Mammalian serum amyloid A (SAA) is a major acute phase protein that shows a massive increase in plasma concentration during inflammation. In the present study, we demonstrate that the expression of mouse SAA1 in serum was increased when infected with Candida albicans, a major human fungal pathogen, in a systemic infection model. We then set out to investigate the antifungal activity of SAA proteins against C. albicans.
KEYWORDS: Candida albicans, SAA1, serum amyloid A, antifungal activity
ABSTRACT
Mammalian serum amyloid A (SAA) is a major acute phase protein that shows a massive increase in plasma concentration during inflammation. In the present study, we demonstrate that the expression of mouse SAA1 in serum was increased when infected with Candida albicans, a major human fungal pathogen, in a systemic infection model. We then set out to investigate the antifungal activity of SAA proteins against C. albicans. Recombinant human and mouse SAA1 (rhSAA1 and rmSAA1) were expressed and purified in Escherichia coli. Both rhSAA1 and rmSAA1 exhibited a potent antifungal activity against C. albicans. We further demonstrate that rhSAA1 binds to the cell surface of C. albicans, disrupts cell membrane integrity, and induces rapid fungal cell death in C. albicans. Our finding expands the known functions of SAA1 and provides new insight into host-Candida interactions during fungal infection.
INTRODUCTION
Antimicrobial peptides and proteins (AMPs) produced by the human innate immune system are the first line of defense against bacterial and fungal infections (1). AMPs have been identified in a range of human bodily fluids, including blood, saliva, urine, milk, and sweat, and are known to defend against microbial pathogens in corresponding host niches. Candida albicans often exists on the mucosal surface of healthy people as a harmless commensal organism, but it can cause both superficial and life-threatening systemic infections when the host immune system is compromised (2). Several human AMPs, such as the cathelicidin LL-37 and histatin Hst-5, have been reported as potent antifungal peptides against C. albicans (3–5). However, our understanding of AMPs-mediated host-pathogen interactions is still limited.
Serum amyloid A (SAA) proteins represent a conserved and major inflammatory acute-phase family in humans and most mammals and are thought to have antimicrobial activities (6–8). Human SAA1 comprises 122 amino acids and is primarily produced in the liver by hepatocytes (9). Its concentration in plasma increases by as much as 1,000-fold during inflammation (6). The expression of SAA1 has also been detected in several epithelial components of extrahepatic tissues, including the breast, kidney, lung, and gut (10). SAA1 is also closely linked to diseases such as rheumatoid arthritis, atherosclerosis, and diabetes (11–13). In addition to its roles in cholesterol transport and lipid metabolism, SAA1 is also involved in the regulation of inflammation and immunity by promoting the production of proinflammatory cytokines and chemotaxis of immune cells. These functions are mediated by SAA1 receptors or binding proteins, such as FPR2 (formyl peptide receptor 2), FPRL1 (formyl peptide receptor like-1), and TLR2 (Toll-like receptor 2) (14, 15).
Several studies have demonstrated that SAA proteins exhibit potent antibacterial activity (6–8). Recombinant human SAA1 is able to form ion channels in planar lipid bilayer membranes at physiologic concentrations, and the recombinant expression of SAA1 in Escherichia coli causes cell lysis (7). Eckhardt et al. have reported that SAA1 derived from mouse intestinal epithelial cells show strong bactericidal activity against cocultured E. coli cells (8). SAA1 can also function as an opsonin for macrophages and neutrophils and modulate their capacity to clear invading bacterial cells (16). Outer membrane protein A (OmpA) of Gram-negative bacteria is one of the known targets (17). Moreover, SAA proteins may play roles in defense against fungal infections by inducing chemotaxis of immune cells. For example, recombinant SAA protein functions as an activator of polymorphonuclear cells and enhances its anti-Candida activity (18).
In the present study, we report that human and mouse SAA1 proteins have a potent antifungal activity against C. albicans. Recombinantly expressed human SAA1 (rhSAA1) binds to the surface of C. albicans cells, impairs the integrity of the fungal cell membrane, and induces rapid cell death. Our study suggests that SAA1 may act as an innate antifungal against the common human fungal pathogen C. albicans.
