Highlights
-
•
Roles for IRF8, IRF4, NOTCH, ZEB2, KLF4 and TBet in cDC1 and cDC2 fate specification.
-
•
Switch from +41 kb to +32 kb enhancer in IRF8 is required for cDC1 development.
-
•
NFIL3 functions to inhibit ZEB2 enabling ID2 expression in cDC1s.
-
•
cDC2 heterogeneity across tissues remains incompletely understood.
-
•
Role of tissue microenvironment in cDC2 heterogeneity remains to be studied.
Keywords: Dendritic cells, Transcription Factors, DC fate, Irf4, Irf8, Zeb1, Zeb2, Notch, Tbet, Klf4
Abstract
Dendritic cells function in the immune system to instruct adaptive immune cells to respond accordingly to different threats. While conventional dendritic cells can be subdivided into two main subtypes, termed cDC1s and cDC2s, it is clear that further heterogeneity exists within these subtypes, particularly for cDC2s. Understanding the signals involved in specifying each of these lineages and subtypes thereof is crucial to (i) enable us to determine their specific functions and (ii) put us in a position to be able to target these cells to promote or prevent a specific function in any given disease setting. Although we still have much to learn regarding the specification of these cells, here we review the most recent advances in our understanding of this and highlight some of the next questions for the future.
1. Introduction
Dendritic cells (DCs) are professional antigen-presenting cells with nonredundant functions in both innate and adaptive immunity. While patrolling peripheral tissues, DCs sample antigens and integrate environmental cues regarding the nature of the threats they encounter to initiate appropriate immune responses. Dendritic cells develop in the bone marrow (BM) through different progenitor stages in a tightly regulated transcriptional process (Sichien et al., 2017) and reviewed elsewhere in this issue) giving rise to plasmacytoid DCs (pDCs) and pre-conventional DCs (pre-cDCs). However, the DC nature of pDCs is currently challenged as they have also been shown to differentiate from lymphoid cell progenitors (Dekker et al., 2018; Dress et al., 2019; Rodrigues et al., 2018). As these cells are discussed in detail in other reviews in this issue, we will focus this review on conventional DCs (cDCs). Upon their egress from the BM, pre-cDCs seed lymphoid and most non-lymphoid tissue where they further differentiate into cDCs driven by environmental cues and the concerted actions of transcription factors (TFs) (Sichien et al., 2017). Both the development, homeostasis and function of cDCs requires Fms-related tyrosine kinase 3 (Flt3) receptor signaling (Fujita et al., 2019; Ginhoux et al., 2009; Onai et al., 2006; Waskow et al., 2008). Based on differences in transcriptional control, specialized function, phenotype and ontogeny, mature cDCs are further subdivided into cDC1s and cDC2s (Guilliams et al., 2014).
Transcriptionally, cells of the cDC1 and cDC2 lineages are commonly distinguished based on their mutually exclusive expression of interferon-regulatory factor 8 (IRF8) and IRF4 respectively (Guilliams et al., 2016; Murphy et al., 2016). Functionally, cDC1s excel in antigen cross-presentation to CD8 T cells, whereas cDC2s typically drive CD4 T cells toward distinct effector subsets. While the cDC1 subset is precisely delineated by the expression of specific markers (such as XCR1) and relies on a distinct set of TFs, cDC2s seem more heterogenous on a transcriptional level and express surface markers (such as SIRPα and CD11b) that often overlap with macrophages and monocyte-derived cells (MCs) (Guilliams et al., 2016).
Already in the BM pre-cDCs become gradually committed to the cDC1 or cDC2 fate and enter the bloodstream as pre-cDC1s and pre-cDC2s respectively (Breton et al., 2016; Grajales-Reyes et al., 2015; Ma et al., 2019; Schlitzer et al., 2015; See et al., 2017; Villani et al., 2017). Guided by tissue-specific signals, such as those derived from the local niche or invading pathogens, pre-cDCs and cDCs alter their transcriptional profile that dictates a functional module best suited to deal with the requirements of the current microenvironment (Roquilly et al., 2017). This can result in a change of surface markers and/or dependency on certain TFs to exert these functions, and as such cDCs may obtain a maturation state different from the classically defined subset. In this review we will focus on recent findings regarding the factors that regulate this fate decision and the functional adaptations of the different cDC subsets.
2. Irf8 and Batf3
Irf8, formerly known as interferon consensus binding protein (ICSBP) plays an important lineage-determining role in the BM at several developmental stages of monocytes, pDCs and cDC1s (Bagadia et al., 2019; Durai et al., 2019; Grajales-Reyes et al., 2015; Kurotaki et al., 2018; Murphy et al., 2016; Sichien et al., 2016). The action of IRF8 at the macrophage and DC precursor (MDP) and common monocyte/macrophage precursor (cMoP) stages is crucial for monocyte development and the exclusion of granulocyte potential (Kurotaki et al., 2014; Sichien et al., 2016). Beyond the cMoP stage, Irf8 is redundant for the survival of monocytes or monocyte-derived cells (MCs) (Kurotaki et al., 2013; Sichien et al., 2016), yet the functional role of IRF8 in these cells remains largely unknown.
By conditionally deleting Irf8 at different stages of DC development, Irf8 has been shown to be a terminal selector for cDC1s as it is required for the specification of pre-cDCs toward terminally differentiated cDC1s (Bajaña et al., 2016; Grajales-Reyes et al., 2015; Luda et al., 2016; Sichien et al., 2016) and that its continuous high expression through BATF3-mediated autoactivation is required to maintain the identity of cDC1s (Grajales-Reyes et al., 2015). Two recent reports by the Murphy lab, deleting specific enhancers within Irf8 and employing a genetic epistasis strategy, (Bagadia et al., 2019; Durai et al., 2019) have offered insights in the mechanism and functional hierarchy of TFs involved in Irf8 expression and subsequent DC fate specification. The Irf8 gene carries three distinct enhancers bound by specific TFs that regulate Irf8 at different stages during development (Durai et al., 2019). Binding of the TF PU.1 to the −50 kb (relative to the Irf8 transcriptional start site) enhancer was shown to regulate Irf8 levels in mature monocytes and some macrophages populations (Durai et al., 2019), but was not required for Irf8 expression in MDPs or for the downstream DC populations as previously reported (Schönheit et al., 2013). The +41 kb Irf8 enhancer likely activated by E2A (encoded by Tcf3) is required to specify common DC precursors (CDPs) to pre-cDC1s by promoting IRF8 expression. In addition, the +41 kb Irf8 enhancer is also required for normal pDC phenotype with Irf8 +41kb-/- pDCs having a similar phenotype to Irf8-/- pDCs (Durai et al., 2019; Sichien et al., 2016) potentially through binding another E protein, E2-2 (encoded by Tcf4), the lineage-determining TF of pDCs (Cisse et al., 2008). Once IRF8 expression is induced in cDC1-committed progenitors, the +32 kb Irf8 enhancer, which is activated by BATF3 and IRF8 itself (Grajales-Reyes et al., 2015), becomes active in the pre-cDC1 stage, promoting both their transition to mature cDC1s and their maintenance by sustaining IRF8 expression. This essential switch from the E2A-dependent +41 kb Irf8 enhancer in DC progenitors to the BATF3-dependent +32 kb enhancer in mature cDC1s depends on a NFIL3-ZEB2-ID2 regulatory circuit in which NFIL3 enforces ID2 expression by suppressing ZEB2 (see below) (Bagadia et al., 2019). Ultimately, ID2 inhibits the E2A activity at the +41 kb Irf8 enhancer from the pre-cDC1 stage onwards, thereby mediating the switch to the BATF3-controlled +32 kb Irf8 enhancer in mature cDC1s.
As the +32 kb enhancer is absolutely required for cDC1 development, the +32 kb−/− mice generated as part of this study (Durai et al., 2019) represent a nice tool to further study cDC1 function, without compensatory mechanisms at play as previously observed for the loss of Batf3, Nfil3 and Id2 (Seillet et al., 2013). Nevertheless, studies using Batf3 or Irf8 KO mice have put forward the cDC1 branch as a major cellular source of IL-12 in several models of infection with intracellular pathogens (Everts et al., 2016; Hildner et al., 2008; Martínez-López et al., 2015; Mashayekhi et al., 2011; Scharton-Kersten et al., 1997; Schiavoni et al., 2002). This may result from the constitutive expression of Irf8 in cDC1s (Aliberti et al., 2003; Everts et al., 2016; Sichien et al., 2016), as Irf8 has previously been suggested to play a role in the expression of Il12-genes in macrophages, (Liu et al., 2004; Scharton-Kersten et al., 1997; Wang et al., 2000). However, as it is not possible to uncouple Irf8 expression and IL-12 production by cDC1s (cDC1s do not develop in the absence of Irf8) this remains to tested. As cDC2s can also express Il12b (Kinnebrew et al., 2012; Satpathy et al., 2013; unpublished data) and IFN-γ driven IL-12 production by MCs is important at sites of infection (Goldszmid et al., 2012), the dependence on IRF8 for IL12 production in these cells should also be examined. Interestingly, it has been proposed that IL-12 produced by peripheral DCs upon infection can activate NK cells in the BM, that in response produce IFN-γ locally (Askenase et al., 2015). One could envisage that such an altered milieu in the BM could also induce epigenetic or even transcriptional changes in the DC precursors biasing the generation toward specific progeny on demand of the biological need. As recent reports in both mice and humans have demonstrated that IRF8 expression as early as the multipotent progenitor stage marked cells biased toward the cDC1 lineage (Kurotaki et al., 2019; Lee et al., 2017), it would be interesting to further investigate whether such priming of precursors actually takes place under different settings.
