Abstract
Aim: Isocitrate dehydrogenase 1 (IDH1) is key enzyme involved in cellular metabolism and DNA repair. Mutations in IDH1 occur in up to 25% of cholangiocarcinomas. The present study aimed to explore the features of cellosaurus REB cells with mutant and wide-type IDH1.
Methods: To compare the features of IDH1 knockout and mutation in cholangiocarcinoma, we firstly constructed the IDH1 knockout and IDH1 mutation cell lines. We then evaluated the viability of these cell lines using the cell count assay and MTT assay. Next, we determined cell migration and invasion using the Transwell assay. Additionally, to evaluate the effects of IDH1 on cellular metabolism, the levels of α-ketoglutarate (α-KG) and nicotinamide adenine dinucleotide phosphate (NADPH) were determined using enzyme-linked immunosorbent assay. We then applied ChIPbase dataset to explore the genes that were regulated by IDH1.
Results: High frequency of mutated IDH1 was observed in the cholangiocarcinoma and IDH1 R132C was presented in more than 80% of mutations. The results showed that IDH1 knockout decreased cell proliferation, migration and invasion, whereas the overexpression of IDH1 in IDH1 knockout cell line recovered its proliferation, migration and invasion capacities. Additionally, IDH1 mutation reduced the levels of NADPH and α-KG. Furthermore, investigation into the underlying mechanisms revealed that IDH1 overexpression induced the expression of aldehyde dehydrogenase 1 thereby promoting cell proliferation, migration and invasion.
Conclusion: IDH1 plays an important role in cholangiocarcinoma and its mutation impairs tumor progression in part by inhibition of isocitrate metabolism.
Keywords: cholangiocarcinoma, isocitrate dehydrogenase 1, aldehyde dehydrogenase 1, α-ketoglutarate, NADPH
Introduction
Cholangiocarcinoma is an adenocarcinoma of epithelial cells that occurs in any areas of bile duct (1). Although it is a rare type of cancer, its occurrence has increased in the past three decades (2). Cholangiocarcinoma can be classified into intrahepatic, perihilar and distal types based on their clinical features. However, surgery and liver transplantation are the recommended therapeutic options for all types of cholangiocarcinoma in a minority of patients (1, 3). The 5-years survival rate is 15% for patients with early-stage intrahepatic cholangiocarcinoma. However, if the intrahepatic cholangiocarcinoma has spread to a distant part of the body, the 5-years survival rate is reduced to only 2% (4). Many patients are diagnosed with cholangiocarcinoma in an advanced stage. Due to the fact that surgery cannot completely remove metastatic tumors, additional treatment is recommended (2, 5).
Genetic alternations are thought to play a critical role in the occurrence and development of cholangiocarcinoma (2, 6, 7). Mutations in several oncogenes and tumor suppressor genes have been reported to be involved in the carcinogenesis of cholangiocarcinoma (6). For instance, activated KRAS has been reported to be associated with the development of cholangiocarcinoma (8). Inactivated p53, a tumor suppressor gene, is observed in the tumor tissue of cholangiocarcinoma (8, 9). In addition to those genes, other genes including SMAD4, PTEN, EGFR, and PDGFB have also been described to be associated with the occurrence and development of cholangiocarcinoma (10).
Isocitrate dehydrogenase 1 (IDH1) is an enzyme encoded by IDH1, which is responsible for catalyzing isocitrate to produce α-ketoglutarate (α-KG) and generating nicotinamide adenine dinucleotide phosphate (NADPH) (11, 12). α-KG and NADPH are components of the tricarboxylic acid (TCA) cycle, which are thought to detoxify against oxidative stress (12, 13). Therefore, IDH1 indirectly regulates oxidative damage. Additionally, IDH1 has also been reported to play an important role in the process of glucose and lipid metabolism (14). Recently, alternations of IDH1 have been implicated in many types of cancer (11). In 2008, for the first time, Parsons and colleagues have demonstrated IDH1 mutations in the human genome related to the glioblastoma multiforme (15). The following studies performed by other groups have further revealed that mutations in IDH1 are associated with leukemia, colon cancer and prostate cancer (11, 12, 14). In 2012, Borger and colleagues have revealed IDH1 mutations in cholangiocarcinoma (16). Interestingly, mutations in IDH1 have been frequently observed in poorly differentiated tumors (16). These results support that IDH1 might be used as a potential biomarker for the detection of cholangiocarcinoma. In 2018, Khurshed et al. have reported that mutations in IDH1 are associated with improved response to irradiation and chemotherapy in colon carcinoma and glioblastoma cells (17). More recently, they found that IDH mutation in gliomas depended on lactate and the neurotransmitter glutamate as metabolic substrates to rescue cells from the metabolic stress (18). These results suggested that IDH mutation might affect tumor progression by regulating metabolic stress. In the present study, we aimed to explore the effects of IDH1 mutation on cholangiocarcinoma. Furthermore, we revealed the mechanisms of IDH1 mutation underlying the tumor progression of cholangiocarcinoma.
