fig. 3A and B). Consistently, although EnvZ/OmpR and CpxAR belong to the same “OmpR subfamily” as PhoP/PhoQ (30), the two selected compounds showed negligible effects on the β-galactosidase activity expressed from the EnvZ/OmpR or CpxAR-dependent reporter genes (Fig. 3C and D), indicating high selectivity of these pharmacophores with respect to the PhoP/PhoQ TCS.
FIG 3.
Dose-response inhibition effect of selected molecules on the activity of S. Typhimurium PhoP-activated reporter genes. β-Galactosidase activity from (A) virK::lacZ, (B) pagC::lacZ, (C) tppB::lacZ, or (D) cpxP::lacZ transcriptional fusions was measured in cells grown overnight in LB with the indicated final concentration of the corresponding compound. β-Galactosidase activity was measured as described in Materials and Methods. Results represent averages from three independent assays performed in duplicate, and error bars correspond to SD.
Each PhoP-controlled gene has been shown to display differential PhoP-binding sequence features, including orientation, coregulatory cis-acting motifs located at different distances of the DNA PhoP box, and/or the need to overcome silencing by nucleoid-associated proteins such as H-NS (31). These differences imply distinct requirements of activated PhoP levels required for induction or repression and also characteristic expression timings for each PhoP-dependent gene in the presence of a given PhoP/PhoQ input signal (32, 33). This accounts for the different extents of decrease of β-galactosidase activity levels mediated by the compounds for each of the six PhoP-regulated reporters, in spite of the fact that all of them harbor a conserved PhoP recognition site (27, 33, 34). Moreover, although the downregulation of the activity levels driven by the six reporters strongly suggested an inhibitory effect on the TCS, we cannot at this point rule out the possibility that the compounds might exert an indirect effect on PhoP/PhoQ.
GI262866A and GI261520A target PhoQ HK activity.
Our results indicate that GI262866A and GI261520A were the most effective compounds in the downregulation of the activity levels of PhoP-dependent transcriptional reporters. To examine whether these compounds exert a direct effect on PhoP/PhoQ and taking into account their original design as kinase inhibitors, we sought to explore whether PhoQ autophosphorylation activity, the first phosphotransfer checkpoint in the PhoP/PhoQ signal transduction cascade, was targeted by the selected quinazolines. We isolated PhoQ-enriched membrane vesicles derived from the S. Typhimurium phoQ strain that expresses PhoQ from the pUHE-21–2::phoQ plasmid, as we described previously (27) (also see Materials and Methods). Salmonella-derived vesicles were prepared from bacteria grown in LB with or without the addition of the indicated concentration of each compound. To determine PhoQ autophosphorylation activity, vesicles were incubated with a [γ-32P]ATP-containing reaction medium, samples were analyzed by SDS-PAGE followed by autoradiography, and the intensity of the bands was quantified by densitometric analysis as described previously (27). As shown in Fig. 4 (see also Fig. S2 [please see supplemental material at http://www.ibr-conicet.gov.ar/wp-content/uploads/2019/10/Carabajal-et-al-AAC01744-19-Supplementary-Information-Combined.pdf]), PhoQ autophosphorylation was inhibited by either GI262866A or GI261520A in a dose-dependent manner over the 6 to 50 μM concentration range tested, demonstrating that the chosen compounds target PhoQ autokinase activity. Consistent with the results determined for the PhoP-regulated reporters, compound GI261590A showed a reduced inhibitory effect compared with the selected hits, while GW320571X exerted no effect as a repressor of PhoQ autokinase activity (Fig. 4; see also Fig. S2).
FIG 4.
Autophosphorylation inhibitory activity of PhoQ of candidate molecules. (A) Increasing concentrations of GI261520A, GI262866A, GI261590A, or GW320571X were added to the Salmonella growth medium. Membranes harboring PhoQ, obtained by cell fractionation, were incubated for 10 min at 37°C in a reaction mixture containing [γ-32P]ATP, as described in Materials and Methods. The phosphorylation levels and the protein expression levels of PhoQ under each condition assayed were determined by densitometry, and the ratio (labeled against immunodetected) was plotted, taking the values obtained with the sole addition of DMSO (compound vehicle; 0.5% [vol/vol]) as 100%. Data shown represent results from three independent experiments, and error bars correspond to SD. All groups were compared by the use of the Kruskal-Wallis test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Structure-activity relationship analysis.
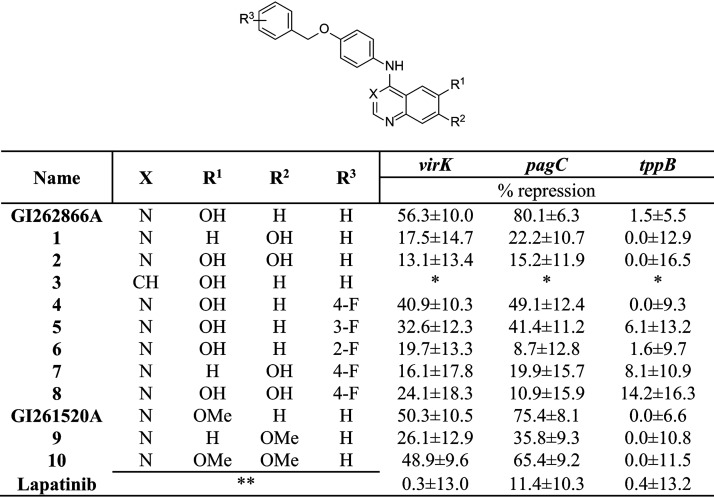
Additional quinazolines were designed to define structure-activity relationships. We synthesized a series of compounds, including compound 1 to compound 21, based on GI261520A and GI262866A through nucleophilic aromatic displacement of 4-chloroquin(az)olines. We were able to furnish products in excellent yields (55% to 91%) that were consistent with previous reports and without protection of the alcohol-substituted quinazoline starting material (please see supplemental material at http://www.ibr-conicet.gov.ar/wp-content/uploads/2019/10/Carabajal-et-al-AAC01744-19-Supplementary-Information-Combined.pdf [Fig. S1, supplemental information—compound characterization, and supplementary methods]) (35–38).
GI262866A from the screening was resynthesized, tested, and verified to produce virK and pagC activity level repression consistent with our previous results (Fig. 5). Moving the alcohol to the 7 position (compound 1) reduced inhibition by 3-fold for both virK and pagC repression. This repression was not rescued by restoring the alcohol to form the 6,7-diol (compound 2), which showed activity similar to that seen with compound 1. The switch to the quinoline of GI262866A (compound 3) yielded a compound with toxicity for the bacterium rather than inhibition of the TCS.
FIG 5.
