Here, we report an NDM-5-producing sequence type 35 (ST35) hypervirulent Klebsiella pneumoniae strain, isolated from the blood of a male patient. It showed a remarkable resistance to serum killing and neutrophil phagocytosis and high virulence in a mouse peritonitis infection model.
Keywords: carbapenem-resistant hypervirulent K. pneumoniae, NDM-5, ICEKp1, ST35
ABSTRACT
Here, we report an NDM-5-producing sequence type 35 (ST35) hypervirulent Klebsiella pneumoniae strain, isolated from the blood of a male patient. It showed a remarkable resistance to serum killing and neutrophil phagocytosis and high virulence in a mouse peritonitis infection model. Instead of carrying a pLVPK-like virulence plasmid, chromosomal integration of ICEKp1 (∼76 kb) was identified in a specific asparagine-tRNA gene, harboring the iron acquisition system salmochelin genes (iroBCDN), a yersiniabactin gene, and a variant of the rmpA gene.
INTRODUCTION
The emergence and global dissemination of carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKp) pose a significant therapeutic challenge to public health, leaving few treatment options for the severe infections caused by these strains (1). Previous studies indicated that such strains were developed through two mechanisms. The first is acquisition by an hvKp strain of a conjugal plasmid that contains carbapenem resistance determinants (2, 3), while the second is acquisition of the hvKp virulence plasmid by carbapenem-resistant K. pneumoniae (CRKP) strains (4, 5). However, the species-specific integrative and conjugative element ICEKp, harboring biosynthetic genes for siderophores, plays an important role in the pathogenesis and evolutionary history of K. pneumoniae (6, 7). The horizontal transfer of ICEKp provides an important mechanism for virulence factors to spread within the general population of K. pneumoniae (6, 7). Here, we report an NDM-5-producing CR-hvKp strain, RJY9645, with chromosomal integration of ICEKp1, harboring the iron acquisition system salmochelin genes (iroBCDN), a yersiniabactin gene, and a variant of the rmpA gene.
A total of 345 clinical CRKP isolates were recovered in our hospital over 3 years (2016 to 2018). All isolates were screened for carbapenemases (blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA-48) and virulence genes (iucABCDiutA, iroBCDN, rmpA, and rmpA2) (see Table S1 in the supplemental material) (8, 9). Only one strain, K. pneumoniae RJY9645, displayed positive detection of blaNDM-5, and the virulence genes iroBCDN and rmpA were also detected (Table 1). K. pneumoniae RJY9645 was resistant to ceftazidime, ceftriaxone, cefepime, piperacillin-tazobactam, imipenem, and mempenem, as determined using the CLSI reference broth microdilution method (10). In contrast, it displayed susceptibility to aztreonam, amikacin, gentamicin, ciprofloxacin, levofloxacin, tigecycline, and polymyxin B (Table S2).
TABLE 1.
Microbiological characteristics of K. pneumoniae RJY9645 and the control strains
| Strain | Isolation source | Carbapenemase | ST | Serotype | String test | Serum killing (grade)a | Phagocytosis rateb | Virulence genes present |
LD50 (CFU) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| iucABCDiutA | iroBCDN | rmpA | rmpA2 | |||||||||
| RJY9645 | Blood | NDM-5 | ST35 | K108 | + | 4 | R | − | + | + | − | ∼104 |
| HAP-Kp11 | Sputum | KPC-2 | ST11 | K47 | − | 1 | S | − | − | − | − | >106 |
| LA-Kp23 | Liver pus | ST23 | K1 | + | 5 | R | + | + | + | + | ∼102 | |
A strain was defined as serum sensitive at grades of 1 and 2, intermediately resistant at 3 and 4, and resistant at 5 and 6.
R, phagocytic rate of <50% at 30 min; S, phagocytic rate of ≥50% at 30 min.