Materials and Methods
Cell Line and Cell Viabilities
Cholangiocarcinoma RBE cell line was purchased from the First Affiliated Hospital of Anhui Medical University and cultured in complete Dulbecco modified eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% antibiotics under 37°C in the presence of 5% CO2 at constant humidity.
Cell viability of RBE cell line and RBE IDH1 knockout or mutation cells was determined using the MTT assay and cell count assay. For MTT assay, an MTT solution (Sigma, St. Louis, MO, USA) was added into each well and the plate was incubated at 37°C. After 4 h, DMSO solution was added and the optical density was read at 570 nm using a microplate reader (Molecular Devices, Sunnyvale, CA, United States). For cell count assay, trypan blue staining solution was added to the cells and then the cell viabilities were calculated by counting live and dead cells.
Construction of IDH1 Knockout and IDH1 Mutation Cell Line
The IDH1 knockout (IDH1 KO) cell line was constructed using CRISPR-Cas9 (Shanghai Liangtai Biotech Company, Shanghai, China). In brief, when the IDH1 cells reached 70% confluency, the cells were transfected with CRISPR-Cas9 knockout plasmids containing guide RNA sequence of IDH1 and sequence of Cas9 protein.
The IDH1 R132C mutation cell line was constructed by transfecting the IDH1 KO cell line with IDH1 R132C mutation plasmids.
Cell Invasion and Migration Assays
Cell invasion and migration assays were performed according to previously reported methods (19, 20). Transwell chamber consisted of a membrane filter coated with Matrigel was used in this study. In brief, the cells were detached using trypsin and then resuspended in serum-free DMEM medium. The cells were then seeded into the upper chamber and the complete DMEM medium was added into the lower chamber. The invaded cells which located in lower chamber were counted after incubation for 12 h. Crystal violet solution was used to stain the cells and then a microscope was used to count the cells.
Detection of α-KG and NADPH
The levels of α-KG and NADPH were determined using relevant qualification kits, according to the documents of the manufacturer (BioVision, Milpitas, CA, United States).
Xenograft Mouse Model
Male BALB/c nude mice (6 weeks old) were purchased from Guangzhou Laboratory Animal Center (Guangzhou, China). The mice were housed in a room on a 12-h light-dark cycle and fed under experimental conditions with a temperature of 22–24°C and humidity of 50 ± 5%. Animal procedures used in this study were approved by the First Affiliated Hospital of Anhui Medical University' Animal Care and Use Committee.
After 1 week acclimatization, the mice were subcutaneously injected with 2 × 106 REB IDH1 KO cells, or REB IDH1 KO cells transfected with wild-type IDH1 or IDH1 R132C mutation plasmids. After 1 week, the tumor volume was measured every 3 days, according to the formula: length × width2 × 0.52.
Immunohistochemistry
After the mice were sacrificed, the tumor tissue was removed and washed with cold PBS, then fixed in 4% paraformaldehyde. The tissues were embedded into paraffin and the sections were then cut at 4-μm thickness. Ki67 (KeyGEN BioTECH, Nanjing, China) staining was performed according to a previously reported method (21) and the sections were then observed under microscopy.
Quantitative Polymerase Chain Reaction (qPCR)
Trizol reagent was used to extract total RNA from the cells, according to the instructions of the manufacturer (TaKaRa, Dalian, China). Isolation of total RNA was performed under sterile condition. To remove DNA contamination, RNase-free DNase I was used. Primers for CDH1, TJP1, VIM, FN1, Snail, Twist, ZEB1, and β-actin were used for amplification of these genes. To analyze the accuracy of the PCR reaction, the melt curves were used. To evaluate the expressions of genes, 2−ΔΔCt values were calculated. The mRNA expression values of CDH1, TJP1, VIM, FN1, Snail, Twist, and ZEB1 were normalized to that of β-actin.
Western Blotting
Extraction and qualification of proteins were performed according to a previously reported method (22). Radioimmunoprecipitation buffer was used to lysis the cells. Next, the lysate was centrifuged at 13,000 g to remove insoluble materials. The concentrations of protein were qualified using the BCA protein assay kit (Beyotime, Jiangsu, China).
Protein samples were loaded at equal amounts for each sample and 10% sodium dodecyl sulfate (SDS) gel was used to separate the samples. Next, the SDS gel was then transferred to a polyvinylidene fluoride membrane and 5% non-fat milk was used to block the membrane. The membrane was probed with primary antibody followed by an appropriated secondary antibody. An imaging system (Bio-Rad, CA, USA) was used to examine chemiluminescence. Expression values of protein were normalized to that of β-actin.
Statistical Analysis
The TCGA cholangiocarcinoma dataset (MSK, Clin Cancer Res 2018) was downloaded and analyzed in cBioportal (www.cbioportal.org). SPSS 13.0 (SPSS, Chicago, IL, USA) was used to statistical analysis. All Data were expressed as mean ± standard deviation (S.D.). To evaluate the significance, one-way analysis of variance with multiple comparisons and Student-Newman-Keuls (SNK) test were performed. A P < 0.05 was considered as a statistical significance.
