Abstract
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic is a global public health emergency, and new therapeutics are needed. This article reports the potential drug target and mechanism of action of Arbidol (umifenovir) to treat coronavirus disease 2019 (COVID-19). Molecular dynamics and structural analysis were used to show how Arbidol targets the SARS-CoV-2 spike glycoprotein and impedes its trimerization, which is key for host cell adhesion and hijacking, indicating the potential of Arbidol to treat COVID-19. It is hoped that knowledge of the potential drug target and mechanism of action of Arbidol will help in the development of new therapeutics for SARS-CoV-2.
Keywords: Coronavirus, Antiviral, Spike glycoprotein, Molecular dynamics, COVID-19
1. Introduction
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic has had a major impact on the health of millions of people and the global economy [1]. To date, more than 126,212 deaths and nearly 2 million confirmed cases have been reported globally, making SARS-CoV-2 an urgent public health concern. As well as using neutralizing antibodies that target spike glycoproteins, which are involved in host cell adhesion [2], several antiviral drugs and other drugs (e.g. hydroxychloroquine) are being evaluated to repurpose as possible treatments for coronavirus disease 2019 (COVID-19) [3]. The different classes of antivirals under evaluation include 3CL protein inhibitors (ribavirin, lopinavir/ritonavir), RNA synthesis inhibitors (remdesivir, tenofovir disoproxil fumarate and 3TC), neuraminidase inhibitors (oseltamivir and peramivir ) [4] and other small molecule drugs which target the ability of SARS-CoV-2 to interact with host cells (ACE2 inhibitors) [3,5]. However, the potential drug target and mechanism of action of several candidate drugs remain elusive, and further structural and biophysical studies are needed to determine how these drugs bind and impact on SARS-CoV-2. Arbidol (umifenovir) (Fig. 1 A) is also being screened for use against SARS-CoV-2 [6]. However, the potential drug target and mechanism of action of Arbidol against SARS-CoV-2 are not known. Considering the current public health crisis, this study aimed to determine the potential drug target, molecular interactions and mechanism of action of Arbidol on SARS-CoV-2. It is hoped that knowledge of the mechanism of action of Arbidol will help in the development of new therapeutics for SARS-CoV-2.
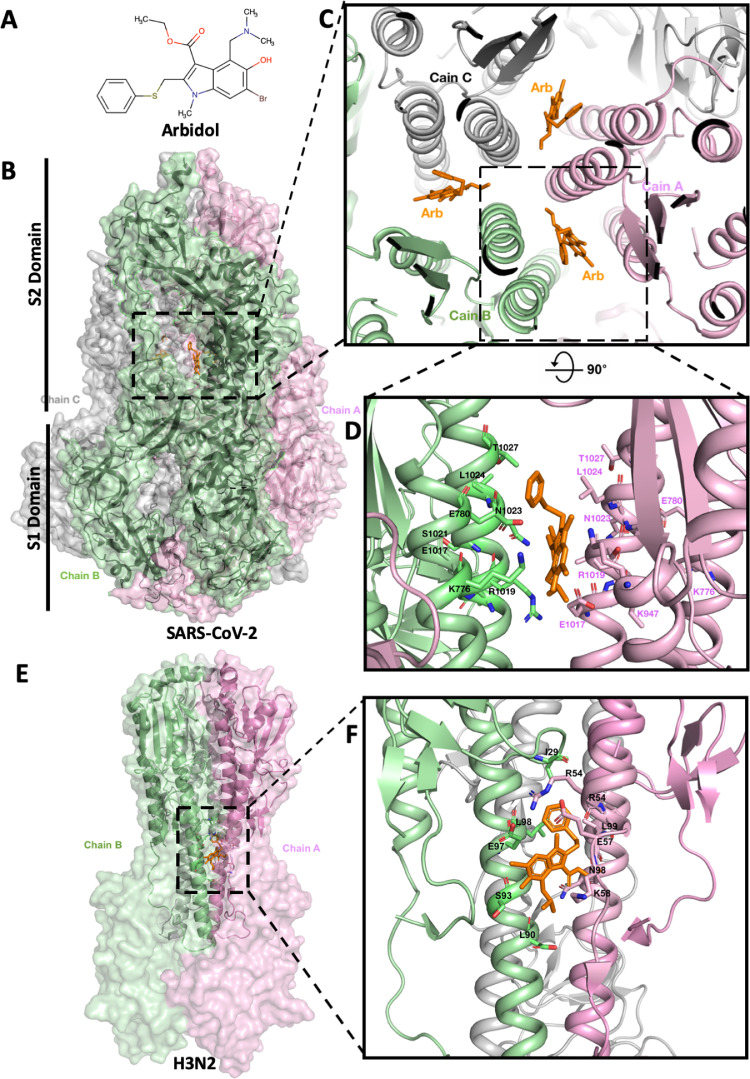
Fig. 1.