RESULTS
C. albicans induces the expression of mouse SAA1 in serum in a systemic infection model.
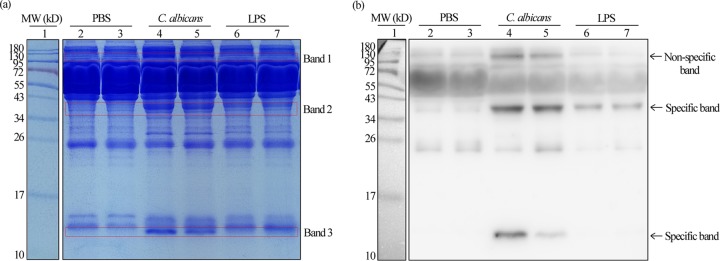
SAA1 is a well-known biomarker for acute infections and can be used as a clinical indicator of infection (6). C. albicans is a major cause of mucosal and systemic candidiasis and often triggers inflammation (2). We set out to reveal whether C. albicans infections could induce the elevated expression of SAA1 using a mouse systemic infection model. As shown in Fig. 1, SDS-PAGE and Western blot analyses were performed using an anti-mouse SAA1 antibody. Compared to the phosphate-buffered saline (PBS) treatment controls, three bands with significantly elevated expression levels (indicated in the boxes) were observed in the serum samples of C. albicans-infected mice. One band with an enhanced signal was observed in the samples from lipopolysaccharide (LPS)-treated mice, which served as positive controls. To confirm that these signals were not due to nonspecific reactions, we performed mass spectrometry analysis using proteins in the indicated band regions. Peptides derived from mouse SAA1 protein were found in the samples from bands 2 and 3 but not from band 1 (see Data Set S1 in the supplemental material), suggesting that the signals from the band 1 region could be due to nonspecific reactions. Of note, the molecular weights (MWs) of proteins in band 2 were larger than the predicted MW according to the amino acid sequences of mouse SAA1. This band might represent a trimeric form of the protein or might be due to the binding of high-density lipoprotein or heparin (19). These results indicate that C. albicans cells, like bacteria and viruses, are able to induce the expression of SAA1 in the mammalian systemic infection model.
FIG 1.
Expression of mSAA1 in serum samples of mice treated with PBS or LPS or infected with C. albicans cells. (a) Coomassie brilliant blue-stained SDS-PAGE analysis. (b) Western blot analysis. Lane 1, markers; lanes 2 and 3, serum samples from mice treated with PBS (negative control); lanes 4 and 5, serum samples from mice infected with C. albicans cells; lanes 6 and 7, serum samples from mice treated with LPS (positive control). Three band regions (indicated by the red boxes) from the samples of C. albicans-infected mice were excised and used for nanoLC-MS/MS analysis (see Data Set S1 in the supplemental material for details).
Potent anti-Candida effect of recombinant human and mouse SAA1 (rhSAA1 and rmSAA1).
Since C. albicans infection induces the elevated expression of SAA1 in mice, we next recombinantly expressed human and mouse SAA1 in E. coli and examined the antifungal activity of purified protein against C. albicans. His6-tagged rhSAA1 was purified using a nickel-chelated agarose column (see Fig. S1 in the supplemental material). To further verify the purity and specificity of rhSAA1 (Fig. S1) and rmSAA1 (data not shown), we performed SDS-PAGE and Western blot analyses with anti-human SAA1 and anti-mouse SAA1 antibodies. To evaluate the antifungal activity of rhSAA1 and rmSAA1 against C. albicans, purified proteins were tested in both PBS and Leeʼs glucose medium. As shown in Fig. 2 and Fig. S2, rhSAA1 exhibited obvious killing activity against C. albicans in a temperature- and dosage-dependent manner. It showed a higher killing effect on C. albicans at 37°C than at 30°C. At 100 mg/liter, approximately 80% of C. albicans cells were killed after 3 h of treatment with rhSAA1 in Leeʼs glucose medium.
FIG 2.
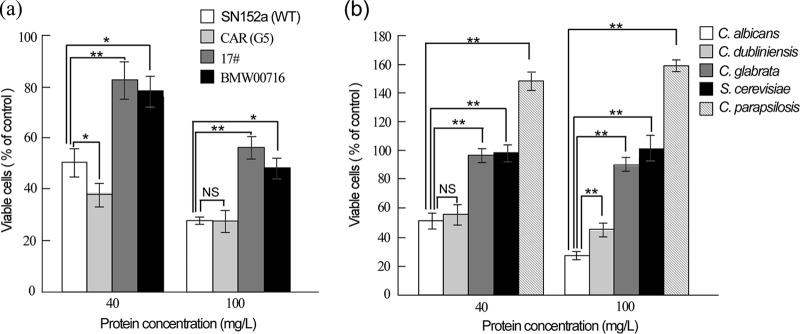
Killing of C. albicans cells by rhSAA1 at 30°C. C. albicans cells of strain SC5314 were initially cultured in liquid YPD medium to logarithmic phase at 30°C, collected, washed, and resuspended in liquid Lee’s glucose medium or PBS for killing assays. C. albicans cells (2 × 105 cells/ml) were treated with 10 mg/liter (triangles), 40 mg/liter (circles), or 100 mg/liter (squares) rhSAA1 protein for 1 to 5 h at 30°C in PBS (a) or Lee’s glucose medium (b). The percentages of viable cells were determined using plating assays. The percentage of viable cells is calculated as follows: (number of colonies of the rhSAA1-treated sample/average number of colonies of the PBS-treated sample) × 100%. PBS treatment served as a negative control. Three biological repeats were performed (n = 3). Values are presented as means ± the SD.