Results
IDH1 Mutation and Frequency in Cholangiocarcinoma
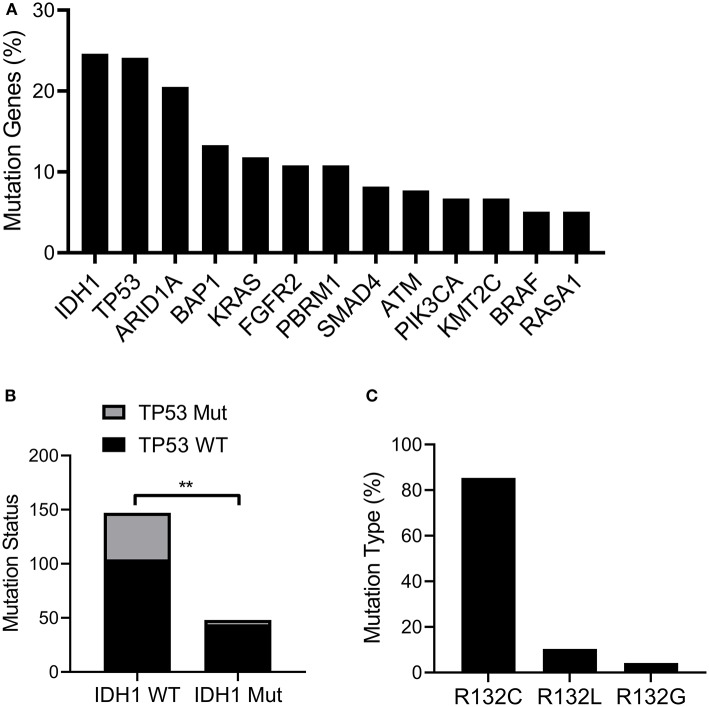
First, we explored the frequency of the mutated genes in cholangiocarcinoma. The results indicated that IDH1 was mutated at the highest frequency among all mutated genes including TP53, ARID1A, BAP1, and KRAS (Figure 1A). We also noticed that mutated TP53 also appeared at a high frequency. It is known that TP53 mutations are frequently observed in the occurrence and development of many types of cancer and are implicated to be an important risk factor for tumorigenesis (23). Therefore, we further analyzed the frequency of mutated TP53 in patients with wild-type (WT) or mutated IDH. Interestingly, we found mutated TP53 with higher frequency in patients with WT IDH than patients with mutated IDH (Figure 1B). Additionally, we also detected the proportions of different mutation types of IDH1. The results showed IDH1 R132C was presented in more than 80% of mutation types (Figure 1C).
Figure 1.
IDH1 was mutated in cholangiocarcinoma. The TCGA cholangiocarcinoma dataset (MSK, Clin Cancer Res 2018) was downloaded and analyzed in cBioportal (www.cbioportal.org). (A) The frequency of mutated genes in Cholangiocarcinoma. (B) The proportion of TP53 mutated patients in IDH1 wildtype (WT) or mutated group. (C) The proportion of IDH1 with different mutation types. The data were represented as mean ± S.D. **P < 0.01.
IDH1 Promoted Cell Proliferation of Cholangiocarcinoma
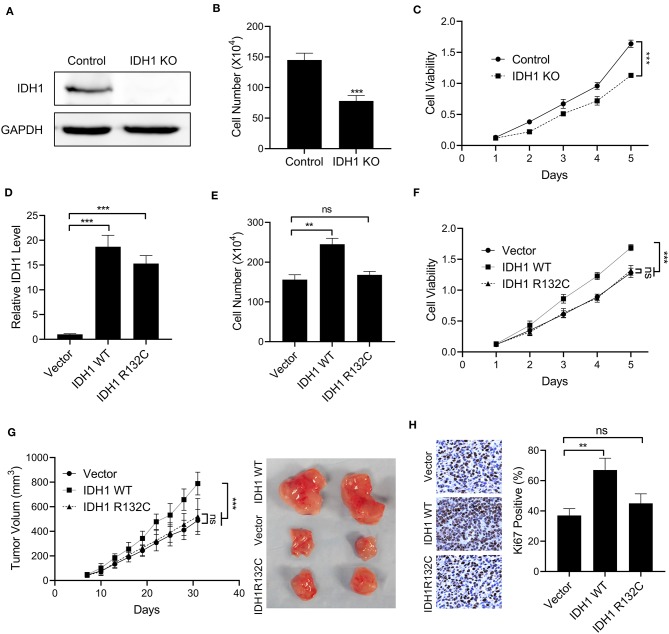
To explore the effects of IDH1 on cell proliferation, the IDH1 KO and WT cell lines were constructed. The IDH1 WT cell line was created in the IDH1 KO cells transfected with WT IDH1 expressing plasmid. As shown in Figure 2A, we did not find expression of IDH1 in the IDH1 KO cells, indicating IDH1 was successfully knocked out. Cell count assay and MTT assay demonstrated that cell number and viability were significantly decreased after IDH1 was knocked out (Figures 2B,C). Next, the IDH1R132C cell line was created in the IDH1 KO cells with IDH1 R132C expressing plasmid. As shown in Figure 2D, the IDH1 R132C mutation and the IDH WT cells were successfully constructed. The results showed that cell number and viability were significantly increased in the IDH WT cells when compared with those in cells transfected with vector control (Figures 2E,F). However, we did not find a significant difference in cell number and viability between IDH R132C mutation cells and cells transfected with vector control. Furthermore, a xenograft animal model was constructed. As shown in Figure 2G, tumor volume in animals transplanted with the IDH WT cells were significantly increased when compared with that in animals transplanted with the IDH R132C mutation cells or cells transfected with vector control. Additionally, we found more Ki67 positive cells in tumor tissues from animals transplanted with the IDH WT cells (Figure 2H).
