Abstract
Precise regulation of cellular proliferation is critical to tissue homeostasis and development, but misregulation leads to diseases of excess proliferation or cell loss. To achieve precise control, cells utilize distinct mechanisms of growth arrest such as quiescence and senescence. The decision to enter these growth arrested states or proliferate is mediated by the core cell-cycle machinery that responds to diverse external and internal signals. Recent advances have revealed the molecular underpinnings of these cell-cycle decisions, highlighting the unique nature of cell-cycle entry from quiescence, identifying endogenous DNA damage as a quiescence-inducing signal, and establishing how persistent arrest is achieved in senescence.
Introduction
One of the most fundamental decisions that a cell continuously makes is whether to proliferate or exist in a nondividing state. Cells integrate a wide variety of internal and external stimuli to determine if the cell-cycle machinery will be initiated. In multicellular organisms, defects in the transition between proliferation and arrest lead to diseases of excess proliferation (cancer) or cell loss (aging and degeneration). To address the complex organismal needs for proper tissue development and homeostasis, there exists great diversity in the forms and functions of growth arrest. Recent work has improved our understanding on how cells integrate signaling information to regulate cell-cycle arrest.
Broadly, growth arrest is categorized as either quiescence, terminal differentiation, or senescence. Quiescence is a non-proliferative state induced by a variety of stimuli including mitogen starvation, contact inhibition, and loss of adhesion. A defining characteristic of quiescence is the ability of cells to re-enter the cell cycle following the reversal of these signals. Quiescent cells are resistant to stress and toxicity, a trait that is critical to the maintenance of tissue stem cells. While some aspects of the quiescence program are shared, there are unique properties that distinguish the response to different quiescence-inducing signals. Contrary to the reversible nature of cell-cycle exit in quiescence, terminal differentiation entails permanent exit from the cell cycle. Classically, cellular senescence entails permanent exit combined with persistent stress (e.g., oncogene activation, telomere shortening). Initially thought to function as an important safeguard against uncontrolled cellular proliferation and cancer, cellular senescence now has many appreciated roles in development (e.g., renal morphogenesis), tissue homeostasis (e.g., wound healing), and aging [1].
In this review, we focus on three regulatory aspects of quiescence and senescence as they relate to mammalian cells. Terminal differentiation has many similarities to quiescence but with more permanent repressive mechanisms that are not discussed here. First, we discuss how ongoing studies have clarified the molecular mechanisms and systems properties underlying the classical concept of the restriction point in quiescence. Next, we consider how cells utilize transient quiescence to tolerate the inherently error prone nature of replicating the genome. Finally, we compare the quiescence and senescence growth arrest programs, evaluating the mechanisms that promote and maintain the irreversible arrest characteristic of senescence. For each section, we summarize the relevant arrest-inducing stimuli and describe the core machinery functioning in induction of arrest, maintenance of arrest, and re-entry into the cell cycle.
Quiescence Decisions and the Restriction Point
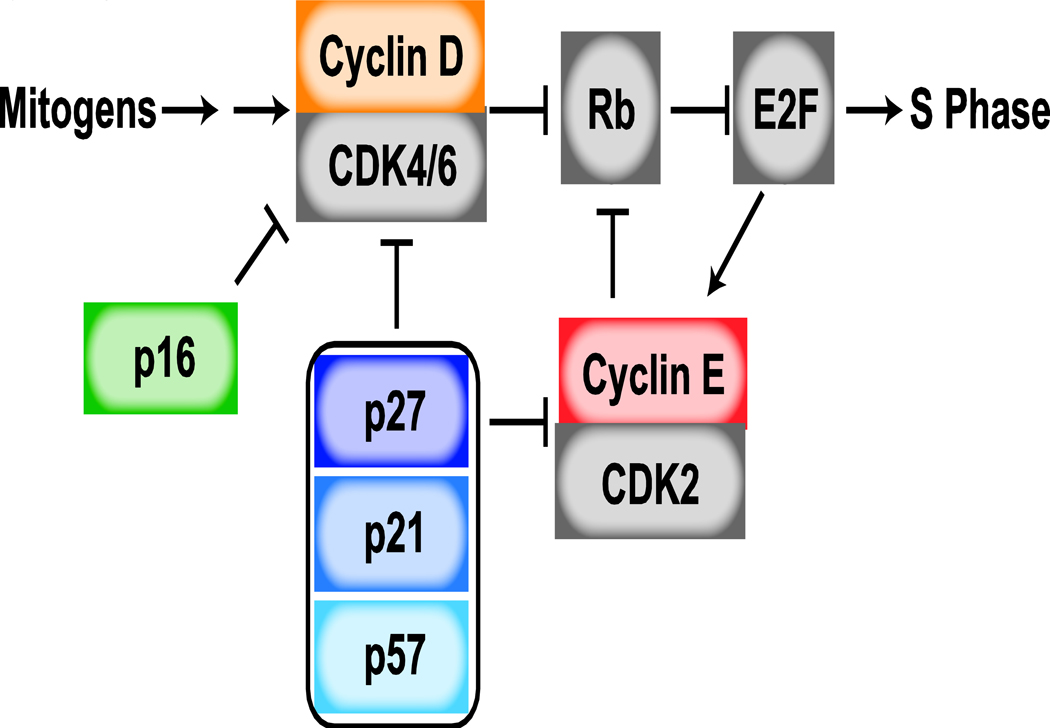
The decision to enter quiescence versus proliferate relies on the funneling of diverse and complex signals into a core cell-cycle machinery [2,3]. Critical to cell-cycle entry is the phosphorylation of the pocket protein family – including p107, p130, and retinoblastoma protein (Rb) – of which we will focus on the regulation of Rb in this review. When unphosphorylated, Rb binds to chromatin and prevents E2F transcription factors from inducing genes essential for proliferation (Figure 1). Under pro-growth conditions, Cyclin-dependent kinases 4 and 6 (CDK4/6) are activated by the binding of cyclin D (D1, D2, D3) and phosphorylate Rb, releasing it from chromatin and enabling E2F target transcription. Activation of Cyclin-dependent kinase 2 (CDK2) following binding by E2F-target cyclin E (E1, E2) helps to reinforce and maintain Rb phosphorylation and cell-cycle progression. The pro-proliferative effects of CDK4/6-cyclin D and CDK2-cyclin E complexes are opposed by anti-proliferative proteins. The CDK interacting protein/kinase inhibitory protein (CIP/KIP) family of inhibitors, including p21, p27, and p57, inhibit both CDK4/6 and CDK2 complexes, while the inhibitor of kinase 4 (INK4) family of inhibitors, including p16, specifically inhibit CDK4/6. Discovery of how this core molecular machinery contributes to cell-cycle regulation is the foundation for recent work investigating the different forms of cellular growth arrest.
