Abstract
Coronaviruses (CoV) are a large family of viruses causing a spectrum of disease ranging from the common cold to more severe diseases as Middle East Respiratory Syndrome (MERS-CoV) and Severe Acute Respiratory Syndrome (SARS-CoV). The recent outbreak of coronavirus disease 2019 (COVID-19) has become a public health emergency worldwide. SARS-CoV-2, the virus responsible for COVID-19, is spread by human-to-human transmission via droplets or direct contact. However, since SARS-CoV-2 (as well as other coronaviruses) has been found in the fecal samples and anal swabs of some patients, the possibility of fecal-oral (including waterborne) transmission need to be investigated and clarified.
This scoping review was conducted to summarize research data on CoV in water environments. A literature survey was conducted using the electronic databases PubMed, EMBASE, and Web Science Core Collection. This comprehensive research yielded more than 3000 records, but only 12 met the criteria and were included and discussed in this review.
In detail, the review captured relevant studies investigating three main areas: 1) CoV persistence/survival in waters; 2) CoV occurrence in water environments; 3) methods for recovery of CoV from waters.
The data available suggest that: i) CoV seems to have a low stability in the environment and is very sensitive to oxidants, like chlorine; ii) CoV appears to be inactivated significantly faster in water than non-enveloped human enteric viruses with known waterborne transmission; iii) temperature is an important factor influencing viral survival (the titer of infectious virus declines more rapidly at 23°C–25 °C than at 4 °C); iv) there is no current evidence that human coronaviruses are present in surface or ground waters or are transmitted through contaminated drinking-water; v) further research is needed to adapt to enveloped viruses the methods commonly used for sampling and concentration of enteric, non enveloped viruses from water environments.
The evidence-based knowledge reported in this paper is useful to support risk analysis processes within the drinking and wastewater chain (i.e., water and sanitation safety planning) to protect human health from exposure to coronavirus through water.
Keywords: Coronavirus, SARS-CoV-2, Water disinfection, Survival, Occurrence, Method
Highlights
-
•
SARS Coronavirus has been detected in wastewater but not as infectious particles.
-
•
Temperature is an important environmental factor affecting CoV survival in water.
-
•
CoV show limited environmental stability and sensitivity to oxidants as chlorine.
-
•
There is no evidence of CoV transmission through contaminated water.
-
•
Methods for CoV concentration from waters should be optimized.
1. Introduction
Faecal contamination of water supplies has been historically recognised as a risk for human health: water can provide a vehicle for pathogen spread, creating the conditions for outbreaks or sporadic cases of infection. Human pathogenic viruses are often detected in water environments and are deemed to be responsible for a considerable proportion of waterborne diseases (Hamza and Bibby, 2019; Haramoto et al., 2018; La Rosa et al., 2012; Moreira and Bondelind; Rusinol, and Girones, 2017; WHO, 2017). Viruses of concern for their potential waterborne transmission belong mainly to the group of enteric viruses, a diverse group of non-enveloped viruses, which can multiply in the gastrointestinal tract of humans. They can be mostly responsible of gastrointestinal illness, but also of a wide spectrum of other diseases, such as conjunctivitis, respiratory symptoms, viral hepatitis, infections of the central nervous system.
The most important waterborne enteric viruses belong to the families Caliciviridae (Norovirus), Picornaviridae (Enterovirus and Hepatitis A virus) and Adenoviridae (Adenovirus) (WHO, 2017). These viruses are often excreted at high titres in the feces (and occasionally, at lower concentrations, in urines) of infected humans (Rusinol and Girones, 2017). They have also been detected from virtually all types of water: wastewater, seawater, fresh waters, groundwater and drinking water and have been associated with drinking and recreational water outbreaks (Bonadonna and La Rosa, 2019; Gall et al., 2015; La Rosa et al., 2012; Moreira and Bondelind; Rusinol, and Girones, 2017).
Conversely, enveloped viruses, are structurally dissimilar to the enteric (non-enveloped) viruses, and are believed to behave differently in water environments (Wigginton et al., 2015). This group of viruses includes families such as Orthomyxoviridae (es. Influenza viruses), Paramyxoviridae (measles virus, mumps virus, respiratory syncytial virus, etc.), Herpesviridae, Coronaviridae and several others viruses. Among the enveloped viruses, coronaviruses (CoV) (order Nidovirales, family Coronaviridae, subfamily Coronavirinae) are single-stranded positive-sense RNA viruses.