Figure 2.
The overexpression of IDH1 promoted cell proliferation of cholangiocarcinoma. (A) Western blot was used to determine the expressions of IDH1 in the IDH knockout (KO) REB cells. Cell count assay (B) and MTT assay (C) were used to determine the cell viabilities of REB and IDH1 KO REB cells. (D) qPCR was used to determine the relative expressions of IDH1 in IDH1 KO REB cells transfected with vector, plasmids containing wildtype IDH1 or plasmids containing IDH1 R132C mutation sequence. Cell count assay (E) and MTT assay (F) were used to determine cell viabilities of IDH1 KO cells transfected with vector, plasmids containing wildtype IDH1 or plasmids containing IDH1 R132C mutation type. (G) The tumor volume of xenograft mouse transplanted with IDH1 KO REB cells transfected with vector, wildtype IDH1 or IDH1 R132C mutation plasmid. (H) Immunohistochemistry demonstrated percentage of Ki67 positive in tumor tissue from each group. Data are shown as mean ± S.D. **P < 0.01, ***P < 0.001; ns indicates no significance.
IDH1 Promoted Cell Migration and Invasion of Cholangiocarcinoma
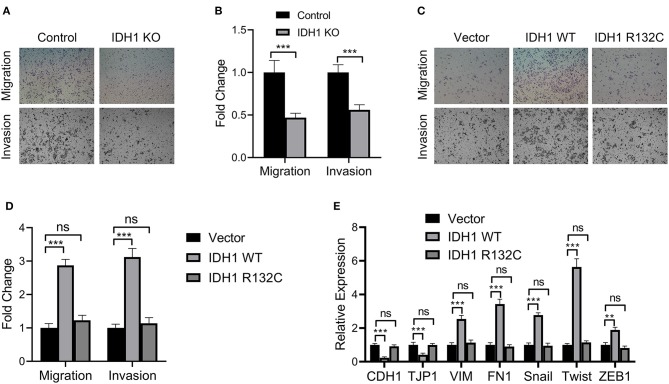
We then investigated the effects of IDH1 on cell migration and invasion. In the IDH1 KO cells, we found significant reductions in cell migration and invasion (Figures 3A,B). However, in the cells transfected with plasmids containing WT IDH1, we observed increased cell migration and invasion when compared with those in the cells transfected with vector control (Figures 3C,D). Additionally, no significant difference for cell migration and invasion between cells transfected with plasmids containing IDH1 R132C and cells transfected with vector control (Figures 3C,D). Moreover, the results showed the relative expression of a panel of epithelial-mesenchymal transition (EMT) markers including CDH1, TJP1, VIM, FN1, Snail, Twist, and ZEB1 were significantly changed after the cells were transfected with plasmids containing wild-type IDH1. We did not find cells transfected with plasmids containing IDH1 R132C with those changes when compared with cells transfected with vector control (Figure 3E).
Figure 3.
IDH1 promoted cell migration and invasion and IDH1 knockout decreased cell migration and proliferation. (A) Cell migration and invasion of the REB and IDH1 KO REB cells were determined using Transwell and (B) the values in each group were shown as mean ± S.D. (C) Cell migration and invasion of the IDH1 KO REB cells transfected with vector, wildtype IDH1, or IDH1 R132C mutation plasmid and (D) the values in each group were shown as mean ± S.D. (E) qPCR was used to analyze the relative expressions of a panel of EMT relevant markers in IDH1 KO REB cells transfected with vector, wildtype IDH1 or IDH1 R132C mutation plasmid. Data were shown as mean ± S.D. **P < 0.01, ***P < 0.001; ns indicates no significance.