As cDC1s typically do not develop in Irf8 and/or Batf3 deficient mice, it is challenging to assess any additional functional role(s) for these TFs in mature cDC1s. However, the requirement for BATF3 to sustain IRF8 expression can be compensated for by Batf and Batf2 binding the same +32 kb Irf8 enhancer (Durai et al., 2019) in certain conditions (Seillet et al., 2013; Tussiwand et al., 2012). The BATF3 dependency can also be artificially bypassed by crossing Batf3 knock-out (Batf3−/−) mice with transgenic Irf8VENUS mice, which have an increased copy number of endogenous Irf8 (Grajales-Reyes et al., 2015; Theisen et al., 2019). By comparing Batf3+/+ Irf8VENUS+ and Batf3-/- Irf8VENUS+ cDC1s, a role for BATF3 in cDC1-mediated tumor rejection was uncovered, as BATF3 was found to specifically target a small set of genes required by cDC1s to carry out this function (Theisen et al., 2019). Additionally, administration of exogenous IL-12 in Batf3-/- mice has also been shown to restore cDC1 development and the capacity to cross-present antigen and mediate tumor rejection (Tussiwand et al., 2012). This suggests that IL-12 provides additional signals, absent in Batf3-/- Irf8VENUS+ mice, that can compensate for impaired tumor rejection (Theisen et al., 2019), although the transcriptional program that dictates this compensation remains elusive.
3. Irf4
IRF4 is preferentially expressed by cDC2s (Guilliams et al., 2016) and was originally proposed to be required for the specification of BM progenitors toward splenic CD4+ cDCs or CD11bhi cDCs (Suzuki et al., 2004; Tamura et al., 2005). Although full Irf4 deficient mice have a reduced number of cDC2s in most tissues (Guilliams et al., 2016), conditional deletion of the Irf4 gene in CD11c-expressing cells resulted in an altered surface marker expression profile and functional deficits (Bajaña et al., 2016, 2012; Krishnaswamy et al., 2017; Persson et al., 2013; Schlitzer et al., 2013) of the cDC2 lineage but does not result in the loss of the entire population. Thus while IRF4 controls some aspects of cDC2 specification, it, unlike IRF8 in cDC1s, is not absolutely required for the presence of the entire cDC2 lineage. Phenotypically, loss of Irf4 from the pre-cDC stage on in Irf4fl/fl mice driven by an ‘early-acting’ Itgax-Cre (Caton et al., 2007) resulted in a reduction (although not complete ablation) of specific cDC2 subtypes expressing CD24 in the lung (Ainsua-Enrich et al., 2019; Bajaña et al., 2016; Schlitzer et al., 2013) and the heart (Van der Borght et al., 2017), both CD24 and CD103 in the intestine (Persson et al., 2013; Schlitzer et al., 2013), CD103b (MGL2) in the skin (Gao et al., 2013) and CD4 in the spleen (Bajaña et al., 2016; Suzuki et al., 2004; Tamura et al., 2005), although whether this reduction is due to a lack of development or impaired survival (see below) remains unclear. Functionally, IRF4 appears to control CCR7-mediated migration of peripheral cDC2s to the draining lymph nodes (LNs) (Ainsua-Enrich et al., 2019; Akbari et al., 2014; Bajaña et al., 2016, 2012; Deckers et al., 2017; Van der Borght et al., 2018; Worbs et al., 2017) and favors a transcriptional module that governs MHCII-dependent antigen presentation required for T cell priming (Roquilly et al., 2017; Vander Lugt et al., 2014). IRF4 promotes the expression of cathepsin S, H2-Oa and H2-DM, which are involved in antigen loading into the peptide-binding pocket of MHC class II molecules (Busch et al., 2000; Vander Lugt et al., 2014). In addition, IRF4 was found to negatively regulate March1, preventing MHCII surface expression (Shin et al., 2006), and Cystatin C, which in turn inhibits the activity of cathepsin S (Pierre and Mellman, 1998; Vander Lugt et al., 2014). Therefore is tempting to speculate that a drop in IRF4 levels in murine lung cDC1s and cDC2s induced upon resolution of pneumonia, could be causally associated with the impaired innate and adaptive immune responses against a secondary bacterial infection (Roquilly et al., 2017). Moreover, in vitro cDC2-like cells and MCs induced by GM-CSF and IL-4 required IRF4 to prime CD8+ T cells (Briseño et al., 2016), opposed to in vivo cDC1s that rely on IRF8 and BATF3 for efficient cross-presentation. Aside from roles in antigen presentation and migration, IRF4 in cDC2s may also antagonize apoptosis (Persson et al., 2013; Schlitzer et al., 2013; Sichien et al., 2017). Thus, it remains unclear whether Irf4-deficient cDC2s fail to migrate because Ccr7 is directly regulated by IRF4 and hence they die in the tissue unable to migrate or if they die prematurely before they can acquire CCR7 expression. Evidently, the induction of cytotoxic or T helper (Th) responses requires efficient MHCI or MHCII presentation of transferred antigen in the LN. Therefore the impaired survival and/or migration of specific subsets of Irf4-deficient cDC2s could, at least partially, explain the deficient priming of CD4 T cells to Th2 (Gao et al., 2013; Williams et al., 2013; Zhou et al., 2014), Th17 (Persson et al., 2013; Schlitzer et al., 2013), T regulatory (Treg) (Ainsua-Enrich et al., 2019; Vander Lugt et al., 2017) or T follicular helper (Tfh) subtypes (Flores-Langarica et al., 2018a, 2018b; Krishnaswamy et al., 2017) that has been reported. Aside from CD4 T cell priming, upon viral infection or specific PRR signals, cDC2s can also function in CD8 T cell priming (unpublished and Ainsua-Enrich et al., 2019; Ballesteros-Tato et al., 2010; Desch et al., 2014; Kim et al., 2014; Shin et al., 2016), however, the underlying transcriptional program that licenses cDC2s to initiate CD8 T cell responses remains to be elucidated. Next to its role in survival and/or migration, some reports indicate that IRF4 could also intrinsically control a functional gene module in cDCs that drives Th specification under certain circumstances (Ainsua-Enrich et al., 2019; Akbari et al., 2014; Calabro et al., 2016; Flores-Langarica et al., 2018a, 2018b; Krishnaswamy et al., 2017; Laidlaw et al., 2015; Mayer et al., 2017; Moon et al., 2018; Negishi et al., 2005; Persson et al., 2013; Schlitzer et al., 2013; Scott et al., 2015; Vander Lugt et al., 2017; Williams et al., 2013).
4. Cooperation with Irf4 and Irf8
It was recently shown in humans that the discrimination between pre-cDC1s and pre-cDC2s correlated with the ratio of IRF8 to IRF4 expression, indicating that the combinatorial dose of these TFs determines fate decision (Ma et al., 2019; Villani et al., 2017). Furthermore, reciprocal expression of IRF4 and IRF8 in pre-cDC subsets might directly prime distinctive molecular programming of the progeny, thereby promoting the presentation of antigens via the MHCII or I pathway, respectively (Vander Lugt et al., 2014). Since antagonism between IRF4 and IRF8 has been demonstrated in activated B cell differentiation (Xu et al., 2015), it is tempting to assume that competition between both TFs would also contribute to the fate decision between cDC1s and cDC2s. Further supporting this hypothesis, IRF4 is strongly increased in uncommitted pre-cDCs and committed pre-cDC2s in the absence of IRF8 which in turn might accelerate cDC2 development (Sichien et al., 2016). In pDCs an increase in IRF4 expression compensated for the loss of IRF8 as IRF4−/−IRF8−/− mice completely lacked pDCs (Sichien et al., 2016). Next to the functional dichotomy of IRF8 and IRF4 in cDCs, both TFs similarly correlated with the expression of genes encoding MHCII, CD80, CD86 and CCR7 in cDC1 and cDC2 subsets, indicating that IRF8 and IRF4 also induce comparable aspects of DC maturation (Schiavoni et al., 2004; Vander Lugt et al., 2014). Even though cDC2s are not quantitatively affected by Irf8 or Batf3 deficiency in steady state, expression of costimulatory molecules and MHCII dependent antigen presentation was found to be impaired in cDC2s derived from Irf8−/− mice (Aliberti et al., 2003; Mattei et al., 2006). Therefore, further research is warranted to investigate whether the loss of these subset-associated TFs could intrinsically affect the functional maturation of cDC2s under certain conditions, and vice versa.
5. Klf4
Another TF associated with cDC2s is Kruppel-like factor 4 (KLF4). Interestingly, KLF4 has been shown to be required for a subpopulation of murine cDC2s. As such, Itgax-Cre Klf4fl/fl mice show a reduction of cDC2s across lymphoid and non-lymphoid tissues, although the missing cDC2 subsets differed in their surface expression pattern (Tussiwand et al., 2015). For instance, loss of Klf4 selectively eliminated cDC2s expressing CD24 and MGL2 (CD301b) in the lung, while compromising the development of migratory CD11b−CD24− cDC2s in the skin. Functionally, these KLF4-dependent cDC2s appear essential for mounting Th2 responses to certain stimuli (Tussiwand et al., 2015), consistent with reduced Th2 responses observed in reports using Itgax-Cre Irf4fl/fl and Mgl2-DTR mice (Gao et al., 2013; Kumamoto et al., 2013). Notably, KLF4 is not alone in regulating a subset of cDC2s (see below).