Arbidol binding site on SARS-CoV-2 spike glycoprotein. (A) Two-dimensional molecular structure of Arbidol. (B) Side view and overall view of the three-dimensional structure of Arbidol in complex with SARS-CoV-2 spike glycoprotein (surface model). The homotrimer structure of the spike glycoprotein is shown as a transparent surface (Chains A, B and C coloured in pink, green and grey, respectively), and the secondary structure in backbone traces. Arbidol is shown in orange. S1 and S2 domains are labelled. (C) Arbidol binding region in SARS-CoV-2 spike glycoprotein (top view). Three identical Arbidol binding sites are shown, viewed along the three-fold symmetry axis of the trimer. Individual monomers are coloured as above and labelled accordingly. (D) Cartoon model showing the Arbidol binding site and the key side chain residues (labelled accordingly) of SARS-CoV-2 spike glycoprotein involved in the interaction with Arbidol (orange). (E) Side view and overall view of the three-dimensional structure of Arbidol in complex with H3N2 haemagglutinin (HA) (surface model). Colour coding and labelling as above. (F) Cartoon model showing the Arbidol binding site and the key side chain residues (labelled accordingly) of H3N2 HA involved in the interaction with Arbidol (orange).
2. Rationale and sequence comparison
Arbidol is used to treat influenza [1,7] and acts by binding to haemagglutinin (HA) protein. Any sequence or structural similarities between SARS-CoV-2 spike glycoprotein and influenza virus (H3N2) HA could have a positive drug effect. Comparative protein sequence analysis showed that a short region of the trimerization domain (S2) (aa947–aa1027) of SARS-CoV-2 spike glycoprotein resembles that of H3N2 HA (Fig. S1A, see online supplementary material). The outer membrane of SARS-CoV-2 spike glycoprotein is essential for host cell adhesion via human ACE2 and CD26 receptors [2,8], and its trimerization is imperative for host membrane fusion. This study aimed to determine if Arbidol could bind to H3N2 HA and SARS-CoV-2 spike glycoprotein in a similar way. Finding the potential drug target and mechanism of action of Arbidol has great implications, and could help in the development of new therapeutics for SARS-CoV-2.
3. Molecular dynamics, docking and structure refinement
Molecular dynamics and structure-guided drug-binding analysis were undertaken to screen Arbidol binding sites in SARS-CoV-2 spike glycoprotein through two independent servers – HADDOCK2.2 (https://haddock.science.uu.nl/) and SwissDock (http://swissdock.ch/docking) – using the spike glycoprotein trimer (PDB: 6VSB) [2]. The predictions from both servers were consistent and showed six positions where Arbidol could potentially interact with SARS-CoV-2 spike glycoprotein (Fig. S1B and S1C, see online supplementary material): a single false-positive site (C1), a single true site (C2) and four unimportant/surface binding regions (C3–6). These were assessed and corroborated based on solvent accessibility surface area, C-score (confidence score) and Z-score (clash score) of the binding location and exposed residues of SARS-CoV-2 spike glycoprotein. Further refinement was completed using Coot (www.mrc-imb.cam.uk/) to ensure appropriate docking and no clashes in the side chain residues. Binding free energies were taken into consideration to select the best-possible docking site. The final model with Aribidol docked in the homotrimer structure of SARS-CoV-2 spike glycoprotein (Fig. 1B) was visualized in PyMol.