Figure 1: Cell-Cycle Regulation in G1 phase.
Progression through the cell cycle depends upon the activation of cyclin-dependent kinases (CDK), whose activity requires the binding of specific regulatory subunits, known as cyclins. CDK4/6 is activated by the binding of D-type cyclins, whose expression are controlled by mitogenic growth factors. CDK2 is activated by E-type cyclins, known transcriptional targets of E2F. The activity of both CDK4/6 and CDK2 complexes are opposed by CIP/KIP CDK inhibitors, including p21, p27, and p57, while INK4 inhibitors like p16 exclusively bind CDK4/6, preventing cyclin D-CDK4/6 interaction. CDK activity promotes cell-cycle entry through the phosphorylation and inactivation of the transcriptional repressor, Rb. Following phosphorylation of Rb, E2F transcription is activated, promoting cell-cycle progression.
Cell-cycle exit occurs via the induction of CDK inhibitors and/or the downregulation of cyclins and CDKs. For example, serum starvation results in an upregulation of p27 and a suppression of the G1 cyclins, cyclin D and cyclin E. Thus, the quiescent state is defined by low CDK activity and hypo-phosphorylation of Rb protein. Despite their defined roles in cell-cycle entry, previous genetic knockout experiments in mice have shown that the majority of cells proliferate normally in mouse embryos lacking all three cyclin D proteins (D1−/−D2−/−D3−/−) or lacking cyclin E1 and E2 (E1−/−E2−/−) [4]. These results led to the current model that both types of G1 cyclins can adequately promote Rb phosphorylation and that a single G1 cyclin is sufficient for mammalian cell proliferation. Recently, this model has been tested by genetic ablation of all D-type and E-type cyclins. While some cell types (e.g., mouse embryonic fibroblasts) failed to proliferate, other cell types, including embryonic stem cells, proliferated normally in the absence of all G1 cyclins, likely by utilizing cyclin A2 [5]. Nevertheless, the loss of G1 cyclins attenuated the pluripotent characteristics of the stem cells. This result is reminiscent of previous findings that prolonged overexpression of CDK inhibitors results in the differentiation of human embryonic stem cells [6]. However, the ablation of all G1 cyclins by Liu et al illuminates that pluripotency is not primarily affected by a lack of proliferation but rather by a lack of G1 CDK activity, which they show directly phosphorylates pluripotency factors Nanog, Oct4, and Sox2 to promote their stability. In contrast to direct inhibition of CDKs by CDK inhibitor overexpression, cell-cycle arrest by quiescence-inducing signals (contact inhibition, serum starvation) suppresses terminal differentiation [7]. This contrast indicates unique signaling mechanisms present in quiescence to promote the stability of the pluripotency factors or inhibit differentiation transcriptional programs, and may imply that the mechanism of quiescence induction could dictate the sensitivity of cells toward terminal differentiation.
Before cells can exit quiescence, there must be a reversal of the quiescence-inducing signals, for example by the reintroduction of growth factor signaling or lowering of cell density. In classic work, Pardee tested the requirement for mitogen signaling in cell-cycle entry from a serum-starved, quiescent state by restimulating with mitogens for various amounts of time. A point termed the “restriction point” was identified where cells will progress through to division even if growth factors are subsequently removed [8]. Later, use of E2F transcriptional reporters demonstrated that the restriction point approximately coincides with the activation of E2F [9]. More recently, the effects of prolonged arrest in quiescence have been investigated in both contact inhibited or serum starved quiescence cells: cells that were arrested for progressively longer time periods require stronger growth factor stimulation and more time to reach the restriction point [10]. Moreover, cellular re-entry into the cell cycle from quiescence is heterogeneous, whereby individual cells exhibit significantly different rates of restarting the cell cycle. The application of live-cell sensors, including the development of CDK2, E2F, and APC/CCDH1 activity reporters, have been critical to observing the heterogeneous nature of cellular re-entry [9–15]. One factor that may contribute to this heterogeneity is memory of previous cell growth conditions at quiescence induction [16]. Other inherent properties of quiescence may also contribute to this heterogeneity, and more work is required to understand how and why quiescence exit is so distinct from re-entry into the cell cycle for continuously cycling cells.
While cells coming out of quiescence have a G1 restriction point, recent work using single-cell imaging has challenged whether this model is true in continuously cycling cells. Following mitosis, cells either enter a reversible, CDK2-activity low, mitogen-sensitive state or immediately begin to increase CDK2 activity, entering G1 in a mitogen-insensitive state [12]. The cells born with high CDK2 activity have largely committed to the cell cycle when tested by mitogen removal. Thus, rather than a restriction point in G1, rapidly cycling cells can commit to the cell cycle in the G2 phase of mother cells [17,18]. However, whether primary cells are subject to a G1 restriction point is still debated [17,18]. Although cells are no longer sensitive to mitogens past the restriction point, recent work has shown that acute cellular stress can still cause cell-cycle exit after the mitogen restriction point [15]. Treatment of cells with multiple stress-inducing conditions, including DNA damage, hyper-osmotic stress, and reactive oxygen stress, in combination with live cell imaging using the APC/CCDH1 activity reporter, identified that the molecular basis for the point of no return for stress sensitivity is the rapid, bi-stable inactivation of anaphase promoting complex (APC/CCDH1) [15,19]. APC/CCDH1 inactivation is mediated by a dual negative feedback loop between APC/CCDH1 and Emi1, an E2F target gene [20].