Initial optimization with GI262866A and GI261520A. The chemical structure shows X, R1, R2, R3, and R4 positions that were substituted by the functional groups indicated below. The inhibition action was calculated on the basis of the β-galactosidase activity from lacZ transcriptional fusions to two different PhoP-activated genes (virK and pagC) and one PhoP-unrelated control reporter gene (tppB). Cells were grown overnight in LB plus DMSO (0.25% [vol/vol]) or the indicated compound at a concentration of 25 μM. Repression percentages were calculated, taking the values obtained with the sole addition of DMSO (0.25% [vol/vol]) as 100%. Results represent averages from three independent assays performed in duplicate. Single asterisks (*) represent compounds that show bacterial growth inhibition. Double asterisks (**) represent the chemical structure of lapatinib (provided in Table S2).
As shown in Fig. 5, a fluorine substitution at either the para position (compound 4) or the meta position (compound 5) of the pendant benzyl of GI262866A reduced activity for both virK and pagC repression by up to half. The fluorine substitution at the ortho position (compound 6) had the most defined effect, showing highly diminished activity as a repressor of the expression levels of either the virK or pagC reporter. The combination of para-fluorine and the alcohol at the 7 position (compound 7) and the addition of the alcohol to form the 6,7-diol (compound 8) did not change the activity of the respective unsubstituted benzyl compounds, compound 1 and compound 2. Methylation of the alcohol of GI262866A to form GI261520A resulted in equipotent repression of the levels of activity of the virK and pagC reporters. The 7-methoxy position analog (compound 9) resulted in diminished levels of the two reporters´ activity repression similar to those seen with compound 1. The most intriguing result was that the dimethoxy substitution (compound 10) was able to restore the repression to the original levels, something not observed with the 6,7-diol of compound 2 or compound 8.
Assessment of structural boundaries.
Given these observations, we sought to explore the limits of the pharmacophore with additional structural alterations. GI261590A and GW320571X (Fig. 2A) were structurally related to lapatinib and were likely generated in the same medicinal chemistry program. We incorporated fragments from this clinical inhibitor into our quinazoline series, along with substructures from erlotinib (Fig. 5; see also Fig. S3). Both of these inhibitors showed levels of inhibition of virK or pagC expression that were lower than 12.2%. Modification of the erlotinib quinazoline to the 6-position alcohol (compound 11) yielded a 15-fold rise in virK repression and a slight increase in pagC repression (Fig. S3). There was no loss in repression from relocating the alcohol from the 6 position to the 7 position (compound 12). However, the 6,7-diol derivative (compound 13) showed up to a 5-fold decrease in virK or pagC repression compared to compound 12. In accordance with the importance of a 6-position alcohol, while the quinoline analog with the alcohol in the 7 position (compound 15) was inactive, a switch to the 6-position quinoline (compound 14) modestly restored repressive activity. Capping the 6-position alcohol of compound 11 to produce the methoxy (compound 16) yielded up to a 10-fold reduction of repression levels. Both the 7-methoxy analog (compound 17) and the 6,7-dimethoxy analog (compound 18) as well as the switch to the quinoline (compound 19) showed inactivity (Fig. S3). Interestingly, the clipped aniline of GI262866A with a simple para-methoxy (compound 20) showed good repression of β-galactosidase activity levels driven from the virK reporter (33%) or the pagC reporter (45%), representing about two-thirds of the level seen with GI262866A. The truncated aniline of GI261520A with a para-methoxy group (compound 21) showed a >5-fold drop in repression potency (Fig. S3). All these results point to the pendant benzyl being an important contributor to activity, and the partial inhibition recovery in compounds 20 and 21 supports the idea of the involvement of the 6-position quinazoline alcohol.
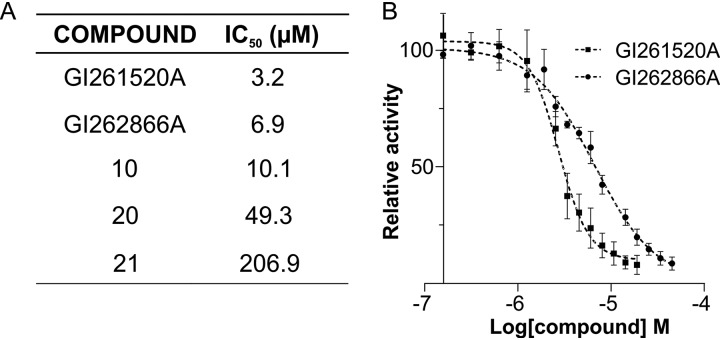
We performed dose-response analyses of pagC reporter expression levels with GI262866A and GI261520A and compounds 10, 20, and 21 to assess the 50% inhibitory concentration (IC50) values for these compounds (Fig. 6). In accordance to our results described above (Fig. 4 and 5), GI262866A, GI261520A, and compound 10 all showed IC50 values of ≤10 μM, indicating that an alcohol or a methoxy group is required at the 6-position quinazoline for the inhibitory action. Compound 20 showed an estimated 7-fold increase relative to GI262866A in the IC50 values, while compound 21 showed a 64-fold increase relative to GI261520A, substantiating the importance of the pendant benzyl group. In sum, the analysis of these structural analogs helped confirm that GI262866A and GI261520A were indeed the most active analogs and further defined the activity of this chemotype.
FIG 6.
Assessment of the IC50 values for compound optimization of GI262866A and GI261520A. (A) IC50 values corresponding to GI262866A and GI261520A and of compounds 10, 20, and 21 were calculated on the basis of the β-galactosidase activity from a pagC::lacZ transcriptional fusion measured in cells grown in LB supplemented with 0.25% DMSO or with the corresponding compound. (B) Dose-response curve of inhibitory action of GI261520A and GI262866A on the expression of S. Typhimurium gene pagC. IC50 values were calculated by the use of Prism 6.1 (GraphPad Software). β-Galactosidase activity was measured as described in Materials and Methods. Results represent averages from three independent assays performed in duplicate.
Molecular mechanism of action for PhoQ HK activity.