K. pneumoniae RJY9645 was recovered in the Department of Liver Surgery from the blood of a male patient. The patient was discharged after liver transplantation, but he had intermittent fever 3 weeks later and was hospitalized for further treatment. He had been on combination therapy with imipenem and levofloxacin for 3 weeks after the isolation of strain RJY9645 and was then discharged. However, he was admitted to the hospital again for the recurrent bloodstream infection only 1 week later, with the positive blood culture of K. pneumoniae RJY9699, which displayed identical pulsed-field gel electrophoresis (PFGE) patterns with RJY9645 and belonged to the same clone (Fig. S1). Since RJY9645 was resistant to imipenem (MIC, 16 μg/ml), the combination therapy of imipenem and levofloxacin is suboptimal and may contribute to the reoccurrence of the bloodstream infection. For the second episode of bloodstream infection, the patient received 3 weeks of antimicrobial therapy of aztreonam and levofloxacin and was discharged after 4 weeks of hospitalization. From then on, the patient was seen for follow-up visits once every 3 months, and he had no recent history of foreign travel.
K. pneumoniae RJY9645 showed a hypermucoviscous phenotype, with a positive string test. Strain virulence was estimated using serum killing, neutrophil phagocytosis, and mouse lethality assays. Serum resistance and neutrophil phagocytosis assays were performed as previously described (11, 12). The mouse lethality assay was performed for estimation of the in vivo virulence, and the 50% lethal dose (LD50) was determined with pathogen-free, 6- to 8-week-old, female BALB/c mice. A 10-fold serial dilution of CFU was made from a starting concentration of 106 CFU/ml to 102 CFU/ml, and BALB/c mice were infected intraperitoneally with 0.1 ml of each concentration. A total of six mice were used as a sample population for each bacterial concentration. Symptoms and mortality rates were observed for 14 days. The exact inoculation dose was confirmed on LB agar, and the LD50 was calculated using the SigmaPlot program (version 11.0). One KPC-2-producing K. pneumoniae strain, HAP-Kp11, which was associated with hospital-acquired pneumonia and belonged to sequence type 11 (ST11), was used as the low-virulence control (Table 1). In contrast, one ST23 K. pneumoniae strain, LA-Kp23, isolated from the abscess drainage of a patient with community-acquired liver abscess, was used as the hypervirulence control (Table 1).
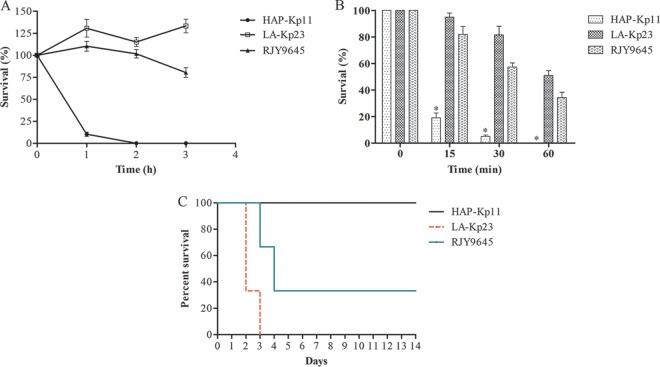
K. pneumoniae RJY9645 exhibited grade 4 response to serum killing, with viable counts of >100% of the inoculum after both 1 and 2 h but <100% after 3 h (Fig. 1 and Table 1). Also, RJY9645 had a survival rate of >50% after incubation with human neutrophils for 30 min, and the survival of both RJY9645 and LA-Kp23 was significantly higher than that of HAP-Kp11 at each time point (P < 0.05 by one-way analysis of variance [ANOVA], Fig. 1). The mouse lethality assay indicated that RJY9645 showed high virulence, with an LD50 of 104 CFU (Table 1). When infected intraperitoneally with 5 × 104 CFU, RJY9645 killed four of the six mice tested within 4 days (Fig. 1).
FIG 1.