Coronavirinae, includes four genera, Alpha-, Beta-, Gamma- and Delta-coronavirus, of which the first two host viruses infecting humans (Human Coronavirus, HCoV): HCoV-229E and HCoV-NL63 (alphacoronaviruses) and HCoV-HKU1, HCoV-OC43, Middle East respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory syndrome coronavirus (SARS-CoV) (betacoronaviruses) (Cui et al., 2019). Moreover, several coronavirus are reported to infect wildlife, pets or livestock, such as in the case of bat coronaviruses (BatCoV), porcine enteric diarrhoea CoV (PEDV) and transmissible gastroenteritis virus (TGEV), feline infectious peritonitis virus (FIPV), bovine coronavirus (BCoV) and others (reviewed in Wong et al., 2019; Wang et al., 2019; Tekes and Thiel, 2016; Amer, 2018).
HCoV are respiratory pathogens and their primary transmission mode is person-to-person contact through respiratory droplets generated by breathing, sneezing, coughing, etc., and contact (direct contact with an infected subject or indirect contact, trough hand-mediated transfer of the virus from contaminated fomites to the mouth, nose, or eyes). Waterborne transmission has never been demonstrated in humans, however detection of HCoV in the feces of infected patients has been reported (Esper et al., 2010; Jevšnik et al., 2013; Risku et al., 2010; Vabret et al., 2006), suggesting the fecal-oral route may contribute to HCoV transmission. In 2003, the SARS-CoV was detected in the feces of infected patients (Isakbaeva et al., 2004) and, during an outbreak in a residential complex of Amoy Garden in Hong Kong, transmission by aerosolized wastewater was suspected (McKinney et al., 2006).
In late 2019, a new acute respiratory disease known as COVID-19, sustained by a novel coronavirus, SARS-CoV-2 (Gorbalenya et al., 2020), emerged in Wuhan, China and following global spread of the disease. The outbreak was declared a Public Health Emergency of International Concern on 30 January 2020 and the World Health Organization (WHO) on 11 February 2020 announced a name for the new coronavirus disease: COVID-19. On March 11, WHO upgraded the status of the COVID-19 outbreak from epidemic to pandemic.
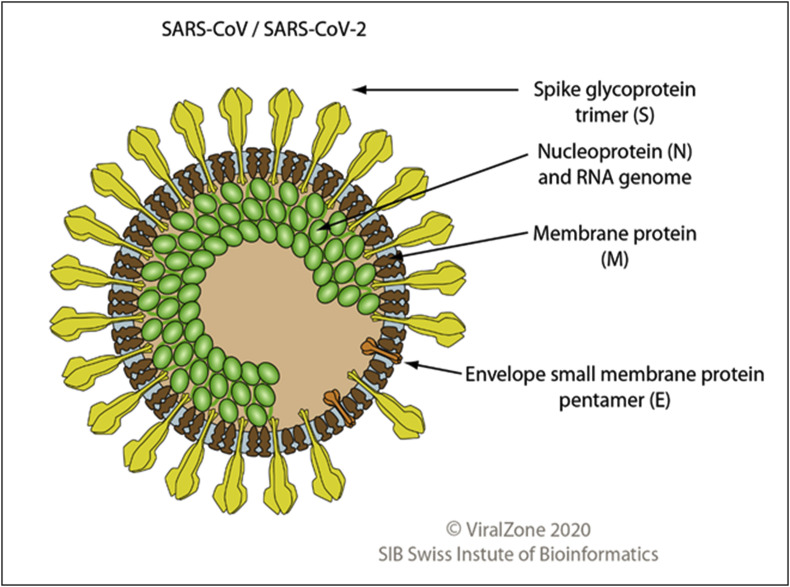
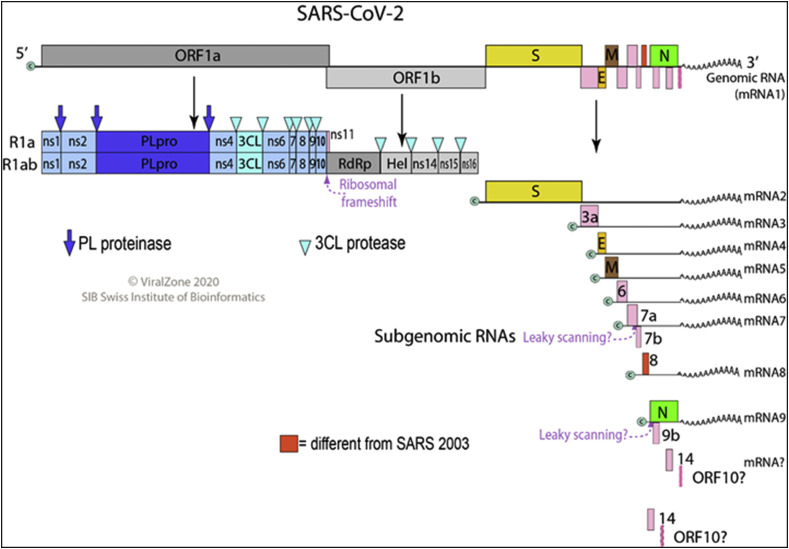
Coronavirus virion is enveloped, spherical, and about 120 nm in diameter. Envelope proteins are involved in several aspects of the virus life cycle, such as assembly, envelope formation, and pathogenesis. Inside the envelope is the helical capsid containing nucleoprotein and the RNA genome. Fig. 1 shows the virion structure of SARS-CoV/SARS-CoV-2. The 25–32 kb genome of SARS-CoV-2 is organized in two large open reading frames (ORF1a and ORF1b, located at the 5’ end) coding for replicase polyproteins, followed, in the terminal one-third of the genome, by a region encoding for the structural proteins (spike, envelope, membrane, and nucleocapsid protein). Fig. 2 shows the linear ssRNA(+) genome of SARS-CoV2.