6. Zeb1 and Zeb2
The ZEB proteins are a family TFs consisting of two members, ZEB1 and ZEB2 (Scott and Omilusik, 2019). While Zeb1 is expressed across DC subsets in mice, Zeb2 expression is found in murine pDCs and cDC2s, but absent in cDC1s (Miller et al., 2012; Scott and Omilusik, 2019). Despite its wide expression across the cDC lineage, the role of ZEB1 in cDCs and their progenitors remains largely unclear. In vitro Zeb1 knock down in a murine tumor-derived DC cell line or FLT3L-induced BMDCs reportedly results in impaired activation and cytokine secretion and a Th2 biased response following DC:T cell coculture (Smita et al., 2018). Thus it will be interesting to determine if this is also the case in vivo. The other ZEB family member, ZEB2, was recently proposed to act as a switch at the bifurcation of cDC1 and pDC fate specification (Bagadia et al., 2019; Wu et al., 2016). However, it is currently questioned whether pDCs and cDC1s actually share a common progenitor, as it was proposed that pDCs may primarily develop from lymphoid progenitor cells independently of the myeloid cDC lineage (Dress et al., 2019). Nevertheless, it is clear that the presence of both cDC1s and pDCs rely on the mutually exclusive repression of either ZEB2 or ID2 in addition to their use of distinct Irf8 enhancers (see above). Repression of ZEB2 by NFIL3 leading to increased ID2 expression is essential for cDC1 specification at the CDP stage (Bagadia et al., 2019; Wu et al., 2016), while in pDCs, ZEB2 is required for their development and maintenance (Scott et al., 2016; Wu et al., 2016), presumably through inhibition of ID2 which in turn antagonizes Tcf4 expression (Ghosh et al., 2010). Zeb2 is also consistently expressed throughout the development and maturation of the cDC2 lineage (Schlitzer et al., 2015). Although loss of Zeb2 only slightly affected cDC2 numbers in a cell-intrinsic manner, this effect varied between cDC2 subsets in different tissues (Scott et al., 2016; Wu et al., 2016), while Zeb2fl/fl mice crossed with a “late-acting” Itgax-Cre did not have a defect in cDC2 numbers. Taken together, these data suggest that ZEB2 may act early in cDC2 development, in a specific subset manner (Scott et al., 2016). In the future, the use of cDC subset-specific Zeb2 deletion may help to unravel the cell-intrinsic effects of Zeb2 deficiency on DC function.
7. Notch
Apart from cell-intrinsic effects of certain TFs, tissue-specific signals, such as NOTCH ligands or lymphotoxin β (LTβ), can inform DCs of their surroundings and thereby guide their differentiation and function (Cheng et al., 2007; Fasnacht et al., 2014). NOTCH receptors transmit signals from membrane-bound ligands of the Delta-like (DL) and Jagged (Jag) families through the common TF RBPjk. It has been shown that NOTCH2-RBPjk signaling is involved in the terminal differentiation of a cDC2 subset expressing ESAM localized in the marginal zone bridging channels of the spleen and CD103 in the small intestine (Caton et al., 2007; Lewis et al., 2011; Satpathy et al., 2013) without actually affecting the development of cDC precursors. Mice devoid of TF RUNX3 displayed a similar loss of ESAM+ cDC2s, potentially through direct interactions with RBPjk (Dicken et al., 2013). Functionally, NOTCH2-signaling in cDCs is required for IL-23 production in the innate defense against C. rodentium (Satpathy et al., 2013). Moreover a reduced fraction of intestinal Th17 cells, previously associated with the lack of CD103+ intestinal cDC2s (Lohoff et al., 2002; Persson et al., 2013; Schlitzer et al., 2013), was found in Itgax-Cre Notch2fl/fl mice in steady state (Lewis et al., 2011), although others have shown that NOTCH2 signaling was dispensable to mount Th17 responses to certain bacteria (Linehan et al., 2015; Panea et al., 2015). Since CD103− cDC2s are not quantitively affected upon loss of Notch2, it is possible that the CCR2+CD103− cDC2 subset in the intestine compensates for Th17 induction (Scott et al., 2015), although the Notch independency of this subset remains to be shown. Moreover, NOTCH2-signaling appeared to be dispensable for mounting Th2 responses against S. mansoni (Tussiwand et al., 2015) or differentiation of peripheral Tregs (Nutsch et al., 2016). A recent study implied a role for Notch2-dependent cDC2s in mediating Tfh differentiation and germinal center (GC) responses in the spleen, but not in Peyer Patches (Briseño et al., 2018). Next to the quantitative defect of splenic cDCs (i.e. ESAM+ subsets), in Itgax-Cre Notch2fl/fl mice (Satpathy et al., 2013) this effect was further ascribed to qualitative changes in gene expression of the Notch2-deficient cDC2s (Briseño et al., 2018). However, the functional module targeted by NOTCH2 in cDC2s that favors Tfh priming (Briseño et al., 2018) or determines the correct anatomical localization in the spleen (Lu et al., 2017; Satpathy et al., 2013) remains elusive. One concern regarding this latter study (Briseño et al., 2018) is that the Itgax-Cre system reportedly causes leakiness in B and T cells (Krishnaswamy et al., 2017; Schlitzer et al., 2013) thus Tfh differentiation may be impaired due to targeting T cell-intrinsic NOTCH2-signaling (Auderset et al., 2013; Dell’aringa and Lee Reinhardt, 2018). Therefore, future studies will have to examine the DC-intrinsic role of Notch2 in these observations by using more specific conditional deletion models, chimeras and/or ex vivo assays with purified subsets. Moreover, it remains to be shown whether cDC2s are involved in a Notch2-dependent way in Tfh differentiation in humans as observed in mice (Durand et al., 2019). Next to NOTCH instruction, splenic ESAM+ and intestinal CD103+ cDC2s also rely on downstream LTβR signaling through ligands provided by the local environment (Kabashima et al., 2005; Satpathy et al., 2013; Wang et al., 2005), but how both pathways interact remains to be elucidated. Interestingly, in order to correctly localize in the marginal zone bridging channels NOTCH2- and LTβR-dependent cDC2s require expression of chemokine receptor Epstein-Barr virus-induced G-protein coupled receptor 2 (EBI2, encoded by Gpr183) (Gatto et al., 2013; Yi and Cyster, 2013). Since the loss of splenic CD4+ cDC2s in Gpr183-deficient mice could be rescued by LTβR-agonism (Yi and Cyster, 2013), it is tempting to speculate that EBI2 drives the positioning of cDC2s into a niche that provides NOTCH and LTβR ligands. Therefore it will be interesting to assess the role of NOTCHsignaling in the anatomical positioning of DCs in future research.
Next to cDC2s, Notch2-deficiency also affected the terminal maturation of cDC1s in the spleen (Lewis et al., 2011; Satpathy et al., 2013). Given the key role of NOTCH signaling in cDC progenitors for terminal differentiation, an alternative in vitro ‘cDC1′ culture system for both mouse and human was recently developed that uses the FLT3L system (Naik et al., 2005) in combination with a stromal cell line (OP9) expressing DLL1 (Kirkling et al., 2018). While pDC development was largely abolished, these in vitro generated cDC1s more closely aligned with their primary counterparts (Kirkling et al., 2018), further emphasizing the key role of NOTCH signaling in terminal cDC differentiation. Another in vitro differentiation method similarly showed that NOTCH signaling in a GM-CSF system promoted cDC1 generation from human CD34+ precursors (Balan et al., 2018).
8. Tbet
Although typically thought of as a TF associated with T helper cell and innate lymphoid cell (ILC) subsets, recently, Tbet expression, reported through the use of Tbx21RFP−Cre mice, has also been suggested to delineate two subsets of cDC2s (Brown et al., 2019), with those cDC2s expressing Tbet being termed cDC2As, while cDC2Bs lacked expression of this TF (Brown et al., 2019). Notably, this is not the first time Tbet expression has been reported in cDC2s (Lugo-Villarino et al., 2003; Wang et al., 2006). Functionally, these two populations were also suggested to be distinct, with cDC2As being more anti-inflammatory than their cDC2B counterparts (Brown et al., 2019). However, this appears somewhat at odds with the original studies suggesting that lack of Tbet expression in cDCs protected mice from collagen antibody induced arthritis (Wang et al., 2006) and that Tbet expression was required for IFN-γ production by cDCs (Lugo-Villarino et al., 2003). While IFN-γ production from cDC2As and cDC2Bs was not assessed in the recent study, preventing the direct comparison, the authors found that cDC2Bs were better at inducing IFN-γ+ Th1 cells in non-polarizing conditions than cDC2As (although levels of IFN-γ measured were relatively low) (Brown et al., 2019), which was previously attributed to the Tbet-dependent DCs (Lugo-Villarino et al., 2003; Wang et al., 2006). However, Brown and colleagues also observed that IFN-γ promoted Tbet expression in cDC2s (Brown et al., 2019). As the original studies used total CD11c+MHCII+ cells from WT or Tbet KO mice and did not discriminate Tbet+ from Tbet- cDCs, one possible hypothesis to explain this discrepancy, could be that Tbet-expressing cDC2As may promote IFN-γ production from cDC2Bs which in turn promote cDC2As creating a loop to ensure sufficient IFN-γ production from cDC2s when required. However, this remains to be formally investigated.
Although the proportions of cDC2As and cDC2Bs differed, both subsets were identified across several murine tissues. Focusing on those found in the murine spleen, despite an enrichment for binding sites of Rbpj and Runx in cDC2As suggesting a role for NOTCH signaling, the authors concluded that Tbet expression did not perfectly correlate with ESAM expression, although further investigation may be warranted based on mRNA expression profiles (Brown et al., 2019). Additionally, while CLEC10A and CLEC12A could be used to further distinguish cDC2Bs into different fractions, no perfect markers were reported to accurately distinguish between cDC2As and cDC2Bs (Brown et al., 2019). The presence of further subsets of cDC2Bs, also suggests there could be further heterogeneity within the population not accounted for by Tbet expression. For example, despite being expressed across cDC2As and cDC2Bs (Brown et al., 2019), loss of KLF4 selectively affects CLEC12A+ cDC2Bs in the spleen (Tussiwand et al., 2015 and reviewed in detail in Murphy et al., 2016). In addition, it remains to be seen how traditional markers used to delineate cDC2 subsets in other tissues, for example CD103 in the intestine, fair when it comes to dividing the Tbet+ and Tbet− fractions.