There are clear differences in the regulation of G1 in cells coming out of quiescence compared to continuously cycling cells. The delayed and heterogeneous cell-cycle entry from quiescence may function to ensure resources for continued cellular division before leaving the protection of the quiescent state. It also may function to control the rate of tissue replenishment. Multiple mechanisms could account for this delay, including reversal of the global changes (transcription, metabolism, or epigenetics) that occur in quiescent cells [7,21–23]. The requirement for significant changes in the expression of the cell-cycle machinery, including decreases in CDK inhibitors and increases in cyclins as well as the CDKs themselves, may also contribute to this delay upon quiescence exit. Future work interrogating the differences between cycling cells and cell-cycle re-entry from quiescence will help determine how these regulatory mechanisms underlie cellular proliferation.
DNA Replication Stress-Mediated Quiescence
In addition to external signaling cues, quiescence can be initiated by internal cellular stress. One form of endogenous cellular stress is the intrinsically error prone nature of DNA replication. Historically, the DNA damage response has been well characterized for exogenous stress, in which DNA damage has been shown to mediate cell-cycle exit through the stabilization of p53 and the upregulation of its transcriptional targets, including p21 [24]. Recent work, outlined in this section, has shown that this same pathway senses and responds to endogenous DNA replication stress [25–28], which is caused by stalled replication forks that fail to restart or collapse [29–31]. It has been estimated that one to two forks stall every S phase in unperturbed cells [32,33]. Thus, endogenous DNA replication stress significantly impacts cellular proliferation.
In unperturbed cycling cells, cells either immediately increase CDK activity and enter the next cell cycle following mitosis, or exit the cell cycle into a quiescent state, defined by low CDK activity and significantly higher levels of p21. Quiescence entry depends on p21 expression, as p21−/− cells rarely enter the CDK low state [12,34]. Using single-cell imaging techniques coupled to tools like endogenously tagged p53 or p21 cell lines and fluorescently-tagged 53BP1, recent work has demonstrated that endogenous DNA replication stress generates this heterogeneity in p21 levels [26–28]. Higher levels of p53 and p21 are observed in cells that enter quiescence compared to cells that continue to cycle. These increased levels correlate with increased markers for DNA damage, including 53BP1 and γH2AX.
While endogenous DNA damage during S phase results in increased p53, it is not until G2 of mother cells and G1 of daughter cells that heterogeneity in p21 levels is observed. Both modeling and experimental testing of the timing of DNA damaging drugs revealed that the sensitivity of cells to DNA stress weakens as cells progress through G1 [35]. Critical for tolerating and repairing replication stress, E2F transcriptional activity regulates a variety of checkpoint and replication repair proteins [36]. If not repaired, memory of DNA damage incurred in S phase of the mother cell is passed on to daughter cells, either through DNA damage signaling molecule p53 [28] or through persistence of the damage itself [26,27].
Endogenous DNA replication stress doesn’t solely dictate the proliferation versus quiescence decision. Rather, Rb phosphorylation is ultimately controlled by a stoichiometric competition between the induction of p21 by endogenous DNA damage stress and the induction of cyclin D controlled by mitogen signaling [28]. This stoichiometric competition converts two graded responses to DNA damage stress and mitogen signaling into an ultrasensitive activation of CDK4/6 [28,35]. Cells are maintained in quiescence as long as p21 levels exceed cyclin D1 levels. To activate CDK4/6 and exit quiescence, either the DNA damage is resolved, decreasing p21 levels, or mitogen signaling is strengthened, increasing cyclin D1 levels above p21 levels. While previous reports have identified positive roles for CDK inhibitors in the cell cycle (reviewed previously) [37], the recent stoichiometric competition observed implies that CDK4/6-cyclin D-p21 complexes are mostly inactive.