Fig. 1.
Virion structure of SARS-CoV/SARS-CoV-2
(permission obtained from Philippe Le Mercier,
ViralZone,
SIB Swiss Institute of Bioinformatics).
Fig. 2.
Genome structure of SARS-CoV-2
(permission obtained from Philippe Le Mercier, ViralZone,SIB Swiss Institute of Bioinformatics).
As for other respiratory HCoV, the main vehicle of transmission of SARA-CoV-2 are droplets generated by breathing, sneezing, coughing, etc., and contact (direct contact with an infected subject or indirect contact, trough hand-mediated transfer of the virus from contaminated fomites to the mouth, nose, or eyes). In the rapidly evolving picture of the scientific knowledge on COVID-19 and SARS-CoV-2, some studies have reported the presence fragments of viral RNA in feces or anal swab of infected patients (Holshue et al., 2020; Xiao et al., 2020). Transmission of COVID-19 through the fecal-oral route, however, has not been demonstrated, nor occurrence of SARS-CoV-2 in water environments has been proved to date. Information on the presence, quantitative levels, and survival in water environments of coronaviruses of interest for human health are, indeed, limited, and few studies approached development and optimization of methods to concentrate CoV or other enveloped viruses from wastewater, biosolids, surface waters or other water types (see Table 3 ).
Table 3.
Occurrence of Coronavirus of interest for human health in water environments.
| Reference | Virus | Water matrix | Country | Year | Main findings |
|---|---|---|---|---|---|
| Wang et al., 2005b | Severe acute respiratory syndrome Coronavirus (SARS-CoV) | Sewage water from two hospitals receiving SARS patients | Beijing, China | 2003 |
|
|
Benchmark Stools (n = 11) from symptomatic patients in the two hospitals |
|
||||
| Blanco et al. (2019) |
Alphacoronavirus Betacoronavirus |
Surface water (water channels) | Central Saudi Arabia | 2015 |
|
|
Benchmark Hepatitis A virus |
|
||||
| Bibby et al. (2011) | Human coronavirus 229E a Human coronavirus HKU1 a |
Class B biosolids from wastewater treatment facility b | USA | unk |
|
| Benchmark virome |
|
||||
| Bibby& Peccia (2013) | Human Coronavirus HKU1 a Human coronavirus 229E a |
Influent and effluent sludge c | USA | unk |
|
| Benchmark virome |
|
||||
| Alexyuk et al. (2017) | Coronaviridae a | Surface water (river, water reservoir, lake) | Ile-Balkhash, Kazakhstan | 2017 |
|
| Benchmark virome |
|
Note: For comparison purposes, other microorganisms detected in the studies were reported under ‘benchmark’.
Metagenomic study.
Solid residuals by primary sedimentation and secondary activated sludge clarification, treated by mesophilic anaerobic digestion, and partially dewatered by belt pressing.
Influent and effluent sludge from mesophilic anaerobic digesters from domestic wastewater treatment plants. Influent samples were mixtures of primary and secondary sludge; effluent samples were of a class B product, prior to dewatering.