Despite the potential for further heterogeneity within the cDC2s, intriguingly, ATAC-Seq analysis revealed that cDC2As were enriched for binding sites for ID2 and NFIL3 when compared with cDC2Bs (Brown et al., 2019), thus given the relationship identified by the lab of Ken Murphy between these TFs and ZEB2 (Bagadia et al., 2019) it is tempting to speculate that Tbet expression could delineate the subset of cDC2s existing independently of ZEB2 expression (Scott et al., 2016; Wu et al., 2016), as NFIL3 would inhibit ZEebBEBEB2 expression which would in turn enable ID2 to be expressed. Potentially fitting with this hypothesis, when the authors compared human and murine splenic cDC2s to find counterparts, ZEB2 was one of the defining TFs found to overlap between human and murine cDC2Bs (Brown et al., 2019). Additionally Zeb2 expression level does seem to distinguish between two cDC2s subsets in the murine spleen (unpublished data) however both subsets express Zeb2 compared with cDC1s (Scott and Omilusik, 2019 and unpublished data), perhaps this could be explained by a lower expression of NFIL3 in cDC2s than cDC1s. Indeed, this requires a further, more in-depth examination.
As mentioned above cDC2A and cDC2B counterparts were also identified in human splenic tissue, expressing CLEC4A and CLEC10A respectively, where they appeared to have similar functional properties to those in the mouse (Brown et al., 2019). Notably, CLEC10A−CLEC4A+ cDC2As were only found in the spleen and not in human PBMCs highlighting the need to also examine human tissue (Brown et al., 2019). While Tbet+ cDC2 equivalents were not found in human blood in this study, many recent studies have also highlighted a significant amount of heterogeneity in the cDC2 populations present within PBMCs (Dutertre et al., 2019; Günther et al., 2019; See et al., 2017; Villani et al., 2017). This suggests that, similar to the mouse, Tbet expression alone is likely not sufficient to explain all cDC2 heterogeneity in humans. The most recent study, upon bringing together all this data, proposed that there are two main cDC2 subsets present within the PBMC population based on expression of CD5 and CD163 which were termed cDC2 (CD5-) and cDC3 (CD5+CD163+). Here, the cDC3 population were identified to be the most inflammatory and fitting with this were also increased in proportion in SLE patients (Dutertre et al., 2019). While NOTCH2 was more highly expressed than KLF4 in the cDC3s (Dutertre et al., 2019), which TFs may specify these populations remains an open question for the future. However, with their shared expression of NOTCH, the link between the cDC3s and cDC2As also requires closer attention.
9. Conclusion
In the last decade significant progress has been made in our understanding of DC development and the different mature cDC populations across species and tissues (Fig. 1). Although cDCs are commonly divided in two main lineages (cDC1s and cDC2s) based on ontogeny (Guilliams et al., 2014) and relating to function and phenotype, it has become apparent that the cDC2s, still represent a heterogeneous population. This heterogeneity is furthermore reflected by the differential reliance on certain TFs, depending on the tissue and/or event that disrupts homeostasis. While a set of TFs appear to be required for the development of the entire cDC1 (e.g. Irf8 Sichien et al., 2016) or pDC (e.g. Tcf4, Zeb2, Cisse et al., 2008; Scott et al., 2016; Wu et al., 2016) lineages, no single transcriptional regulator has yet been identified that is essential for cDC2 development or maintenance. However, this is perhaps not surprising given the level of heterogeneity observed within this populations. Future research will need to further consider and define the contextual signals that influence the transcriptional program of different subsets of cDC2s in any specific location as perhaps similar to what is seen for macrophages (Lavin et al., 2014), local environmental features may be key in understanding these populations. Moreover, whether the heterogeneity in transcriptional program within the cDC subsets, and in particular cDC2s, relates to specific differentiation programs induced in the primed T cells will have to be determined by future studies. A better understanding of the transcriptional and functional adaptation cDC subtypes will ultimately provide a basis for better therapeutic strategies in humans.
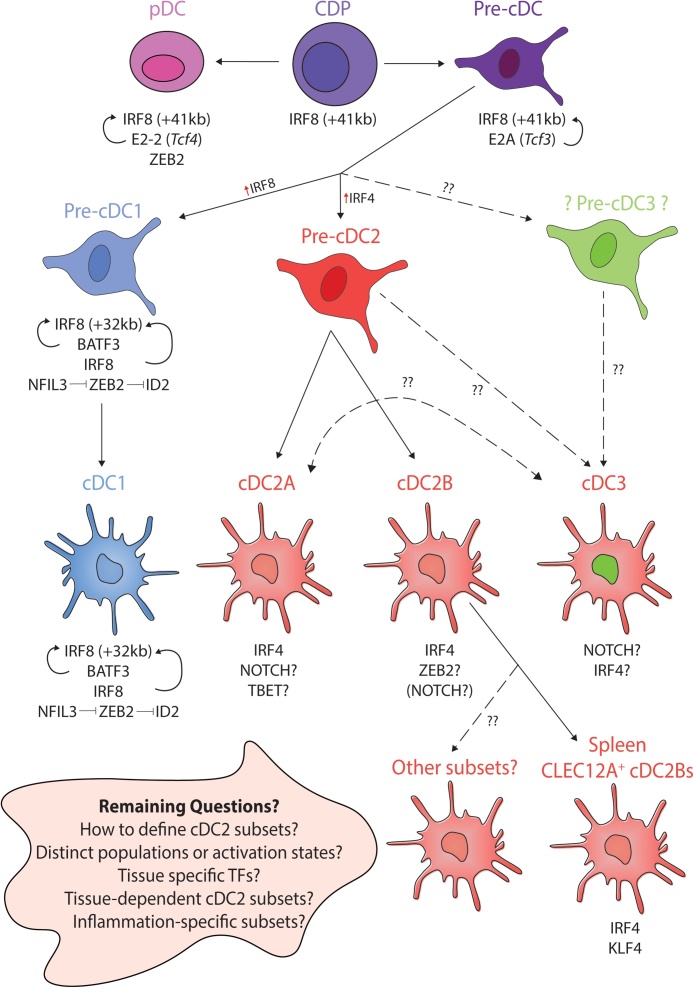
Fig. 1.
Recent advances in cDC fate specification by Transcription Factors.
Summary figure detailing recent advances in our understanding of transcriptional control of DC fate, highlighting remaining unknowns and main questions for the future. cDC1s develop from CDPs through pre-cDC1 intermediates. In mice, progression to cDC1 is achieved through increasing expression of IRF8 and switching from the +41 kb enhancer to the +32 kb enhancer at the pre-cDC stage. cDC1s express IRF8, BATF3, ID2, NFIL3 (red arrows) and absolutely require IRF8 (maintained through BATF3 and IRF8 autoactivation) for their development and maintenance. Expression of NFIL3, inhibits ZEB2 expression which in turn enables ID2 expression, normally repressed by ZEB2. Like cDC1s, cDC2s develop from CDPs but via a pre-cDC2 intermediate. There is still much to learn regarding cDC2s. Typically IRF4 is thought of as the main cDC2 TF in mice, however, it is not required for the development of all subsets, but may instead be important for survival and/or migration of specific subsets. In recent years, considerable heterogeneity has been demonstrated within the cDC2 population including the recent description of Tbet+ and Tbet− cDC2s termed cDC2As and cDC2Bs respectively identified in mouse and human spleen. To date no distinct precursors for these subsets have been identified. In mice, cDC2As appear to also express and require NOTCH signaling being enriched for Rbpj and Runx binding sites. Whether Tbet is absolutely required for cDC2As remains to be studied. Some evidence suggests murine cDC2Bs (or a subset thereof), may require ZEB2 for their development but this remains to be directly investigated. Clec12A+ cDC2Bs, one subset of cDC2Bs, have been shown to require KLF4 in mice. Requirements for the other subsets remain to be studied. In human PBMCs, a pro-inflammatory cDC3 population has also recently been described, the TFs mediating this population and the precise progenitor remain to be identified however, they were shown to express higher NOTCH2 than KLF4. Thus the relationship of these cDC3s to cDC2As requires further investigation. Key questions for the future, especially related to heterogeneity within the cDC2 lineage, including understanding the role of the tissue microenvironment in cDC2 biology. How many distinct populations of cDC2s are there? Are there different subsets based on tissue? Do distinct TFs play a role in each tissue to generate the final cDC2 phenotype? Are cDC2 subsets arising from distinct developmental pathways or do they represent alternative activation states? Understanding this, will be a main goal in the coming years.
Acknowledgements
CB is supported by a FWO PhD student fellowship (1138019N). CLS is a Francqui Professor and her lab is also supported by an ERC starting grant (MyeFattyLiver, #851908) and FWO project grants.