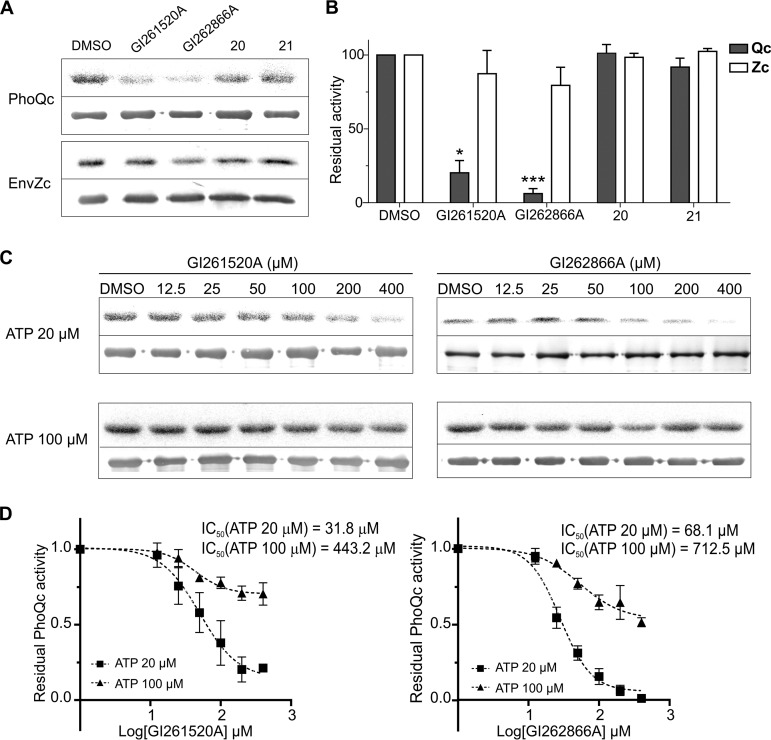
We sought to assess whether GI262866A and GI261520A directly interfered with the activity of the PhoQ catalytic domain. An autokinase assay was performed by using a soluble purified protein combined the maltose binding protein (MBP) with PhoQ cytoplasmic domain that harbors the HK catalytic domain and that was previously demonstrated to retain both PhoQ autokinase and PhoP-phosphotransfer activities (PhoQc) (39). The cytoplasmic domain of EnvZ, purified as a histidine-tagged fusion protein (EnvZc), was used as control. GI261520A and GI262866A inhibited PhoQc autokinase activity by 80% and 94%, respectively, while they did not significantly affect EnvZc autophosphorylation capacity, reinforcing the idea of the specificity of the compounds toward PhoQ (Fig. 7A and B). Consistent with their failure to downregulate the expression of the PhoP-activated reporters, neither compound 20 nor compound 21 was able to alter PhoQc or EnvZc autokinase activity (Fig. 7A and B). Both GI262866A and GI261520A inhibited PhoQc autokinase activity in a dose-dependent manner, within the 12.5 to 400 μM concentration range (Fig. 7C).
FIG 7.
Autophosphorylation inhibitory activity of PhoQ cytoplasmic domain mediated by GI262866A and GI261520A. (A) Autophosphorylation of catalytically active PhoQc or EnvZc in the presence of compound GI261520A, GI262866A, 20, or 21 at 200 μM. Purified protein was incubated for 10 min at 37°C in a reaction mixture containing [γ-32P]ATP. The autophosphorylation reactions were analyzed by SDS-PAGE (12% polyacrylamide) and transferred to nitrocellulose, followed by autoradiography (top panel in each pair of panels) or by immunodetection analysis (bottom panel in each pair) developed with anti-PhoQCyt (top panels) or anti-EnvZCyt (bottom panels) polyclonal antibodies. (B) The phosphorylation levels and the protein expression levels of PhoQc (Qc) or EnvZc (Zc) under each condition assayed were determined by densitometry (as described in Materials and Methods), and the ratio (labeled against immunodetected) was plotted, taking the values obtained with the sole addition of 0.5% DMSO as 100%. All groups were compared by the use of the Kruskal-Wallis test. *, P < 0.05; ***, P < 0.001. (C) Autoradiography (top panel in each pair of panels) and immunodetection analysis (bottom panel in each pair) (D) and determinations of relative activity levels of catalytically active PhoQc in the presence of increasing amounts of compounds GI261520A (left) and GI262866A (right) were performed in one-time-point (10-min) experiments using ATP 20 μM (■) or ATP 100 μM (▲). IC50 values were calculated by the use of Prism 6.1 (GraphPad Software). Data shown represent results from five independent experiments, and error bars correspond to SD.
To test whether GI262866A or GI261520A acts as an allosteric or competitive inhibitor of PhoQc, we compared the IC50 values obtained for the two compounds at two ATP concentrations (20 and 100 μM). The IC50 of an allosteric inhibitor should remain unchanged and independent of the ATP concentration, whereas in the case of a competitive inhibitor, the IC50 value would be influenced by the ATP concentration used. The reduction of the calculated IC50 values seen when the concentration of ATP in the substrate was lowered from 100 to 20 μM in the reaction medium indicated that the two molecules acted as competitive inhibitors of ATP in the PhoQ autophosphorylation reaction (Fig. 7C and D).
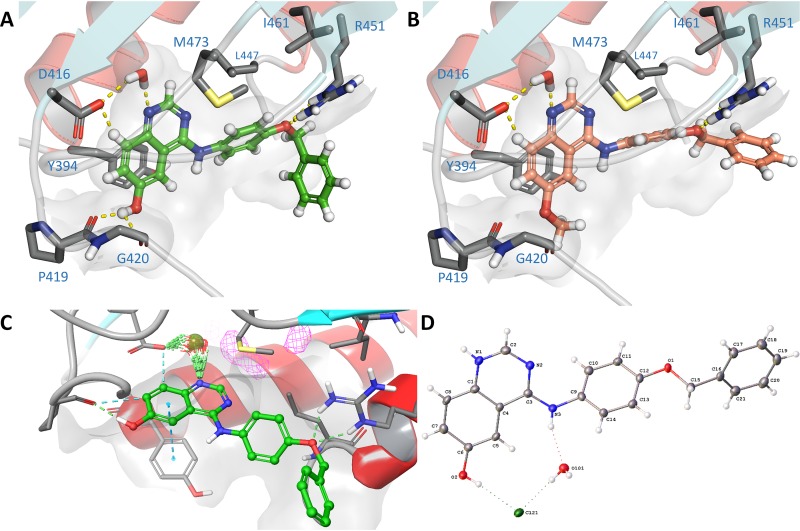
To rationalize the molecular basis of binding, as shown in Fig. 8A and B, we docked the two key compounds (GI261520A and GI262866A) into the ATP-binding domain of PhoQ (PDB identifier [ID] 3CGY) (19). The compounds were able to act as ATP mimics in a nontraditional hinge binder conformation, where a key water-mediated bridge with Asp416 forms the strongest interaction with the quinazoline nitrogen. This locks the molecule in place, with an additional direct interaction with the alcohol and Gly420. We also found the benzyl was required for extended pi-pi aromatic interactions, supporting our observation of the pronounced loss of functional activity of the benzyl clipped compounds 20 and 21. To further investigate the binding, we employed WaterMap (see Materials and Methods) and located the key water network interactions, confirming that a water bridge was formed between the quinazoline nitrogen and Asp416 on PhoQ (Fig. 8C) (40). We also solved the small-molecule crystal structure of GI262866A as a monoclinic structure with each unit found to be in a conformation similar to that predicted for the bound inhibitor (Fig. 8D; Cambridge Crystallographic Data Centre, deposition number 1953813 [see also the supplemental material]). These results demonstrate the key interactions occurring with the bound ligand and the wider protein structure and reinforce the idea of the mechanistic action of the quinazoline molecules that resemble substrate binding and hinder ATP interaction, precluding the PhoQ autocatalytic phosphorylation reaction.