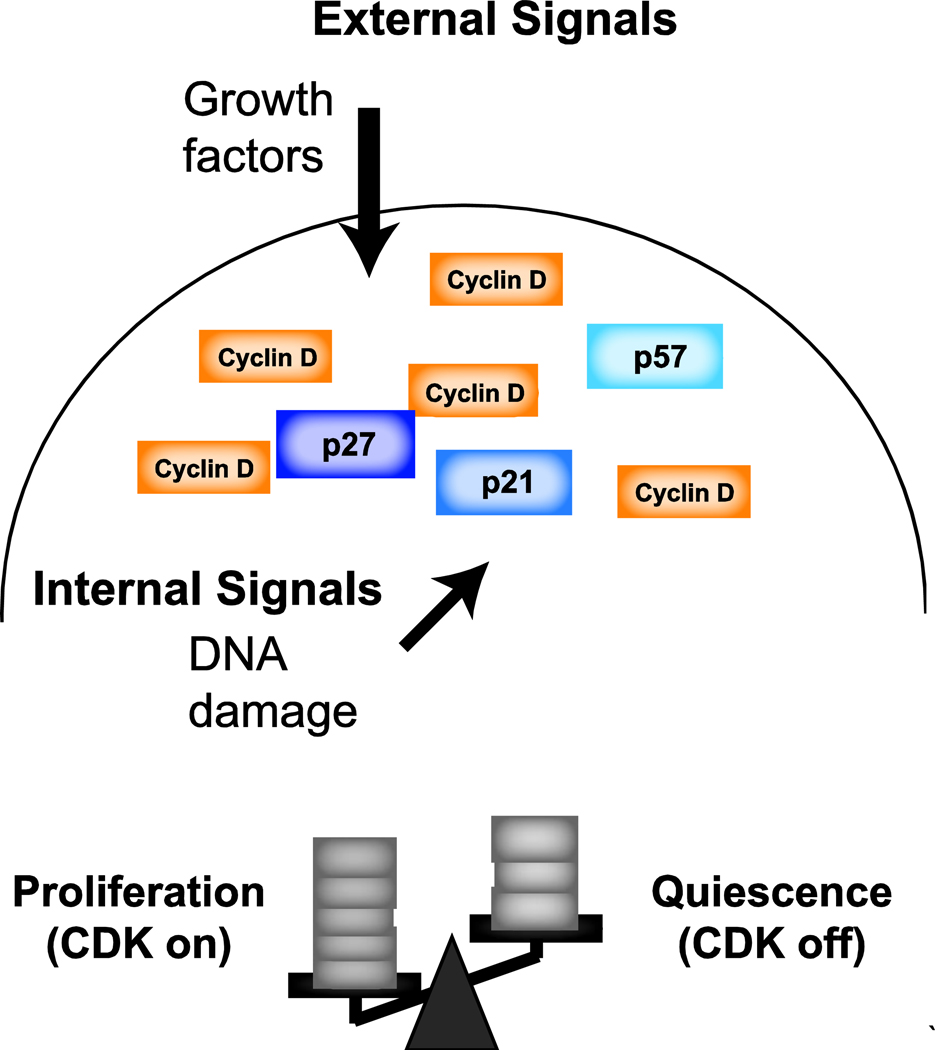
It has previously been reported that most cell types in p21-null, p27-null, and p21- p27- double knockout mice lack severe phenotypes [38]. Moreover, while p57 knock-out mice are inviable, mice in which p57 is replaced by p27 are viable and lack most developmental defects characteristic of p57-null mice [39]. These results contrast the model that any one CIP/KIP inhibitor is required in proliferation decisions. We suggest that cell-cycle entry decisions do not rely on a single cell-cycle inhibitor, but these decisions rely on the expression levels of all CDK inhibitors. An attractive model is that different pro- and anti-growth signals are integrated by their combined effects on the expression of CDK activators and inhibitors (Figure 2). We hypothesize that besides DNA damage, other extrinsic and intrinsic anti-growth signals could similarly induce transient quiescence. However, these signals may regulate cell-cycle entry through control of alternative cell-cycle regulators, not necessarily p21.
Figure 2: Model for Integration of Cell-Cycle Inputs.
External and internal signals regulate the core cell-cycle machinery, including cyclins and CDK inhibitors. While mitogenic signals are known to regulate cyclin D expression, recent work has shown that endogenous DNA damage leads to the upregulation of p21. Additionally, extrinsic and intrinsic growth signals contribute to cell-cycle decisions. We propose a model in which the cell cycle decisions rely on the combined effects of pro-and anti-growth signals and their integration by their combined effects on the expression of CDKs, cyclins, and CDK inhibitors. This model is consistent with an ultrasensitive activation of CDKs.
Mechanisms of cell-cycle arrest in senescence
The senescence phenotype is heterogeneous, with many described stimuli, changes to cellular physiology, and tissue/organismal effects that are beyond the scope of this review but are covered in detail in many excellent recent articles [1,40,41]. An extensive number of stimuli have been reported to trigger cellular senescence: oncogene activation [42,43], ionizing radiation and other DNA damaging agents, mitochondrial dysfunction [44,45], chromatin alterations [46–48], and impairment of ribosome biogenesis [49,50], for example. However, a fundamental trend does appear to link together these diverse stimuli: a potent cellular stressor induces persistent damage signaling, causing permanent cell-cycle arrest.
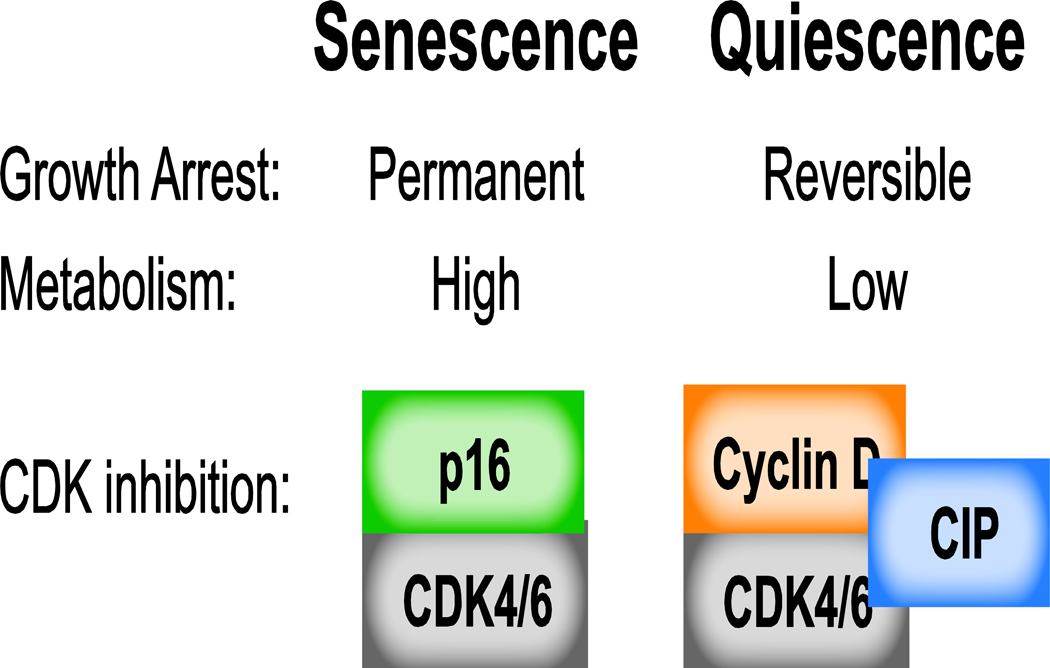
Activation of both the DNA damage response p53-p21 pathway and the Rb cell-cycle inhibitory pathway is a long-recognized theme of senescence. As in several forms of quiescence, activation of the DNA damage response pathway is the initial stimulus triggering cell-cycle arrest. However, senescence appears to be unique from quiescence in the persistence of the DNA damage response: while the DNA damage response (DDR) is often transient in quiescence, recent evidence suggests that the DDR is persistently activated in senescence [51–53]. In both senescence and quiescence, upregulation of p21 enables robust cell-cycle exit via CDK4/6 inhibition. Unique to senescence is an eventual decline in p21 levels several days following the senescence-inducing stimulus combined with a concomitant increase in p16 protein levels, presumably to maintain CDK4/6 inactivation [46,54,55]. Extremely high affinity binding of p16 to CDK4/6 ensures suppression of CDK4/6 activity and contributes to the irreversible nature of cell-cycle arrest in senescence (Figure 3) [56,57]. Interestingly, in contrast to quiescence, G1 cyclin levels frequently remain high in senescent cells, though the significance of this is unknown.