References
- Ainsua-Enrich E., Hatipoglu I., Kadel S., Turner S., Paul J., Singh S., Bagavant H., Kovats S. IRF4-dependent dendritic cells regulate CD8+ T-cell differentiation and memory responses in influenza infection. Mucosal Immunol. 2019;12:1025–1037. doi: 10.1038/s41385-019-0173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari M., Honma K., Kimura D., Miyakoda M., Kimura K., Matsuyama T., Yui K. IRF4 in dendritic cells inhibits IL-12 production and controls Th1 immune responses against leishmania major. J. Immunol. 2014;192:2271–2279. doi: 10.4049/jimmunol.1301914. [DOI] [PubMed] [Google Scholar]
- Aliberti J., Schulz O., Pennington D.J., Tsujimura H., Reis e Sousa C., Ozato K., Sher A. Essential role for ICSBP in the in vivo development of murine CD8α+ dendritic cells. Blood. 2003;101:305–310. doi: 10.1182/blood-2002-04-1088. [DOI] [PubMed] [Google Scholar]
- Askenase M.H., Han S.-J., Byrd A.L., Morais da Fonseca D., Bouladoux N., Wilhelm C., Konkel J.E., Hand T.W., Lacerda-Queiroz N., Su X., Trinchieri G., Grainger J.R., Belkaid Y. Bone-Marrow-Resident nk cells prime monocytes for regulatory function during infection. Immunity. 2015;42:1130–1142. doi: 10.1016/j.immuni.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auderset F., Schuster S., Fasnacht N., Coutaz M., Charmoy M., Koch U., Favre S., Wilson A., Trottein F., Alexander J., Luther S.A., MacDonald H.R., Radtke F., Tacchini-Cottier F. Notch signaling regulates follicular helper t cell differentiation. J. Immunol. 2013;191:2344–2350. doi: 10.4049/jimmunol.1300643. [DOI] [PubMed] [Google Scholar]
- Bagadia P., Huang X., Liu T.-T., Durai V., Grajales-Reyes G.E., Nitschké M., Modrusan Z., Granja J.M., Satpathy A.T., Briseño C.G., Gargaro M., Iwata A., Kim S., Chang H.Y., Shaw A.S., Murphy T.L., Murphy K.M. An Nfil3–Zeb2–Id2 pathway imposes Irf8 enhancer switching during cDC1 development. Nat. Immunol. 2019;20:1174–1185. doi: 10.1038/s41590-019-0449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaña S., Roach K., Turner S., Paul J., Kovats S. IRF4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. J. Immunol. 2012;189:3368–3377. doi: 10.4049/jimmunol.1102613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaña S., Turner S., Paul J., Ainsua-Enrich E., Kovats S. IRF4 and IRF8 Act in CD11c + cells to regulate terminal differentiation of lung tissue dendritic cells. J. Immunol. 2016;196:1666–1677. doi: 10.4049/jimmunol.1501870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balan S., Arnold-Schrauf C., Abbas A., Couespel N., Savoret J., Imperatore F., Villani A.-C., Vu Manh T.-P., Bhardwaj N., Dalod M. Large-scale human dendritic cell differentiation revealing notch-dependent lineage bifurcation and heterogeneity. Cell Rep. 2018;24:1902–1915. doi: 10.1016/j.celrep.2018.07.033. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros-Tato A., León B., Lund F.E., Randall T.D. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8+ T cell responses to influenza. Nat. Immunol. 2010;11:216–224. doi: 10.1038/ni.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton G., Zheng S., Valieris R., Tojal da Silva I., Satija R., Nussenzweig M.C. Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs. J. Exp. Med. 2016;213:2861–2870. doi: 10.1084/jem.20161135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briseño C.G., Haldar M., Kretzer N.M., Wu X., Theisen D.J., Wumesh K.C., Durai V., Grajales-Reyes G.E., Iwata A., Bagadia P., Murphy T.L., Murphy K.M. Distinct transcriptional programs control cross-priming in classical and monocyte-derived dendritic cells. Cell Rep. 2016;15:2462–2472. doi: 10.1016/j.celrep.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briseño C.G., Satpathy A.T., Davidson J.T., Ferris S.T., Durai V., Bagadia P., O’Connor K.W., Theisen D.J., Murphy T.L., Murphy K.M. Notch2-dependent DC2s mediate splenic germinal center responses. Proc. Natl. Acad. Sci. 2018;115:10726–10731. doi: 10.1073/pnas.1809925115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.C., Gudjonson H., Pritykin Y., Deep D., Lavallée V.-P., Mendoza A., Fromme R., Mazutis L., Ariyan C., Leslie C., Pe’er D., Rudensky A.Y. Transcriptional basis of mouse and human dendritic cell heterogeneity. Cell. 2019;179:846–863. doi: 10.1016/j.cell.2019.09.035. e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch R., Doebele R.C., Patil N.S., Pashine A., Mellins E.D. Accessory molecules for MHC class II peptide loading. Curr. Opin. Immunol. 2000;12:99–106. doi: 10.1016/S0952-7915(99)00057-6. [DOI] [PubMed] [Google Scholar]
- Calabro S., Gallman A., Gowthaman U., Liu D., Chen P., Liu J., Krishnaswamy J.K., Nascimento M.S.L., Xu L., Patel S.R., Williams A., Tormey C.A., Hod E.A., Spitalnik S.L., Zimring J.C., Hendrickson J.E., Stowell S.R., Eisenbarth S.C. Bridging channel dendritic cells induce immunity to transfused red blood cells. J. Exp. Med. 2016;213:887–896. doi: 10.1084/jem.20151720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton M.L., Smith-Raska M.R., Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., Nefedova Y., Corzo C.A., Gabrilovich D.I. Regulation of dendritic-cell differentiation by bone marrow stroma via different Notch ligands. Blood. 2007;109:507–515. doi: 10.1182/blood-2006-05-025601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse B., Caton M.L., Lehner M., Maeda T., Scheu S., Locksley R., Holmberg D., Zweier C., den Hollander N.S., Kant S.G., Holter W., Rauch A., Zhuang Y., Reizis B. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckers J., Sichien D., Plantinga M., Van Moorleghem J., Vanheerswynghels M., Hoste E., Malissen B., Dombrowicz D., Guilliams M., De Bosscher K., Lambrecht B.N., Hammad H. Epicutaneous sensitization to house dust mite allergen requires interferon regulatory factor 4–dependent dermal dendritic cells. J. Allergy Clin. Immunol. 2017;140:1364–1377. doi: 10.1016/j.jaci.2016.12.970. [DOI] [PubMed] [Google Scholar]
- Dekker J.D., Rhee C., Hu Z., Lee B.-K., Lee J., Iyer V.R., Ehrlich L.I.R., Georgiou G., Tucker H.O., Ippolito G.C. Lymphoid origin of a lineage of intrinsically activated plasmacytoid dendritic cell in mice and humans. bioRxiv. 2018 doi: 10.1101/310680. [DOI] [Google Scholar]
- Dell’aringa M., Lee Reinhardt R. Notch signaling represents an important checkpoint between follicular T-helper and canonical T-helper 2 cell fate article. Mucosal Immunol. 2018;11:1079–1091. doi: 10.1038/s41385-018-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desch A.N., Gibbings S.L., Clambey E.T., Janssen W.J., Slansky J.E., Kedl R.M., Henson P.M., Jakubzick C. Dendritic cell subsets require cis-activation for cytotoxic CD8 T-cell induction. Nat. Commun. 2014;5:4674. doi: 10.1038/ncomms5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicken J., Mildner A., Leshkowitz D., Touw I.P., Hantisteanu S., Jung S., Groner Y. Transcriptional reprogramming of CD11b+Esamhi dendritic cell identity and function by loss of Runx3. PLoS One. 2013;8 doi: 10.1371/journal.pone.0077490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dress R.J., Dutertre C.-A., Giladi A., Schlitzer A., Low I., Shadan N.B., Tay A., Lum J., Kairi M.F.B.M., Hwang Y.Y., Becht E., Cheng Y., Chevrier M., Larbi A., Newell E.W., Amit I., Chen J., Ginhoux F. Plasmacytoid dendritic cells develop from Ly6D+ lymphoid progenitors distinct from the myeloid lineage. Nat. Immunol. 2019;20:852–864. doi: 10.1038/s41590-019-0420-3. [DOI] [PubMed] [Google Scholar]
- Durai V., Bagadia P., Granja J.M., Satpathy A.T., Kulkarni D.H., Davidson J.T., Wu R., Patel S.J., Iwata A., Liu T.-T., Huang X., Briseño C.G., Grajales-Reyes G.E., Wöhner M., Tagoh H., Kee B.L., Newberry R.D., Busslinger M., Chang H.Y., Murphy T.L., Murphy K.M. Cryptic activation of an Irf8 enhancer governs cDC1 fate specification. Nat. Immunol. 2019;20:1161–1173. doi: 10.1038/s41590-019-0450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand M., Walter T., Pirnay T., Naessens T., Gueguen P., Goudot C., Lameiras S., Chang Q., Talaei N., Ornatsky O., Vassilevskaia T., Baulande S., Amigorena S., Segura E. Human lymphoid organ cDC2 and macrophages play complementary roles in T follicular helper responses. J. Exp. Med. 2019;216:1561–1581. doi: 10.1084/jem.20181994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre C.-A., Becht E., Irac S.E., Khalilnezhad A., Narang V., Khalilnezhad S., Ng P.Y., van den Hoogen L.L., Leong J.Y., Lee B., Chevrier M., Zhang X.M., Yong P.J.A., Koh G., Lum J., Howland S.W., Mok E., Chen J., Larbi A., Tan H.K.K., Lim T.K.H., Karagianni P., Tzioufas A.G., Malleret B., Brody J., Albani S., van Roon J., Radstake T., Newell E.W., Ginhoux F. Single-cell analysis of human mononuclear phagocytes reveals subset-defining markers and identifies circulating inflammatory dendritic cells. Immunity. 2019;51:573–589. doi: 10.1016/j.immuni.2019.08.008. e8. [DOI] [PubMed] [Google Scholar]