FIG 8.
Docking of GI261520A and GI262866A compounds into the PhoQ binding site. (A and B) Docking of GI262866A (A) and GI262866A (B) in PDB 3CGY using the Schrödinger glide suite, highlighting key binding interactions. (C) WaterMap simulation of GI262866A in PhoQ binding site, proving that there is a pivotal interaction with a bridging water molecule and the quinoline nitrogen. (D) Small-molecule crystal structure of GI262866A, including Cl-counterion and solvent water. ADP ellipsoids are shown at 50% probability.
Effect on Salmonella intramacrophage survival.
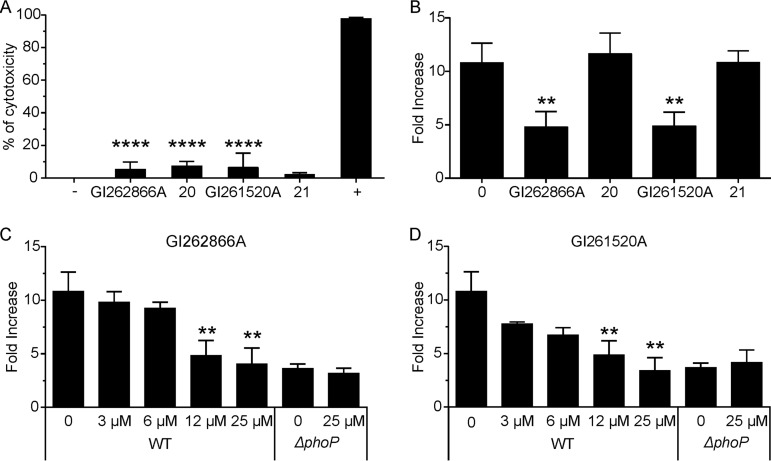
PhoP/PhoQ governs the capacity of Salmonella to survive and replicate inside macrophages, and the control of bacteria within these cells is critical to overcome infection. Therefore, we examined the effect of the selected compounds as antivirulence agents (41). To first investigate their potential cytotoxicity, we performed a tetrazolium dye reduction assay in cultured RAW264.7 macrophages by the use of 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) (42). This assay measures NAD(P)H-dependent cellular oxidoreductase activity (43). Neither GI261520A nor GI262866A affected the viability of the cultured cells (Fig. 9A). Additionally, compounds 20 and 21 were included and similarly showed no cytotoxicity in RAW264.7 cells (Fig. 9A).
FIG 9.
Inhibitory effect on S. Typhimurium intracellular growth. (A) Cytotoxicity effect assay. RAW264.7 macrophages were incubated for 18 h with a 25 μM concentration of either GI261520A or GI262866A and of either compound 20 or 21 as a control, cells were washed, and MTT was added. After incubation, violet formazan crystals were dissolved and absorbance was read at λ = 570 nm. +, cells incubated with 0.01% Triton X-100 as a positive control; −, cell incubated with 0.25% DMSO (vol/vol) as a negative control. Results represent averages from five independent assays performed in triplicate, and error bars correspond to SD. (B) Recovery of intracellular Salmonella at 1.5 and 18 h postinfection from RAW264.7 macrophages incubated with 12 μM GI261520A, GI262866A, compound 20, or compound 21. (C and D) Dose-response effect on intracellular Salmonella recovery at 1.5 and 18 h postinfection from RAW264.7 macrophages incubated with GI262866A (C) or GI261520A (D). All groups were compared by the use of one-way ANOVA. **, P < 0.01; ****, P < 0.0001.
Next, we measured levels of S. Typhimurium intramacrophage survival and replication capacity by performing a conventional gentamicin protection invasion assay (41). In the same media used for the MTT reduction assay, we tested both wild-type and the otherwise isogenic phoP mutant strains. After 30 min postinvasion, 12.5 μM GI262866A, GI261520A, compound 20, or compound 21 was added to RAW264.7 macrophages culture medium. The Salmonella intramacrophage CFU fold increase was calculated by dividing the values at 18 h by the values at 1.5 h postinfection. Both compounds lowered by 66% the intramacrophage replication of Salmonella (Fig. 9B). As expected, neither compound 20 nor compound 21 showed any effect on the capacity of Salmonella to survive or replicate inside macrophages (Fig. 9B). GI262866A or GI261520A exerted its effect in a dose-dependent manner within the 3.0 to 25 μM concentration range assayed. Moreover, the use of a 25 μM concentration of either GI262866A or GI261520A phenocopied the mutant phoP strain impairment in intracellular survival (Fig. 9C and D). Those last results indicate that GI262866A and GI261520A are able to successfully permeate the eukaryotic plasmatic and vacular cell membranes and reach intravacuolar Salmonella to block intramacrophage proliferation, one of the key PhoP/PhoQ-dependent phenotypes that confers to the bacteria the capacity to spread systemically, a pathogenic trait of the bacteria crucial for remaining viable within the mammalian host.
In addition, as shown in Fig. S4A, we determined that GI261520A downregulated the expression of the representative PhoP-activated virK gene in β-galactosidase activity assays performed in the S. Typhimurium STM23, S. Enteritidis Pt4, and S. Dublin Sdu5 strain backgrounds (linoleic acid, a known PhoP/PhoQ inhibitor, was used as control [27]). Accordingly, GI261520A was also able to inhibit intramacrophage proliferation when the macrophage infection assay was carried out using these Salmonella strains (Fig. S4B). These results demonstrated the activity of the quinazoline compound in Salmonella enterica serovars beyond the initially tested S. Typhimurium 14028 strain.
We also assessed the potential utility of these compounds in an in vivo context. A common limiting factor for in vivo compound exposure is P450-mediated oxidative metabolism in the liver. Accordingly, we coincubated GI261520A and GI262866A with mouse liver microsomes and quantified the amount of compound remaining as a function of time. The GI261520A compound demonstrated moderate stability, with a half-life (t1/2) of 71.6 min and a ratio with propranolol control of 10.6. The GI262866A compound was marginally less stable (t1/2 of 19.6 min with a ratio to propranolol control of 2.4). GI262866 was 5-fold less stable than GI261520A once normalized to the control. For comparison, the marketed drug erlotinib had a t1/2 of 22.7 min with a ratio of 2.9. These results suggest that first-pass metabolism of these compounds would not be a prohibitive liability for in vivo use.
Concluding remarks.