Interestingly, at 100 mg/liter, rhSAA1 exhibited potent antifungal activity against C. albicans azole-resistant strains CAR (G5), 17, and BMW00716 (20, 21). However, the survival rates of strains 17 and BMW00716 were higher than those of the wild-type and azole-resistant CAR (G5) strains, especially when treated with of rhSAA1 at 40 mg/liter (Fig. 3a).
FIG 3.
Killing effect of rhSAA1 on the cells of C. albicans antifungal-resistant strains (a) and C. dubliniensis, C. glabrata, C. parapsilosis, and S. cerevisiae strains (b). Fungal cells were initially cultured in liquid YPD medium to logarithmic phase at 30°C, collected, washed, and resuspended in liquid Lee’s glucose medium for killing assays. Cells (2 × 105 cells/ml) were treated with 40 or 100 mg/liter rhSAA1 protein for 3 h at 30°C. The percentages of viable cells were determined using plating assays. PBS treatment served as a negative control. The percentage of viable cells was calculated as follows: (number of colonies of the rhSAA1-treated sample/average number of colonies of the PBS-treated sample) × 100%. Three biological repeats were performed (n = 3). Values are presented as means ± the SD. (a) C. albicans antifungal-susceptible strain SN152a (fluconazole-susceptible control) and fluconazole-resistant strains CAR (G5), 17, and BMW00716. Student t tests (two-tailed) were performed between SN152 and each fluconazole-resistant strain as indicated in the figure. *, P < 0.05; **, P < 0.01; NS, no significant difference. (b) Student t tests (two-tailed) were performed between C. albicans and C. dubliniensis, C. albicans and C. glabrata, C. albicans and C. parapsilosis, and C. albicans and S. cerevisiae, as indicated in the figure. **, P < 0.01; NS, no significant difference. No significant difference was observed between the survival rate of the PBS treatment (100%) and the rhSAA1 treatments in C. glabrata and S. cerevisiae. The survival rates for the rhSAA1 treatments were significantly higher than that for the PBS treatment in C. parapsilosis (P < 0.01).
We next examined the antifungal activity of rhSAA1 against several other fungal species, including Candida dubliniensis, Candida glabrata, Candida parapsilosis, and Saccharomyces cerevisiae, at 30°C. As shown in Fig. 3b, rhSAA1 only exhibited a killing effect on cells of C. dubliniensis, a closely related species to C. albicans (22), suggesting that the antifungal activity of SAA1 is species specific.
Using a similar strategy, we examined the antifungal activity of mouse SAA1 against C. albicans using recombinant proteins (rmSAA1). As expected, rmSAA1 exhibited a notable anti-C. albicans activity in a dosage-dependent manner (Fig. S3), although its killing activity was less potent than that of rhSAA1 (Fig. S3b). These results suggest that human and mouse SAA1 have a conserved antifungal activity.
rhSAA1 has no effect on the intracellular levels of ROS in C. albicans.
Antifungals often kill fungal cells through the increased production of intracellular reactive oxygen species (ROS) (23). To explore the anti-C. albicans mechanism of rhSAA1, we first tested whether the treatment of rhSAA1 increased the intracellular levels of ROS in fungal cells. To our surprise, no obvious fluorescence signals of ROS were detected both in the rhSAA1-untreated and rhSAA1-treated C. albicans cells, whereas the signal was strong in cells treated with a commercially available oxidative reagent mixture (used as a positive control) (Fig. S4). These results suggest that the antifungal activity of rhSAA1 against C. albicans is independent of the production of ROS.
rhSAA1 binds to the cell surface and impairs the membrane integrity of C. albicans.
