Second-generation HIV-1 integrase strand transfer inhibitors (INSTIs) dolutegravir (DTG), bictegravir (BIC), and cabotegravir (CAB) showed a high genetic barrier to resistance and limited cross-resistance with first-generation INSTIs raltegravir (RAL) and elvitegravir (EVG).
KEYWORDS: HIV-1 resistance, integrase inhibitors, phenotypic assay, dolutegravir, cabotegravir, bictegravir
ABSTRACT
Second-generation HIV-1 integrase strand transfer inhibitors (INSTIs) dolutegravir (DTG), bictegravir (BIC), and cabotegravir (CAB) showed a high genetic barrier to resistance and limited cross-resistance with first-generation INSTIs raltegravir (RAL) and elvitegravir (EVG). In this study, DTG, BIC, and CAB demonstrated a comparable activity on a panel of INSTI-resistant strains isolated from patients exposed to RAL, EVG, and/or DTG, with a significantly reduced susceptibility only with the pathway Q148H/K/R plus one to two additional INSTI mutations.
INTRODUCTION
Exposure to first-generation HIV-1 integrase strand transfer inhibitors (INSTIs), such as raltegravir (RAL) and elvitegravir (EVG), is frequently associated with the selection of resistance-associated mutations (RAMs) at virological failure. Recently developed second-generation INSTIs, such as dolutegravir (DTG; approved by FDA and European Medicines Agency [EMA] in 2013 and 2014, respectively), bictegravir (BIC; approved by FDA and EMA in 2018 as part of a single tablet regimen, including tenofovir alafenamide [TAF] and emtricitabine [FTC]), and cabotegravir (CAB; currently under phase III clinical investigation) have demonstrated a superior genetic barrier to resistance and a variable activity against viruses harboring INSTI resistance mutations selected by RAL and EVG. However, predicting the activity of second-generation INSTIs on HIV-1 variants with different combination of INSTI RAMs is not straightforward. This study aimed at clarifying cross-resistance to DTG, BIC, and CAB in a panel of INSTI-resistant strains isolated from patients previously exposed to RAL, EVG, and/or DTG.
Plasma samples from 19 patients were collected during routine drug resistance testing under virological failure of an INSTI regimen, based on the detection of at least one major INSTI RAM (T66A, E92Q, E138K/T, G140A/C/S, Y143C/R, S147G, Q148H/R, N155H, and R263K). Virological failure was defined at the individual clinics as confirmed viral load of >50 HIV-1 RNA copies/ml or one viral load of >1,000 HIV-1 RNA copies/ml. Access to residual samples for research purposes was regulated by informed consent approved by the Southeast Tuscany Ethics Committee. Plasma RNA was extracted by the DSP virus kit using the EZ1 workstation (Qiagen, Hilden, Germany). Population sequencing of the integrase region was performed by homebrew technology using primers previously described (1). For in vitro susceptibility testing, the integrase coding region was amplified through a 2-step PCR protocol, and the resulting amplicon was used for the generation of chimeric viruses by homologous recombination with an integrase-deleted pNL4-3 vector in 293T cells (2). Integrase sequences of recombinant viruses were confirmed by Sanger sequencing.
Susceptibility to DTG, BIC, and CAB was assessed through a TZM-bl cell line-based phenotypic assay, shown to correlate well with the de facto reference PhenoSense assay by Monogram Biosciences and expressed as fold change (FC) with respect to the reference wild type NL4-3 virus (2). Differences in FC values among groups were tested by Friedman’s ANOVA, and multiple comparisons were performed by the Dunn’s test when appropriate.
