In the present study, we employed untargeted metabolomics to investigate the synergistic killing mechanism of polymyxin B in combination with an aminoglycoside, amikacin, against a polymyxin-susceptible isolate of Pseudomonas aeruginosa, FADDI-PA111 (MIC = 2 mg/liter for both polymyxin B and amikacin), and a polymyxin-resistant Liverpool epidemic strain (LES), LESB58 (the corresponding MIC for both polymyxin B and amikacin is 16 mg/liter).
KEYWORDS: Pseudomonas aeruginosa, amikacin, metabolomics, polymyxin
ABSTRACT
In the present study, we employed untargeted metabolomics to investigate the synergistic killing mechanism of polymyxin B in combination with an aminoglycoside, amikacin, against a polymyxin-susceptible isolate of Pseudomonas aeruginosa, FADDI-PA111 (MIC = 2 mg/liter for both polymyxin B and amikacin), and a polymyxin-resistant Liverpool epidemic strain (LES), LESB58 (the corresponding MIC for both polymyxin B and amikacin is 16 mg/liter). The metabolites were extracted 15 min, 1 h, and 4 h following treatment with polymyxin B alone (2 mg/liter for FADDI-PA111; 4 mg/liter for LESB58), amikacin alone (2 mg/liter), and both in combination and analyzed using liquid chromatography-mass spectrometry (LC-MS). At 15 min and 1 h, polymyxin B alone induced significant perturbations in glycerophospholipid and fatty acid metabolism pathways in FADDI-PA111 and, to a lesser extent, in LESB58. Amikacin alone at 1 and 4 h induced significant perturbations in peptide and amino acid metabolism, which is in line with the mode of action of aminoglycosides. Pathway analysis of FADDI-PA111 revealed that the synergistic effect of the combination was largely due to the inhibition of cell envelope biogenesis, which was driven initially by polymyxin B via suppression of key metabolites involved in lipopolysaccharide, peptidoglycan, and membrane lipids (15 min and 1 h) and later by amikacin (4 h). Overall, these novel findings demonstrate that the disruption of cell envelope biogenesis and central carbohydrate metabolism, decreased levels of amino sugars, and a downregulated nucleotide pool are the metabolic pathways associated with the synergistic killing of the polymyxin-amikacin combination against P. aeruginosa. This mechanistic study might help optimize synergistic polymyxin B combinations in the clinical setting.
INTRODUCTION
The World Health Organization (WHO) recently classified multidrug-resistant (MDR) Pseudomonas aeruginosa as a top-priority critical pathogen that urgently requires new antibiotic therapies (1). MDR P. aeruginosa often causes life-threatening nosocomial infections such as pneumonia and bloodstream infections, in particular in immunocompromised and critically ill patients (2, 3). P. aeruginosa is often responsible for the colonization of the lungs of adult cystic fibrosis (CF) patients and is associated with high mortality rates (4, 5). The large genome (5.9 to 6.3 Mb) of P. aeruginosa encodes complex regulation systems and remarkable metabolic flexibility, endowing it with the ability to rapidly adapt to diverse conditions, such as antimicrobial treatment (6). The known mechanisms of antimicrobial resistance in P. aeruginosa include induced efflux pumping, altering target binding sites, and enzymatic inactivation of antibiotics (7, 8). Most worryingly, P. aeruginosa can rapidly develop resistance to all current antibiotics, including the last-resort lipopeptide antibiotics, the polymyxins (polymyxin B and colistin, also known as polymyxin E) (9–11).
Polymyxins are nonribosomal polycationic decapeptides that consist of a cyclic heptapeptide linked to a tripeptide linear chain and an N-terminal fatty acyl tail. Polymyxins are amphipathic molecules owing to their five basic l-α-γ-diaminobutyric acid (Dab) residues, two hydrophobic amino acids at positions 6 and 7, and the N-terminal fatty acyl chain (12). A model for their mode of action entails that the cationic Dab residues of the polymyxin molecule interact electrostatically with the negatively charged phosphate groups of the lipid A component of lipopolysaccharide (LPS), followed by the displacement of divalent cations (Mg2+ and Ca2+) that bridge and stabilize the LPS leaflet of the outer membrane (OM) (13). This then enables the insertion of the N-terminal fatty acyl group and the hydrophobic amino acid side chain at positions 6 to 7 of the polymyxin into the OM fatty acyl layer, leading to outer membrane destabilization, self-promoted uptake, osmotic imbalance, and cell death (12, 14). However, there are still significant gaps in the exact mechanism(s) of polymyxin activity and resistance in P. aeruginosa (15). Treatment failure due to suboptimal polymyxin plasma concentrations or the presence of heteroresistant subpopulations highlights the need to optimize antibiotic combination therapies (e.g., polymyxin B combined with the aminoglycoside amikacin) (16, 17). The most common mechanism of polymyxin resistance in P. aeruginosa is mainly related to LPS and involves the addition of phosphoethanolamine (pEtN) and 4-amino-4-deoxy-l-arabinose (l-Ara4N) or deacylation, hydroxylation, and palmitoylation to its lipid A component (18–20). As a result, the overall net negative charge of the OM is reduced, which attenuates the binding of the polycationic polymyxin molecule (12). Currently used polymyxin combination therapies are empirical, and most combination studies focus only on phenotypic killing (21). The lack of a fundamental understanding of the mechanistic synergy underlying polymyxin combination therapies hinders their clinical utility. Systems pharmacology is a powerful approach for deciphering the complex interplay between cellular pathways in response to antibiotic treatments (22). Metabolomics is the combination of state-of-the-art bioanalytical techniques and bioinformatics, which are of considerable utility for elucidating the complex modes of action and bacterial cellular processes in response to antibiotic killing (23, 24).
To the best of our knowledge, we are the first to conduct a metabolomics analysis of the synergistic killing mechanism of a polymyxin in combination with amikacin. We profiled the response of polymyxin-susceptible P. aeruginosa (FADDI-PA111) and polymyxin-resistant P. aeruginosa (LESB58) following treatment with the polymyxin B-amikacin combination. This study was the first to reveal that the synergistic bactericidal effect of the combination involves the disruption of bacterial cell envelope biogenesis and inhibition of central carbohydrate metabolism and the pyridine nucleotide cycle (PNC).
RESULTS
Metabolic impact of polymyxin B, amikacin, and their combination on polymyxin-susceptible P. aeruginosa FADDI-PA111.
Lipopolysaccharide biosynthesis, the pyridine nucleotide cycle, central carbon metabolism, and peptidoglycan biosynthesis are the main pathways influenced by polymyxin B and amikacin monotherapies and their combination.
Lipid metabolism.
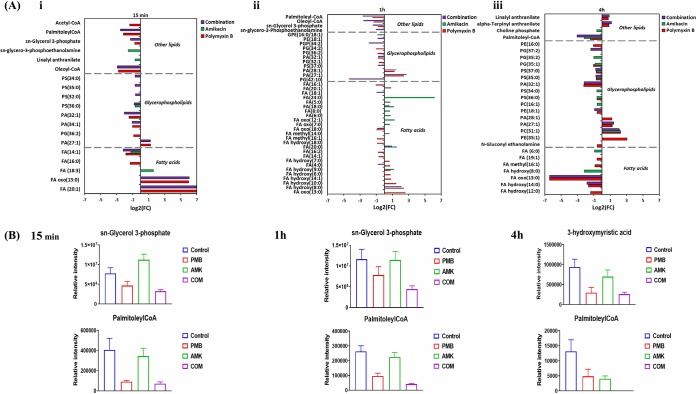
Lipid levels were markedly perturbed in P. aeruginosa FADDI-PA111 following polymyxin B monotherapy and the combination at 15 min, 1 h, and 4 h. Polymyxin B treatment predominantly perturbed bacterial membrane lipids across all time points, including fatty acids (FAs) and glycerophospholipids (GPLs) (≥0.58553-log2-fold; P ≤ 0.05; false discovery rate [FDR] ≤ 0.1) (Fig. 1A). The impact of amikacin on the levels of these lipid metabolites at 15 min was marginal; however, amikacin induced significant perturbations of lipid levels at 1 h and 4 h. Intriguingly, the combination produced a differential pattern of lipid perturbation across all three time points; notably, more GPL perturbations were evident than FA perturbations (Fig. 1A and B). The combination significantly reduced the concentration of palmitoleyl-CoA, an essential cellular intermediate that maintains the LpxC stability (≥−2.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05), across all three time points (Fig. 1B) (25). Furthermore, the level of a key precursor of membrane phospholipids, sn-glycerol-3-phosphate, was significantly decreased following the combination treatment at 15 min and 1 h (≥−1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05). The levels of essential bacterial membrane lipids involved in phospholipid and LPS biosynthesis were significantly reduced following the combination treatment, including sn-glycero-3-phosphoethanolamine, FA hydroxy(14:0) (3-hydroxymyristic acid), and FA(14:1) (myristoleic acid) (≥−1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05) (Fig. 1A and B). Our results suggest that the combination of polymyxin B and amikacin reduced the main precursors of bacterial membrane lipids, particularly those related to LPS and GPL biosynthesis.