Figure 3: Comparing Senescence and Quiescence.
Contrary to the reversible nature of quiescence, senescence is a permanent growth arrest. Fundamental differences between these two types of growth arrest include continued high metabolism in senescent cells compared to the low metabolic state of quiescent cells. Moreover, senescent cells rely on p16 to inhibit CDK4/6, while quiescent cells utilize the CIP/KIP inhibitors to bind and inhibit CDK cyclin complexes.
Recently, numerous other changes to cellular physiology have been discovered. Changes to cellular chromatin architecture help to reinforce the senescent state. Senescence-associated heterochromatic foci transcriptionally silence E2F target genes, decreasing cellular sensitivity to mitogenic stimuli [58,59]. Recent work employing Hi-C to investigate genome architecture demonstrated that senescent cells undergo broad changes in global chromatin organization [60,61]. This change appears to be due in part to histone and nuclear lamin depletion [62–64], as well as decreased tethering of chromatin to the nuclear lamin [65,66]. Alterations in chromatin dynamics may contribute to increased transcription of inflammatory cytokines in senescent cells [67,68]. Several recent studies employing both cellular and mouse models of radiation- or oncogene-induced senescence demonstrated that cytoplasmic chromatin fragments accumulate in senescent cells, activating the pro-inflammatory cGAS-STING pathway and contributing to a pro-inflammatory phenotype [69–71]. Secretion of inflammatory proteins, termed the senescence associated secretory phenotype (SASP), functions as a pro-senescence stimulus in both an autocrine and paracrine manner [1]. cGAS activation and the SASP phenotype appear to be unique to senescence and not found in quiescent cells. Moreover, while quiescent cells reside in a state of low metabolic activity, senescent cells remain metabolically active. However, metabolic profiling and functional studies of senescent cells indicate that senescent cell metabolism differs significantly from proliferating cells [45,72,73]. These multitude of changes to cellular physiology appear to be important in reinforcing the growtharrest of senescent cells.
Although senescence is generally considered to be an irreversible cell-cycle arrest, several studies have suggested mechanisms to alter the fate of senescent cells. It was previously known that acute genetic deletion of Rb and Rb-related proteins induces cell-cycle re-entry in fibroblasts [74]. Inactivation of p53 was also shown to promote senescence escape, highlighting the role for a persistent DNA damage response in senescence maintenance [75,76]. More recent work has demonstrated that senescent cells exist on the verge of apoptosis, spurring efforts to pharmacologically eliminate senescent cells by triggering the apoptotic pathway [54,77,78]. Finally, genetically reprogramming senescent cells through an iPSC state has recently been demonstrated to ameliorate features of senescence and aging both in vitro and in vivo, suggesting that increased stemness may be a route out of the senescent cell state [79,80]. Future work identifying mechanisms of cellular rejuvenation and cell-cycle re-entry will provide greater insight into the mechanisms of senescence maintenance and may ultimately have therapeutic potential.
Conclusions
The similarities between quiescence and the first several days following a senescence-inducing stimulus suggest that there may be a critical cellular decision window during which a cell can either enter a temporary quiescent state or initiate permanent cell-cycle exit via a senescence program. Moreover, important factors have been identified that are critical in protecting quiescent cells from senescence and differentiation [81]. Future work will focus on identifying the core decision-making machinery and the signals that a cell integrates when making this decision. Moreover, to date, the majority of studies of cellular senescence have examined phenotypes at the population level. Future studies examining cell-to-cell heterogeneity within a senescent population will help to unravel the complexities underlying senescence.
While the stimuli and mechanisms of cell-cycle arrest are diverse, common principles have emerged. Essential to any form of arrest is inhibition of cyclin-dependent-kinase activity, especially the G1 kinases CDK4 and CDK6, which play a critical role in promoting cell-cycle entry. Control over CDK activity therefore appears to be the ultimate determinant of cell-cycle arrest. Future work uncovering additional mechanisms of CDK regulation, and how these mechanisms dictate which form of cell-cycle arrest cells enter, will continue to advance the field.