The present review summarizes the current state of knowledge on coronaviruses of interest for human health in water environments, with an emphasis on their occurrence and persistence, and on concentration methods for their detection in different water matrices. The reported outcomes are aimed to improve knowledge on transmission pathways and possible infection hazards related to poor drinking water and sanitation management; additionally, research gaps on methodologies for detection (with focus on concentration methods) enveloped viruses are specifically examined, to strengthen their monitoring in water media.
2. Methods
An electronic search of available literature was run on 23 February 2020. Search was conducted using the electronic databases PubMed, EMBASE, and Web Science Core Collection with no restriction for publication date or language. The search strategy included terms related to the virus group and the environmental matrices of interest (see Table 1).
Table 1.
Literature search Strategy.
| Search | Field | |
|---|---|---|
| #1 | Coronavirus | Coronavirus OR “Human Coronavirus” OR “Middle East Respiratory Syndrome Coronavirus” OR “Human Coronavirus NL63” OR “Porcine Respiratory Coronavirus” OR “Human Coronavirus OC43” OR “Human Coronavirus 229E″ OR “Coronavirus Infections” OR “Rat Coronavirus” OR “Canine Coronavirus” OR “Bovine Coronavirus” OR “Feline Coronavirus” OR “Turkey Coronavirus” OR “Severe acute respiratory syndrome” OR “SARS Virus” OR “COVID-19” OR HCoV OR 229E OR OC43 OR NL63 OR HKU1 OR SARS OR MERS OR 2019-nCoV OR HCoV-229E OR HCoV-OC43 OR HCoV-NL63 OR HCoV-HKU1 OR SARS-CoV OR SARS-CoV-2 OR MERS-CoV |
| #2 | Water environments | Water OR “Waste Water” OR Sewage OR Wastewater OR River OR “Surface water” OR Groundwater OR “brackish water” OR Seawater OR “sea water” OR “wastewater treatment plant” OR influent OR effluent OR “drinking water” OR “tap water” OR “potable water” OR lake OR “fresh water” OR freshwater OR “marine water” |
| #3 | #1 AND #2 |
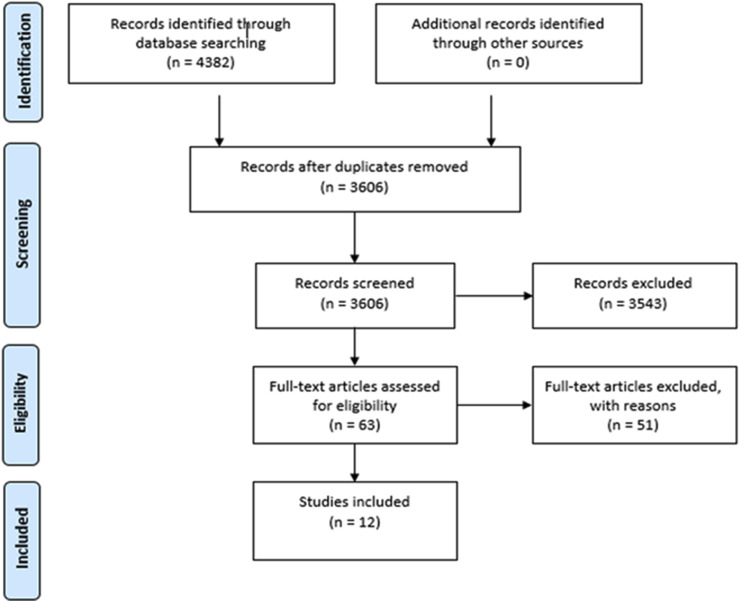
A total of 4382 articles were retrieved by the search and duplicates (n = 776) were automatically removed using the EndNote Reference Manager software online. Using the Rayyan Review platform (https://rayyan.qcri.org/welcome), titles and abstracts of the retained 3606 articles were screened and assessed for eligibility by two independent reviewers (GLR and ES) and the disagreements were resolved by discussion between the reviewers and a third referee (LB). Based on the objective of the study 3543 records were eliminated as not relevant. Full text screening was undertaken on the retained 63 articles and further 51 articles were excluded as either i) unrelated to CoV in water environments, ii) records duplicating results retrieved by earlier articles (linked articles), iii) articles related only to inactivation of surrogate viruses other than CoV, iv) reviews not including data relevant to the study, v) non-relevant erratum. For one of the 63 retained records, full text was not available for screening but the article was assessed as relevant based on abstract content.