FIG 1.
Perturbations of bacterial lipids. (A) Significantly perturbed lipids in P. aeruginosa FADDI-PA111 following treatment with polymyxin B, amikacin, and the combination at 15 min (i), 1 h (ii), and 4 h (iii). Lipid names are putatively assigned based on accurate mass. FC, fold change. (B) Bar charts showing the depletion of essential bacterial membrane lipids after treatment with polymyxin B (PMB), amikacin (AMK), and the combination (COM) across all three time points (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.1). Control, untreated samples; PE, phosphoethanolamines; PG, glycerophosphoglycerols; PS, glycerophosphoserines; PC, glycerophosphocholines; PA, glycerophosphates; GPE, glycerophosphatidylethanolamine; PGP, glycerophosphoglycerophosphates; FA, fatty acids.
LPS biosynthesis.
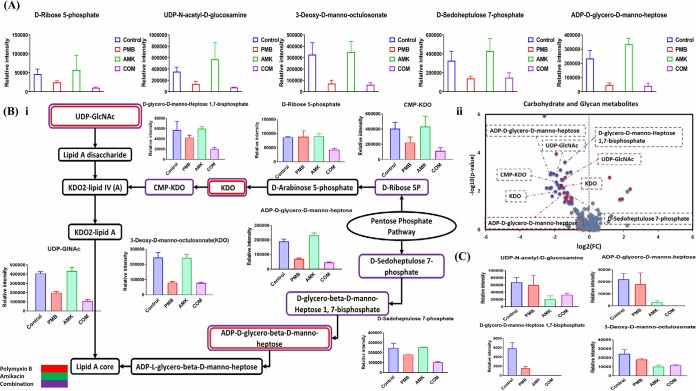
As alluded to above, the combination treatment substantially decreased the levels of intermediates involved in LPS biosynthesis, particularly at 1 h. At 15 min, the polymyxin-amikacin combination caused a prominent decline in the concentrations of three essential pentose phosphate pathway (PPP) metabolites (≥−2.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05), namely, d-ribose-5-phosphate, erythrose 4-phosphate, and d-sedoheptulose-7-phosphate. The levels of three precursors of LPS biosynthesis underwent a remarkable decrease (≥−2.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05) after combination treatment at 15 min, including ADP-d-glycero-d-manno-heptose, 3-deoxy-d-manno-octulosonate (KDO), and UDP-N-acetyl-d-glucosamine (UDP-GlcNAc) (Fig. 2A). Polymyxin B monotherapy had a similar impact on LPS biosynthesis, wherein four key metabolites underwent a significant decline (≥−1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05), including d-sedoheptulose-7-phosphate, ADP-d-glycero-d-manno-heptose, KDO, and UDP-GlcNAc (Fig. 2A). On the other hand, the effect of amikacin treatment on the FADDI-PA111 LPS biosynthetic pathway was unremarkable. Interestingly, the combination at 1 h induced a significant decrease (≥−1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05) in the levels of seven metabolites that are crucial for the formation of LPS, namely, d-ribose-5-phosphate, d-sedoheptulose-7-phosphate, CMP-KDO, KDO, UDP-GlcNAc, ADP-d-glycero-d-manno-heptose, and d-glycero-d-manno-heptose 1,7-bisphosphate (Fig. 2Bi and Bii). Similarly, but to a lesser extent, polymyxin B monotherapy induced a significant drop in the levels of three key intermediates of the LPS biosynthetic pathway, including UDP-GlcNAc, KDO, and ADP-d-glycero-β-d-manno-heptose. Similar to the 15-min time point, we did not observe any impact on LPS biosynthesis at 1 h following amikacin monotherapy (Fig. 2Bii). The combination also induced a significant decrease (≥−1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05) in the levels of key precursors of LPS biogenesis at 4 h, namely, ADP-d-glycero-d-manno-heptose, KDO, UDP-GlcNAc, and d-glycero-d-manno-heptose 1,7-bisphosphate (Fig. 2C). Amikacin monotherapy reduced the levels of four key intermediates of LPS biosynthesis, namely, ADP-d-glycero-d-manno-heptose, d-glycero-d-manno-heptose 1,7-bisphosphate, KDO, and UDP-GlcNAc, at 4 h (≥−1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05) (Fig. 2C). On the other hand, the influence of polymyxin B monotherapy on LPS biosynthesis in FADDI-PA111 at 4 h was unremarkable.
FIG 2.
Impact of treatment on lipopolysaccharide biosynthesis. (A) Bar charts for significantly perturbed intermediates of the PPP and downstream LPS in P. aeruginosa FADDI-PA111 following treatment with polymyxin B (PMB), amikacin (AMK), and the combination (COM) at 15 min. (B and C) Schematic diagram and bar charts (i) and volcano plots (ii) for the significantly impacted intermediates of LPS biogenesis in P. aeruginosa FADDI-PA111 after treatment with polymyxin B, amikacin, and the combination at 1 h (B) and at 4 h (C) (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05). Control, untreated samples.
Central carbon metabolites.
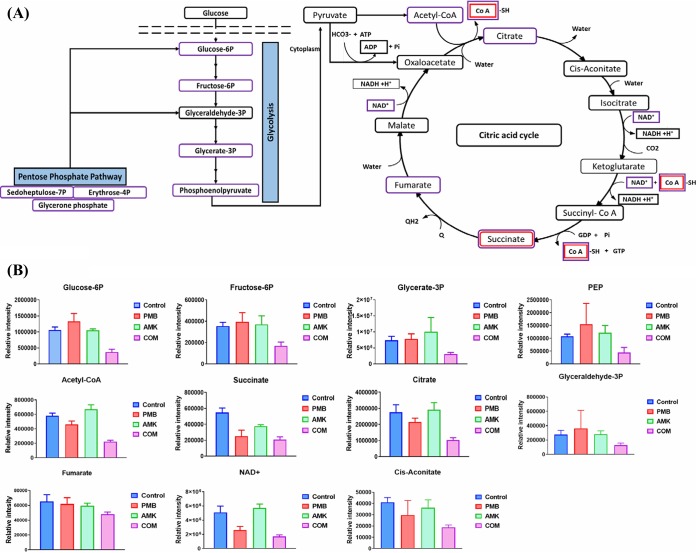
Besides the significant impact of the polymyxin B-amikacin combination on the PPP of FADDI-PA111 at 15 min and 1 h, it also caused a remarkable suppression of the levels of five key intermediates of the glycolysis pathway at 1 h, including glycerate-3-phosphate, d-fructose-6-phosphate, d-glucose-6-phosphate, glyceraldehyde-3-phosphate, and phosphoenolpyruvate (≥−1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05) (Fig. 3A and B). Moreover, the combination significantly reduced the concentrations of eight tricarboxylic acid (TCA) cycle metabolites, namely, acetyl-CoA, citrate, cis-aconitate, isocitrate, succinate, fumarate, NAD+, and CoA (≥−1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05) (Fig. 3A and B). Polymyxin treatment induced a significant reduction in the concentrations of only two intermediates of this pathway at 1 h, namely, succinate (log2-fold change = −1.13) and CoA (log2-fold change = −1.48); amikacin monotherapy had no detectable impact on the central carbon metabolites at 1 h (Fig. 3B).
FIG 3.
Changes in central carbon metabolism. (A) Schematic diagram for the significantly perturbed intermediates of the PPP, glycolysis, and the interrelated tricarboxylic acid cycle in P. aeruginosa FADDI-PA111 following treatment with polymyxin B, amikacin, and the combination at 1 h. (B) Bar graphs for the main intermediates of the PPP, glycolysis, and the TCA cycle after treatment with polymyxin B (PMB), amikacin (AMK), and the combination (COM) at 1 h (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05). PEP, phosphoenolpyruvate. Control, untreated samples.