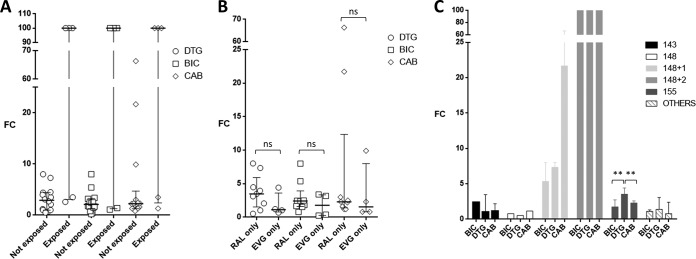
The patients had been exposed to RAL only (n = 9), EVG only (n = 4), RAL and DTG (n = 5), or RAL and EVG (n = 1). By querying the COMET HIV-1 subtyping tool (3), sequences were assigned to subtype B (n = 13), subtype F1 (n = 2), subtype G (n = 2), CRF03_AB (n = 1), and CRF06_cpx (n = 1). Sequences were grouped according to the presence of major INSTI RAMs at codon 143, 148, or 155, alone or in combination with other INSTI RAMs. Four samples with INSTI RAMs E92Q (n = 2), T66A plus S147SG, and R263K were grouped together as “others” (Table 1). Median (interquartile range [IQR]) FC values for DTG, BIC, and CAB were 3.5 (1.2 to 7.3), 2.4 (1.4 to 5.4), and 2.3 (1.3 to 21.7), respectively, without statistically significant differences between groups. Median (IQR) DTG, BIC, and CAB FC values from patients not exposed to DTG were 2.9 (1.0 to 4.4), 2.1 (1.3 to 3.3), and 2.3 (1.2 to 4.7), respectively, while exposure to RAL and then DTG resulted in a FC of >100 in three cases (Q148H/K/R plus 2 INSTI RAMs) and FC of <3.5 in two cases (samples with E92Q and with R263K) for all three drugs (Fig. 1A). Median (IQR) DTG, BIC, and CAB FC values were consistently, but not significantly, higher following exposure to RAL only versus EVG only (3.5 [1.6 to 5.9], 2.4 [1.6 to 3.9], and 2.3 [1.4 to 12.3] versus 1.1 [0.8 to 3.6], 1.8 [0.3 to 3.3], and 1.6 [0.8 to 8.0], respectively) (Fig. 1B). According to the three major INSTI resistance pathways, median (IQR) DTG, BIC, and CAB FC values were 2.3 (1.0 to 3.5), 2.5 (2.4 to 2.5), and 1.7 (1.2 to 2.2), respectively, with Y143R/C alone (n = 2); 3.6 (2.6 to 4.4), 1.8 (1.7 to 2.7), and 2.3 (1.5 to 2.6) with N155H alone (n = 6); 0.5, 0.8, and 1.2 with Q148R alone (n = 1); 7.3 (1.0 to 8.0), 5.4 (3.2 to 8.0), and 21.7 (9.9 to 66.3) with Q148R plus one additional INSTI RAM (n = 3); and >100 for all the three drugs with Q148H/K/R plus two additional INSTI RAMs (n = 3) (Fig. 1C). In the N155H group, the BIC and CAB FC values were significantly lower than the DTG FC values (P = 0.005). Among samples with other INSTI RAMs (two with E92Q, one with R263K, and one with T66A plus S147S/G), FC values of ≤3.5 were measured for all the three drugs. In particular, one sample from a failing DTG-based regimen harboring R263K had DTG and CAB FCs of 3.5, while BIC FC was 1.1.
TABLE 1.
Stanford HIVdb major and accessory INSTI RAMs and DTG, BIC, and CAB IC50 and FC values in a panel of 19 samples from patients failing INSTI-based therapya
| Sample | Subtype | INSTI exposureb | Major mutation(s) | Accessory mutation | DTG |
BIC |
CAB |
Group | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IC50 ± SD nM | FC | IC50 ± SD nM | FC | IC50 ± SD nM | FC | ||||||
| NL4-3 | B | Wild type | None | None | 0.8 ± 0.4 | 1 | 0.5 ± 0.1 | 1 | 1.8 ± 0.9 | 1 | Wild type |
| 151277 | F1 | RAL | Y143R | T97A | 0.8 ± 0.1 | 1.0 | 1.1 ± 0.4 | 2.4 | 2.1 ± 1.0 | 1.2 | Y143C/H/R |