Finally, 12 articles were included in the study, corresponding to original studies whose main findings are presented in Table 2, Table 3, Table 4 (Abd-Elmaksoud et al., 2014; Alexyuk et al., 2017; Bibby et al., 2011; Bibby and Peccia, 2013; Blanco et al., 2019; Casanova et al., 2009; Collomb et al., 1986; Gundy et al., 2019; Wang et al., 2005a; Wang et al., 2005b; Wang et al., 2005c; Ye et al., 2016).
Table 2.
Persistence and survival of Coronavirus in water environments.
| Reference | Virus | Water matrix | Main findings |
|---|---|---|---|
| Wang et al., 2005a |
|
|
|
|
Benchmark Escherichia coli Enterobacteria phage f2 (non-enveloped) |
|
||
| Casanova et al. (2009) |
|
|
|
| Gundy et al., 2019 |
|
|
|
|
Benchmark Poliovirus-1 (PV-1), strain LSc-2ab (non-enveloped) |
In tap water (both filtered and unfiltered) at 23 °C, PV-1 survived six times longer than coronaviruses; in wastewater (primary and secondary sludge) PV-1 survived 2 to 3 times longer than coronaviruses | ||
| Ye et al. (2016) |
|
|
|
|
Benchmark Pseudomonas phage φ6 (enveloped) Enterobacteria phage MS2, ATCC 15597-B1 (non-enveloped) Enterobacteria phage T3, ATCC 11303-B4 (non-enveloped) |
|
Note: Findings were reported differentiating experimental results (reduction ‘reached’) and results obtained by predictive modelling (reduction ‘expected’). For comparison purposes, other microorganisms used in the experimental plans were reported under ‘benchmark’.
Table 4.
Concentration methods for Coronavirus in water matrices.
| Reference | Virus | Water matrix | Concentration method | Volume | Main findings |
|---|---|---|---|---|---|
| Collomb et al., 1986a | Bovine enteric coronavirus | Adsorption-elution: adsorption on glass-powder at acid-pH followed by alkaline-pH elution |
|
||
| Wang et al. (2005c) | Severe acute respiratory syndrome Coronavirus (SARS-CoV) |
|
Adsorption-elution-PEG precipitation: adsorption on positive charged filter media particle (silica gel plus Al(OH)3), elution with neutral buffer, PEG precipitation | 100 ml |
|
|
Benchmark Enterobacteria phage f2 (non-enveloped) |
|
||||
| Ye et al. (2016) | Murine hepatitis virus, strain A59 (MHV) | Municipal wastewater |
|
|
∼5% with ultracentrifugation 25.1% with ultrafiltration
|
|
|
||||
|
|
||||
|
Benchmark Enterobacteria phage MS2, ATCC 15597-B1 (non-enveloped) |
∼5% with ultracentrifugation 55.6% with ultrafiltration |
||||
| Abd-Elmaksoud et al. (2014) | Bovine Coronavirus (BoCoV) | Dechlorinated tap water from groundwater source | Adsorption-elution: adsorption on glass wool, elution with alkaline buffer, PEG precipitation | 20 L |
|
|
Benchmark - Bovine rotavirus gr. A (BoRV gr.A) - Bovine viral diarrhea virus types 1 (BVDV1) - Bovine viral diarrhea virus types 2 (BVDV2) - Poliovirus 3 (Sabin) - E. coli O157:NM - Campylobacter jejuni |
|
||||
| Blanco et al. (2019) | Transmissible gastroenteritis virus (TGEV), strain PUR46-MAD | – | Adsorption-elution adsorption on glass wool, elution with alkaline buffer, PEG precipitation | 5L 50L |
|
|
Benchmark Hepatitis A virus, strain HM175 43c |
|
Note: For comparison purposes, other microorganisms used in the experimental plans were reported under ‘benchmark’.
the full text of this paper was not recovered, therefore information was retrieved from the abstract.
3. Results and discussion
The twelve retrieved records were divided according to their content in studies related to the investigation of CoV persistence and survival in water environments (n = 4, Table 2), occurrence of CoV, pathogenic or potentially pathogenic to humans, in water environments (n = 5, Table 3), and analytical methods for concentration of CoV from water (n = 5, Table 4). The flow chart of the systematic literature review is illustrated in Fig. 3 .
Fig. 3.
Flow chart for the systematic literature search.