- Everts B., Tussiwand R., Dreesen L., Fairfax K.C., Huang S.C.C., Smith A.M., O’Neill C.M., Lam W.Y., Edelson B.T., Urban J.F., Murphy K.M., Pearce E.J. Migratory CD103 + dendritic cells suppress helminth-driven type 2 immunity through constitutive expression of IL-12. J. Exp. Med. 2016;213:35–51. doi: 10.1084/jem.20150235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasnacht N., Huang H.-Y., Koch U., Favre S., Auderset F., Chai Q., Onder L., Kallert S., Pinschewer D.D., MacDonald H.R., Tacchini-Cottier F., Ludewig B., Luther S.A., Radtke F. Specific fibroblastic niches in secondary lymphoid organs orchestrate distinct Notch-regulated immune responses. J. Exp. Med. 2014;211:2265–2279. doi: 10.1084/jem.20132528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Langarica A., Cook C., Müller Luda K., Persson E.K., Marshall J.L., Beristain-Covarrubias N., Yam-Puc J.C., Dahlgren M., Persson J.J., Uematsu S., Akira S., Henderson I.R., Lindbom B.J., Agace W., Cunningham A.F. Intestinal CD103+CD11b+ cDC2 conventional dendritic cells are required for primary CD4+ t and B cell responses to soluble flagellin. Front. Immunol. 2018;9(2409) doi: 10.3389/fimmu.2018.02409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Langarica A., Müller Luda K., Persson E.K., Cook C.N., Bobat S., Marshall J.L., Dahlgren M.W., Hägerbrand K., Toellner K.M., Goodall M.D., Withers D.R., Henderson I.R., Johansson Lindbom B., Cunningham A.F., Agace W.W. CD103+CD11b+ mucosal classical dendritic cells initiate long-term switched antibody responses to flagellin. Mucosal Immunol. 2018;11:681–692. doi: 10.1038/mi.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K., Chakarov S., Kobayashi T., Sakamoto K., Voisin B., Duan K., Nakagawa T., Horiuchi K., Amagai M., Ginhoux F., Nagao K. Cell-autonomous FLT3L shedding via ADAM10 mediates conventional dendritic cell development in mouse spleen. Proc. Natl. Acad. Sci. 2019;116:14714–14723. doi: 10.1073/pnas.1818907116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Nish S., Jiang R., Hou L., Licona-Limón P., Weinstein J., Zhao H., Medzhitov R. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity. 2013;39:722–732. doi: 10.1016/j.immuni.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto D., Wood K., Caminschi I., Murphy-Durland D., Schofield P., Christ D., Karupiah G., Brink R. The chemotactic receptor EBI2 regulates the homeostasis, localization and immunological function of splenic dendritic cells. Nat. Immunol. 2013;14:446–453. doi: 10.1038/ni.2555. [DOI] [PubMed] [Google Scholar]
- Ghosh H.S., Cisse B., Bunin A., Lewis K.L., Reizis B. Continuous Expression of the Transcription Factor E2-2 Maintains the Cell Fate of Mature Plasmacytoid Dendritic Cells. Immunity. 2010;33:905–916. doi: 10.1016/j.immuni.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Liu K., Helft J., Bogunovic M., Greter M., Hashimoto D., Price J., Yin N., Bromberg J., Lira S.A., Stanley E.R., Nussenzweig M., Merad M. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldszmid R.S., Caspar P., Rivollier A., White S., Dzutsev A., Hieny S., Kelsall B., Trinchieri G., Sher A. NK Cell-Derived Interferon-γ Orchestrates Cellular Dynamics and the Differentiation of Monocytes into Dendritic Cells at the Site of Infection. Immunity. 2012;36:1047–1059. doi: 10.1016/j.immuni.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajales-Reyes G.E., Iwata A., Albring J., Wu X., Tussiwand R., Kc W., Kretzer N.M., Briseño C.G., Durai V., Bagadia P., Haldar M., Schönheit J., Rosenbauer F., Murphy T.L., Murphy K.M. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α + conventional DC clonogenic progenitor. Nat. Immunol. 2015;16:708–717. doi: 10.1038/ni.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M., Ginhoux F., Jakubzick C., Naik S.H., Onai N., Schraml B.U., Segura E., Tussiwand R., Yona S. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol. 2014;14:571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M., Dutertre C.A., Scott C.L., McGovern N., Sichien D., Chakarov S., Van Gassen S., Chen J., Poidinger M., De Prijck S., Tavernier S.J., Low I., Irac S.E., Mattar C.N., Sumatoh H.R., Low G.H.L., Chung T.J.K., Chan D.K.H., Tan K.K., Hon T.L.K., Fossum E., Bogen B., Choolani M., Chan J.K.Y., Larbi A., Luche H., Henri S., Saeys Y., Newell E.W., Lambrecht B.N., Malissen B., Ginhoux F. Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity. 2016;45:669–684. doi: 10.1016/j.immuni.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther P., Cirovic B., Baßler K., Händler K., Becker M., Dutertre C.A., Bigley V., Newell E., Collin M., Ginhoux F., Schlitzer A., Schultze J.L. A rule-based data-informed cellular consensus map of the human mononuclear phagocyte cell space. bioRxiv. 2019 doi: 10.1101/658179. [DOI] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., Schreiber R.D., Murphy T.L., Murphy K.M. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic t cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashima K., Banks T.A., Ansel K.M., Lu T.T., Ware C.F., Cyster J.G. Intrinsic Lymphotoxin-β receptor requirement for homeostasis of lymphoid tissue dendritic cells. Immunity. 2005;22:439–450. doi: 10.1016/j.immuni.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Kim T.S., Gorski S.A., Hahn S., Murphy K.M., Braciale T.J. Distinct Dendritic Cell Subsets Dictate the Fate Decision between Effector and Memory CD8+T Cell Differentiation by a CD24-Dependent Mechanism. Immunity. 2014;40:400–413. doi: 10.1016/j.immuni.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnebrew M.A., Buffie C.G., Diehl G.E., Zenewicz L.A., Leiner I., Hohl T.M., Flavell R.A., Littman D.R., Pamer E.G. Interleukin 23 production by intestinal CD103+CD11b+ dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkling M.E., Cytlak U., Lau C.M., Lewis K.L., Resteu A., Khodadadi-Jamayran A., Siebel C.W., Salmon H., Merad M., Tsirigos A., Collin M., Bigley V., Reizis B. Notch signaling facilitates in vitro generation of cross-presenting classical dendritic cells. Cell Rep. 2018;23:3658–3672. doi: 10.1016/j.celrep.2018.05.068. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy J.K., Gowthaman U., Zhang B., Mattsson J., Szeponik L., Liu D., Wu R., White T., Calabro S., Xu L., Collet M.A., Yurieva M., Alsén S., Fogelstrand P., Walter A., Heath W.R., Mueller S.N., Yrlid U., Williams A., Eisenbarth S.C. Migratory CD11b + conventional dendritic cells induce T follicular helper cell–dependent antibody responses. Sci. Immunol. 2017;2 doi: 10.1126/sciimmunol.aam9169. eaam9169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto Y., Linehan M., Weinstein J.S., Laidlaw B.J., Craft J.E., Iwasaki A. CD301b+ dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity. 2013;39:733–743. doi: 10.1016/j.immuni.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotaki D., Osato N., Nishiyama A., Yamamoto M., Ban T., Sato H., Nakabayashi J., Umehara M., Miyake N., Matsumoto N., Nakazawa M., Ozato K., Tamura T. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood. 2013;121:1839–1849. doi: 10.1182/blood-2012-06-437863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotaki D., Yamamoto M., Nishiyama A., Uno K., Ban T., Ichino M., Sasaki H., Matsunaga S., Yoshinari M., Ryo A., Nakazawa M., Ozato K., Tamura T. IRF8 inhibits C/EBPα activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nat. Commun. 2014;5:4978. doi: 10.1038/ncomms5978. [DOI] [PubMed] [Google Scholar]
- Kurotaki D., Nakabayashi J., Nishiyama A., Sasaki H., Kawase W., Kaneko N., Ochiai K., Igarashi K., Ozato K., Suzuki Y., Tamura T. Transcription factor IRF8 governs enhancer landscape dynamics in mononuclear phagocyte progenitors. Cell Rep. 2018;22:2628–2641. doi: 10.1016/j.celrep.2018.02.048. [DOI] [PubMed] [Google Scholar]
- Kurotaki D., Kawase W., Sasaki H., Nakabayashi J., Nishiyama A., Morse H.C., Ozato K., Suzuki Y., Tamura T. Epigenetic control of early dendritic cell lineage specification by the transcription factor IRF8 in mice. Blood. 2019;133:1803–1813. doi: 10.1182/blood-2018-06-857789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw B.J., Cui W., Amezquita R.A., Gray S.M., Guan T., Lu Y., Kobayashi Y., Flavell R.A., Kleinstein S.H., Craft J., Kaech S.M. Production of IL-10 by CD4+ regulatory T cells during the resolution of infection promotes the maturation of memory CD8+ T cells. Nat. Immunol. 2015;16:871–879. doi: 10.1038/ni.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Zhou Y.J., Ma W., Zhang W., Aljoufi A., Luh T., Lucero K., Liang D., Thomsen M., Bhagat G., Shen Y., Liu K. Lineage specification of human dendritic cells is marked by IRF8 expression in hematopoietic stem cells and multipotent progenitors. Nat. Immunol. 2017;18:877–888. doi: 10.1038/ni.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K.L., Caton M.L., Bogunovic M., Greter M., Grajkowska L.T., Ng D., Klinakis A., Charo I.F., Jung S., Gommerman J.L., Ivanov I.I., Liu K., Merad M., Reizis B. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan J.L., Dileepan T., Kashem S.W., Kaplan D.H., Cleary P., Jenkins M.K. Generation of Th17 cells in response to intranasal infection requires TGF-β1 from dendritic cells and IL-6 from CD301b + dendritic cells. Proc. Natl. Acad. Sci. 2015;112:12782–12787. doi: 10.1073/pnas.1513532112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Guan X., Tamura T., Ozato K., Ma X. Synergistic activation of Interleukin-12 p35 gene transcription by interferon regulatory Factor-1 and interferon consensus sequence-binding protein. J. Biol. Chem. 2004;279:55609–55617. doi: 10.1074/jbc.M406565200. [DOI] [PubMed] [Google Scholar]