IDH1 Mutation Reduced Levels of α-KG and NADPH
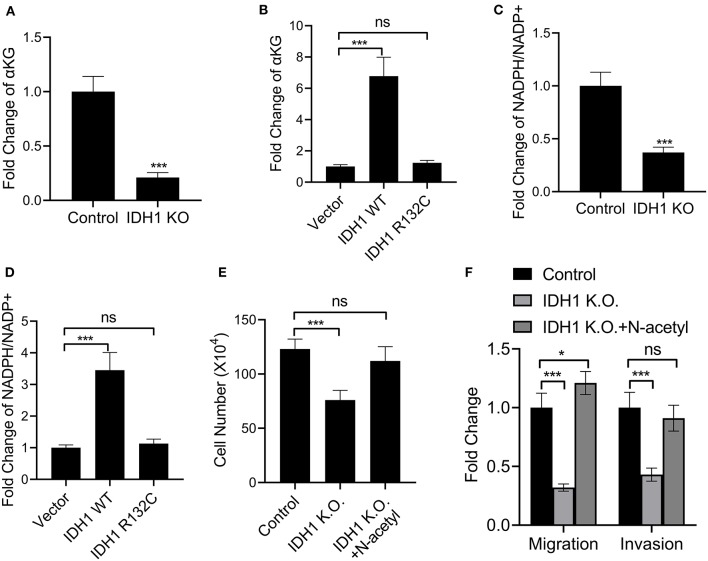
We next explored the mutation effects of IDH1 on α-KG and NADPH. First, we determined the levels of α-KG and NADPH in RBE cells and IDH KO RBE cells. The results demonstrated that the levels of α-KG and NADPH were significantly decreased in the IDH KO RBE cells when compared with those in the RBE cells (Figures 4A,C). However, in the IDH KO cells transfected with plasmids containing WT IDH1, the levels of α-KG and NADPH were increased, indicating IDH affected the levels of α-KG and NADPH (Figures 4B,D). Interestingly, when we transfected the IDH KO cells with plasmids containing IDH1 R132C, reductions in α-KG and NADPH were observed (Figures 4B,D). Moreover, N-acetyl cysteine treatment could significantly rescue the cell loss and impairments in migration and invasion in the IDH1 KO cells (Figures 4E,F). These results supported that IDH1 mutation reduced levels of α-KG and NADPH.
Figure 4.
IDH1 mutation reduced NADPH and α-KG levels. (A,B) ELISA was used to determine the levels of α-KG in REB cells, IDH1 KO REB cells, IDH1 KO REB cells transfected with vector, wildtype IDH1 or IDH1 R132C mutation plasmid. (C,D) ELISA was used to determine the levels of NADPH in REB cells, IDH1 KO REB cells, IDH1 KO REB cells transfected with vector, wildtype IDH1 or IDH1 R132C mutation plasmid. (E) Cell count assay were used to determine the cell viabilities of REB cells or REB IDH1 K.O. cells treated with or without N-acetyl cysteine. (F) Cell migration and invasion assay of the REB cells or REB IDH1 K.O. cells treated with or without N-acetyl cysteine. Data were shown as mean ± S.D. *P < 0.05, ***P < 0.001; ns indicates no significance.
IDH1 Regulated the Expressions of ALDH1 in Cholangiocarcinoma
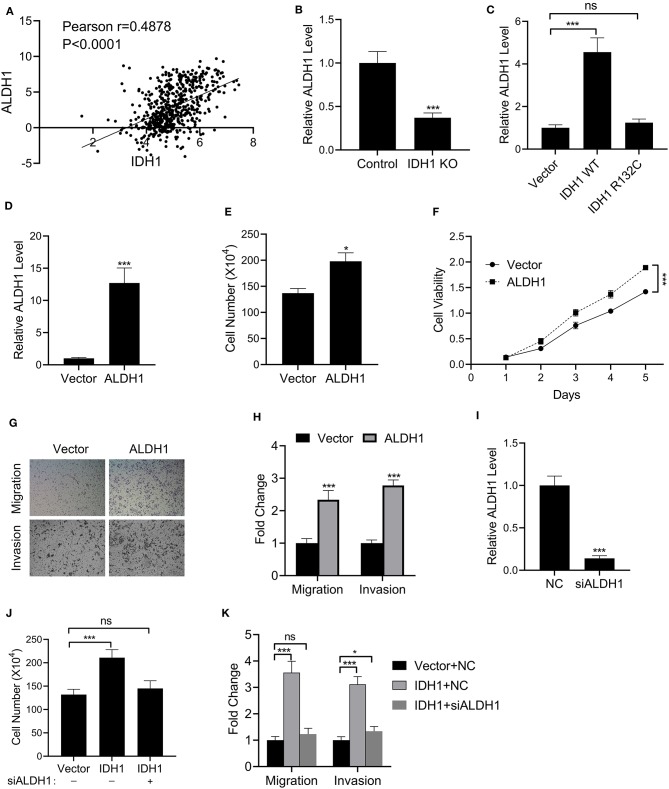
We further investigated the effects of IDH1 on ALDH1. First, we found that IDH was correlated to the expression of ALDH1 in patients with cholangiocarcinoma (Figure 5A). We then found that The levels of ALDH1 were significantly decreased in IDH KO RBE cells (Figure 5B). Additionally, the levels of ALDH1 were significantly increased in IDH KO cells transfected with plasmids containing WT IDH1, whereas a reduction in ALDH1 was observed in the IDH KO cells transfected with plasmids containing IDH1 R132C (Figure 5C). Next, we successfully transfected the IDH KO cells with plasmids containing ALDH1 (Figure 5D). Interestingly, the cell number and viability were increased in the IDH KO cells transfected with plasmids containing ALDH1 (Figures 5E,F). We also observed increased cell migration and invasion in the IDH KO cells with plasmids containing ALDH1 (Figures 5G,H). To further confirm if IDH1 regulated ALDH1 in such events, ALDH1 siRNA was employed to knock down the level of ALDH1 in REB cells (Figure 5I). It was found that ALDH1 siRNA could obviously reduce the viability of REB cells transfected with vector control or IDH1 expression plasmid (Figure 5J), as well as significantly decrease cell migration and invasion (Figure 5K).