Acknowledgements
We would like to thank Chad Liu and Yilin Fan and other members of the Meyer Lab for their insights and thoughtful comments during the preparation of this article. L.R.P. was supported by National Institute of General Medical Sciences of the National Institutes of Health F32 Ruth L. Kirschstein fellowship F32GM125246. L.H.D. was supported by NIH training grant T32GM007365 and National Institute of Aging of the NIH F32 Ruth L. Kirschstein fellowship F30AG060634. T.M. was supported by NIH grants GM118377, GM030179, and P50GM107615.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hernandez-Segura A, Nehme J, Demaria M: Hallmarks of Cellular Senescence. Trends Cell Biol 2018, 28:436–453. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DG, Walker CL: Cyclins and cell cycle checkpoints. Annu Rev Pharmacol Toxicol 1999, 39:295–312. [DOI] [PubMed] [Google Scholar]
- 3.Massague J: G1 cell-cycle control and cancer. Nature 2004, 432:298–306. [DOI] [PubMed] [Google Scholar]
- 4.Sherr CJ, Roberts JM: Living with or without cyclins and cyclin-dependent kinases. Genes Dev 2004, 18:2699–2711. [DOI] [PubMed] [Google Scholar]
- 5.Liu L, Michowski W, Inuzuka H, Shimizu K, Nihira NT, Chick JM, Li N, Geng Y, Meng AY, Ordureau A, et al. : G1 cyclins link proliferation, pluripotency and differentiation of embryonic stem cells. Nat Cell Biol 2017, 19:177–188.**Embryonic stem cells continue to proliferate in the absence of all G1 cyclins, but have compromised pluripotent characteristics.
- 6.Ruiz S, Panopoulos AD, Herrerias A, Bissig KD, Lutz M, Berggren WT, Verma IM, Izpisua Belmonte JC: A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr Biol 2011, 21:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coller HA, Sang L, Roberts JM: A new description of cellular quiescence. PLoS Biol 2006, 4:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardee AB: A restriction point for control of normal animal cell proliferation. Proc Natl Acad Sci U S A 1974, 71:1286–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao G, Lee TJ, Mori S, Nevins JR, You L: A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol 2008, 10:476–482. [DOI] [PubMed] [Google Scholar]
- 10.Kwon JS, Everetts NJ, Wang X, Wang W, Della Croce K, Xing J, Yao G: Controlling Depth of Cellular Quiescence by an Rb-E2F Network Switch. Cell Rep 2017, 20:3223–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn AT, Jones JT, Meyer T: Quantitative analysis of cell cycle phase durations and PC12 differentiation using fluorescent biosensors. Cell Cycle 2009, 8:1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spencer SL, Cappell SD, Tsai FC, Overton KW, Wang CL, Meyer T: The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 2013, 155:369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong P, Zhang C, Parker BT, You L, Mathey-Prevot B: Cyclin D/CDK4/6 activity controls G1 length in mammalian cells. PLoS One 2018, 13:e0185637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. : Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008, 132:487–498. [DOI] [PubMed] [Google Scholar]
- 15.Cappell SD, Chung M, Jaimovich A, Spencer SL, Meyer T: Irreversible APC(Cdh1) Inactivation Underlies the Point of No Return for Cell-Cycle Entry. Cell 2016, 166:167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Fujimaki K, Mitchell GC, Kwon JS, Della Croce K, Langsdorf C, Zhang HH, Yao: Exit from quiescence displays a memory of cell growth and division. Nat Commun 2017, 8:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moser J, Miller I, Carter D, Spencer SL: Control of the Restriction Point by Rb and p21. Proc Natl Acad Sci U S A 2018, 115:E8219–E8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz C, Johnson A, Koivomagi M, Zatulovskiy E, Kravitz CJ, Doncic A, Skotheim JM: A Precise Cdk Activity Threshold Determines Passage through the Restriction Point. Mol Cell 2018, 69:253–264 e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barr AR, Heldt FS, Zhang T, Bakal C, Novak B: A Dynamical Framework for the All-or-None G1/S Transition. Cell Syst 2016, 2:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cappell SD, Mark KG, Garbett D, Pack LR, Rape M, Meyer T: EMI1 switches from being a substrate to an inhibitor of APC/C(CDH1) to start the cell cycle. Nature 2018, 558:313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evertts AG, Manning AL, Wang X, Dyson NJ, Garcia BA, Coller HA: H4K20 methylation regulates quiescence and chromatin compaction. Mol Biol Cell 2013, 24:3025–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kieffer-Kwon KR, Nimura K, Rao SSP, Xu J, Jung S, Pekowska A, Dose M, Stevens E, Mathe E, Dong P, et al. : Myc Regulates Chromatin Decompaction and Nuclear Architecture during B Cell Activation. Mol Cell 2017, 67:566–578 e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HJ, Jedrychowski MP, Vinayagam A, Wu N, Shyh-Chang N, Hu Y, Min-Wen C, Moore JK, Asara JM, Lyssiotis CA, et al. : Proteomic and Metabolomic Characterization of a Mammalian Cellular Transition from Quiescence to Proliferation. Cell Rep 2017, 20:721–736.*This study compares the proteomic and metabolomics changes of pro-B lymphocytes transitioning from quiescence to proliferation. Their comparisons identify an upregulation of glycolysis, lipid metabolism, and amino acid and nucleotide synthesis and a down regulation of oxidative phosphorylation in proliferating compared to quiescent cells.
- 24.Fei P, El-Deiry WS: P53 and radiation responses. Oncogene 2003, 22:5774–5783. [DOI] [PubMed] [Google Scholar]
- 25.Daigh LH, Liu C, Chung M, Cimprich KA, Meyer T: Stochastic Endogenous Replication Stress Causes ATR-Triggered Fluctuations in CDK2 Activity that Dynamically Adjust Global DNA Synthesis Rates. Cell Syst 2018, 7:17–27 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arora M, Moser J, Phadke H, Basha AA, Spencer SL: Endogenous Replication Stress in Mother Cells Leads to Quiescence of Daughter Cells. Cell Rep 2017, 19:1351–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barr AR, Cooper S, Heldt FS, Butera F, Stoy H, Mansfeld J, Novak B, Bakal C: DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat Commun 2017, 8:14728.*Using live single cell measurements of p21 protein, Barr et al demonstrate that endogenous DNA damage replication stress in S phase causes accumulation of p21 in G2 of mother cells and G1 of daughter cells, which can lead to G1 arrest.