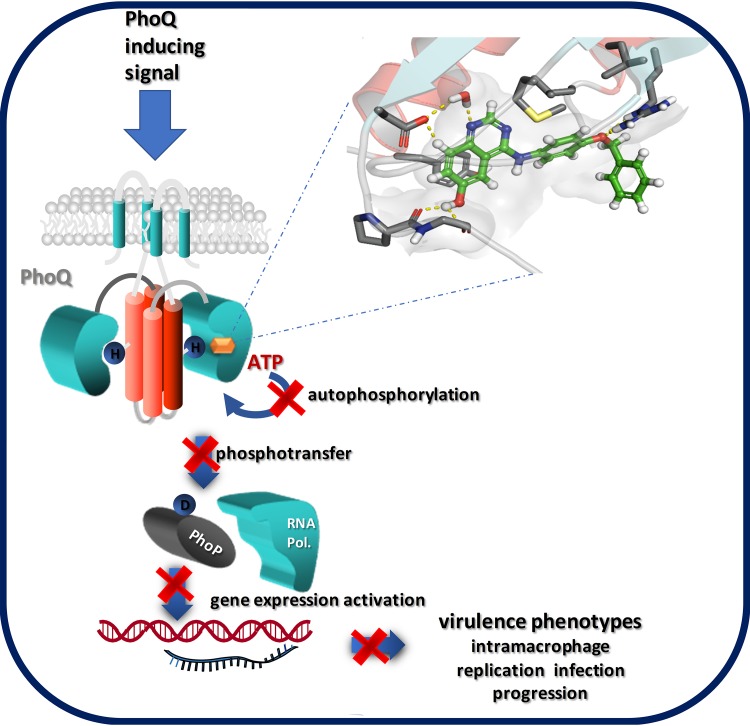
We have identified two key quinazoline-based lead compounds, GI262866A and GI261520A, which downregulate PhoP-activated genes in S. Typhimurium by the selective inhibition of PhoQ histidine kinase activity (see Fig. 10 for a model that depicts the mechanistic action of the compounds on the Salmonella PhoP/PhoQ signal transduction system). Profiling a focused series of designed analogs allowed initial definition of the pharmacophore and highlighted that small structural changes had a significant impact on antibacterial activity. These quinazolines have properties that are significantly improved over those of currently available antivirulence compounds directed against Salmonella identified in the literature.
FIG 10.
Model of action for the quinazoline-based antivirulence compounds. Compounds GI262866A and GI261520A (orange hexagon) bind to the catalytic site of PhoQ and compete for the binding of substrate ATP, precluding the phosphotransfer reaction from ATP to the conserved His (H) residue located in the DHp domain of PhoQ (protein domain in red). This inhibition hinders the downstream steps in the signal transduction reaction: His (H)-Asp (D) phosphotransfer from phosphorylated PhoQ to the response regulator PhoP and its subsequent binding to the target promoters of PhoP-regulated genes. This blockage prevents the induction of expression of virulence genes, including those required for Salmonella intramacrophage replication and disease progression in the infected host.
The components of the PhoP/PhoQ signal transduction system reside inside the bacteria. By employing a whole-cell screening approach, we were able to circumvent or exclude potential problems derived from accessibility of the drugs to the desired bacterial target. The strategy allowed us to select only those molecules that were competently permeative and that were not completely extruded by efflux pumps nor consumed by inactivating enzymes. The selectivity for PhoQ over other HKs suggests the possibility of a therapeutic intervention directed to inhibit Salmonella pathogenic traits with minimal impact on beneficial microbiota.
The ability of the compounds to block intramacrophage survival and the replication capacity of S. Typhimurium without cytotoxic effects supports the idea of their capacity to reach and exert their effect on intravacuolar Salmonella, being innocuous for host cells. Moreover, the hit molecules were able to inhibit the intramacrophage replication of Salmonella enterica serovars differently from that of S. Typhimurium, indicating an antivirulence effect useful for potential treatment of salmonellosis in diverse warm-blooded vertebrates.
The pathoblocker compounds identified here could be used as standalone therapeutic agents to treat Salmonella-borne infections. Alternatively, they might be employed as adjuvants, administered in conjunction with classic antimicrobials, lowering the required dose of usage and reducing the prospects for development of resistance. The in vitro and in vivo activity of the compounds demonstrates an exciting possibility for the development of a new generation of antivirulence drugs against Salmonella.
MATERIALS AND METHODS
Bacterial strains and cell culture and growth conditions.
Bacterial strains used in this work are listed in Table S1 (please see supplemental material at http://www.ibr-conicet.gov.ar/wp-content/uploads/2019/10/Carabajal-et-al-AAC01744-19-Supplementary-Information-Combined.pdf). Overnight cultures of bacteria were inoculated using a single colony and routinely grown in LB media (10 g liter−1 NaCl, 10 g liter−1 Tryptone, 5 g liter−1 yeast extract) supplemented with antibiotics as appropriate (kanamycin, 50 μg ml−1; ampicillin, 100 μg ml−1) and 0.7 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when required and/or with the addition of the compound of interest at the concentrations indicated for each assay.
Compound screening and dose-response inhibition assays.
The potential inhibitory effect of the PKIS library was evaluated by growing S. Typhimurium in LB broth containing either the compound vehicle dimethyl sulfoxide (DMSO; 0.5% [vol/vol]) as a control or the corresponding compound solution at a final concentration of 25 or 50 μM, dissolved in DMSO. The assays were carried out in sterile 96-well microtiter plates, the reaction mixtures were incubated for 18 h with agitation at 37°C, and optical density at 600 nm was measured every 60 min with a BioTek Synergy 2 multimode microplate reader. β-Galactosidase activity from PhoP-modulated reporter genes was determined for the compounds that did not exhibit alterations in growth curves (44). Dose-dependent inhibition was performed using final concentration of the compounds in the 0 to 50 μM range. Samples were processed in duplicate, and proper blank controls were used in each microplate.
General procedure for the synthesis of 4-anilinoquin(az)olines.
A 4-chloroquin(az)oline derivative (1.0 equivalent), an aniline derivative (1.1 equivalent), and iPr2NEt (2.5 equivalent) were suspended in ethanol (10 ml) and subjected to refluxing for 18 h. The crude mixture was purified by flash chromatography using EtOAc-hexane followed by 1% to 5% methanol–EtOAc. After solvent removal under reduced pressure, the product was obtained as solid or was recrystallized from ethanol–water. SMILES and Labbook codes for the compounds are provided in Table S2. The chemical structure, the mass spectrometry method, the chart of spectra for the compounds, and the crystal and experimental data for GI261520A (2017ncs0878q) are provided in the supplemental material (supplementary information, compound characterization, and supplementary methods).
Preparation of PhoQ-enriched membranes.