Figure 5.
IDH1 regulated the expressions of ALDH1. (A) The correlation of IDH1 and ALDH1 expression in the patients with cholangiocarcinoma was analyzed using ChIPbase dataset. (B,C) qPCR was used to determine the levels of ALDH1 in REB cells, IDH1 KO REB cells, and IDH1 KO REB cells transfected with vector, wildtype IDH1 or IDH1 R132C mutation plasmid. (D) qPCR was used to determine the relative expressions of ALDH1 in REB cells transfected with ALDH1 expression plasmid. (E,F) Cell count assay and MTT assay were used to determine the cell viabilities of REB cells transfected with vector or ALDH1 expression plasmid. (G,H) Cell migration and invasion assay of REB cells transfected with vector or ALDH1 expression plasmid were determined using Transwell and the values in each group were shown as mean ± S.D. (I) qPCR was used to determine the relative expressions of ALDH1 in REB cells transfected with ALDH1 expression plasmid. (J) Cell count assay were used to determine the cell viabilities of REB cells transfected with vector or IDH1 expression plasmid in the presence or absence of ALDH1 siRNA. (K) Cell migration and invasion assay of the REB cells transfected with vector or IDH1 expression plasmid in the presence or absence of ALDH1 siRNA. *P < 0.05, ***P < 0.001; ns indicates not significance.
Discussion
In the present study, for the first time, IDH1 was identified as a high frequency mutated gene in patients with cholangiocarcinoma. In our in vitro study, IDH promoted cell proliferation, invasion and migration, whereas those cellular events were inhibited in the IDH R132C mutation cells. In the in vivo study, tumor volume in mice transplanted with the IDH WT cells was significantly increased when compared with that in mice transplanted with the IDH R132C mutation cells. And more Ki67 positive cells were observed in tumor tissue from animals transplanted with the IDH WT cells. We further revealed that IDH1 mutation reduced the levels of α-KG and NADPH, and IDH1 regulated the expressions of ALDH1 in cholangiocarcinoma. These results supported that IDH1 promoted the development of cholangiocarcinoma in part by inhibiting isocitrate metabolism.
IDH1 mutations have been identified in many types of cancers including glioma, hepatoma, leukemia, colon cancer and prostate cancer (15, 16, 24). As a key metabolic enzyme, IDH1 is responsible for catalyzing isocitrate to produceα-KG and generating NADPH (11, 24). In the present study, we identified a high frequency of mutated IDH1 in cholangiocarcinoma.
Besides, we also found a high frequency of TP53 mutation in patients with IDH WT, whereas low frequency of TP53 mutation in patients was accompanied with mutated IDH. Adam and colleagues have demonstrated that IDH mutation occurs earlier in the development of glioma and its mutation precedes TP53 mutation (25). Therefore, we speculate that IDH mutation and TP53 mutation occur at different stages in cholangiocarcinoma. By analyzing the proportions of different mutation types of IDH1, we revealed that IDH1 R132C was presented in more than 80% of mutation types. These results are consistent with a previous finding, in which IDH1 mutation was found in 16.88% of patients with cholangiocarcinoma and IDH1 R132C was presented in 12.55% of patients (26).
In the present study, we further verified the effects of IDH1 on cholangiocarcinoma. IDH1 promoted cell proliferation, migration and invasion in REB cells, whereas those cellular events were not significantly changed in the IDH1 R132C REB cells. Furthermore, we found that IDH1 regulated the expression of EMT markers including CDH1, TJP1, VIM, FN1, Snail, Twist, and ZEB1. These results suggested that IDH1 promoted the development of cholangiocarcinoma and its mutation impaired the progression of cholangiocarcinoma. Many previous studies have demonstrated that IDH1 plays an important role in the cellular redox state in part by regulating cellular α-KG and NADPH generation within the TCA cycle (27). It is well-known that TCA cycle is essential for cell growth, because many intermediates are required for production of lipids, proteins and nucleic acids (28). Additionally, NADPH production in cancer cell mitochondria is critical to maintain glutathione and other scavenging molecules in a reduced state, thereby preventing oxidative damage to mitochondrial structures (29). However, the accumulation of α-KG and NADPH may activate cancer-related proteins including mammalian target of rapamycin 1, thereby promoting tumorigenesis (30). Regulation of α-KG levels is also essential in modulating the epigenetic landscape of cancer cells (13). In our study, the levels of α-KG and NADPH were increased in the IDH KO cells transfected with plasmids containing WT IDH1. However, when we transfected the IDH KO cells with plasmids containing IDH1 R132C, reduced α-KG and NADPH were observed. Therefore, we verified that IDH1 was associated with the levels of α-KG and NADPH in cholangiocarcinoma. The mutations in IDH1 affected the expression of α-KG and NADPH, thereby impacting mitochondrial functions in cholangiocarcinoma.