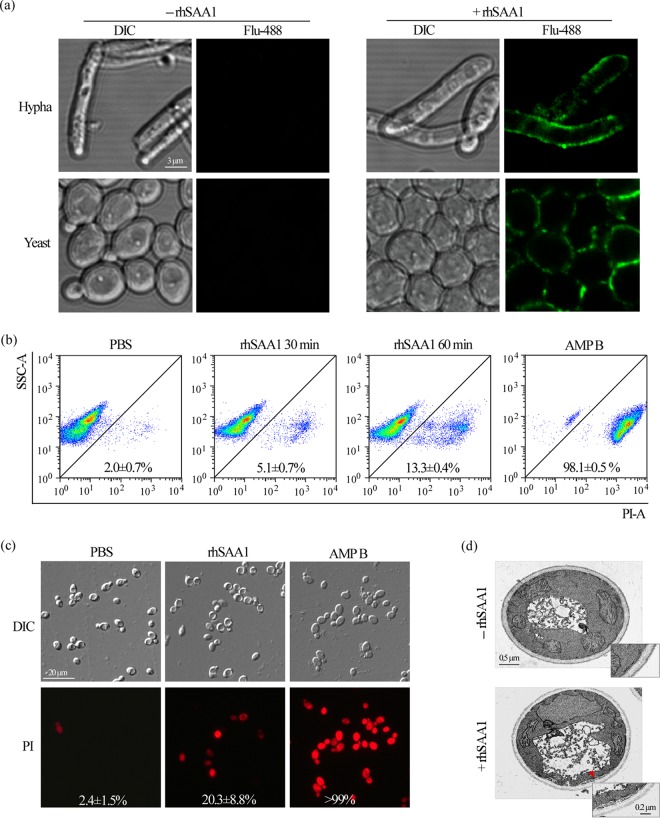
To uncover its mechanism of antifungal activity, we next treated C. albicans cells with rhSAA1 and performed immunofluorescence assays to test its subcellular localization. As demonstrated in Fig. 4a, rhSAA1 had been detected on the surfaces of both yeast-form and filamentous cells of C. albicans, but not on the surfaces of C. glabrata and S. cerevisiae cells (Fig. S5). Flow cytometry and microscopy analyses demonstrated that the treatment of rhSAA1 led to the uptake of the dye propidium iodide (PI) in C. albicans cells (Fig. 4b and c); amphotericin B treatment served as a positive control (Fig. 4b and c). PI staining assays demonstrated that the treatment of rhSAA1 also led to the uptake of the dye and the death of C. albicans filamentous cells (Fig. S6). These findings suggest that rhSAA1 effects the cell surface and impairs the integrity of the cell membrane. To validate this hypothesis, we next performed transmission electron microscopy (TEM) assays and observed obvious impairments of the integrity of the fungal cell membrane in rhSAA1-treated C. albicans cells (Fig. 4d). However, other cell organelles, such as the nucleus, vacuole, and mitochondria, remained intact. The disruption of cell membrane integrity may cause the leakage of cellular contents from the cytoplasm and thus result in cell death of C. albicans cells.
FIG 4.
rhSAA1 binds to the cell surface of C. albicans and causes cell membrane damage. (a) Immunofluorescence staining assays. Yeast-form and filamentous cells of C. albicans (SC5314) were treated with or without 200 mg/liter rhSAA1. Anti-human SAA1 antibody and Alexa Fluor 488-conjugated anti-mouse IgG were used for immunofluorescence staining assays. (b) Fluorescence-activated cell sorting analysis of PI uptake. Yeast-form cells of C. albicans (SC5314, 5 × 106 cells/ml) were treated with 200 mg/liter rhSAA1 for 30 min or 60 min and stained with the red dye PI. The cells were then subjected to flow cytometry analysis. The PBS and amphotericin B (AMP B) treatments served as negative and positive controls, respectively. SSC-A, side scatter area; PI-A, propidium iodide area. The percentages of cells stained with PI are expressed as means ± the SD of three independent experiments. (c) Cellular images of PI staining assays. C. albicans yeast-form cells (5 × 106 cells/ml) were treated with 200 mg/liter rhSAA1 for 60 min and stained with PI. The PBS and amphotericin B (AMP B) treatments served as negative and positive controls, respectively. The percentages of stained cells are shown in the corresponding images. (d) TEM assays. C. albicans cells treated with or without 200 mg/liter rhSAA1 for 3 h were used. The red arrow indicates membrane damage.
DISCUSSION
Host defense peptides and proteins are an integral part of the innate immune system (1). The limited choice of antifungal drugs and the emergence of resistant strains in the clinic support the medical need to discover and develop alternative antifungal agents such as AMPs. In the present study, we report the antifungal activity of recombinant human and mouse SAA1 against C. albicans by targeting the cell surface membrane.