4. Key findings and mechanism of Arbidol binding to SARS-CoV-2 spike glycoprotein
To determine the binding mode and mechanism of action of Arbidol, the most likely amino acid residues of SARS-CoV-2 spike glycoprotein involved in establishing the interaction with Arbidol were considered. K776, E780, K947, E1017, R1019, S1021, N1023, L1024 and T1027 of the S2 trimerization domain were found to interact with Arbidol via van der Waals forces or hydrogen bonding (Fig. 1D and Fig. S1E, see online supplementary material). Arbidol is sandwiched between the trimerization helices of two protomers: K776, R1019, N1023, L1024, T1027 (Chain A) and E780, K947, E1017, S1021, L1024 (Chain B) (Fig. 1C and 1D) with a total buried interface of ~900Å2 for three Arbidol binding regions (~300Å2/Arbidol), as calculated from the PISA server (https://www.ebi.ac.uk/pdbe/pisa/). Next, the interaction between Arbidol and H3N2 HA (PDB: 5T6N) was examined [7]. Interestingly, this interaction is mediated with Arg, Lys, Leu and Glu as the key interacting residues (R54, E57, K58, L90, E97, L98 and L99) (Fig. 1E and 1F). Furthermore, Arbidol is sandwiched between the trimerization helices of two protomers (Chains A and B, Fig. 1F). The positions of H3N2 HA binding residues E57, R54, E97, L98, L99, K307, T59 and K58 were similar to those of SARS-CoV-2 spike glycoprotein residues K776, E780, E1017, R1019, L1024 and Y1027. In order to compare structural similarities between the SARS-CoV-2 and H3N2 trimerization domains involved in Arbidol binding, the binding regions of SARS-CoV-2 and H3N2 were superimposed, and the root-mean-square deviation for Cα atoms was 0.82Å. This suggests possible structural similarities in the drug binding region of SARS-CoV-2 spike glycoprotein and H3N2 HA, and shows how an influenza drug (Arbidol) could be a potential drug to treat COVID-19.
5. Conclusions
The sequence and structural similarities between the Arbidol binding sites for SARS-CoV-2 spike glycoprotein and H3N2 HA seem promising and suggest that Arbidol may have efficacy to treat COVID-19. Molecular dynamics and structural analysis showed that SARS-CoV-2 spike glycoprotein is the drug target for Arbidol, and suggested the potential drug binding mode with key interacting residues and mechanism of action, whereby Arbidol can effectively block or impede the trimerization of SARS-CoV-2 spike glycoprotein, which is key to cell adherence and entry [9]. Blocking the trimerization of SARS-CoV-2 spike glycoprotein also leads to the formation of naked or immature virus which is less infectious. This study has significant implications, but the efficacy and safety of Arbidol against SARS-CoV-2 still require clinical investigation. Although current drug options are mainly focused on ACE2 inhibitors and viral RNA polymerase, potential drugs to impede the trimerization of SARS-CoV-2 spike glycoprotein are equally essential as a combination of drugs may have a profound effect on the battle against SARS-CoV-2. It is hoped that knowledge of the potential drug target and mechanism of action of Arbidol will help in the development of new therapeutics for SARS-CoV-2.
Acknowledgements
The author wishes to thank Monash University Software Platform for access to the software licence, and Joseph Polidano of the University of Melbourne and Vijaya Kumar Pidugu of the National Cancer Institute for editing and proofreading the manuscript.
Funding: None.
Competing interests: None declared.
Ethical approval: Not required.
Editor: Jean-Marc Rolain
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijantimicag.2020.105998.
Appendix. Supplementary materials
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 4.Baron SA, Devaux C, Colson P, Raoult D, Rolain JM. Teicoplanin: an alternative drug for the treatment of coronavirus COVID-19? Int J Antimicrob Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Q, Xiong HR, Lu L, Liu YY, Luo F, Hou W, et al. Antiviral and anti-inflammatory activity of arbidol hydrochloride in influenza A (H1N1) virus infection. Acta Pharmacol Sin. 2013;34:1075–1083. doi: 10.1038/aps.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Zhou L, Yang Y, Peng W, Wang W, Chen X. Therapeutic and triage strategies for 2019 novel coronavirus disease in fever clinics. Lancet Respir Med. 2020;8:e11–e12. doi: 10.1016/S2213-2600(20)30071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadam RU, Wilson IA. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc Natl Acad Sci USA. 2017;114:206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vankadari N, Wilce JA. Emerging Wuhan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg Microb Infect. 2020;9:601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.