ALDH1 is a group of detoxifying enzymes capable of catalyzing aldehydes into carboxylic acids (31, 32). It is also a marker of stem cells and is widely expressed in many types of cancers including gastric cancer, ovarian cancer and lung cancer (33, 34). Additionally, ALDH1 is involved in cell differentiation and detoxification [32; Our results demonstrated that IDH1 regulated the levels of α-KG and NADPH, which are thought to detoxify against oxidative stress. Interestingly, we also revealed that IDH was correlated with the expression of ALDH1 in patients with cholangiocarcinoma by analyzing the ChIPbase dataset. In our in vitro study, the levels of ALDH1 were significantly increased in IDH KO cells transfected with plasmids containing WT IDH1, whereas a reduction in ALDH1 was observed in the IDH KO cells transfected with plasmids containing IDH1 R132C. We further evaluated the cellular functions including cell proliferation, migration and invasion. Interestingly, in the cells transfected with ALDH1 expression plasmids, cellular functions including cell proliferation, migration and invasion were recovered. These results supported that the effects of IDH on cholangiocarcinoma was mediated in part by regulating ALDH1.
Conclusion
The present study has identified IDH1 as a high frequency mutated gene in patients with cholangiocarcinoma. In our in vitro study, IDH promotes cell proliferation, invasion and migration. In the in vivo study, IDH increases the tumor volume in mice transplanted with the IDH WT cells. Further study reveals that IDH1 regulates the levels of α-KG and NADPH and the expression of ALDH1. These results support that IDH1 promotes the development of cholangiocarcinoma in part by inhibiting the isocitrate metabolism.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
Animal procedures used in this study were approved by the First Affiliated Hospital of Anhui Medical University' Animal Care and Use Committee.
Author Contributions
PL conceived and designed research and wrote the manuscript. LS, XZ, LZ, MW, and ZZ conducted experiments and analyzed data. All authors read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The work was supported by National Natural Science Foundation of China (81673908).
References
- 1.Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD. Cholangiocarcinoma. Lancet. (2005) 366:1303–14. 10.1016/S0140-6736(05)67530-7 [DOI] [PubMed] [Google Scholar]
- 2.Blechacz B, Gores GJ. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology. (2008) 48:308–21. 10.1002/hep.22310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. (2014) 383:2168–79. 10.1016/S0140-6736(13)61903-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazzaferro V, Gorgen A, Roayaie S, Droz Dit Busset M, Sapisochin G. Liver resection and transplantation for intrahepatic cholangiocarcinoma. J Hepatol. (2020) 72:364–77. 10.1016/j.jhep.2019.11.020 [DOI] [PubMed] [Google Scholar]
- 5.Khan SA, Davidson BR, Goldin RD, Heaton N, Karani J, Pereira SP, et al. Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut. (2012) 61:1657–69. 10.1136/gutjnl-2011-301748 [DOI] [PubMed] [Google Scholar]
- 6.Chiang NJ, Shan YS, Hung WC, Chen LT. Epigenetic regulation in the carcinogenesis of cholangiocarcinoma. Int J Biochem Cell Biol. (2015) 67:110–4. 10.1016/j.biocel.2015.06.012 [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Chen Z. Mutation detection and molecular targeted tumor therapies. STEMedicine. (2020) 1:e11 10.37175/stemedicine.v1i1.11 [DOI] [Google Scholar]
- 8.Hsu M, Sasaki M, Igarashi S, Sato Y, Nakanuma Y. KRAS and GNAS mutations and p53 overexpression in biliary intraepithelial neoplasia and intrahepatic cholangiocarcinomas. Cancer. (2013) 119:1669–74. 10.1002/cncr.27955 [DOI] [PubMed] [Google Scholar]
- 9.Khan SA, Thomas HC, Toledano MB, Cox IJ, Taylor-Robinson SD. p53 Mutations in human cholangiocarcinoma: a review. Liver Int. (2005) 25:704–16. 10.1111/j.1478-3231.2005.01106.x [DOI] [PubMed] [Google Scholar]