Virulence characteristics of K. pneumoniae RJY9645. (A) Serum resistance assay. K. pneumoniae RJY9645 exhibited a grade 4 response, with viable counts of >100% of the inoculum after both 1 and 2 h but <100% after 3 h. The hypervirulence control LA-Kp23 and the low-virulence control HAP-Kp11 exhibited grade 5 and grade 1 responses, respectively. (B) Human neutrophil phagocytosis assay. The data are presented as the means ± standard deviation. The survival of RJY9645 and LA-Kp23 was significantly higher than that of HAP-Kp11 at each time point (*, P < 0.05, one-way ANOVA). (C) Survival curves for K. pneumoniae RJY9645-infected mice. Mice were intraperitoneally infected with 5 × 104 CFU of each strain. Six mice were used as a sample population for each strain. Symptoms and mortality rates were observed for 14 days.
Strain RJY9645 was subjected to whole-genome sequencing using a MiSeq platform (Illumina, USA) and long-read PromethION platform (Nanopore, Oxford, United Kingdom). Nanopore reads were assembled using CANU version 1.7, which returned four contigs. The sequences were polished by Illumina reads to decrease the error rates. The genome size was 5,587,671 bp, including a circular chromosome of 5,269,438 bp, with an average GC content of 57.6% and three circular plasmids, pY9645-166 (166,344 bp), pY9645-105 (105,728 bp), and pY9645-NDM5 (46,161 bp). The sizes of the plasmids were confirmed using S1-PFGE (Fig. S1). K. pneumoniae RJY9645 belonged to ST35 and capsular type K108 (wzi_allele 194), as determined using the assembled contigs to query the multilocus sequence typing (MLST) and wzi allele (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html) databases. Phylogenetic analysis of RJY9645 was performed using RAxML version 8.0 (13) with the whole genomes of other representative K. pneumoniae strains available in GenBank (Table S3). RJY9645 was grouped with classical K. pneumoniae (cKp) strains, such as OXA-232-producing UCLAOXA232KP and KPC-2-producing HS11286 (Fig. S2). In contrast, RJY9645 was distant from the clades grouped by ST23 or K2 (ST66 or ST86) hvKp strains.
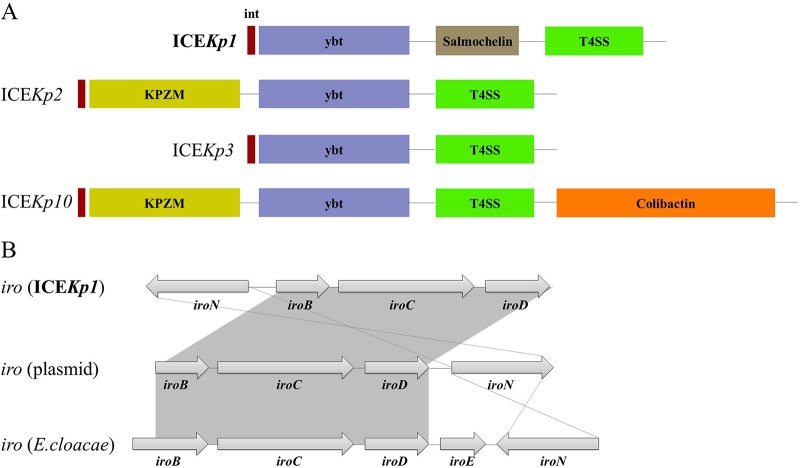
Unlike the previously reported CR-hvKp strains, no pLVPK-like virulence plasmid was identified in the genome of K. pneumoniae RJY9645. Instead, the chromosomal integration of the integrative and conjugative element ICEKp1 (∼76 kb) was spotted in a specific asparagine-tRNA gene. ICEKp1 was divided into three regions, with one similar to the high-pathogenicity island of Yersinia species encoding the siderophore yersiniabactin, followed by a middle region similar to a part of the large virulence plasmid pLVPK (GenBank accession no. AY378100), encoding the siderophore salmochelin (iroBCDN) and the capsular polysaccharide regulator RmpA, while the third region encoded the type IV secretion system (T4SS) (Fig. 2A). The genetic context of salmochelin located on ICEKp1 was distinct from pLVPK-carried iro lineage, with an inverted and preamble iroN sequence in gene cluster (Fig. 2B). In order to exclude genome misassembly events, PCRs verifying the junctions were performed between the virulence cluster (iroBCDN and rmpA) and ICEKp1 and between the intact ICEKp1 and chromosome, respectively (Table S1). Apart from ICEKp1, another four chromosomally borne ICE variants were identified by VRprofile, with default parameters (14), including two different type VI secretion system (T6SS) loci and two prophages encoding the prophage core component proteins (Table S4). Other chromosomally borne virulence genes, such as type 1 fimbrial genes (fimACDGH) and an enterobactin gene cluster (entABCEF), have also been identified.