Unlike most membrane-disrupting AMPs that contain a high ratio of hydrophobic residues, human SAA1 is hydrophilic and negatively charged (pI = 6.3) (24). Crystal structure analysis indicates that native SAA1 often exists as a hexamer, which may favor amyloid formation and conformational changes (19). Here, we demonstrate that rhSAA1 is able to bind to the cell surfaces of C. albicans and its closely related species C. dubliniensis but not C. glabrata and S. cerevisiae (Fig. 4 and Fig. S5). This result is consistent with the killing effect on these species. The reasons for this species-specific effect of rhSAA1 on the killing of C. albicans remain to be investigated. One possibility is that C. albicans and C. dubliniensis are commensals of humans, whereas the other species are not, and thus SAA1 could represent a newly evolved interaction between C. albicans and its human host that has recently arisen over evolutionary time. Another possibility is that species-specific cell wall proteins (or motifs of such proteins, e.g., Als proteins) could be involved in this interaction. Since the genomes of C. glabrata and S. cerevisiae do not contain ALS genes, this could explain why there is no effect of rhSAA1 with these species. We further demonstrate that rhSAA1 causes damage to the fungal cell membrane (Fig. 4) that could lead to leakage of cellular components and cell death. It has been suggested that SAA1 could be able to form stable oligomers that escape lysosomal degradation and disrupt cellular membranes (25). Antifungals often induce the generation of high levels of intracellular ROS and thus promote the cell death of fungal pathogens (23). However, we found that the treatment of rhSAA1 did not increase the intracellular ROS in C. albicans cells (Fig. S4). It is possible that the binding of the rhSAA1 protein to the fungal surface may alter fungal cell membrane integrity and affect the overall functionality of the cell membrane.
Plasma proteins have multiple roles in controlling infections. A recent study demonstrated that human plasma protein serum amyloid P component (SAP) was able to bind to surface amyloid-like structures of C. albicans and resulted in poor recognition and ingestion of yeast cells by host macrophages (26). Combined with this previous finding, our study implies that this innate immune protein could play a critical role in defense against C. albicans infections either through indirect induction of degranulation and phagocytosis or directly by killing the pathogen. During the evolutionary “arms race,” both C. albicans cells and host cells have developed a range of strategies to persist and defend against one another. Given the general function of SAA1 in systemic inflammatory conditions and its broad-spectrum activity against bacteria, it is reasonable that mammalian SAA1 also has antifungal activity. Our findings uncover a novel innate defense strategy of the mammalian host against fungal infections and also provide a potential avenue for the development of new antifungal agents.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this study are listed in Table S1 in the supplemental material. YPD medium (2% glucose, 2% peptone, 1% yeast extract) was used for routine growth of Candida albicans, Candida dubliniensis, Candida glabrata, Saccharomyces cerevisiae, and Candida parapsilosis strains. YPD plus 10% fetal bovine serum (Gibco, Grand Island, NY) medium was used for the induction of C. albicans filamentation. Luria-Bertani (LB) medium (1% peptone, 1% NaCl, 0.5% yeast extract) was used for bacterial growth.
C. albicans systemic infection assay.
Cells of C. albicans (SC5314) were cultured in liquid YPD medium to exponential phase at 30°C, collected, and washed with PBS. C. albicans cells (2.6 × 106) in 200 μl of PBS were intravenously injected into a 6- to 8-week-old female C57BL/6J mouse. Also, 200 μl of PBS only and 4 μg of LPS (Innaxon, Bristol, UK) in 200 μl of PBS served as the negative and positive controls for the induction of mouse SAA1 protein. Two mice were used for each treatment. After 24 h of infection, blood was collected from the eyeballs of mice and used for serum preparation. Equal volume of each serum sample (40 μl) was analyzed by SDS-PAGE and Western blot assays. Anti-mouse SAA1 antibody (Abcam, Cambridge, UK) and anti-rabbit IgG (secondary anti-body) coupled to horseradish peroxidase (Santa Cruz, Dallas, TX) were used for Western blot assays.
NanoLC-MS/MS analysis.
Protein bands of interest were excised from Coomassie blue-stained SDS-PAGE gels and subjected to in-gel tryptic digestion. The peptide samples were subjected to an EASY-nLC 1000 system interfaced via a Nanospray Flex ion source to an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Tandem mass spectrometry (MS/MS) data were analyzed using the Mascot database search engine. Peptide sequences were interpreted from the MS/MS spectra by searching the Swiss-Prot protein database. Carbamidomethylation of cysteines and methionine oxidation were used as a fixed and a variable modification, respectively. The peptide mass tolerance was set to 10 ppm, and the fragment mass tolerance to 0.5 Da.
Recombinant expression and purification of human and mouse SAA1 in E. coli.