| 151648 | G | RAL | Y143C | S230R | 2.6 ± 1.7 | 3.5 | 1.2 ± 0.1 | 2.5 | 4.0 ± 0.5 | 2.2 | |
| 150678 | B | RAL | Q148R | None | 0.4 ± 0.3 | 0.5 | 0.4 ± 0.3 | 0.8 | 2.1 ± 0.1 | 1.2 | Q148H/K/R |
| 146943 | B | RAL | G140C, Q148R | None | 6.0 ± 4.9 | 8.0 | 3.8 ± 2.4 | 8.0 | 121.3 ± 40.9 | 66.3 | Q148H/K/R plus one RAM |
| 147026 | B | EVG | E138K, Q148R | None | 0.8 ± 0.9 | 1.0 | 1.5 ± 0.3 | 3.2 | 18.2 ± 8.0 | 9.9 | |
| 147086 | B | RAL | G140A, Q148R | None | 5.5 ± 5.7 | 7.3 | 2.5 ± 0.5 | 5.4 | 39.6 ± 28.8 | 21.7 | |
| 149357 | B | RAL, DTG | E138K, G140S, Q148H, N155H | None | >100 | >100 | >100 | >100 | >200 | >100 | Q148H/K/R plus two RAMs |
| 151377 | B | RAL, DTG | E138T, G140S, Q148H | T97A | >100 | >100 | >100 | >100 | >200 | >100 | |
| 151236 | B | RAL, DTG | E138K, G140A, S147SG, Q148R | T97A | >100 | >100 | >100 | >100 | >200 | >100 | |
| 150953 | CRF03_AB | RAL | N155H | E157Q | 1.6 ± 1.2 | 2.1 | 0.9 ± 0.2 | 1.8 | 3.0 ± 0.7 | 1.6 | N155H |
| 150336 | G | RAL | N155H | T97A | 3.0 ± 0.2 | 4.0 | 0.7 ± 0.2 | 1.5 | 5.4 ± 4.2 | 3.0 | |
| 149789 | B | RAL, EVG | N155H | None | 2.0 ± 0.9 | 2.7 | 0.8 ± 0.4 | 1.7 | 2.3 ± 0.9 | 1.3 | |
| 150077 | B | EVG | N155H | None | 3.3 ± 0.7 | 4.4 | 1.6 ± 0.8 | 3.4 | 4.3 ± 1.2 | 2.3 | |
| 149865 | B | RAL | N155H | None | 3.4 ± 1.4 | 4.5 | 1.2 ± 0.3 | 2.5 | 4.5 ± 2.8 | 2.5 | |
| 150348 | B | RAL | N155H | None | 2.3 ± 2.0 | 3.1 | 0.8 ± 0.4 | 1.7 | 4.2 ± 0.2 | 2.3 | |
| 149570 | CRF06_cpx | EVG | E92Q | None | 0.9 ± 0.1 | 1.2 | 0.2 ± 0.1 | 0.4 | 1.4 ± 0.5 | 0.8 | Others |
| 148640 | B | RAL, DTG | E92Q | None | 1.9 ± 0.3 | 2.6 | 0.7 ± 0.6 | 1.4 | 2.3 ± 1.0 | 1.3 | |
| 150129 | B | EVG | T66A, S147SG | G163K | 0.5 ± 0.1 | 0.7 | 0.1 ± 0.1 | 0.2 | 1.4 ± 0.4 | 0.8 | |
| 151422 | F1 | RAL, DTG | R263K | None | 2.7 ± 0.2 | 3.5 | 0.5 ± 0.1 | 1.1 | 6.5 ± 2.3 | 3.5 | |
| Median (IQR) | 2.6 (1.4–3.9) | 3.5 (1.2–7.3) | 1.1 (0.7–1.8) | 2.4 (1.4–5.4) | 4.3 (2.3–39.6) | 2.3 (1.3–21.7) | |||||
INSTI, integrase strand transfer inhibitor; RAM, resistance-associated mutation; DTG, dolutegravir; BIC, bictegravir; CAB, cabotegravir; IC50, half maximal inhibitory concentration; FC, fold change; SD, standard deviation; IQR, interquartile range; HIVdb, HIV database.
Where two INSTIs are indicated, the former was used before and the latter was in use at the time of genotyping; therapy based on the first INSTI failed in all these cases except for cases 149789 and 149357, where the second INSTI-based therapy was started as simplification while the patient was virologically suppressed. Where only one INSTI is indicated, this was in use at the time of genotyping.
FIG 1.
Fold change (FC) dolutegravir (DTG), bictegravir (BIC), and cabotegravir (CAB) susceptibility values stratified by exposure to DTG (A), exposure to RAL only or EVG only (B), and mutational pathway (C). ns, P value not significant; **, P = 0.005.