- 28.Yang HW, Chung M, Kudo T, Meyer T: Competing memories of mitogen and p53 signalling control cell-cycle entry. Nature 2017, 549:404–408.*Cell-cycle entry is ultimately dictated by a stoichiometric competition between the induction of p21 by endogenous DNA damage stress and the induction of cyclin D controlled by mitogen signaling
- 29.Saldivar JC, Cortez D, Cimprich KA: The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol 2017, 18:622–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA: Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 2005, 19:1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, Vindigni A, Lopes M: Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol 2015, 208:563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al Mamun M, Albergante L, Moreno A, Carrington JT, Blow JJ, Newman TJ: Inevitability and containment of replication errors for eukaryotic genome lengths spanning megabase to gigabase. Proc Natl Acad Sci U S A 2016, 113:E5765–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreno A, Carrington JT, Albergante L, Al Mamun M, Haagensen EJ, Komseli ES, Gorgoulis VG, Newman TJ, Blow JJ: Unreplicated DNA remaining from unperturbed S phases passes through mitosis for resolution in daughter cells. Proc Natl Acad Sci U S A 2016, 113:E5757–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overton KW, Spencer SL, Noderer WL, Meyer T, Wang CL: Basal p21 controls population heterogeneity in cycling and quiescent cell cycle states. Proc Natl Acad Sci U S A 2014, 111:E4386–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heldt FS, Barr AR, Cooper S, Bakal C, Novak B: A comprehensive model for the proliferation-quiescence decision in response to endogenous DNA damage in human cells. Proc Natl Acad Sci U S A 2018, 115:2532–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertoli C, Herlihy AE, Pennycook BR, Kriston-Vizi J, de Bruin RAM: Sustained E2F-Dependent Transcription Is a Key Mechanism to Prevent Replication-Stress-Induced DNA Damage. Cell Rep 2016, 15:1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherr CJ, Roberts JM: CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 1999, 13:1501–1512. [DOI] [PubMed] [Google Scholar]
- 38.Nakayama K, Nakayama K: Cip/Kip cyclin-dependent kinase inhibitors: brakes of the cell cycle engine during development. Bioessays 1998, 20:1020–1029. [DOI] [PubMed] [Google Scholar]
- 39.Susaki E, Nakayama K, Yamasaki L, Nakayama KI: Common and specific roles of the related CDK inhibitors p27 and p57 revealed by a knock-in mouse model. Proc Natl Acad Sci U S A 2009, 106:5192–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munoz-Espin D, Serrano M: Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol 2014, 15:482–496. [DOI] [PubMed] [Google Scholar]
- 41.Sharpless NE, Sherr CJ: Forging a signature of in vivo senescence. Nat Rev Cancer 2015, 15:397–408. [DOI] [PubMed] [Google Scholar]
- 42.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA: Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 2005, 436:660–665. [DOI] [PubMed] [Google Scholar]
- 43.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. : Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006, 444:638–642. [DOI] [PubMed] [Google Scholar]
- 44.Tezze C, Romanello V, Desbats MA, Fadini GP, Albiero M, Favaro G, Ciciliot S, Soriano ME, Morbidoni V, Cerqua C, et al. : Age-Associated Loss of OPA1 in Muscle Impacts Muscle Mass, Metabolic Homeostasis, Systemic Inflammation, and Epithelial Senescence. Cell Metab 2017, 25:1374–1389 e1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiley CD, Velarde MC, Lecot P, Liu S, Sarnoski EA, Freund A, Shirakawa K, Lim HW, Davis SS, Ramanathan A, et al. : Mitochondrial Dysfunction Induces Senescence with a Distinct Secretory Phenotype. Cell Metab 2016, 23:303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito T, Teo YV, Evans SA, Neretti N, Sedivy JM: Regulation of Cellular Senescence by Polycomb Chromatin Modifiers through Distinct DNA Damage- and Histone Methylation-Dependent Pathways. Cell Rep 2018, 22:3480–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rapisarda V, Borghesan M, Miguela V, Encheva V, Snijders AP, Lujambio A, O’Loghlen A: Integrin Beta 3 Regulates Cellular Senescence by Activating the TGF-beta Pathway. Cell Rep 2017, 18:2480–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sen P, Lan Y, Li CY, Sidoli S, Donahue G, Dou Z, Frederick B, Chen Q, Luense LJ, Garcia BA, et al. : Histone Acetyltransferase p300 Induces De Novo Super-Enhancers to Drive Cellular Senescence. Mol Cell 2019, 73:684–698 e688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lessard F, Igelmann S, Trahan C, Huot G, Saint-Germain E, Mignacca L, Del Toro N, Lopes-Paciencia S, Le Calve B, Montero M, et al. : Senescence-associated ribosome biogenesis defects contributes to cell cycle arrest through the Rb pathway. Nat Cell Biol 2018, 20:789–799. [DOI] [PubMed] [Google Scholar]
- 50.Nishimura K, Kumazawa T, Kuroda T, Katagiri N, Tsuchiya M, Goto N, Furumai R, Murayama A, Yanagisawa J, Kimura K: Perturbation of ribosome biogenesis drives cells into senescence through 5S RNP-mediated p53 activation. Cell Rep 2015, 10:1310–1323. [DOI] [PubMed] [Google Scholar]
- 51.Feringa FM, Raaijmakers JA, Hadders MA, Vaarting C, Macurek L, Heitink L, Krenning L, Medema RH: Persistent repair intermediates induce senescence. Nat Commun 2018, 9:3923.*This study provided evidence that unrepaired DNA breaks causing persistent DNA damage response activation can induce senescence
- 52.Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, Kaplunov JM, Bucci G, Dobreva M, Matti V, Beausejour CM, et al. : Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol 2012, 14:355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hewitt G, Jurk D, Marques FD, Correia-Melo C, Hardy T, Gackowska A, Anderson R, Taschuk M, Mann J, Passos JF: Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat Commun 2012, 3:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baar MP, Brandt RMC, Putavet DA, Klein JDD, Derks KWJ, Bourgeois BRM, Stryeck S, Rijksen Y, van Willigenburg H, Feijtel DA, et al. : Targeted Apoptosis of Senescent Cells Restores Tissue Homeostasis in Response to Chemotoxicity and Aging. Cell 2017, 169:132–147 e116.**The authors demonstrated that p53 sequestration by FOXO4 prevents apoptosis following induction of senescence by ionizing radiation. Designed a peptide to block the FOXO4-p53 interaction, leading to increased senescent cell apoptosis and improved aging-related phenotypes in naturally-aged mice following peptide treatment.