FIG 2.
Structures of the representative ICEKp variants and salmochelin (iro) locus. (A) The representative ICEKp structures. ybt, yersiniabactin synthesis locus; T4SS, type 4 secretion system; KPZM, Zn2+/Mn2+ metabolism module. (B) Salmochelin (iro) locus. The gray area indicates that this region was identical between different sequences. The iro lineages were based on the sequences of ICEKp1, plasmid pLVPK, and Enterobacter cloacae iroBCDEN under GenBank accession numbers AB298504, AY378100, and CP032832.
The blaNDM-5 gene was located on an IncX3 plasmid of 46,161 bp, designated pY9645-NDM5, which could be transferred to Escherichia coli strain J53 through a conjugation assay. The blaNDM-5-carrying plasmid could be transferred at a frequency of 10−4 transconjugants per donor cell. This plasmid displayed 100% coverage and 99% identity to the epidemic blaNDM-5-carrying plasmid pP768-NDM5 in China (GenBank accession no. MF547510) (15), and only two nucleotide substitutions were identified. The pY9645-NDM5 plasmid did not carry resistance genes other than blaNDM-5. In addition, most of the blaNDM-5-carrying plasmids reported in China belonged to the IncX3 type and showed highly identical nucleotide sequences (15, 16). The IncX3-type plasmids facilitate the rapid dissemination of blaNDM-5 among Enterobacteriaceae in China. Other resistance genes of K. pneumoniae RJY9645 were predicted using ResFinder (Center for Genomic Epidemiology, http://genomicepidemiology.org/). A chromosomally borne broad-spectrum β-lactamase gene (blaSHV-33), a fosfomycin resistance determinant gene (fosA), and efflux pump genes (oqxAB) were identified, and strain RJY9645 was found to display resistance to fosfomycin through antimicrobial susceptibility testing using a disk diffusion method.
Instead of carrying a pLVPK-like virulence plasmid, chromosomal integration of ICEKp1 was identified in K. pneumoniae RJY9645. ICEKp is the most common virulence-associated mobile genetic element of K. pneumoniae, and 14 different structural variants have been reported to date (6). ICEKp1 was first described in hvKp strain NTUH-K2044 and was more prevalent in hvKp than in cKp strains, suggesting a role in hvKp pathogenesis (17). The sequence of ICEKp1 in RJY9645 displayed 100% coverage and 99% identity to that of strain NTUH-K2044. Though all ICEKp variants carried yersiniabactin, only ICEKp1 harbored the salmochelin biosynthesis gene cluster (6). Yersiniabactin has been shown to be an important factor for cKp in mouse infection models (18). Salmochelin is hvKp specific and does not just act as a siderophore. In conjunction with microcin E492, salmochelin is also a critical mediator of hvKp colonization (19). Together with the rmpA gene, the chromosomal integration of ICEKp1 may facilitate genetic diversity and contribute significantly to the hypervirulence in K. pneumoniae RJY9645.
ST35 K. pneumoniae is an international multidrug-resistant clone. It has worldwide distribution and has been reported in Tunisia (20), Spain (21), Denmark (22), France (23), and Yemen (24). Various extended-spectrum β-lactamases (ESBLs) have been detected in ST35 K. pneumoniae strains, such as CTX-M-15 and SHV-12 (20, 21). Recently, one NDM-1-producing ST35 K. pneumoniae strain was recovered from the blood of a 5-year-old boy in a pediatric intensive care unit (24). However, little attention has been paid to the pathogenicity of ST35 K. pneumoniae. To the best of our knowledge, this is the first report of ST35 NDM-5-producing CR-hvKp with chromosomal integration of ICEKp1.