To assess the autokinase activity of sensor PhoQ, membranes were prepared from overnight cultures of S. Typhimurium strains that express PhoQ from pUHE-21-2::phoQ, as described previously (27). Briefly, an overnight culture of S. Typhimurium strain PB4663 was used to inoculate LB containing the corresponding compound at the final concentration indicated in each experiment. These were then grown at 37°C to the logarithmic phase (A630 = 0.6), and protein expression was induced by the addition of 0.7 mM IPTG for an additional 3 h with shaking. Cells were collected and treated as detailed previously (27). Cells were collected and resuspended in a solution containing 20 mM Tris-HCl (pH 8.0), 20% sucrose, 5 mM EDTA, and 150 mg μl−1 lysozyme. After 40 min of incubation at 4°C, 20 mM MgCl2 was added and the pellet was resuspended in ice-cold 10 mM Tris-HCl (pH 8.0) and subjected to sonication. Membrane fractions were recovered and washed subsequently using a solution containing 10 mM Tris-HCl (pH 8.0), 2 M KCl, 10 mM Tris-HCl, 5 mM EDTA, and 10 mM Tris HCl and 40-min centrifugations rounds at 21,000 × g. The supernatant was discarded each time. Finally, the membranes were resuspended in 25 mM Tris-HCl (pH 8.0)–50 mM KCl. All procedures were carried out at 4°C. Protein concentrations were determined by the bicinchoninic acid assay (Sigma) using bovine serum albumin as the standard.
Autokinase activity assays and immunodetection analysis.
To test the autokinase activity of the sensor, PhoQ-enriched membranes (50 μg of total protein) were obtained as described previously (27). Membranes were incubated 10 min at 37°C in a 30-μl mixture containing 25 mM Tris-HCl (pH 8.0), 50 mM KCl, 1 mM MgCl2, 50 μM nonlabeled ATP, and 0.16 μCi μl−1 of [γ-32P]ATP. For the identification of the molecular mechanism of action on PhoQ, nonlabeled ATP (20 and 100 μM) and trace amounts (0.03 and 0.32 μCi μl−1) of [γ-32P]ATP were incubated with purified PhoQc (10 μg) or EnvZc (2 μg) fusion proteins and increasing amounts of compounds GI261520A, GI262866A, 20, and 21, as indicated. Reactions were started by addition of the reaction mixture and stopped by addition of 6 μl of 5 × SDS-PAGE sample buffer (2.5% β-mercaptoethanol, 9% glycerol, 10% SDS, 600 mM Tris-HCl [pH 6.8], 0.006% bromphenol blue). All reaction mixtures were analyzed by SDS-PAGE (12% polyacrylamide), transferred to nitrocellulose, and then subjected to Western blotting or to autoradiography analysis. Western blot membranes were incubated with rabbit anti-PhoQCyt or anti-EnvZCyt polyclonal antibodies (27) and developed by incubation with protein A conjugated with phosphatase, coupled to a chromogenic reaction using nitro blue tetrazolium and 5 bromo-4-chloro-3-indolyl phosphate as substrates. Autoradiography images and Western blot membranes were scanned for densitometry using Adobe Photoshop CS to perform quantitative determinations (27).
Gentamicin protection assay.
Salmonella invasion of RAW264.7 macrophages was tested by gentamicin assays as previously described (45). Briefly, monolayers of RAW264.7 macrophages cultured in 48-well tissue plates containing Dulbecco’s modified Eagle’s medium (DMEM) plus 10% fetal calf serum (FCS) medium were infected at a multiplicity of infection (MOI) of 10:1 (bacterial to eukaryotic cells). The cells were incubated for 30 min at 37°C and rinsed with gentamicin-containing medium 100 μg ml−1 and the selected compound up to 25 μM or DMSO 0.5% (vol/vol) as the control (compound vehicle). After 1.5 h, cells were incubated at a lower gentamicin concentration (30 μg ml−1) for a total of 18 h postinfection. The cells were washed with gentamicin-free medium and lysed with 0.1% Triton X-100. Intracellular bacteria were collected and enumerated on Luria broth agar plates, and the CFU count per milliliter was calculated. The results for each experiment represent averages from at least three independent assays performed in duplicate.
Data analysis.
Data were prepared and analyzed in GraphPad Prism (version 6.01; GraphPad Software, San Diego, CA, USA). For all dose-response curves (β-galactosidase and autokinase activity assays), data were fitted the following equation:
Some compounds exhibited incomplete dose-response curves because going to higher concentrations would have caused a solubility problem; therefore, the IC50 values were estimated for purposes of comparison to other compounds by constraining the bottom of the curve to a value of zero, as described previously (25).
Statistical analysis.
Statistical analysis was performed using one-way analysis of variance (ANOVA) and the Kruskal-Wallis multiple-comparison test with an overall significance level of 0.05. Asterisks in the plots denote the values among the treatment groups in which a statistically significant difference was determined.
Molecular modeling.
Molecular modeling was performed using the Schrödinger Maestro software package. Structures of small molecules were prepared using the LigPrep module of the Schrödinger suite, employing OPLS3 force for all computations. The X-ray crystal structure for PhoQ (PDB ID 3CGZ and 3CGY [19]) was preprocessed using the protein preparation wizard of the Schrödinger suite in order to optimize the hydrogen bonding network. Prior to Glide docking, the grid box was centered using the corresponding X-ray ligand as the template. The ligand docking was performed using default SP settings of Schrödinger Glide with additional hydrogen bond constraints to NH of CYS126 (hinge residue). Graphical illustrations were generated using MOE software, Maestro (WaterMap), or PyMOL (The PyMOL Molecular Graphics System, ver. 1.8; Schrödinger LLC).
Hydration site analysis.
Hydration site analysis calculations were performed with WaterMap (Schrödinger release 2016-3; Schrödinger LLC, New York, NY, USA, 2016). The structures (PDB ID 3CGZ and 3CGY [19]) were prepared with the Protein Preparation Wizard (as described above). Water molecules were analyzed within 5 Å of the cocrystallized ligand, and the 2-ns simulation was conducted with the OPLS3 force field (46).
Additional details for Materials and Methods are provided in the supplemental material.
ACKNOWLEDGMENTS
E.G.V. is a Career Investigator of Consejo de Investigaciones Cientificas y Tecnológicas (CONICET), Argentina. M.A.C. has fellowships from CONICET. This work was supported by grants PICT 2017-1627 and PICT-O-2013-0041 from Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), Argentina, to E.G.V. and by grant Proyecto Unidad Ejecutora P-UE 22920160100039CO from CONICET to Instituto de Biología Molecular y Celular de Rosario (IBR). The Structural Genomics Consortium (SGC) is a registered charity (registration number 1097737) that receives funds from AbbVie; Bayer Pharma AG; Boehringer Ingelheim; Canada Foundation for Innovation; Eshelman Institute for Innovation; Genome Canada; Innovative Medicines Initiative (EU/EFPIA) (ULTRA-DD grant no. 115766); Janssen; Merck KGaA, Darmstadt, Germany; MSD; Novartis Pharma AG; Ontario Ministry of Economic Development and Innovation; Pfizer; São Paulo Research Foundation (FAPESP); Takeda; and Wellcome (106169/ZZ14/Z). We thank Biocenter Finland/DDCB for financial support.