Pyridine nucleotide cycle.
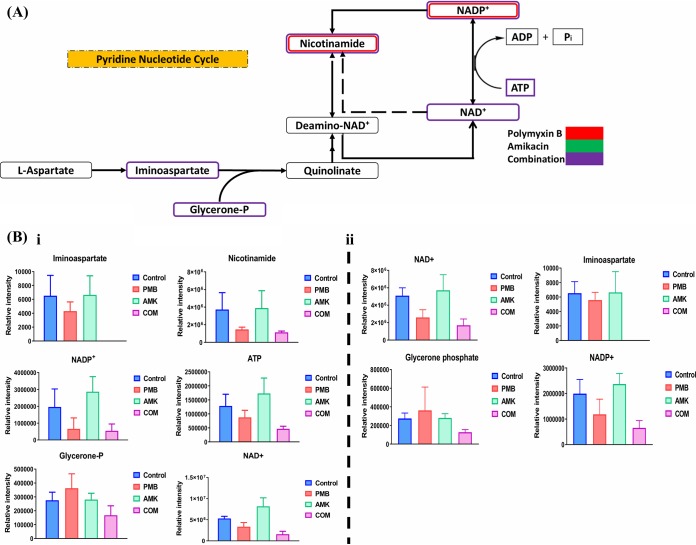
Treatment with polymyxin B alone and the combination (Fig. 4A) markedly disrupted the PNC pathway, particularly at 15 min and, to a lesser extent, at 1 h (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05). Polymyxin B treatment decreased the levels of two key metabolites, namely, nicotinamide and NADP+, at 15 min, whereas the combination reduced the levels of six key metabolites compromising the backbone of the PNC, namely, iminoaspartate, nicotinamide, NADP+, NAD, ATP, and glycerone phosphate, at 15 min (Fig. 4Bi). At 1 h, the combination reduced the levels of four metabolites, namely, iminoaspartate, NADP+, NAD, and glycerone phosphate (Fig. 4Bii). The influence of amikacin monotherapy on the PNC was inconsequential across all time points. Based on these findings, we purport that the perturbations in PNC metabolites by the combination were largely driven by polymyxin B.
FIG 4.
Perturbations of the pyridine nucleotide cycle (PNC). (A) Schematic diagram for the significantly perturbed intermediates of the PNC in P. aeruginosa FADDI-PA111 following treatment with polymyxin B, amikacin, and the combination at 15 min. (B) Bar graphs for the main depleted precursors of the PNC after treatment with polymyxin B (PMB), amikacin (AMK), and the combination (COM) at 15 min (i) and 1 h (ii) (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.1). Control, untreated samples.
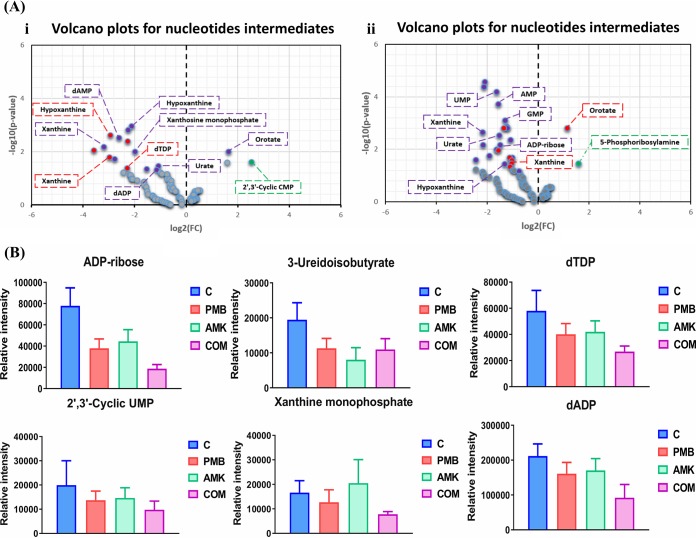
Nucleotide levels.
Polymyxin B monotherapy induced a marked decline in the levels of nucleotides across all the time points, and this effect was even more pronounced following combination treatment (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.1) (Fig. 5A and B; see also Table S2 in the supplemental material). At 15 min, a marked decline in the levels of five nucleotides (hypoxanthine, xanthine, dTMP, dCMP, and dTDP) was observed following polymyxin B monotherapy, whereas amikacin monotherapy had negligible effects. Nine nucleotides underwent a remarkable decrease in their levels after combination treatment (except for an increment in orotate levels) (Fig. 5Ai). At 1 h, the combination profoundly reduced (≥−1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05) the concentrations of 16 nucleotide intermediates (Fig. 5B; Table S2). Similarly to the 15-min time point, polymyxin B monotherapy caused a reduction of 6 nucleotides and conversely induced an increase in the levels of orotate (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.1). Amikacin monotherapy increased (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.1) the concentrations of 5-phosphoribosylamine at 1 h (Fig. 5Aii). Key nucleotides were also significantly depleted in FADDI-PA111 following the combination treatment at 4 h (Fig. 5B).
FIG 5.
Induction of nucleotide turnover and DNA damage. (A) Volcano plots for the significantly perturbed nucleotides at 15 min (i) and 1 h (ii) in P. aeruginosa FADDI-PA111 following treatment with polymyxin B, amikacin, and the combination. (B) Bar graphs for the nucleotides after treatment with polymyxin B (PMB), amikacin (AMK), and the combination (COM) at 4 h (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.1). Control, untreated samples.
Amino acid, peptide, and peptidoglycan metabolism.
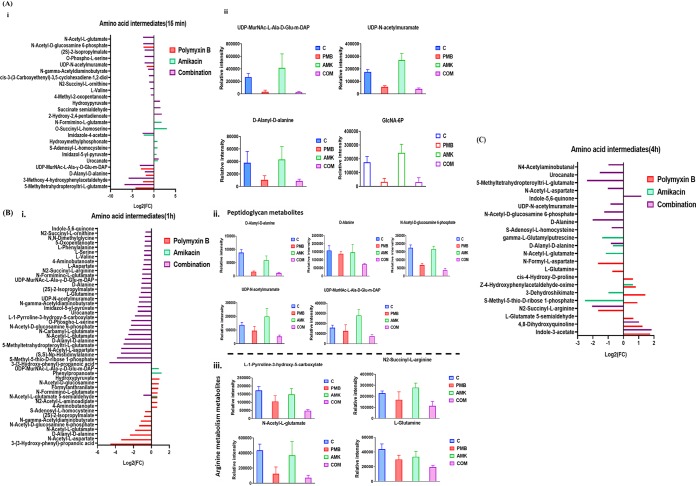
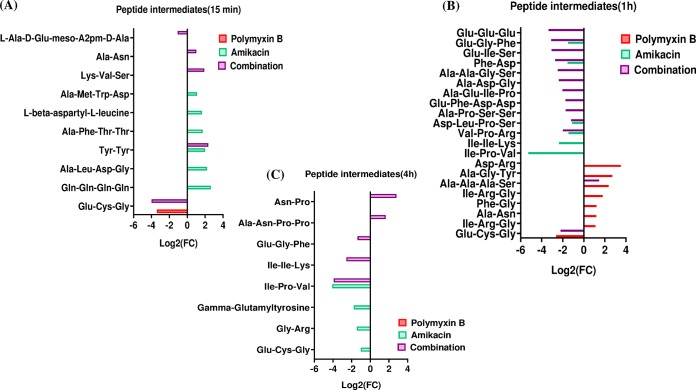
Differential perturbation patterns in amino acid metabolism were identified under each treatment condition across all time points; the most notable difference was evident in the main precursors of peptidoglycan biosynthesis and arginine metabolism (Fig. 6). A dramatic decrease in the levels of four essential metabolites of the peptidoglycan biosynthetic pathway was observed following polymyxin B monotherapy at 15 min (≥−1.5-log2-fold; P ≤ 0.05; FDR ≤ 0.05), including d-alanyl-d-alanine, UDP-N-acetylmuramoyl-l-alanyl-d-γ-glutamyl-meso-2,6-diaminopimelate (UDP-MurNAc-l-Ala-γ-d-Glu-m-DAP), UDP-N-acetylmuramate (UDP-MurNAc), and N-acetyl-d-glucosamine-6-phosphate (Fig. 6Ai and Aii). This effect was less pronounced at the latter time points of 1 and 4 h (Fig. 6B and C). Amikacin monotherapy caused a moderate increase (≥0.58553-log2-fold; P ≤ 0.05; FDR ≤ 0.1) in the levels of four intermediates of histidine metabolism at 15 min (except for a decline in imidazole-4-acetate levels), namely, imidazol-5-yl-pyruvate, N-formimino-l-glutamate, and urocanate (Fig. 6Ai). On the other hand, the combination treatment resulted in a significant suppression of the levels of key intermediates of peptidoglycan biosynthesis at 15 min, including d-alanyl-d-alanine, UDP-MurNAc-l-Ala-γ-d-Glu-m-DAP, UDP-MurNAc, and N-acetyl-d-glucosamine-6-phosphate (≥−2.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05) (Fig. 6Ai, and Aii). This effect on peptidoglycan biosynthesis persisted over the latter time points, in particular at 1 h, causing a dramatic decrease in the levels of five metabolites, d-alanyl-d-alanine, alanine, UDP-MurNAc-l-Ala-γ-d-Glu-m-DAP, UDP-MurNAc, and N-acetyl-d-glucosamine-6-phosphate (≥−1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05) (Fig. 6Bi and Bii). This effect tapered at 4 h, at which the combination therapy reduced the concentrations of only two important intermediates of peptidoglycan biosynthesis, namely, d-alanine (log2-fold change = −2.02) and N-acetyl-glucosamine-6-phosphate (log2-fold change = −1.26) (Fig. 6C). The effect of the polymyxin-amikacin combination on the intermediates of arginine metabolism was most pronounced at 15 min and 1 h, reducing the levels of N2-succinyl-l-ornithine and N-acetyl-l-glutamate (at 15 min) and of N-acetyl-l-glutamate, l-1-pyrroline-3-hydroxy-5-carboxylate, l-glutamine, and N2-succinyl-l-arginine (at 1 h) (Fig. 6A, Bi, and Biii). Amikacin treatment and the combination induced major perturbations in peptides. The combination treatment significantly decreased the levels of two essential peptides, glutathione and l-Ala-d-Glu-meso-A2pm-d-Ala (muropeptide), at 15 min (≥−1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05) (Fig. 7A). Twelve peptide metabolites (including glutathione) (≥−2.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05) were remarkably diminished after 1 h of the combination treatment (Fig. 7B). The combination treatment produced a marginal effect on peptide metabolism at 4 h (Fig. 7C). Amikacin monotherapy increased the levels of six peptide metabolites at 15 min and then induced a significant reduction in their levels at 1 and 4 h (Fig. 7A to C). Polymyxin B monotherapy did not show a significant impact on peptide metabolism across all time points except for 1 h (Fig. 7B). Furthermore, the glutathione level underwent a significant decline after polymyxin B treatment at 15 min and 1 h (≥−2.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05). It should be noted that peptide identifications were based on accurate mass and isomeric forms of the putative peptides where possible.