A previous comparative analysis of 2,498 whole-genome sequences of K. pneumoniae indicated that independent ICEKp integration events were detected, affecting hundreds of phylogenetically distinct lineages, including CRKP and hvKp strains (6). The horizontal gene transfer of ICEKp1 provided an important mechanism for the yersiniabactin, salmochelin, and rmpA virulence genes to spread within K. pneumoniae populations. Also, NDM-5-positive K. pneumoniae strains are distributed across a number of sequence types (STs), with no predominant lineages (25). Furthermore, RJY9645 exhibited a cKp genetic background distant from the clades formed by K1 (ST23) or K2 (ST66 or ST86) hvKp strains. Taken together, the development of CR-hvKp strain RJY9645 might be attributed to chromosomal integration of ICEKp1 and acquisition of the blaNDM-5-carrying plasmid.
All procedures performed in this study involving human participants and animals were in accordance with the ethical standards of the institutional review board ethics committee of Renji Hospital, School of Medicine, Shanghai Jiaotong University. For this type of retrospective study, formal consent is not required.
Data availability.
The complete sequences of the chromosome of strain RJY9645 and plasmids pY9645-166, pY9645-105, and pY9645-NDM5 have been deposited in GenBank under accession numbers CP041353, CP044029, CP044030, and MN064714, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grant no. 81802065), Shanghai Sailing Program (grant no. 18YF1413300), and a research fund from Renji Hospital for young scholars (grant no. PYIII-17-010).
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, Jeong BC, Lee SH. 2017. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol 7:483. doi: 10.3389/fcimb.2017.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siu LK, Huang DB, Chiang T. 2014. Plasmid transferability of KPC into a virulent K2 serotype Klebsiella pneumoniae. BMC Infect Dis 14:176. doi: 10.1186/1471-2334-14-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, Zeng J, Liu W, Zhao F, Hu Z, Zhao C, Wang Q, Wang X, Chen H, Li H, Zhang F, Li S, Cao B, Wang H. 2015. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect 71:553–560. doi: 10.1016/j.jinf.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 5.Huang YH, Chou SH, Liang SW, Ni CE, Lin YT, Huang YW, Yang TC. 2018. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother 73:2039–2046. doi: 10.1093/jac/dky164. [DOI] [PubMed] [Google Scholar]
- 6.Lam M, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney A, Brisse S, Holt KE. 2018. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom 4: . doi: 10.1099/mgen.0.000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcoleta AE, Berrios-Pasten C, Nunez G, Monasterio O, Lagos R. 2016. Klebsiella pneumoniae asparagine tDNAs are integration hotspots for different genomic islands encoding microcin E492 production determinants and other putative virulence factors present in hypervirulent strains. Front Microbiol 7:849. doi: 10.3389/fmicb.2016.00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Ye M, Tu J, Jiang J, Bi Y, You W, Zhang Y, Ren J, Zhu T, Cao Z, Yu Z, Shao C, Shen Z, Ding B, Yuan J, Zhao X, Guo Q, Xu X, Huang J, Wang M. 2016. Clinical and genomic analysis of liver abscess-causing Klebsiella pneumoniae identifies new liver abscess-associated virulence genes. Front Cell Infect Microbiol 6:165. doi: 10.3389/fcimb.2016.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed. CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Siu LK, Fung CP, Chang FY, Lee N, Yeh KM, Koh TH, Ip M. 2011. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol 49:3761–3765. doi: 10.1128/JCM.00977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deleo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, Chavda KD, Jacobs MR, Mathema B, Olsen RJ, Bonomo RA, Musser JM, Kreiswirth BN. 2014. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A 111:4988–4993. doi: 10.1073/pnas.1321364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Tai C, Deng Z, Zhong W, He Y, Ou HY. 2018. VRprofile: gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Brief Bioinform 19:566–574. doi: 10.1093/bib/bbw141. [DOI] [PubMed] [Google Scholar]