We thank Dolores Campos and Marina Avecilla for technical assistance. We thank Gastón Viarengo for preliminary assays performed with the compounds. We thank CSC—IT Center for Science Ltd. Finland for the use of their facilities, software licenses, and computational resources. We are also grateful to Brandie Ehrmann for LC-MS/HRMS support provided by the Mass Spectrometry Core Laboratory at the University of North Carolina at Chapel Hill. We are grateful to the SALMOIBER CYTED Network (Control de salmonelosis en Iberoamérica) for travel exchange support and for useful suggestions from all its members (in alphabetical order, not including authors E.G.V., L.Y., and J.A.C. of the present article): Laura Betancor, Coralith García, Francisco García del Portillo, Theresa J. Ochoa, José Pedraza, M. Graciela Pucciarelli, José Luis Puente, Griselda Ruiz, Claudia Silva, Lorena Soleto, and Fernando Soncini. In addition, we thank the EPSRC UK National Crystallography Service for funding and for the collection of the crystallographic data of GI261520A. We also thank Zengbiao Li and Lianyong Su (Drumetix Laboratories, Greensboro, NC, USA) for their useful discussions and microsome assay assistance. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM; International Collaboration on Enteric Disease ‘Burden of Illness’ Studies. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 2.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. 2012. Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guerrant RL, Van Gilder T, Steiner TS, Thielman NM, Slutsker L, Tauxe RV, Hennessy T, Griffin PM, DuPont H, Sack RB, Tarr P, Neill M, Nachamkin I, Reller LB, Osterholm MT, Bennish ML, Pickering LK. 2001. Practice guidelines for the management of infectious diarrhea. Clin Infect Dis 32:331–351. doi: 10.1086/318514. [DOI] [PubMed] [Google Scholar]
- 4.Hohmann EL. 2001. Nontyphoidal salmonellosis. Clin Infect Dis 32:263–269. doi: 10.1086/318457. [DOI] [PubMed] [Google Scholar]
- 5.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 6.Calvert MB, Jumde VR, Titz A. 2018. Pathoblockers or antivirulence drugs as a new option for the treatment of bacterial infections. Beilstein J Org Chem 14:2607–2617. doi: 10.3762/bjoc.14.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickey SW, Cheung GYC, Otto M. 2017. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov 16:457–471. doi: 10.1038/nrd.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasko DA, Sperandio V. 2010. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov 9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 9.Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu Rev Microbiol 63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groisman EA, Hollands K, Kriner MA, Lee E-J, Park S-Y, Pontes MH. 2013. Bacterial Mg2+ homeostasis, transport, and virulence. Annu Rev Genet 47:625–646. doi: 10.1146/annurev-genet-051313-051025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamond G, Beckloff N, Weinberg A, Kisich K. 2009. The roles of antimicrobial peptides in innate host defense. Curr Pharm Des 15:2377–2392. doi: 10.2174/138161209788682325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaRock CN, Nizet V. 2015. Cationic antimicrobial peptide resistance mechanisms of streptococcal pathogens. Biochim Biophys Acta 1848:3047–3054. doi: 10.1016/j.bbamem.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst RK, Guina T, Miller SI. 1999. How intracellular bacteria survive: surface modifications that promote resistance to host innate immune responses. J Infect Dis 179:S326–S330. doi: 10.1086/513850. [DOI] [PubMed] [Google Scholar]
- 14.Núñez-Hernández C, Tierrez A, Ortega ÁD, Pucciarelli MG, Godoy M, Eisman B, Casadesús J, García-del Portillo F. 2013. Genome expression analysis of nonproliferating intracellular Salmonella enterica serovar Typhimurium unravels an acid pH-dependent PhoP-PhoQ response essential for dormancy. Infect Immun 81:154–165. doi: 10.1128/IAI.01080-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueira R, Holden DW. 2012. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology 158:1147–1161. doi: 10.1099/mic.0.058115-0. [DOI] [PubMed] [Google Scholar]
- 16.Drewry DH, Wells CI, Andrews DM, Angell R, Al-Ali H, Axtman AD, Capuzzi SJ, Elkins JM, Ettmayer P, Frederiksen M, Gileadi O, Gray N, Hooper A, Knapp S, Laufer S, Luecking U, Muller S, Muratov E, Denny RA, Saikatendu KS, Treiber DK, Zuercher WJ, Willson TM. 2017. Progress towards a public chemogenomic set for protein kinases and a call for contributions. bioRxiv https://www.biorxiv.org/content/10.1101/104711v1. [DOI] [PMC free article] [PubMed]
- 17.Ryan Q, Ibrahim A, Cohen MH, Johnson J, Ko CW, Sridhara R, Justice R, Pazdur R. 2008. FDA drug approval summary: lapatinib in combination with capecitabine for previously treated metastatic breast cancer that overexpresses HER-2. Oncologist 13:1114–1119. doi: 10.1634/theoncologist.2008-0816. [DOI] [PubMed] [Google Scholar]
- 18.Cohen MH, Johnson JR, Chen YF, Sridhara R, Pazdur R. 2005. FDA drug approval summary: erlotinib (Tarceva) tablets. Oncologist 10:461–466. doi: 10.1634/theoncologist.10-7-461. [DOI] [PubMed] [Google Scholar]
- 19.Guarnieri MT, Zhang L, Shen J, Zhao R. 2008. The Hsp90 inhibitor radicicol interacts with the ATP-binding pocket of bacterial sensor kinase PhoQ. J Mol Biol 379:82–93. doi: 10.1016/j.jmb.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 20.Boibessot T, Zschiedrich CP, Lebeau A, Bénimèlis D, Dunyach-Rémy C, Lavigne J-P, Szurmant H, Benfodda Z, Meffre P. 2016. The rational design, synthesis, and antimicrobial properties of thiophene derivatives that inhibit bacterial histidine kinases. J Med Chem 59:8830–8847. doi: 10.1021/acs.jmedchem.6b00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilmour R, Foster JE, Sheng Q, McClain JR, Riley A, Sun PM, Ng WL, Yan D, Nicas TI, Henry K, Winkler ME. 2005. New class of competitive inhibitor of bacterial histidine kinases. J Bacteriol 187:8196–8200. doi: 10.1128/JB.187.23.8196-8200.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilliard JJ, Goldschmidt RM, Licata L, Baum EZ, Bush K. 1999. Multiple mechanisms of action for inhibitors of histidine protein kinases from bacterial two-component systems. Antimicrob Agents Chemother 43:1693–1699. doi: 10.1128/AAC.43.7.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephenson K, Hoch JA. 2004. Developing inhibitors to selectively target two-component and phosphorelay signal transduction systems of pathogenic microorganisms. Curr Med Chem 11:765–773. doi: 10.2174/0929867043455765. [DOI] [PubMed] [Google Scholar]