FIG 6.
Changes in amino acid metabolism in FADDI-PA111. (A) Fold changes for the significantly affected amino acids (i) and bar charts of peptidoglycan intermediates (ii) following treatment with polymyxin B, amikacin, and the combination at 15 min. (B) Fold changes of significantly perturbed amino acids (i) and bar graphs for intermediates of peptidoglycan biosynthesis (ii) and arginine metabolism (iii) after treatment with polymyxin B (PMB), amikacin (AMK), and the combination (COM) at 1 h (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.1). (C) Fold changes of significant amino acid metabolites at 4 h (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.1). Control, untreated samples.
FIG 7.
Significantly affected metabolites of peptide metabolism in FADDI-PA111. Shown are fold changes for the significantly affected peptides following treatment with polymyxin B, amikacin, and the combination at 15 min (A), 1 h (B), and 4 h (C) (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.1).
Impact of polymyxin B, amikacin, and their combination on the metabolome of polymyxin-resistant CF P. aeruginosa isolate LESB58.
The Liverpool epidemic strain LESB58 is intrinsically resistant to polymyxin B and amikacin, as its resistome carries APH(3′)-IIb aminoglycoside resistance genes, which mediate the inactivation of amikacin via aminoglycoside-modifying enzymes (phosphotransferase) (18, 26). Not surprisingly, the metabolic response of the strain was limited under all treatment conditions except for minor effects on lipid and carbohydrate metabolism; the only significantly perturbed metabolites were comparable in number between polymyxin B monotherapy and combination treatment groups across all time points (Fig. S4B). Polymyxin B monotherapy caused a marked reduction in the levels of metabolites from amino acid, lipid, and carbohydrate metabolism at 15 min and 1 h, including amino acid intermediates involved in glutathione metabolism, such as glutathione disulfide (log2-fold change = −2.07), l-glutamate (log2-fold change = −1.14), and s-glutathionyl-l-cysteine (log2-fold change = −1.44) (Fig. S6B and Table S3). Palmitoleyl-CoA (log2-fold change = −0.82) and oleoyl-CoA (log2-fold change= −0.82) were among the essential lipid membrane intermediates that underwent a marked decline following polymyxin B monotherapy. However, the effect of polymyxin B monotherapy was completely inversed at 4 h, wherein the levels of all intermediates increased. On the other hand, amikacin treatment showed a delayed effect wherein marginal perturbation was evident at 15 min, followed by dramatic changes at 1 and 4 h (Fig. S4B and S6B); amikacin monotherapy imparted a significant perturbation (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05) in the levels of glutathione biosynthesis-related intermediates, such as glutathione disulfide, l-methionine S-oxide, l-methionine, and O-acetyl-l-homoserine (Table S3).
The polymyxin B-amikacin combination treatment mainly caused perturbation to lipids, which were primarily decreased at 15 min and 1 h, while increases were generally observed at 4 h. The main lipid precursors involved in the bacterial outer membrane composition underwent a remarkable perturbation following the combination treatment (≥1.0-log2-fold; P ≤ 0.05; FDR ≤ 0.05), namely, FA(16:0), FA(17:0), FA(18:0), FA(14:1), and sn-glycero-3-phosphoethanolamine (log2-fold change= −0.90) (Table S3). In addition, carbohydrates were also significantly perturbed. There was an elevation in the levels of intermediates (UDP-l-Ara4FN) associated with the lipid A aminoarabinose modification pathway following polymyxin B monotherapy (15 min and 4 h); however, this effect was not observed with the combination treatment at any time points (Table S3).
DISCUSSION
In the present study, we employed metabolomics to characterize the responses of P. aeruginosa to treatment with polymyxin-amikacin. Polymyxins and aminoglycosides exert their primary antibacterial killing activity via disruption of the OM and inhibition of protein synthesis, respectively (12, 27). To the best of our knowledge, this is the first study to investigate the antibacterial killing synergy of the combination of polymyxin B with amikacin using an untargeted metabolomics approach. The most significant findings of the study include (i) differential time-dependent inhibition of key metabolic pathways, (ii) perturbation of central carbon metabolism and suppression of nucleotide pools, (iii) inhibition of the pyridine nucleotide cycle (PNC) (the main pool of NADP), and (iv) inhibition of LPS and cell wall biosynthesis.
The early cellular metabolic perturbations following treatment with polymyxin B monotherapy seen at 15 min and 1 h impacted lipid, nucleotide, amino-nucleotide sugar, and energy metabolism. Similar metabolic changes were evident following amikacin monotherapy (particularly LPS biosynthesis) albeit in a delayed fashion, largely occurring at 4 h posttreatment. Moreover, amikacin monotherapy induced substantial perturbations in peptide metabolism, whereas polymyxin B had little effect on peptide levels (see Fig. S6A in the supplemental material). These mechanistic findings support the use of the polymyxin B-amikacin combination for maintaining a persistent antibacterial effect (i.e., polymyxin B for early and rapid-onset followed by amikacin for delayed-onset bacterial killing) and minimizing potential bacterial regrowth, which can rapidly emerge with monotherapy (28, 29).