- Lohoff M., Mittrucker H.-W., Prechtl S., Bischof S., Sommer F., Kock S., Ferrick D.A., Duncan G.S., Gessner A., Mak T.W. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc. Natl. Acad. Sci. 2002;99:11808–11812. doi: 10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu E., Dang E.V., McDonald J.G., Cyster J.G. Distinct oxysterol requirements for positioning naïve and activated dendritic cells in the spleen. Sci. Immunol. 2017;2 doi: 10.1126/sciimmunol.aal5237. eaal5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luda K.M., Joeris T., Persson E.K., Rivollier A., Demiri M., Sitnik K.M., Pool L., Holm J.B., Melo-Gonzalez F., Richter L., Lambrecht B.N., Kristiansen K., Travis M.A., Svensson-Frej M., Kotarsky K., Agace W.W. IRF8 transcription-factor-dependent classical dendritic cells are essential for intestinal T cell homeostasis. Immunity. 2016;44:860–874. doi: 10.1016/j.immuni.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Lugo-Villarino G., Maldonado-Lopez R., Possemato R., Penaranda C., Glimcher L.H. T-bet is required for optimal production of IFN- and antigen-specific T cell activation by dendritic cells. Proc. Natl. Acad. Sci. 2003;100:7749–7754. doi: 10.1073/pnas.1332767100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Lee J., Backenroth D., Zhou Y.J., Bush E., Sims P., Liu K., Shen Y. Single cell RNA-Seq reveals pre-cDCs fate determined by transcription factor combinatorial dose. BMC Mol. Cell Biol. 2019;20:20. doi: 10.1186/s12860-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-López M., Iborra S., Conde-Garrosa R., Sancho D. Batf3-dependent CD103 + dendritic cells are major producers of IL-12 that drive local Th1 immunity against Leishmania major infection in mice. Eur. J. Immunol. 2015;45:119–129. doi: 10.1002/eji.201444651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashayekhi M., Sandau M.M., Dunay I.R., Frickel E.M., Khan A., Goldszmid R.S., Sher A., Ploegh H.L., Murphy T.L., Sibley L.D., Murphy K.M. CD8α+ dendritic cells are the critical source of Interleukin-12 that controls acute infection by toxoplasma gondii tachyzoites. Immunity. 2011;35:249–259. doi: 10.1016/j.immuni.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei F., Schiavoni G., Borghi P., Venditti M., Canini I., Sestili P., Pietraforte I., Morse H.C., Ramoni C., Belardelli F., Gabriele L. ICSBP/IRF-8 differentially regulates antigen uptake during dendritic-cell development and affects antigen presentation to CD4+ T cells. Blood. 2006;108:609–617. doi: 10.1182/blood-2005-11-4490. [DOI] [PubMed] [Google Scholar]
- Mayer J.U., Demiri M., Agace W.W., MacDonald A.S., Svensson-Frej M., Milling S.W. Different populations of CD11b+ dendritic cells drive Th2 responses in the small intestine and colon. Nat. Commun. 2017;8:15820. doi: 10.1038/ncomms15820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., Brown B.D., Shay T., Gautier E.L., Jojic V., Cohain A., Pandey G., Leboeuf M., Elpek K.G., Helft J., Hashimoto D., Chow A., Price J., Greter M., Bogunovic M., Bellemare-Pelletier A., Frenette P.S., Randolph G.J., Turley S.J., Merad M., Jakubzick C., Best A.J., Knell J., Goldrath A., Koller D., Cohen N., Brennan P., Brenner M., Regev A., Fletcher A., Malhotra D., Jianu R., Laidlaw D., Collins J., Narayan K., Sylvia K., Kang J., Gazit R., Rossi D.J., Kim F., Rao T.N., Wagers A., Shinton S.A., Hardy R.R., Monach P., Bezman N.A., Sun J.C., Kim C.C., Lanier L.L., Heng T., Kreslavsky T., Painter M., Ericson J., Davis S., Mathis D., Benoist C. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 2012;13:888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H.-G., Kim S., Jeong J.J., Han S.-S., Jarjour N.N., Lee H., Abboud-Werner S.L., Chung S., Choi H.S., Natarajan V., Ackerman S.J., Christman J.W., Park G.Y. Airway epithelial cell-derived colony stimulating Factor-1 promotes allergen sensitization. Immunity. 2018;49:275–287. doi: 10.1016/j.immuni.2018.06.009. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T.L., Grajales-Reyes G.E., Wu X., Tussiwand R., Briseño C.G., Iwata A., Kretzer N.M., Durai V., Murphy K.M. Transcriptional control of dendritic cell development. Annu. Rev. Immunol. 2016;34:93–119. doi: 10.1146/annurev-immunol-032713-120204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S.H., Proietto A.I., Wilson N.S., Dakic A., Schnorrer P., Fuchsberger M., Lahoud M.H., O’Keeffe M., Shao Q., Chen W., Villadangos J.A., Shortman K., Wu L. Cutting edge: generation of splenic CD8 + and CD8 − dendritic cell equivalents in fms-like tyrosine kinase 3 ligand bone marrow cultures. J. Immunol. 2005;174:6592–6597. doi: 10.4049/jimmunol.174.11.6592. [DOI] [PubMed] [Google Scholar]
- Negishi H., Ohba Y., Yanai H., Takaoka A., Honma K., Yui K., Matsuyama T., Taniguchi T., Honda K. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc. Natl. Acad. Sci. 2005;102:15989–15994. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutsch K., Chai J.N., Ai T.L., Russler-Germain E., Feehley T., Nagler C.R., Hsieh C.-S. Rapid and efficient generation of regulatory t cells to commensal antigens in the periphery. Cell Rep. 2016;17:206–220. doi: 10.1016/j.celrep.2016.08.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Tussiwand R., Lanzavecchia A., Manz M.G. Activation of the Flt3 signal transduction cascade rescues and enhances type I interferon–producing and dendritic cell development. J. Exp. Med. 2006;203:227–238. doi: 10.1084/jem.20051645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panea C., Farkas A.M., Goto Y., Abdollahi-Roodsaz S., Lee C., Koscsó B., Gowda K., Hohl T.M., Bogunovic M., Ivanov I.I. Intestinal monocyte-derived macrophages control commensal-specific Th17 responses. Cell Rep. 2015;12:1314–1324. doi: 10.1016/j.celrep.2015.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson E.K., Uronen-Hansson H., Semmrich M., Rivollier A., Hägerbrand K., Marsal J., Gudjonsson S., Håkansson U., Reizis B., Kotarsky K., Agace W.W. IRF4 transcription-factor-Dependent CD103+CD11b+ dendritic cells drive mucosal t helper 17 cell differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Pierre P., Mellman I. Developmental regulation of invariant chain proteolysis controls MHC class II trafficking in mouse dendritic cells. Cell. 1998;93:1135–1145. doi: 10.1016/S0092-8674(00)81458-0. [DOI] [PubMed] [Google Scholar]
- Rodrigues P.F., Alberti-Servera L., Eremin A., Grajales-Reyes G.E., Ivanek R., Tussiwand R. Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat. Immunol. 2018;19:711–722. doi: 10.1038/s41590-018-0136-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roquilly A., McWilliam H.E.G., Jacqueline C., Tian Z., Cinotti R., Rimbert M., Wakim L., Caminschi I., Lahoud M.H., Belz G.T., Kallies A., Mintern J.D., Asehnoune K., Villadangos J.A. Local modulation of antigen-presenting cell development after resolution of pneumonia induces long-term susceptibility to secondary infections. Immunity. 2017;47:135–147. doi: 10.1016/j.immuni.2017.06.021. e5. [DOI] [PubMed] [Google Scholar]
- Satpathy A.T., Briseño C.G., Lee J.S., Ng D., Manieri N.A., KC W., Wu X., Thomas S.R., Lee W.-L., Turkoz M., McDonald K.G., Meredith M.M., Song C., Guidos C.J., Newberry R.D., Ouyang W., Murphy T.L., Stappenbeck T.S., Gommerman J.L., Nussenzweig M.C., Colonna M., Kopan R., Murphy K.M. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 2013;14:937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharton-Kersten T., Contursi C., Masumi A., Sher A., Ozato K. Interferon consensus sequence binding protein–deficient mice display impaired resistance to intracellular infection due to a primary defect in interleukin 12 p40 induction. J. Exp. Med. 1997;186:1523–1534. doi: 10.1084/jem.186.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavoni G., Mattei F., Sestili P., Borghi P., Venditti M., Morse H.C., Belardelli F., Gabriele L. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8α+ dendritic cells. J. Exp. Med. 2002;196:1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavoni G., Mattei F., Borghi P., Sestili P., Venditti M., Morse H.C., Belardelli F., Gabriele L. ICSBP is critically involved in the normal development and trafficking of Langerhans cells and dermal dendritic cells. Blood. 2004;103:2221–2228. doi: 10.1182/blood-2003-09-3007. [DOI] [PubMed] [Google Scholar]
- Schlitzer A., McGovern N., Teo P., Zelante T., Atarashi K., Low D., Ho A.W.S., See P., Shin A., Wasan P.S., Hoeffel G., Malleret B., Heiseke A., Chew S., Jardine L., Purvis H.A., Hilkens C.M.U., Tam J., Poidinger M., Stanley E.R., Krug A.B., Renia L., Sivasankar B., Ng L.G., Collin M., Ricciardi-Castagnoli P., Honda K., Haniffa M., Ginhoux F. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A., Sivakamasundari V., Chen J., Sumatoh H.R., Bin Schreuder J., Lum J., Malleret B., Zhang S., Larbi A., Zolezzi F., Renia L., Poidinger M., Naik S., Newell E.W., Robson P., Ginhoux F. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 2015;16:718–728. doi: 10.1038/ni.3200. [DOI] [PubMed] [Google Scholar]
- Schönheit J., Kuhl C., Gebhardt M.L., Klett F.F., Riemke P., Scheller M., Huang G., Naumann R., Leutz A., Stocking C., Priller J., Andrade-Navarro M.A., Rosenbauer F. PU.1 level-directed chromatin structure remodeling at the Irf8 gene drives dendritic cell commitment. Cell Rep. 2013;3:1617–1628. doi: 10.1016/j.celrep.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Scott C.L., Omilusik K.D. ZEBs: novel players in immune cell development and function. Trends Immunol. 2019;40:431–446. doi: 10.1016/j.it.2019.03.001. [DOI] [PubMed] [Google Scholar]