- 15.Ho PL, Wang Y, Liu MC, Lai EL, Law PY, Cao H, Chow KH. 2018. IncX3 epidemic plasmid carrying blaNDM-5 in Escherichia coli from swine in multiple geographic areas in China. Antimicrob Agents Chemother 62:e02295-17. doi: 10.1128/AAC.02295-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Fu Y, Shen M, Huang D, Du X, Hu Q, Zhou Y, Wang D, Yu Y. 2018. Dissemination of blaNDM-5 gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob Resist Infect Control 7:59. doi: 10.1186/s13756-018-0349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin TL, Lee CZ, Hsieh PF, Tsai SF, Wang JT. 2008. Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J Bacteriol 190:515–526. doi: 10.1128/JB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachman MA, Oyler JE, Burns SH, Caza M, Lepine F, Dozois CM, Weiser JN. 2011. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun 79:3309–3316. doi: 10.1128/IAI.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagos R, Baeza M, Corsini G, Hetz C, Strahsburger E, Castillo JA, Vergara C, Monasterio O. 2001. Structure, organization and characterization of the gene cluster involved in the production of microcin E492, a channel-forming bacteriocin. Mol Microbiol 42:229–243. doi: 10.1046/j.1365-2958.2001.02630.x. [DOI] [PubMed] [Google Scholar]
- 20.Elhani D, Bakir L, Aouni M, Passet V, Arlet G, Brisse S, Weill FX. 2010. Molecular epidemiology of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae strains in a university hospital in Tunis, Tunisia, 1999–2005. Clin Microbiol Infect 16:157–164. doi: 10.1111/j.1469-0691.2009.03057.x. [DOI] [PubMed] [Google Scholar]
- 21.Oteo J, Cuevas O, López-Rodríguez I, Banderas-Florido A, Vindel A, Pérez-Vázquez M, Bautista V, Arroyo M, García-Caballero J, Marín-Casanova P, González-Sanz R, Fuentes-Gómez V, Oña-Compán S, García-Cobos S, Campos J. 2009. Emergence of CTX-M-15-producing Klebsiella pneumoniae of multilocus sequence types 1, 11, 14, 17, 20, 35 and 36 as pathogens and colonizers in newborns and adults. J Antimicrob Chemother 64:524–528. doi: 10.1093/jac/dkp211. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen JB, Skov MN, Jorgensen RL, Heltberg O, Hansen DS, Schonning K. 2011. Identification of CTX-M-15-, SHV-28-producing Klebsiella pneumoniae ST15 as an epidemic clone in the Copenhagen area using a semi-automated Rep-PCR typing assay. Eur J Clin Microbiol Infect Dis 30:773–778. doi: 10.1007/s10096-011-1153-x. [DOI] [PubMed] [Google Scholar]
- 23.Marcade G, Brisse S, Bialek S, Marcon E, Leflon-Guibout V, Passet V, Moreau R, Nicolas-Chanoine MH. 2013. The emergence of multidrug-resistant Klebsiella pneumoniae of international clones ST13, ST16, ST35, ST48 and ST101 in a teaching hospital in the Paris region. Epidemiol Infect 141:1705–1712. doi: 10.1017/S0950268812002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alsharapy SA, Gharout-Sait A, Muggeo A, Guillard T, Cholley P, Brasme L, Bertrand X, Moghram GS, Touati A, De Champs C. 2019. Characterization of carbapenem-resistant Enterobacteriaceae clinical isolates in Al Thawra University Hospital, Sana’a, Yemen. Microb Drug Resist, in press. doi: 10.1089/mdr.2018.0443. [DOI] [PubMed] [Google Scholar]
- 25.Wu W, Feng Y, Tang G, Qiao F, McNally A, Zong Z. 2019. NDM metallo-beta-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev 32:e00115-18. doi: 10.1128/CMR.00115-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete sequences of the chromosome of strain RJY9645 and plasmids pY9645-166, pY9645-105, and pY9645-NDM5 have been deposited in GenBank under accession numbers CP041353, CP044029, CP044030, and MN064714, respectively.