It has been shown that the disorganization of the bacterial outer membrane is one of the possible bacterial killing mechanisms caused by polymyxins (12, 30). Coincidently, treatment of FADDI-PA111 with polymyxin B monotherapy caused a marked suppression of several key phospholipids, fatty acids, and lipid intermediates, such as FA(16:0), FA(14:1), oleoyl-CoA, and palmitoleyl-CoA, at 15 min and, to a lesser extent, at 1 h and 4 h (Fig. 1A and Fig. 3B). This finding is in agreement with previous metabolomics studies with P. aeruginosa that revealed that polymyxin B caused perturbations in fatty acids and glycerophospholipids (31, 32). Additionally, it has been found that the genes associated with bacterial OM biosynthesis were differentially expressed as a result of colistin treatment (33). Membrane fatty acid composition and fluidity are vital for the development of antibiotic resistance (34, 35), and an increase in saturated fatty acids is usually associated with a decrease in membrane fluidity and, hence, gives rise to a less permeable bacterial OM (36). It is not surprising that polymyxin B did not show marked perturbations in the lipid intermediates of polymyxin-resistant LESB58 at 15 min; however, a minor reduction in the levels of essential bacterial membrane lipids was evident at 1 h, which was then followed by a significant rise in all significantly affected lipid intermediates at 4 h. These results are likely reflective of the resistance of this strain to polymyxin B.
Amikacin is a semisynthetic aminoglycoside which exerts its bacterial killing activity by interfering with intracellular protein synthesis (37). However, several studies have shown that aminoglycoside-based amphiphiles (like amikacin- and neamine-based amphiphiles) are able to disrupt the negatively charged lipids from bacterial inner membranes in P. aeruginosa, which subsequently leads to membrane permeabilization and depolarization (38, 39). Interestingly, our results are in line with those studies, as amikacin remarkably reduced the levels of several membrane glycerophospholipids in FADDI-PA111, such as PS(36:0) and PS(37:0), at 4 h (Fig. 1Aiii). These glycerophospholipids are commonly involved in the biosynthesis of phosphatidic acid (PA), the key intermediate in the synthesis of all membrane glycerolipids (40). The biogenesis of bacterial OM phospholipids starts via acylation of sn-glycerol-3-phosphate using fatty acyl-acyl carrier protein (acyl-ACP) (25). Importantly, a previous study has shown that the decline in the levels of palmitoleyl-CoA could lead to decreases in the stability of LpxC, which has a vital role in lipid A core formation (25). It is important to note that the combination treatment remained effective in reducing the levels of FADDI-PA111 membrane lipids at 4 h, in which more perturbations in glycerophospholipid metabolism were most prominent (Fig. 1Aiii). In regard to LESB58, the impact of combination treatment was largely related to changes in the levels of fatty acids and glycerophospholipids over 4 h (Table S3). Together, the above-described data highlight that the perturbation of lipid metabolism is a key pathway in killing synergy by the polymyxin B-amikacin combination.
Importantly, our study is the first to report that combining polymyxin B with amikacin caused a significant suppression of intermediates involved in LPS biosynthesis (Fig. 2A to C). This influence may have arisen from the early (15-min and 1-h) inhibition of the pentose phosphate pathway (PPP), which is a key source of precursors for LPS synthesis. A considerable decline in intermediates of LPS formation was observed after polymyxin B monotherapy at 15 min and 1 h but not at 4 h (Fig. 2C). In contrast, amikacin had no impact at early time points (15 min and 1 h), while it caused substantial inhibitory effects on LPS biosynthesis at 4 h. The combination effect on these pathways was not seen in the polymyxin B-resistant strain LESB58; however, the level of the PPP metabolite d-sedoheptulose-7-phosphate was slightly decreased after polymyxin B monotherapy at 15 min (Table S3). This negative impact on LPS biosynthesis is in line with the primary mode of action of polymyxins, which involves disorganizing the bacterial OM through their interaction with LPS (41). It was previously shown that Escherichia coli mutants defective in the biosynthesis of KDO are extremely susceptible to very low concentrations of antibiotics such as novobiocin (42), and others also noticed that inactivation of d-arabinose-5-phosphate isomerase (API), an enzyme that promotes the reversible isomerization of d-ribulose-5-phosphate (Ru5P) to d-arabinose-5-phosphate, a KDO precursor in E. coli, resulted in death of the microorganism (43). Heptose units such as d-sedoheptulose-7-phosphate are crucial building blocks that form the LPS inner core of Gram-negative bacteria (44). Heptose-deficient Haemophilus influenzae mutants, bearing a genetic defect of ADP-glyceromannoheptose isomerase, showed less virulence and more susceptibility to antibiotics (45). Hence, taking data from all of the above-mentioned studies together, a possible mechanism of synergistic killing by the polymyxin B-amikacin combination is strongly related to the inhibition of the PPP and subsequent LPS biosynthesis, and the differential time-dependent inhibition of this pathway by each antibiotic alone is beneficial in terms of pharmacokinetics/pharmacodynamics (PK/PD), due to maintenance of their killing activity over time.
Apart from its potential impact on membrane structure, our pathway analysis illustrated that the polymyxin B-amikacin combination also effectively perturbed the TCA and glycolysis (central carbohydrate metabolism) pathways in FADDI-PA111 (Fig. 3A and B). This effect was not evident for LESB58; only two intermediates (succinate and CoA) from these pathways were affected by polymyxin B monotherapy, with no effect seen for amikacin monotherapy. Bacterial central carbohydrate metabolism is a complex cellular network including various metabolic pathways, such as glycolysis and TCA, and was recently investigated as a new target for the next generation of antibiotics (46). Glycolysis is the main source of acetyl-CoA, which has a direct role in many metabolic processes, such as supply of acetyl groups to TCA, synthesis of fatty acids, and amino acid biosynthesis (47). The TCA cycle has a crucial role in cellular respiration of bacteria and supplies several important intermediates, such as succinate and citrate, which are required for other key metabolic processes (48). It is important to mention that there are two fundamental types of reactions that control the TCA cycle, namely, anaplerotic and cataplerotic reactions, in which the amount of TCA cycle intermediates increases or decreases, respectively (49). Our results demonstrate that polymyxin B-amikacin treatment displayed a cataplerotic effect on the TCA cycle by reduction of fundamental intermediates such as succinate and acetyl-CoA, which are essential for electron transport chain reactions and fatty acid and amino acid metabolism.
The polymyxin B-amikacin combination also caused a dramatic perturbation in the PNC of FADDI-PA111, with a greater effect seen at 15 min and, to a lesser extent, at 1 h (Fig. 4A and B). In contrast, polymyxin B monotherapy was able to reduce the concentrations of only two essential precursors of the PNC at 15 min, namely, nicotinamide and NADP+ (Fig. 4A and Bi), while no effect was seen for amikacin monotherapy. The PNC is an essential network of biochemical transformations that enable bacterial cells to maintain the homeostasis of NADP (50). It has been found that a small change in the concentration of NADP is likely associated with the propagation of various metabolic disruptions, for instance, in the synthesis of proteins and lipids (51). Inhibition of the PNC by polymyxin B-amikacin treatment appears to be a mechanism underlying the synergistic killing activity of the combination.
Polymyxin B-amikacin combination treatment caused a significant reduction in d-ribose-5-phosphate, a key initial intermediate in purine and pyrimidine metabolism, and concomitantly produced a significant depletion in the nucleotide pool of FADDI-PA111 up to 4 h. It was evident that the maximum effect on the nucleotide pool was at 1 h, followed by 4 h. Similarly, polymyxin B monotherapy demonstrated significant perturbations in the levels of nucleotides, especially those related to purine catabolism, such as hypoxanthine and xanthine, at 15 min and 1 h, which then faded away at 4 h (Fig. 5A and B). Amikacin had little impact on the nucleotide pool compared to polymyxin B monotherapy and the combination. No significant effect on nucleotide levels was seen in LESB58. Previous metabolomics studies showed that exposure of Gram-negative (E. coli) and Gram-positive (Staphylococcus aureus) bacteria to different antibiotic treatments (e.g., ampicillin, kanamycin, norfloxacin, and vancomycin) can lead to a depletion of the nucleotide pool, indicative of nucleotide degradation (52, 53).