The coding region of human SAA1 (hSAA1) without the signal peptide (coding for 19 to 122 amino acids [aa]) was amplified from a human liver cDNA library (a generous gift from Zhongshan Hospital, Fudan University) by PCR. The primers were 5′-GGGAATTCCATATGTTCTTTTCGTTCCTTGGCGAG-3′ and 5′-CCGCTCGAGTCAGTATTTCTCAGGCAGGCC-3′. The coding region of mouse SAA1 (mSAA1) without the signal peptide (coding for 19 to 122 aa) was chemically synthesized by WuXi Qinglan Biotech, Inc., Wuxi, China. The PCR products of the coding regions of human and mouse SAA1 were then digested with restriction enzymes NdeI and XhoI and subcloned into plasmid pET28a (4). The expression plasmids pET28-hSAA1 and pET28-mSAA1 were transformed into E. coli strain Rosetta (DE3).
To induce the expression of human or mouse SAA1 in E. coli, bacterial cells were cultured to an optical density at 600 nm of 0.5 at 37°C in LB medium containing 50 mg/liter kanamycin and then 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Sangon Biotech, Shanghai, China) was added to the culture. After 3 h of incubation at 37°C, bacterial cells were harvested by centrifugation at 5,000 × g for 10 min at 4°C and lysed by sonication. The lysate samples were then spun down at 10,000 × g for 15 min at 4°C, and the supernatants were subjected to recombinant SAA1 (rSAA1) rSAA1 purification using a His-Bind purification kit (Millipore, Uppsala, Sweden). The eluted proteins collected at 1 ml per tube were analyzed by 15% SDS-PAGE analysis and stained with Coomassie brilliant blue. To further assess the specificity of rSAA1, samples of IPTG-uninduced and IPTG-induced cell lysates and purified rSAA1 were analyzed by Western blot assays. Anti-human SAA1 monoclonal antibody (Abcam) or mouse-SAA1 polyclonal antibody (Abcam, data not shown) were used for hybridization. The purified recombinant SAA1 was dialyzed with cold PBS, and its concentration was determined using the Bradford method (Bio-Rad, Hercules, CA).
Killing assays of fungal cells.
Antifungal activity of rhSAA1 was determined with a CFU assay as described previously with slight modifications (27). Fungal cells from a single colony were inoculated in liquid YPD medium and cultured to logarithmic phase at 30°C with shaking. Fungal cells were collected, washed, and then diluted with Lee’s glucose or PBS to 2 × 105 cells/ml. Cells were then treated with equal volumes of rhSAA1 at different concentrations and incubated at 30 or 37°C. PBS treatment served as a negative control. The CFU were determined at the time points indicated. Three biological repeats were performed for each test. Independent fungal colonies and liquid cultures were used for each replicate. The percent viable cells was calculated as follows: = (number of colonies of the rhSAA1-treated sample/average number of colonies of the PBS-treated sample) × 100%. Statistical analyses (Student t tests) were performed using Microsoft Excel (v.2010).
Intracellular ROS determination.
The ROS determination assay was performed with a DCFDA-ROS detection kit (Beyotime Company, Shanghai, China) according to our previous publication (28). C. albicans cells (5 × 106 cells/ml; SC5314) were incubated with rhSAA1 at 200 mg/liter for 2 h at 30°C. The cells were harvested, washed with PBS, and then incubated with the DCFDA probe for 30 min at 37°C in dark. Cells treated with the ROS inducer (provided in the kit) served as a positive control. The cells were visualized by fluorescence microscopy (Carl Zeiss Imager A2; Zeiss, Göttingen, Germany).
rhSAA1 binding assays.
Binding assays of rhSAA1 to cells of C. albicans were performed according to previous reports with slight modifications (17). Briefly, fungal cells were grown to logarithmic phase in liquid medium at 30°C, harvested, and washed with PBS. Cells (5 × 106 cells/ml) were mixed with 200 mg/liter rhSAA1 at room temperature for 2.5 h. The binding assays were performed at this relatively low temperature to avoid the high killing rate of rhSAA1. Subsequently, cells were washed three times with PBS and incubated with mouse anti-human SAA1 antibody (Abcam) at 4°C overnight. After a washing step with PBS, Alexa Fluor 488-conjugated goat anti-mouse IgG (Life Technologies, Delhi, India) was added to the cells for visualization by confocal scanning laser microscopy (SP8; Leica, Wetzlar, Germany) or fluorescence microscopy (Carl Zeiss Imager A2) assays.
Propidium iodide staining assay.