- Scott C.L., Bain C.C., Wright P.B., Sichien D., Kotarsky K., Persson E.K., Luda K., Guilliams M., Lambrecht B.N., Agace W.W., Milling S.W.F., Mowat A.M. CCR2 + CD103 - Intestinal dendritic cells develop from DC-committed precursors and induce interleukin-17 production by T cells. Mucosal Immunol. 2015;8:327–339. doi: 10.1038/mi.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C.L., Soen B., Martens L., Skrypek N., Saelens W., Taminau J., Blancke G., Van Isterdael G., Huylebroeck D., Haigh J., Saeys Y., Guilliams M., Lambrecht B.N., Berx G. The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J. Exp. Med. 2016;213:897–911. doi: 10.1084/jem.20151715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See P., Dutertre C.-A., Chen J., Günther P., McGovern N., Irac S.E., Gunawan M., Beyer M., Händler K., Duan K., Sumatoh H.R., Bin Ruffin N., Jouve M., Gea-Mallorquí E., Hennekam R.C.M., Lim T., Yip C.C., Wen M., Malleret B., Low I., Shadan N.B., Fen C.F.S., Tay A., Lum J., Zolezzi F., Larbi A., Poidinger M., Chan J.K.Y., Chen Q., Rénia L., Haniffa M., Benaroch P., Schlitzer A., Schultze J.L., Newell E.W., Ginhoux F. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 2017:356. doi: 10.1126/science.aag3009. eaag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C., Jackson J.T., Markey K.A., Brady H.J.M., Hill G.R., MacDonald K.P.A., Nutt S.L., Belz G.T. CD8α+ DCs can be induced in the absence of transcription factors Id2, Nfil3, and Batf3. Blood. 2013;121:1574–1583. doi: 10.1182/blood-2012-07-445650. [DOI] [PubMed] [Google Scholar]
- Shin J.-S., Ebersold M., Pypaert M., Delamarre L., Hartley A., Mellman I. Surface expression of MHC class II in dendritic cells is controlled by regulated ubiquitination. Nature. 2006;444:115–118. doi: 10.1038/nature05261. [DOI] [PubMed] [Google Scholar]
- Shin H., Kumamoto Y., Gopinath S., Iwasaki A. CD301b+ dendritic cells stimulate tissue-resident memory CD8+ T cells to protect against genital HSV-2. Nat. Commun. 2016;7:13346. doi: 10.1038/ncomms13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichien D., Scott C.L., Martens L., Vanderkerken M., Van Gassen S., Plantinga M., Joeris T., De Prijck S., Vanhoutte L., Vanheerswynghels M., Van Isterdael G., Toussaint W., Madeira F.B., Vergote K., Agace W.W., Clausen B.E., Hammad H., Dalod M., Saeys Y., Lambrecht B.N., Guilliams M. IRF8 transcription factor controls survival and function of terminally differentiated conventional and plasmacytoid dendritic cells. Respectively. Immunity. 2016;45:626–640. doi: 10.1016/j.immuni.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Sichien D., Lambrecht B.N., Guilliams M., Scott C.L. Development of conventional dendritic cells: from common bone marrow progenitors to multiple subsets in peripheral tissues. Mucosal Immunol. 2017;10:831–844. doi: 10.1038/mi.2017.8. [DOI] [PubMed] [Google Scholar]
- Smita S., Ahad A., Ghosh A., Biswas V.K., Koga M.M., Gupta B., Acha-Orbea H., Raghav S.K. Importance of EMT factor ZEB1 in cDC1 “MutuDC line” mediated induction of Th1 immune response. Front. Immunol. 2018;9(2604) doi: 10.3389/fimmu.2018.02604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Honma K., Matsuyama T., Suzuki K., Toriyama K., Akitoyo I., Yamamoto K., Suematsu T., Nakamura M., Yui K., Kumatori A. Critical roles of interferon regulatory factor 4 in CD11b high CD8α – dendritic cell development. Proc. Natl. Acad. Sci. 2004;101:8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T., Tailor P., Yamaoka K., Kong H.J., Tsujimura H., O’Shea J.J., Singh H., Ozato K. IFN regulatory Factor-4 and -8 govern dendritic cell subset development and their functional diversity. J. Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- Theisen D.J., Ferris S.T., Briseño C.G., Kretzer N., Iwata A., Murphy K.M., Murphy T.L. Batf3-dependent genes control tumor rejection induced by dendritic cells independently of cross-presentation. Cancer Immunol. Res. 2019;7:29–39. doi: 10.1158/2326-6066.CIR-18-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R., Lee W.L., Murphy T.L., Mashayekhi M., Kc W., Albring J.C., Satpathy A.T., Rotondo J.A., Edelson B.T., Kretzer N.M., Wu X., Weiss L.A., Glasmacher E., Li P., Liao W., Behnke M., Lam S.S.K., Aurthur C.T., Leonard W.J., Singh H., Stallings C.L., David Sibley L., Schreiber R.D., Murphy K.M. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;290:502–507. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R., Everts B., Grajales-Reyes G.E., Kretzer N.M., Iwata A., Bagaitkar J., Wu X., Wong R., Anderson D.A., Murphy T.L., Pearce E.J., Murphy K.M. Klf4 expression in conventional dendritic cells is required for t helper 2 cell responses. Immunity. 2015;42:916–928. doi: 10.1016/j.immuni.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Borght K., Scott C.L., Nindl V., Bouché A., Martens L., Sichien D., Van Moorleghem J., Vanheerswynghels M., De Prijck S., Saeys Y., Ludewig B., Gillebert T., Guilliams M., Carmeliet P., Lambrecht B.N. Myocardial infarction primes autoreactive t cells through activation of dendritic cells. Cell Rep. 2017;18:3005–3017. doi: 10.1016/j.celrep.2017.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Borght K., Scott C.L., Martens L., Sichien D., Van Isterdael G., Nindl V., Saeys Y., Boon L., Ludewig B., Gillebert T.C., Lambrecht B.N. Myocarditis elicits dendritic cell and monocyte infiltration in the heart and self-antigen presentation by conventional type 2 dendritic cells. Front. Immunol. 2018;9(2714) doi: 10.3389/fimmu.2018.02714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Lugt B., Khan A.A., Hackney J.A., Agrawal S., Lesch J., Zhou M., Lee W.P., Park S., Xu M., Devoss J., Spooner C.J., Chalouni C., Delamarre L., Mellman I., Singh H. Transcriptional programming of dendritic cells for enhanced MHC class II antigen presentation. Nat. Immunol. 2014;15:161–167. doi: 10.1038/ni.2795. [DOI] [PubMed] [Google Scholar]
- Vander Lugt B., Riddell J., Khan A.A., Hackney J.A., Lesch J., DeVoss J., Weirauch M.T., Singh H., Mellman I. Transcriptional determinants of tolerogenic and immunogenic states during dendritic cell maturation. J. Cell Biol. 2017;216:779–792. doi: 10.1083/jcb.201512012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani A.-C., Satija R., Reynolds G., Sarkizova S., Shekhar K., Fletcher J., Griesbeck M., Butler A., Zheng S., Lazo S., Jardine L., Dixon D., Stephenson E., Nilsson E., Grundberg I., McDonald D., Filby A., Li W., De Jager P.L., Rozenblatt-Rosen O., Lane A.A., Haniffa M., Regev A., Hacohen N. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017:356. doi: 10.1126/science.aah4573. eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I.-M., Contursi C., Masumi A., Ma X., Trinchieri G., Ozato K. An IFN-γ-Inducible transcription factor, IFN consensus sequence binding protein (ICSBP), stimulates IL-12 p40 expression in macrophages. J. Immunol. 2000;165:271–279. doi: 10.4049/jimmunol.165.1.271. [DOI] [PubMed] [Google Scholar]
- Wang Y.-G., Kim K.D., Wang J., Yu P., Fu Y.-X. Stimulating lymphotoxin β receptor on the dendritic cells is critical for their homeostasis and expansion. J. Immunol. 2005;175:6997–7002. doi: 10.4049/jimmunol.175.10.6997. [DOI] [PubMed] [Google Scholar]
- Wang J., Fathman J.W., Lugo-Villarino G., Scimone L., Von Andrian U., Dorfman D.M., Glimcher L.H. Transcription factor T-bet regulates inflammatory arthritis through its function in dendritic cells. J. Clin. Invest. 2006 doi: 10.1172/JCI26631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskow C., Liu K., Darrasse-Jèze G., Guermonprez P., Ginhoux F., Merad M., Shengelia T., Yao K., Nussenzweig M. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat. Immunol. 2008;9:676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.W., Tjota M.Y., Clay B.S., Vander Lugt B., Bandukwala H.S., Hrusch C.L., Decker D.C., Blaine K.M., Fixsen B.R., Singh H., Sciammas R., Sperling A.I. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat. Commun. 2013;4:2990. doi: 10.1038/ncomms3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worbs T., Hammerschmidt S.I., Förster R. Dendritic cell migration in health and disease. Nat. Rev. Immunol. 2017;17:30–48. doi: 10.1038/nri.2016.116. [DOI] [PubMed] [Google Scholar]
- Wu X., Briseño C.G., Grajales-Reyes G.E., Haldar M., Iwata A., Kretzer N.M., KC W., Tussiwand R., Higashi Y., Murphy T.L., Murphy K.M. Transcription factor Zeb2 regulates commitment to plasmacytoid dendritic cell and monocyte fate. Proc. Natl. Acad. Sci. 2016;113:14775–14780. doi: 10.1073/pnas.1611408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Chaudhri V.K., Wu Z., Biliouris K., Dienger-Stambaugh K., Rochman Y., Singh H. Regulation of bifurcating B cell trajectories by mutual antagonism between transcription factors IRF4 and IRF8. Nat. Immunol. 2015;16:1274–1281. doi: 10.1038/ni.3287. [DOI] [PubMed] [Google Scholar]
- Yi T., Cyster J.G. EBI2-mediated bridging channel positioning supports splenic dendritic cell homeostasis and particulate antigen capture. Elife. 2013;2(e00757) doi: 10.7554/eLife.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Ho A.W.S., Schlitzer A., Tang Y., Wong K.H.S., Wong F.H.S., Chua Y.L., Angeli V., Mortellaro A., Ginhoux F., Kemeny D.M. GM-CSF–Licensed CD11b + lung dendritic cells orchestrate Th2 immunity to Blomia tropicalis. J. Immunol. 2014;193:496–509. doi: 10.4049/jimmunol.1303138. [DOI] [PubMed] [Google Scholar]