Notably, the polymyxin B-amikacin combination caused a significant inhibition of peptidoglycan biosynthesis only in FADDI-PA111 (Fig. 6A to C). Levels of major metabolites of peptidoglycan biogenesis were dramatically decreased following the combination treatment across all time points, in particular at 1 h (Fig. 6Bii) (54). This effect might be secondary to the marked depletion of amino sugar and nucleotide sugar intermediates that supply key precursors for peptidoglycan biosynthesis (55). Polymyxin B monotherapy also produced a significant depletion of peptidoglycan intermediates in FADDI-PA111 at 15 min (Fig. 6A), while there was no noticeable impact following amikacin monotherapy. Interestingly, our group previously reported transcriptomics and metabolomics studies demonstrating that polymyxins also inhibit peptidoglycan biosynthesis in Acinetobacter baumannii and P. aeruginosa (32, 56, 57). Pathway analysis highlighted the significant impact of the combination on arginine and proline metabolism in FADDI-PA111 at 15 min and 1 h, and there was a little influence of polymyxin B and amikacin monotherapies on this pathway at 1 and 4 h, respectively (Fig. 6A to C). The disruption of amino acid pathways, in particular arginine metabolism, has recently been targeted as a novel approach to manage bacterial infections and subvert pathogenesis (58).
Overall, this study highlights the importance of elucidating the complex and dynamic interactions of multiple cellular metabolic pathways due to antibiotic treatment, which ultimately aids in optimizing the most commonly used combination therapy in clinical practice.
MATERIALS AND METHODS
Drugs and bacterial isolates.
Polymyxin B (batch number 20120204; Beta Pharma, China) and amikacin (Sigma-Aldrich, Saint Louis, MO, USA) solutions were prepared in Milli-Q water (Millipore, Australia) and filtered through 0.22-μm syringe filters (Sartorius, Australia). All other reagents were purchased from Sigma-Aldrich (Australia) and were of the highest commercial grade available. A polymyxin-susceptible P. aeruginosa strain, FADDI-PA111 (polymyxin B MIC = 2 mg/liter; amikacin MIC = 2 mg/liter), and the polymyxin-resistant cystic fibrosis P. aeruginosa isolate LESB58 (Liverpool epidemic strain) (MIC = 16 mg/liter for both polymyxin B and amikacin) were tested. The isolates were stored in tryptone soy broth (Oxoid) with 20% glycerol (Ajax Finechem, Seven Hills, NSW, Australia) in cryovials at −80°C. Before use, both strains (FADDI-PA111 and LESB58) were subcultured onto M9 minimal medium (59).
Bacterial culture preparation for metabolomics experiments.
To investigate the possible molecular mechanisms of the polymyxin B-amikacin combination, we employed untargeted metabolomics to determine the changes in different metabolite levels following 15 min, 1 h, and 4 h of antibiotic treatment.
A single colony of P. aeruginosa grown on nutrient or Mueller-Hinton agar was chosen and grown overnight (16 to 18 h) in 20 ml in M9 minimal medium (59) in 50-ml Falcon tubes (Thermo Fisher, Australia) and incubated in a shaking water bath at 37°C (shaking speed, 180 rpm). P. aeruginosa LES isolates are methionine auxotrophs; therefore, l-methionine was added to M9 minimal medium for growth of the LESB58 strain. Following overnight incubation, each culture was transferred to a 1-liter conical flask with 250 ml of fresh M9 minimal medium at ∼50- to 100-fold dilutions. Flasks were incubated at 37°C with shaking at 180 rpm for ∼3 to 4 h to log phase (optical density at 600 nm [OD600] of ∼0.5). Cultures (50 ml) were transferred to four 500-ml conical flasks, and solutions of polymyxin B, amikacin, or both were added to three of four flasks to give final concentrations of 2 mg/liter for FADDI-PA111, 4 mg/liter for LESB58 for polymyxin B, and 2 mg/liter for amikacin for both strains; the remaining flask acted as a drug-free control. To prevent excessive bacterial killing, preliminary optimization studies were conducted using a high bacterial inoculum (∼108 CFU/ml) and different antibiotic concentrations to ensure no more than a 2-log10 CFU/ml reduction and thereby induce more stress on the microorganism. The flasks were further incubated at 37°C with shaking at 180 rpm. After 2 h, the OD600 reading for each flask was determined and normalized to a value of ∼0.5 with fresh M9 minimal medium, and 10-ml samples were transferred to 15-ml Falcon tubes (Thermo Fisher, Australia) for metabolite extraction. To account for inherent random variation, four biological samples were prepared under each treatment condition for each strain.
Metabolite extraction for metabolomic studies.
Following bacterial culture preparation, extraction of metabolites was immediately carried out in order to decrease further drug effects on metabolite levels. Initially, samples were centrifuged at 3,220 × g at 4°C for 20 min. Supernatants were then removed, and bacterial pellets were washed twice in 1 ml of cold normal saline, followed by centrifugation at 3,220 × g at 4°C for 10 min to remove residual extracellular metabolites and medium components. Next, 300 μl of a cold chloroform-methanol-water (CMW) (1:3:1, vol/vol) extraction solvent containing 1 μM (each) the internal standards {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), N-cyclohexyl-3-aminopropanesulfonic acid (CAPS), piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), and Tris} was added to the washed pellets. The internal standards used are physicochemically different small molecules that do not naturally exist in any microorganism. Samples were then immersed three times in liquid nitrogen, thawed on ice, and vortexed to liberate the intracellular metabolites. The samples were centrifuged for 10 min at 3,220 × g at 4°C after the third freeze-thaw cycle, whereby 300 μl of the supernatants was added to 1.5-ml Eppendorf tubes. Centrifugation at 14,000 × g at 4°C for 10 min was used to detach any particles from samples, and 200 μl was transferred into the injection vials for storage in a −80°C freezer. For liquid chromatography-mass spectrometry (LC-MS) analysis (described below), the samples were taken out from the −80°C freezer to thaw, and 10 μl of each sample was transferred to a vial and used as a pooled quality control (QC) sample, namely, a sample that contains all the analytes that will be encountered during the analysis (60).
LC-MS analysis.
Metabolites were identified by hydrophilic interaction liquid chromatography (HILIC)–high-resolution mass spectrometry (HMS) using a Dionex high-performance liquid chromatography (HPLC) system (RSLC U3000; Thermo Fisher) with a ZIC-pHILIC column (5 μm, polymeric, 150 by 4.6 mm) (SeQuant; Merck). The system was coupled to a Q-Exactive Orbitrap mass spectrometer (Thermo Fisher) operated in both positive and negative electrospray ionization (ESI) modes (rapid switching) at a 35,000 resolution with a detection range of m/z 85 to 1, 275. Two LC solvents, 20 mM ammonium carbonate (solvent A) and acetonitrile (solvent B), were used, which operated via a multistep gradient system. The gradient started at 80% solvent B, which declined to 50% solvent B over 15 min and was then reduced from 50% solvent B to 5% solvent B over 3 min, followed by a wash with 5% solvent B for another 3 min and, finally, an 8-min reequilibration with 80% solvent B at a flow rate of 0.3 ml/min (61). The injection sample volume was 10 μl, and the total run time was 32 min. All samples were analyzed as a single LC-MS batch to avoid batch-to-batch variation. Mixtures of pure standards containing over 300 metabolites were also included in the analysis batch to aid in metabolite identification.
Data processing, bioinformatics, and statistical analyses.
Conversion of LC-MS raw data to metabolite levels was conducted using IDEOM software (http://mzmatch.sourceforge.net/ideom.php) (62), which initially employed ProteoWizard to convert raw LC-MS data to mzXML format and XCMS to pick peaks with Mzmatch.R to convert to peakML files (63, 64). Mzmatch.R was subsequently used for the alignment of samples and the filtering of peaks, using a minimum peak intensity threshold of 100,000, a relative standard deviation (RSD) of <0.5 (reproducibility), and a peak shape (codadw) of >0.8. Mzmatch was also used to retrieve missing peaks and annotate related peaks. Default IDEOM parameters were used to eliminate unwanted noise and artifact peaks. Loss or gain of a proton was corrected in negative or positive ESI mode, respectively, followed by putative identification of metabolites by the exact mass within 2 ppm. Retention times of authentic standards were used to confirm the identification of each metabolite (level 1 identification based on metabolomics standards initiative [MSI] standards). Other metabolites were putatively identified (level 2 identification based on MSI standards) using exact mass and predicted retention time based on the Kyoto Encyclopedia of Genes and Genomes (KEGG), MetaCyc, and LIPIDMAPS databases, with preference for bacterial metabolites annotated in EcoCyc in cases where isomers could not be clearly differentiated by retention time. The raw peak intensity was used to quantify each metabolite. The free online tool MetaboAnalyst 3.0 was used for statistical analysis. Briefly, putative metabolites with a median RSD of ≤0.2 (20%) within the QC group and an IDEOM confidence level of ≥5 were incorporated into a table and uploaded to MetaboAnalyst. Features with >50% missing values were replaced by half of the minimum positive value in the original data. Interquartile ranges (IQRs) were utilized to filter data, and log2 transformation and autoscaling were then used to normalize the data. Principal-component analysis (PCA) was performed to identify and remove outliers. Partial least squares for discriminant analysis is normally used to reduce the dimension of variables from a large data set (65). One-way analysis of variance (ANOVA) was used to identify metabolites with significant level changes between all samples, and Fisher’s least-square difference (LSD) test was used to determine the metabolites with significant level changes between treatment and control groups. Statistically significant metabolites were selected using a false discovery rate of ≤0.1 for one-way ANOVA and a P value of ≤0.05 for Fisher’s LSD test. KEGG mapper was used to determine the pathway modules by statistically significant metabolites containing the KEGG compound numbers.