- 10.Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A, and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. (2013) 45:1470–3. 10.1038/ng.2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yen KE, Schenkein DP. Cancer-associated isocitrate dehydrogenase mutations. Oncologist. (2012) 17:5–8. 10.1634/theoncologist.2011-0429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark O, Yen K, Mellinghoff IK. Molecular pathways: isocitrate dehydrogenase mutations in cancer. Clin Cancer Res. (2016) 22:1837–42. 10.1158/1078-0432.CCR-13-1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vatrinet R, Leone G, De Luise M, Girolimetti G, Vidone M, Gasparre G, et al. The alpha-ketoglutarate dehydrogenase complex in cancer metabolic plasticity. Cancer Metab. (2017) 5:3. 10.1186/s40170-017-0165-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murugan AK, Bojdani E, Xing M. Identification and functional characterization of isocitrate dehydrogenase 1 (IDH1) mutations in thyroid cancer. Biochem Biophys Res Commun. (2010) 393:555–9. 10.1016/j.bbrc.2010.02.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan H, Parsons DW, Jin G, Mclendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. (2009) 360:765–73. 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. (2012) 17:72–9. 10.1634/theoncologist.2011-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khurshed M, Aarnoudse N, Hulsbos R, Hira VVV, Van Laarhoven HWM, Wilmink JW, et al. IDH1-mutant cancer cells are sensitive to cisplatin and an IDH1-mutant inhibitor counteracts this sensitivity. FASEB J. (2018) 32:6344–52. 10.1096/fj.201800547R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenting K, Khurshed M, Peeters TH, Van Den Heuvel C, Van Lith SAM, et al. Isocitrate dehydrogenase 1-mutated human gliomas depend on lactate and glutamate to alleviate metabolic stress. FASEB J. (2019) 33:557–71. 10.1096/fj.201800907RR [DOI] [PubMed] [Google Scholar]
- 19.Moutasim KA, Nystrom ML, Thomas GJ. Cell migration and invasion assays. Methods Mol Biol. (2011) 731:333–43. 10.1007/978-1-61779-080-5_27 [DOI] [PubMed] [Google Scholar]
- 20.Zhang YQ, Yang H, Sun WD, Wang J, Zhang BY, Shen YJ, et al. Ethanol extract of Ilex hainanensis Merr. exhibits anti-melanoma activity by induction of G1/S cell-cycle arrest and apoptosis. Chin J Integr Med. (2018) 24:47–55. 10.1007/s11655-017-2544-8 [DOI] [PubMed] [Google Scholar]
- 21.Yang H, Wang J, Fan JH, Zhang YQ, Zhao JX, Dai XJ, et al. Ilexgenin A exerts anti-inflammation and anti-angiogenesis effects through inhibition of STAT3 and PI3K pathways and exhibits synergistic effects with Sorafenib on hepatoma growth. Toxicol Appl Pharmacol. (2017) 315:90–101. 10.1016/j.taap.2016.12.008 [DOI] [PubMed] [Google Scholar]
- 22.Liu C, Zhao J, Liu Y, Huang Y, Shen Y, Wang J, et al. A novel pentacyclic triterpenoid, Ilexgenin A, shows reduction of atherosclerosis in apolipoprotein E deficient mice. Int Immunopharmacol. (2016) 40:115–24. 10.1016/j.intimp.2016.08.024 [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Sun Q. TP53 mutations, expression and interaction networks in human cancers. Oncotarget. (2017) 8:624–43. 10.18632/oncotarget.13483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. (2010) 465:966. 10.1038/nature09132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. (2013) 13:345. 10.1007/s11910-013-0345-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Consortium APG. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. (2017) 7:818–31. 10.1158/2159-8290.CD-17-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardaci S, Ciriolo MR. TCA cycle defects and cancer: when metabolism tunes redox state. Int J Cell Biol. (2012) 2012:161837. 10.1155/2012/161837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullen AR, Hu Z, Shi X, Jiang L, Boroughs LK, Kovacs Z, et al. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. (2014) 7:1679–90. 10.1016/j.celrep.2014.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciccarese F, Ciminale V. Escaping death: mitochondrial redox homeostasis in cancer cells. Front Oncol. (2017) 7:117. 10.3389/fonc.2017.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy?. Nat Rev Cancer. (2016) 16:635–49. 10.1038/nrc.2016.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perozich J, Nicholas H, Lindahl R, Hempel J. The big book of aldehyde dehydrogenase sequences. An overview of the extended family. Adv Exp Med Biol. (1999) 463:1–7. 10.1007/978-1-4615-4735-8_1 [DOI] [PubMed] [Google Scholar]
- 32.Tomita H, Tanaka K, Tanaka T, Hara A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget. (2016) 7:11018–32. 10.18632/oncotarget.6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Douville J, Beaulieu R, Balicki D. ALDH1 as a functional marker of cancer stem and progenitor cells. Stem Cells Dev. (2009) 18:17–26. 10.1089/scd.2008.0055 [DOI] [PubMed] [Google Scholar]
- 34.Sharrow AC, Perkins B, Collector MI, Yu W, Simons BW, Jones RJ. Characterization of aldehyde dehydrogenase 1 high ovarian cancer cells: towards targeted stem cell therapy. Gynecol Oncol. (2016) 142:341–8. 10.1016/j.ygyno.2016.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.