C. albicans cells (SC5314) were cultured to logarithmic phase in YPD medium at 30°C with shaking. Yeast-form (5 × 106 cells/ml) and filamentous cells were treated with 200 mg/liter rhSAA1 for 30 or 60 min at 30°C and then stained with PI (10 mg/liter; Sigma, St. Louis, MO) for 30 min at 4°C in the dark. To disperse aggregated cells, the yeast samples were treated with trypsin (Solarbio, Beijing, China) for 15 min at 37°C and resuspended in PBS for flow cytometry analysis (BD FACSCalibur, San Jose, CA). C. albicans cells treated with PBS for 60 min served as a negative control, and C. albicans cells treated with amphotericin B (10 μM; Sigma) for 20 min served as a positive control. At least 20,000 events per sample were recorded and analyzed using FlowJo 7.6 software (FlowJo, LLC, Ashland, NC). Three experimental repeats were performed. The averages and standard deviations (SD) of the percentages of PI-stained populations are presented. PI uptake in the yeast-from and filamentous C. albicans cells was also examined using fluorescence microscopy assays (Carl Zeiss Imager A2).
Transmission electron microscopy.
C. albicans cells (SC5314) treated with 200 mg/liter rhSAA1 for 3 h in PBS were used for the TEM assay. TEM assays were performed according to our previous report (28). Briefly, cells were fixed in 0.5% polyoxymethylene and 2.5% glutaraldehyde in a buffer solution (0.2 M PIPES; pH 6.8), 1 mM MgCl2, 1 mM CaCl2, 0.1 M sorbitol), embedded with 1% agarose, and then washed five times with cold deionized water. Cells were then stained with 1.5% KMnO4 for 80 min at 4°C and subjected to dehydration with cold acetone solutions at different concentrations (30, 50, 70, 85, 95, 100, 100, and 100%). C. albicans cells were then infiltrated and embedded in Spurr resin for TEM (JEM-1400; JEOL, Peabody, MA).
Supplementary Material
ACKNOWLEDGMENTS
We thank Lixin Zhang from the Institute of Microbiology, Chinese Academy of Sciences, for the antifungal-resistant strains.
This study was supported by grants from the National Natural Science Foundation of China (31930005 and 31625002 to G.H.) and funding from a National Institutes of Health National Institute of General Medical Sciences award (R35GM124594 to C.J.N.).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bastos P, Trindade F, da Costa J, Ferreira R, Vitorino R. 2018. Human antimicrobial peptides in bodily fluids: current knowledge and therapeutic perspectives in the postantibiotic era. Med Res Rev 38:101–146. doi: 10.1002/med.21435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman J. 2012. Candida albicans. Curr Biol 22:R620–R622. doi: 10.1016/j.cub.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 3.Gyurko C, Lendenmann U, Helmerhorst EJ, Troxler RF, Oppenheim FG. 2001. Killing of Candida albicans by histatin 5: cellular uptake and energy requirement. Antonie Van Leeuwenhoek 79:297–309. doi: 10.1023/a:1012070600340. [DOI] [PubMed] [Google Scholar]
- 4.Orrapin S, Intorasoot S. 2014. Recombinant expression of novel protegrin-1 dimer and LL-37-linker-histatin-5 hybrid peptide mediated biotin carboxyl carrier protein fusion partner. Protein Expr Purif 93:46–53. doi: 10.1016/j.pep.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Tsai PW, Yang CY, Chang HT, Lan CY. 2011. Human antimicrobial peptide LL-37 inhibits adhesion of Candida albicans by interacting with yeast cell-wall carbohydrates. PLoS One 6:e17755. doi: 10.1371/journal.pone.0017755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uhlar CM, Whitehead AS. 1999. Serum amyloid A, the major vertebrate acute-phase reactant. Eur J Biochem 265:501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- 7.Hirakura Y, Carreras I, Sipe JD, Kagan BL. 2002. Channel formation by serum amyloid A: a potential mechanism for amyloid pathogenesis and host defense. Amyloid 9:13–23. doi: 10.3109/13506120209072440. [DOI] [PubMed] [Google Scholar]
- 8.Eckhardt ER, Witta J, Zhong J, Arsenescu R, Arsenescu V, Wang Y, Ghoshal S, de Beer MC, de Beer FC, de Villiers WJ. 2010. Intestinal epithelial serum amyloid A modulates bacterial growth in vitro and proinflammatory responses in mouse experimental colitis. BMC Gastroenterol 10:133. doi: 10.1186/1471-230X-10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorn CF, Lu ZY, Whitehead AS. 2003. Tissue-specific regulation of the human acute-phase serum amyloid A genes, SAA1 and SAA2, by glucocorticoids in hepatic and epithelial cells. Eur J Immunol 33:2630–2639. doi: 10.1002/eji.200323985. [DOI] [PubMed] [Google Scholar]