Supplementary Material
ACKNOWLEDGMENTS
J.L. and T.V. are supported by research grants from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01 AI132681). J.L. and T.V. are also supported by the Australian National Health and Medical Research Council (NHMRC) as principal research and career development level 2 fellows, respectively. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
We thank the Monash Proteomics and Metabolomics Facility for sample analysis and particularly Amanda Peterson for technical assistance. We thank Elena K. Schneider-Futschik for assistance with English grammar editing of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.World Health Organization. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 3.Rice LB. 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. The University of Chicago Press, Chicago, IL. [DOI] [PubMed] [Google Scholar]
- 4.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. doi: 10.1128/cmr.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verkman A, Song Y, Thiagarajah JR. 2003. Role of airway surface liquid and submucosal glands in cystic fibrosis lung disease. Am J Physiol Cell Physiol 284:C2–C15. doi: 10.1152/ajpcell.00417.2002. [DOI] [PubMed] [Google Scholar]
- 6.Moradali MF, Ghods S, Rehm BH. 2017. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol 7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambert PA. 2002. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med 95(Suppl 41):22–26. [PMC free article] [PubMed] [Google Scholar]
- 8.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breidenstein EBM, de la Fuente-Núñez C, Hancock REW. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 11.Nation RL, Li J, Cars O, Couet W, Dudley MN, Kaye KS, Mouton JW, Paterson DL, Tam VH, Theuretzbacher U, Tsuji BT, Turnidge JD. 2015. Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect Dis 15:225–234. doi: 10.1016/S1473-3099(14)70850-3. [DOI] [PubMed] [Google Scholar]
- 12.Velkov T, Roberts KD, Nation RL, Thompson PE, Li J. 2013. Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future Microbiol 8:711–724. doi: 10.2217/fmb.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock RE. 1984. Alterations in outer membrane permeability. Annu Rev Microbiol 38:237–264. doi: 10.1146/annurev.mi.38.100184.001321. [DOI] [PubMed] [Google Scholar]
- 14.Hancock RE, Bell A. 1989. Antibiotic uptake into gram-negative bacteria, p 42–53. In Jackson GG, Schlumberger HD, Zeiler HJ (ed), Perspectives in antiinfective therapy. Springer, Berlin, Germany. [Google Scholar]
- 15.Trimble MJ, Mlynárčik P, Kolář M, Hancock RE. 2016. Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb Perspect Med 6:a025288. doi: 10.1101/cshperspect.a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergen PJ, Bulman ZP, Saju S, Bulitta JB, Landersdorfer C, Forrest A, Li J, Nation RL, Tsuji BT. 2015. Polymyxin combinations: pharmacokinetics and pharmacodynamics for rationale use. Pharmacotherapy 35:34–42. doi: 10.1002/phar.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McPhee JB, Lewenza S, Hancock RE. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol 50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 19.Gellatly SL, Needham B, Madera L, Trent MS, Hancock RE. 2012. The Pseudomonas aeruginosa PhoP-PhoQ two-component regulatory system is induced upon interaction with epithelial cells and controls cytotoxicity and inflammation. Infect Immun 80:3122–3131. doi: 10.1128/IAI.00382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thaipisuttikul I, Hittle LE, Chandra R, Zangari D, Dixon CL, Garrett TA, Rasko DA, Dasgupta N, Moskowitz SM, Malmström L, Goodlett DR, Miller SI, Bishop RE, Ernst RK. 2014. A divergent Pseudomonas aeruginosa palmitoyltransferase essential for cystic fibrosis-specific lipid A. Mol Microbiol 91:158–174. doi: 10.1111/mmi.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim TP, Lee W, Tan TY, Sasikala S, Teo J, Hsu LY, Tan TT, Syahidah N, Kwa AL. 2011. Effective antibiotics in combination against extreme drug-resistant Pseudomonas aeruginosa with decreased susceptibility to polymyxin B. PLoS One 6:e28177. doi: 10.1371/journal.pone.0028177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaddurah-Daouk R, Weinshilboum RM. 2014. Pharmacometabolomics: implications for clinical pharmacology and systems pharmacology. Clin Pharmacol Ther 95:154–167. doi: 10.1038/clpt.2013.217. [DOI] [PubMed] [Google Scholar]
- 23.Mastrangelo A, Armitage EG, García A, Barbas C. 2014. Metabolomics as a tool for drug discovery and personalised medicine. A review. Curr Top Med Chem 14:2627–2636. doi: 10.2174/1568026614666141215124956. [DOI] [PubMed] [Google Scholar]
- 24.Clish CB. 2015. Metabolomics: an emerging but powerful tool for precision medicine. Cold Spring Harb Mol Case Stud 1:a000588. doi: 10.1101/mcs.a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emiola A, Andrews SS, Heller C, George J. 2016. Crosstalk between the lipopolysaccharide and phospholipid pathways during outer membrane biogenesis in Escherichia coli. Proc Natl Acad Sci U S A 113:3108–3113. doi: 10.1073/pnas.1521168113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeukens J, Freschi L, Kukavica-Ibrulj I, Emond-Rheault J-G, Tucker NP, Levesque RC. 2019. Genomics of antibiotic-resistance prediction in Pseudomonas aeruginosa. Ann N Y Acad Sci 1435:5–17. doi: 10.1111/nyas.13358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taber HW, Mueller J, Miller P, Arrow A. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev 51:439–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergen PJ, Bulitta JB, Forrest A, Tsuji BT, Li J, Nation RL. 2010. Pharmacokinetic/pharmacodynamic investigation of colistin against Pseudomonas aeruginosa using an in vitro model. Antimicrob Agents Chemother 54:3783–3789. doi: 10.1128/AAC.00903-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthaiou DK, Michalopoulos A, Rafailidis PI, Karageorgopoulos DE, Papaioannou V, Ntani G, Samonis G, Falagas ME. 2008. Risk factors associated with the isolation of colistin-resistant gram-negative bacteria: a matched case-control study. Crit Care Med 36:807–811. doi: 10.1097/CCM.0B013E3181652FAE. [DOI] [PubMed] [Google Scholar]
- 30.Hancock RE, Chapple DS. 1999. Peptide antibiotics. Antimicrob Agents Chemother 43:1317–1323. doi: 10.1128/AAC.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han M-L, Zhu Y, Creek DJ, Lin Y-W, Anderson D, Shen H-H, Tsuji B, Gutu AD, Moskowitz SM, Velkov T, Li J. 2018. Alterations of metabolic and lipid profiles in polymyxin-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 62:e02656-17. doi: 10.1128/AAC.02656-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hussein M, Han M-L, Zhu Y, Schneider-Futschik EK, Hu X, Zhou QT, Lin Y-W, Anderson D, Creek DJ, Hoyer D, Li J, Velkov T. 2018. Mechanistic insights from global metabolomics studies into synergistic bactericidal effect of a polymyxin B combination with tamoxifen against cystic fibrosis MDR Pseudomonas aeruginosa. Comput Struct Biotechnol J 16:587–599. doi: 10.1016/j.csbj.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry R, Crane B, Powell D, Deveson Lucas D, Li Z, Aranda J, Harrison P, Nation RL, Adler B, Harper M, Boyce JD, Li J. 2015. The transcriptomic response of Acinetobacter baumannii to colistin and doripenem alone and in combination in an in vitro pharmacokinetics/pharmacodynamics model. J Antimicrob Chemother 70:1303–1313. doi: 10.1093/jac/dku536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louesdon S, Charlot-Rougé S, Tourdot-Maréchal R, Bouix M, Béal C. 2015. Membrane fatty acid composition and fluidity are involved in the resistance to freezing of Lactobacillus buchneri R1102 and Bifidobacterium longum R0175. Microb Biotechnol 8:311–318. doi: 10.1111/1751-7915.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikaido H, Hancock R. 1986. Outer membrane permeability of Pseudomonas aeruginosa. Bacteria 10:145–193. [Google Scholar]