- 24.Stephenson K, Yamaguchi Y, Hoch JA. 2000. The mechanism of action of inhibitors of bacterial two-component signal transduction systems. J Biol Chem 275:38900–38904. doi: 10.1074/jbc.M006633200. [DOI] [PubMed] [Google Scholar]
- 25.Wilke KE, Francis S, Carlson EE. 2015. Inactivation of multiple bacterial histidine kinases by targeting the ATP-binding domain. ACS Chem Biol 10:328–335. doi: 10.1021/cb5008019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salazar MO, Viarengo G, Sciara MI, Kieffer PM, Garcia Vescovi E, Furlan RL. 2014. A thin-layer chromatography autographic method for the detection of inhibitors of the Salmonella PhoP-PhoQ regulatory system. Phytochem Anal 25:155–160. doi: 10.1002/pca.2482. [DOI] [PubMed] [Google Scholar]
- 27.Viarengo G, Sciara MI, Salazar MO, Kieffer PM, Furlan RL, Garcia VE. 2013. Unsaturated long chain free fatty acids are input signals of the Salmonella enterica PhoP/PhoQ regulatory system. J Biol Chem 288:22346–22358. doi: 10.1074/jbc.M113.472829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson MM, Ellis EM, Graeme-Cook KA, Higgins CF. 1987. OmpR and EnvZ are pleiotropic regulatory proteins: positive regulation of the tripeptide permease (tppB) of Salmonella typhimurium. Mol Gen Genet 207:120–129. doi: 10.1007/bf00331499. [DOI] [PubMed] [Google Scholar]
- 29.Goh EB, Siino DF, Igo MM. 2004. The Escherichia coli tppB (ydgR) gene represents a new class of OmpR-regulated genes. J Bacteriol 186:4019–4024. doi: 10.1128/JB.186.12.4019-4024.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolanin PM, Thomason PA, Stock JB. 2002. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol 3:REVIEWS3013. doi: 10.1186/gb-2002-3-10-reviews3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zwir I, Yeo WS, Shin D, Latifi T, Huang H, Groisman EA. 2014. Bacterial nucleoid-associated protein uncouples transcription levels from transcription timing. mBio 5:e01485-14. doi: 10.1128/mBio.01485-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harari O, del Val C, Romero-Zaliz R, Shin D, Huang H, Groisman EA, Zwir I. 2009. Identifying promoter features of co-regulated genes with similar network motifs. BMC Bioinformatics 10(Suppl 4):S1. doi: 10.1186/1471-2105-10-S4-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zwir I, Latifi T, Perez JC, Huang H, Groisman EA. 2012. The promoter architectural landscape of the Salmonella PhoP regulon. Mol Microbiol 84:463–485. doi: 10.1111/j.1365-2958.2012.08036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soncini FC, Garcia Vescovi E, Solomon F, Groisman EA. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol 178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asquith CRM, Berger BT, Wan J, Bennett JM, Capuzzi SJ, Crona DJ, Drewry DH, East MP, Elkins JM, Fedorov O, Godoi PH, Hunter DM, Knapp S, Muller S, Torrice CD, Wells CI, Earp HS, Willson TM, Zuercher WJ. 2019. SGC-GAK-1: a chemical probe for cyclin G associated kinase (GAK). J Med Chem 62:2830–2836. doi: 10.1021/acs.jmedchem.8b01213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asquith CRM, Laitinen T, Bennett JM, Godoi PH, East MP, Tizzard GJ, Graves LM, Johnson GL, Dornsife RE, Wells CI, Elkins JM, Willson TM, Zuercher WJ. 2018. Identification and optimization of 4-anilinoquinolines as inhibitors of cyclin G associated kinase. ChemMedChem 13:48–66. doi: 10.1002/cmdc.201700663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asquith CRM, Naegeli KM, East MP, Laitinen T, Havener TM, Wells CI, Johnson GL, Drewry DH, Zuercher WJ, Morris DC. 2019. Design of a cyclin G associated kinase (GAK)/epidermal growth factor receptor (EGFR) inhibitor set to interrogate the relationship of EGFR and GAK in chordoma. J Med Chem 62:4772–4778. doi: 10.1021/acs.jmedchem.9b00350. [DOI] [PubMed] [Google Scholar]
- 38.Asquith CRM, Treiber DK, Zuercher WJ. 2019. Utilizing comprehensive and mini-kinome panels to optimize the selectivity of quinoline inhibitors for cyclin G associated kinase (GAK). Bioorg Med Chem Lett 29:1727–1731. doi: 10.1016/j.bmcl.2019.05.025. [DOI] [PubMed] [Google Scholar]
- 39.Castelli ME, Garcia Vescovi E, Soncini FC. 2000. The phosphatase activity is the target for Mg2+ regulation of the sensor protein PhoQ in Salmonella. J Biol Chem 275:22948–22954. doi: 10.1074/jbc.M909335199. [DOI] [PubMed] [Google Scholar]
- 40.Cappel D, Sherman W, Beuming T. 2017. Calculating water thermodynamics in the binding site of proteins—applications of WaterMap to drug discovery. Curr Top Med Chem 17:2586–2598. doi: 10.2174/1568026617666170414141452. [DOI] [PubMed] [Google Scholar]
- 41.Elsinghorst EA. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol 236:405–420. doi: 10.1016/0076-6879(94)36030-8. [DOI] [PubMed] [Google Scholar]
- 42.Sylvester PW. 2011. Optimization of the tetrazolium dye (MTT) colorimetric assay for cellular growth and viability. Methods Mol Biol 716:157–168. doi: 10.1007/978-1-61779-012-6_9. [DOI] [PubMed] [Google Scholar]
- 43.Lazzaro M, Krapf D, Garcia Vescovi E. 17 May 2019, posting date Selective blockage of Serratia marcescens ShlA by nickel inhibits the pore-forming toxin-mediated phenotypes in eukaryotic cells. Cell Microbiol 21:e13045. doi: 10.1111/cmi.13045. [DOI] [PubMed] [Google Scholar]
- 44.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 45.Thompson JA, Liu M, Helaine S, Holden DW. 2011. Contribution of the PhoP/Q regulon to survival and replication of Salmonella enterica serovar Typhimurium in macrophages. Microbiology 157:2084–2093. doi: 10.1099/mic.0.048926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL, Kaus JW, Cerutti DS, Krilov G, Jorgensen WL, Abel R, Friesner RA. 2016. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]