- 10.Urieli-Shoval S, Cohen P, Eisenberg S, Matzner Y. 1998. Widespread expression of serum amyloid A in histologically normal human tissues: predominant localization to the epithelium. J Histochem Cytochem 46:1377–1384. doi: 10.1177/002215549804601206. [DOI] [PubMed] [Google Scholar]
- 11.Chambers RE, MacFarlane DG, Whicher JT, Dieppe PA. 1983. Serum amyloid A protein concentration in rheumatoid arthritis and its role in monitoring disease activity. Ann Rheum Dis 42:665–667. doi: 10.1136/ard.42.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis KE, Kirk EA, McDonald TO, Wang S, Wight TN, O’Brien KD, Chait A. 2004. Increase in serum amyloid a evoked by dietary cholesterol is associated with increased atherosclerosis in mice. Circulation 110:540–545. doi: 10.1161/01.CIR.0000136819.93989.E1. [DOI] [PubMed] [Google Scholar]
- 13.Jahangiri A, Wilson PG, Hou TF, Brown A, King VL, Tannock LR. 2013. Serum amyloid A is found on ApoB-containing lipoproteins in obese humans with diabetes. Obesity (Silver Spring) 21:993–996. doi: 10.1002/oby.20126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su SB, Gong W, Gao JL, Shen W, Murphy PM, Oppenheim JJ, Wang JM. 1999. A seven-transmembrane, G protein-coupled receptor, FPRL1, mediates the chemotactic activity of serum amyloid A for human phagocytic cells. J Exp Med 189:395–402. doi: 10.1084/jem.189.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen M, Zhou H, Cheng N, Qian F, Ye RD. 2014. Serum amyloid A1 isoforms display different efficacy at Toll-like receptor 2 and formyl peptide receptor 2. Immunobiology 219:916–923. doi: 10.1016/j.imbio.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah C, Hari-Dass R, Raynes JG. 2006. Serum amyloid A is an innate immune opsonin for Gram-negative bacteria. Blood 108:1751–1757. doi: 10.1182/blood-2005-11-011932. [DOI] [PubMed] [Google Scholar]
- 17.Hari-Dass R, Shah C, Meyer DJ, Raynes JG. 2005. Serum amyloid A protein binds to outer membrane protein A of gram-negative bacteria. J Biol Chem 280:18562–18567. doi: 10.1074/jbc.M500490200. [DOI] [PubMed] [Google Scholar]
- 18.Badolato R, Wang JM, Stornello SL, Ponzi AN, Duse M, Musso T. 2000. Serum amyloid A is an activator of PMN antimicrobial functions: induction of degranulation, phagocytosis, and enhancement of anti-Candida activity. J Leukoc Biol 67:381–386. doi: 10.1002/jlb.67.3.381. [DOI] [PubMed] [Google Scholar]
- 19.Lu J, Yu Y, Zhu I, Cheng Y, Sun PD. 2014. Structural mechanism of serum amyloid A-mediated inflammatory amyloidosis. Proc Natl Acad Sci U S A 111:5189–5194. doi: 10.1073/pnas.1322357111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White TC. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother 41:1482–1487. doi: 10.1128/AAC.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franz R, Kelly SL, Lamb DC, Kelly DE, Ruhnke M, Morschhauser J. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob Agents Chemother 42:3065–3072. doi: 10.1128/AAC.42.12.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belenky P, Camacho D, Collins JJ. 2013. Fungicidal drugs induce a common oxidative-damage cellular death pathway. Cell Rep 3:350–358. doi: 10.1016/j.celrep.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang LJ, Gallo RL. 2016. Antimicrobial peptides. Curr Biol 26:R14–R19. doi: 10.1016/j.cub.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 25.Jayaraman S, Gantz DL, Haupt C, Gursky O. 2017. Serum amyloid A forms stable oligomers that disrupt vesicles at lysosomal pH and contribute to the pathogenesis of reactive amyloidosis. Proc Natl Acad Sci U S A 114:E6507–E6515. doi: 10.1073/pnas.1707120114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behrens NE, Lipke PN, Pilling D, Gomer RH, Klotz SA. 2019. Serum amyloid P component binds fungal surface amyloid and decreases human macrophage phagocytosis and secretion of inflammatory cytokines. mBio 10:e00218-19. doi: 10.1128/mBio.00218-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koshlukova SE, Araujo MW, Baev D, Edgerton M. 2000. Released ATP is an extracellular cytotoxic mediator in salivary histatin 5-induced killing of Candida albicans. Infect Immun 68:6848–6856. doi: 10.1128/iai.68.12.6848-6856.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du H, Guan G, Li X, Gulati M, Tao L, Cao C, Johnson AD, Nobile CJ, Huang G. 2015. N-Acetylglucosamine-induced cell death in Candida albicans and its implications for adaptive mechanisms of nutrient sensing in yeasts. mBio 6:e01376. doi: 10.1128/mBio.01376-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.