These data are in line with and yet add further information to those reported with specific laboratory clones and with a few clinical samples (4–7). In the largest data set analyzed comprising 47 patient-derived HIV-1 isolates, BIC appeared to retain more activity against INSTI-resistant mutants than DTG (4). The median FC value for BIC was also lower than that for DTG in our case file; however, the difference was statistically significant only for the N155H pathway and yet not for the data overall. The Q148H/K/R plus G140S pathway was confirmed to be the most challenging for all second-generation INSTIs, with the addition of a third RAM (particularly at codon 138) substantially increasing the FC value, as shown also in previous studies (4–7). Contrary to the results generated with molecular clones (7), we did not detect better activity of DTG than with CAB, likely due to the overrepresentation of N155H single mutants in our data set.
In summary, DTG, BIC, and CAB retain comparable activity against major INSTI RAMs; however, adding one and, particularly, two additional INSTI RAMs to the Q148H/K/R pathway results in a substantial loss of susceptibility for all three drugs. The full spectrum of additional mutations playing such a role in different HIV-1 subtypes remains to be elucidated. More extensive in vivo data are required to fully define the correlation between HIV-1 genotype, FC values, and response to treatment with second-generation INSTIs.
Data availability.
Integrase sequences of study samples were submitted to GenBank under accession numbers MN389749 to MN389767.
ACKNOWLEDGMENTS
We thank the patients for participating in the study and the clinical units for providing information about treatment exposure.
Dolutegravir was kindly provided by ViiV Healtcare, while cabotegravir and bictegravir were purchased from MedChemExpress. The NL4-3 wild-type reference strain and the TZM-bl cell line were obtained through the NIH AIDS Reagent Program.
The authors acknowledge the contribution of CARE Consortium funded by the European Union’s Horizon 2020 program and the Ministry of Science and Higher Education of the Russian Federation. In addition, the study was partially supported by ViiV Healthcare for the project “HIV multidrug resistance pathways in EuResist Integrated DataBase.”
M.Z. reports consultancy for ViiV Healthcare, Gilead Sciences, and Janssen-Cilag and grants for his institution from ViiV Healthcare and Gilead outside the submitted work. All other authors have none to declare.
REFERENCES
- 1.Van Laethem K, Schrooten Y, Covens K, Dekeersmaeker N, De Munter P, Van Wijngaerden E, Van Ranst M, Vandamme AM. 2008. A genotypic assay for the amplification and sequencing of integrase from diverse HIV-1 group M subtypes. J Virol Methods 153:176–181. doi: 10.1016/j.jviromet.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Saladini F, Giannini A, Boccuto A, Vicenti I, Zazzi M. 2018. Agreement between an in-house replication competent and a reference replication defective recombinant virus assay for measuring phenotypic resistance to HIV-1 protease, reverse transcriptase, and integrase inhibitors. J Clin Lab Anal 32:e22206. doi: 10.1002/jcla.22206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Struck D, Lawyer G, Ternes AM, Schmit JC, Bercoff DP. 2014. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res 42:e144. doi: 10.1093/nar/gku739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsiang M, Jones GS, Goldsmith J, Mulato A, Hansen D, Kan E, Tsai L, Bam RA, Stepan G, Stray KM, Niedziela-Majka A, Yant SR, Yu H, Kukolj G, Cihlar T, Lazerwith SE, White KL, Jin H. 2016. Antiviral activity of bictegravir (GS-9883), a novel potent HIV-1 integrase strand transfer inhibitor with an improved resistance profile. Antimicrob Agents Chemother 60:7086–7097. doi: 10.1128/AAC.01474-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassounah SA, Alikhani A, Oliveira M, Bharaj S, Ibanescu RI, Osman N, Xu HT, Brenner BG, Mesplède T, Wainberg MA. 2017. Antiviral activity of bictegravir and cabotegravir against integrase inhibitor-resistant SIVmac239 and HIV-1. Antimicrob Agents Chemother 61:e01695-17. doi: 10.1128/AAC.01695-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang WW, Cheung PK, Oliveira N, Robbins MA, Harrigan PR, Shahid A. 2018. Accumulation of multiple mutations in vivo confers cross-resistance to new and existing integrase inhibitors. J Infect Dis 218:1773–1776. doi: 10.1093/infdis/jiy428. [DOI] [PubMed] [Google Scholar]
- 7.Smith SJ, Zhao XZ, Burke TR Jr, Hughes SH. 2018. Efficacies of cabotegravir and bictegravir against drug-resistant HIV-1 integrase mutants. Retrovirology 15:37. doi: 10.1186/s12977-018-0420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Integrase sequences of study samples were submitted to GenBank under accession numbers MN389749 to MN389767.