- 36.Mansilla MC, Cybulski LE, Albanesi D, de Mendoza D. 2004. Control of membrane lipid fluidity by molecular thermosensors. J Bacteriol 186:6681–6688. doi: 10.1128/JB.186.20.6681-6688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotra LP, Haddad J, Mobashery S. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob Agents Chemother 44:3249–3256. doi: 10.1128/aac.44.12.3249-3256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bera S, Zhanel GG, Schweizer F. 2010. Antibacterial activities of aminoglycoside antibiotics-derived cationic amphiphiles. Polyol-modified neomycin B-, kanamycin A-, amikacin-, and neamine-based amphiphiles with potent broad spectrum antibacterial activity. J Med Chem 53:3626–3631. doi: 10.1021/jm1000437. [DOI] [PubMed] [Google Scholar]
- 39.Sautrey G, El Khoury M, dos Santos AG, Zimmermann L, Deleu M, Lins L, Décout J-L, Mingeot-Leclercq M-P. 2016. Negatively charged lipids as a potential target for new amphiphilic aminoglycoside antibiotics: a biophysical study. J Biol Chem 291:13864–13874. doi: 10.1074/jbc.M115.665364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y-M, Rock CO. 2008. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol 6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 41.Velkov T, Roberts KD, Nation RL, Wang J, Thompson PE, Li J. 2014. Teaching ‘old’ polymyxins new tricks: new-generation lipopeptides targeting Gram-negative ‘superbugs’. ACS Chem Biol 9:1172–1177. doi: 10.1021/cb500080r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brade H. 1999. Endotoxin in health and disease. CRC Press, Boca Raton, FL. [Google Scholar]
- 43.Gourlay LJ, Sommaruga S, Nardini M, Sperandeo P, Deho G, Polissi A, Bolognesi M. 2010. Probing the active site of the sugar isomerase domain from E. coli arabinose-5-phosphate isomerase via X-ray crystallography. Protein Sci 19:2430–2439. doi: 10.1002/pro.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor PL, Blakely KM, De Leon GP, Walker JR, McArthur F, Evdokimova E, Zhang K, Valvano MA, Wright GD, Junop MS. 2008. Structure and function of sedoheptulose-7-phosphate isomerase, a critical enzyme for lipopolysaccharide biosynthesis and a target for antibiotic adjuvants. J Biol Chem 283:2835–2845. doi: 10.1074/jbc.M706163200. [DOI] [PubMed] [Google Scholar]
- 45.Brooke JS, Valvano M. 1996. Molecular cloning of the Haemophilus influenzae gmhA (lpcA) gene encoding a phosphoheptose isomerase required for lipooligosaccharide biosynthesis. J Bacteriol 178:3339–3341. doi: 10.1128/jb.178.11.3339-3341.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murima P, McKinney JD, Pethe K. 2014. Targeting bacterial central metabolism for drug development. Chem Biol 21:1423–1432. doi: 10.1016/j.chembiol.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 47.Wolfe AJ. 2015. Glycolysis for the microbiome generation. Microbiol Spectr 3:MBP-0014-2014. doi: 10.1128/microbiolspec.MBP-0014-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gest H. 1987. Evolutionary roots of the citric acid cycle in prokaryotes. Biochem Soc Symp 54:3–16. [PubMed] [Google Scholar]
- 49.Owen OE, Kalhan SC, Hanson RW. 2002. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem 277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 50.Galeazzi L, Bocci P, Amici A, Brunetti L, Ruggieri S, Romine M, Reed S, Osterman AL, Rodionov DA, Sorci L, Raffaelli N. 2011. Identification of nicotinamide mononucleotide deamidase of the bacterial pyridine nucleotide cycle reveals a novel broadly conserved amidohydrolase family. J Biol Chem 286:40365–40375. doi: 10.1074/jbc.M111.275818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neidhardt FC, Ingraham JL, Schaechter M. 1990. Physiology of the bacterial cell: a molecular approach, vol 20 Sinauer Associates, Sunderland, MA. [Google Scholar]
- 52.Dörries K, Schlueter R, Lalk M. 2014. Impact of antibiotics with various target sites on the metabolome of Staphylococcus aureus. Antimicrob Agents Chemother 58:7151–7163. doi: 10.1128/AAC.03104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belenky P, Ye JD, Porter CBM, Cohen NR, Lobritz MA, Ferrante T, Jain S, Korry BJ, Schwarz EG, Walker GC, Collins JJ. 2015. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep 13:968–980. doi: 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perkins HR, Nieto M. 1973. The significance of D-alanyl-D-alanine termini in the biosynthesis of bacterial cell walls and the action of penicillin, vancomycin and ristocetin. Pure Appl Chem 35:371–382. doi: 10.1351/pac197335040371. [DOI] [Google Scholar]
- 55.Milewski S. 2002. Glucosamine-6-phosphate synthase—the multi-facets enzyme. Biochim Biophys Acta 1597:173–192. doi: 10.1016/S0167-4838(02)00318-7. [DOI] [PubMed] [Google Scholar]
- 56.Maifiah MHM, Creek DJ, Nation RL, Forrest A, Tsuji BT, Velkov T, Li J. 2017. Untargeted metabolomics analysis reveals key pathways responsible for the synergistic killing of colistin and doripenem combination against Acinetobacter baumannii. Sci Rep 7:45527. doi: 10.1038/srep45527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han M-L, Zhu Y, Creek DJ, Lin Y-W, Gutu AD, Hertzog P, Purcell T, Shen H-H, Moskowitz SM, Velkov T, Li J. 2019. Comparative metabolomics and transcriptomics reveal multiple pathways associated with polymyxin killing in Pseudomonas aeruginosa. mSystems 4:e00149-18. doi: 10.1128/mSystems.00149-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiong L, Teng JL, Botelho MG, Lo RC, Lau SK, Woo PC. 2016. Arginine metabolism in bacterial pathogenesis and cancer therapy. Int J Mol Sci 17:363. doi: 10.3390/ijms17030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anonymous. 2016. M9 minimal medium buffer. Cold Spring Harb Protoc doi: 10.1101/pdb.rec088559. [DOI] [Google Scholar]
- 60.Gika HG, Theodoridis GA, Wingate JE, Wilson ID. 2007. Within-day reproducibility of an HPLC-MS-based method for metabonomic analysis: application to human urine. J Proteome Res 6:3291–3303. doi: 10.1021/pr070183p. [DOI] [PubMed] [Google Scholar]
- 61.Zhang T, Creek DJ, Barrett MP, Blackburn G, Watson DG. 2012. Evaluation of coupling reversed phase, aqueous normal phase, and hydrophilic interaction liquid chromatography with Orbitrap mass spectrometry for metabolomic studies of human urine. Anal Chem 84:1994–2001. doi: 10.1021/ac2030738. [DOI] [PubMed] [Google Scholar]
- 62.Creek DJ, Jankevics A, Burgess KE, Breitling R, Barrett MP. 2012. IDEOM: an Excel interface for analysis of LC-MS-based metabolomics data. Bioinformatics 28:1048–1049. doi: 10.1093/bioinformatics/bts069. [DOI] [PubMed] [Google Scholar]
- 63.Scheltema RA, Jankevics A, Jansen RC, Swertz MA, Breitling R. 2011. PeakML/mzMatch: a file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal Chem 83:2786–2793. doi: 10.1021/ac2000994. [DOI] [PubMed] [Google Scholar]
- 64.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. 2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 65.Boulesteix A-L, Strimmer K. 2007. Partial least squares: a versatile tool for the analysis of high-dimensional genomic data. Brief Bioinform 8:32–44. doi: 10.1093/bib/bbl016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.