- 55.Stein GH, Drullinger LF, Soulard A, Dulic V: Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol Cell Biol 1999, 19:2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brenner AJ, Stampfer MR, Aldaz CM: Increased p16 expression with first senescence arrest in human mammary epithelial cells and extended growth capacity with p16 inactivation. Oncogene 1998, 17:199–205. [DOI] [PubMed] [Google Scholar]
- 57.Hallett ST, Pastok MW, Morgan RML, Wittner A, Blundell K, Felletar I, Wedge SR, Prodromou C, Noble MEM, Pearl LH, et al. : Differential Regulation of G1 CDK Complexes by the Hsp90-Cdc37 Chaperone System. Cell Rep 2017, 21:1386–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Narita M, Narita M, Krizhanovsky V, Nunez S, Chicas A, Hearn SA, Myers MP, Lowe SW: A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 2006, 126:503–514. [DOI] [PubMed] [Google Scholar]
- 59.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW: Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 2003, 113:703–716. [DOI] [PubMed] [Google Scholar]
- 60.Criscione SW, De Cecco M, Siranosian B, Zhang Y, Kreiling JA, Sedivy JM, Neretti N: Reorganization of chromosome architecture in replicative cellular senescence. Sci Adv 2016, 2:e1500882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sen P, Shah PP, Nativio R, Berger SL: Epigenetic Mechanisms of Longevity and Aging. Cell 2016, 166:822–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Funayama R, Saito M, Tanobe H, Ishikawa F: Loss of linker histone H1 in cellular senescence. J Cell Biol 2006, 175:869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, et al. : Lysosome-mediated processing of chromatin in senescence. J Cell Biol 2013, 202:129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J: Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol 2010, 17:1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sadaie M, Salama R, Carroll T, Tomimatsu K, Chandra T, Young AR, Narita M, Perez-Mancera PA, Bennett DC, Chong H, et al. : Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev 2013, 27:1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. : Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 2013, 27:1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Capell BC, Drake AM, Zhu J, Shah PP, Dou Z, Dorsey J, Simola DF, Donahue G, Sammons M, Rai TS, et al. : MLL1 is essential for the senescence-associated secretory phenotype. Genes Dev 2016, 30:321–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tasdemir N, Banito A, Roe JS, Alonso-Curbelo D, Camiolo M, Tschaharganeh DF, Huang CH, Aksoy O, Bolden JE, Chen CC, et al. : BRD4 Connects Enhancer Remodeling to Senescence Immune Surveillance. Cancer Discov 2016, 6:612–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dou Z, Ghosh K, Vizioli MG, Zhu J, Sen P, Wangensteen KJ, Simithy J, Lan Y, Lin Y, Zhou Z, et al. : Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 2017, 550:402–406.**This study provided compelling evidence that senescent cells accumulate cytoplasmic chromatin fragments, activating the cGAS-STING pathway and ultimately promoting inflammation via the senescence associated secretory phenotype.
- 70.Gluck S, Guey B, Gulen MF, Wolter K, Kang TW, Schmacke NA, Bridgeman A, Rehwinkel J, Zender L, Ablasser A: Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol 2017, 19:1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang H, Wang H, Ren J, Chen Q, Chen ZJ: cGAS is essential for cellular senescence. Proc Natl Acad Sci U S A 2017, 114:E4612–E4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.James EL, Michalek RD, Pitiyage GN, de Castro AM, Vignola KS, Jones J, Mohney RP, Karoly ED, Prime SS, Parkinson EK: Senescent human fibroblasts show increased glycolysis and redox homeostasis with extracellular metabolomes that overlap with those of irreparable DNA damage, aging, and disease. J Proteome Res 2015, 14:1854–1871. [DOI] [PubMed] [Google Scholar]
- 73.Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, van der Burg SH, Verdegaal EM, Cascante M, Shlomi T, et al. : A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 2013, 498:109–112. [DOI] [PubMed] [Google Scholar]
- 74.Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T: Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 2003, 424:223–228. [DOI] [PubMed] [Google Scholar]
- 75.Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, Campisi J: Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J 2003, 22:4212–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Milanovic M, Fan DNY, Belenki D, Dabritz JHM, Zhao Z, Yu Y, Dorr JR, Dimitrova L, Lenze D, Monteiro Barbosa IA, et al. : Senescence-associated reprogramming promotes cancer stemness. Nature 2018, 553:96–100. [DOI] [PubMed] [Google Scholar]
- 77.Chang J, Wang Y, Shao L, Laberge RM, Demaria M, Campisi J, Janakiraman K, Sharpless NE, Ding S, Feng W, et al. : Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat Med 2016, 22:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu Y, Tchkonia T, Pirtskhalava T, Gower AC, Ding H, Giorgadze N, Palmer AK, Ikeno Y, Hubbard GB, Lenburg M, et al. : The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell 2015, 14:644–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lapasset L, Milhavet O, Prieur A, Besnard E, Babled A, Ait-Hamou N, Leschik J, Pellestor F, Ramirez JM, De Vos J, et al. : Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev 2011, 25:2248–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ocampo A, Reddy P, Martinez-Redondo P, Platero-Luengo A, Hatanaka F, Hishida T, Li M, Lam D, Kurita M, Beyret E, et al. : In Vivo Amelioration of Age-Associated Hallmarks by Partial Reprogramming. Cell 2016, 167: 1719–1733 e1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sang L, Coller HA, Roberts JM: Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 2008, 321:1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]