3.1. Persistence of coronavirus in water environments
Four papers dealing with the persistence or survival of CoV in waters were retrieved (Table 2). The articles were related to seeding experiments in which SARS-CoV, Human CoV (229E) or surrogate animal CoV (TGEV, FIPV, or murine hepatitis virus, MHV) were used to spike different water types (Wang et al., 2005a; Casanova et al., 2009; Gundy et al., 2009; Ye et al., 2016).
Wang and coworkers studied the persistence of SARS-CoV in water (hospital wastewater, domestic sewage and dechlorinated tap water) and in feces and urine (Wang et al., 2005a). In the study, the effect of sodium hypochlorite and chlorine dioxide in inactivating SARS-CoV, Escherichia coli and the Enterobacteria phage f2 spiked in wastewater was evaluated.
SARS-CoV was detected in hospital wastewater, domestic sewage, and tap water for 2 days at 20 °C and up to 14 days at 4 °C, thus demonstrating temperature strongly influences viral persistence. Indeed, it has been universally demonstrated that higher temperatures are associated with rapid inactivation of enteric viruses, and temperature is recognised as the most influential factor for viral survival in water due to increased denaturation of proteins and activity of extracellular enzymes (Pinon and Vialette, 2018).
Wang and coworkers (Wang et al., 2005a) highlighted that SARS-CoV persists 3 days in stools and 17 days in urine stored at 20 °C. On the other hand, at a lower temperature (4 °C) they persist for 17 days. The same study showed that chlorine was more effective than chlorine dioxide in inactivating E. coli, f2 phage and SARS-CoV and a free residual chlorine of 0.5 mg/L from chlorine or 2.19 mg/L from chlorine dioxide in wastewater ensured complete inactivation of SARS-CoV. In the experimental conditions of the study, SARS-CoV was inactivated completely in presence of 10 mg/L chlorine and a minimum contact time of 10 min or in 1 min using 20 mg/L chlorine. Under the same conditions, E. coli and f2 phage were not inactivated effectively. This findings are of specific relevance since, according to the 4th edition of the World Health Organization’s Guidelines for drinking-water quality, viruses are generally more resistant to free chlorine than bacteria (specifically, “moderate” resistance for viruses, and “low” for the vast majority of bacteria) (WHO, 2017). The viruses considered of concern for water in WHO Guidelines, however, are principally enteric viruses (familes Adenoviridae, Astroviridae, Caliciviridae, Hepeviridae, Picornaviridae, and Reoviridae) which are, as previously reported, non-enveloped viruses. It is well known that these viruses are more resistant to environmental conditions, water treatments and disinfectants than enveloped viruses like coronavirus, as lysis of the viral envelope leads to the loss of functional receptors required for infection of susceptible cells (Wigginton et al., 2015). According to the results of Wang (2005a), SARS-CoV resistance to chlorine is lower than for bacteria. It follows that the current water disinfection practices (drinking water, wastewater, water from swimming pool), effective against non-enveloped viruses and bacteria, are expected to be effective also towards enveloped viruses such as coronaviruses.
The study of Casanova et al. (2009) evaluated the survival of two surrogate coronaviruses, TGEV (transmissible gastroenteritis virus, a porcine coronavirus) and MHV (murine hepatitis virus), in reagent-grade water, lake water, and settled human sewage. Two temperatures were evaluated over 6 weeks: room temperature (23–25 °C), and 4 °C. In general, in all the water tested, the titer of infectious virus declined more rapidly at 25 °C than at 4 °C, confirming that temperature is an important factor affecting viral survival in water. At 25 °C, the time required for a 99.9% reduction (T99.9) in reagent-grade water was 33 days and 26 days for TGEV and MHV, respectively, while in pasteurized settled sewage it was 14 days and 10 days, respectively. On the other hand, no significant decrease of TGEV and MHV was reported in reagent-grade water at 4 °C after 49 days, and limited reduction was obtained at the same temperature in lake water after 14 days. Based on these results, the authors suggest that contaminated water may be a potential vehicle for human exposure if aerosols are generated. However, it is important to underline that the surrogate animal coronaviruses used in this study are responsible for gastrointestinal or hepatic diseases in animals and may therefore display a different resistance behaviour compared to respiratory human coronaviruses. This could explain the greater resistance and longer survival displayed by CoV in this work compared to the study of Wang and colleagues (2005a). Moreover, the use of different cell lines and growth media in these persistence studies might have contribute to measurement uncertainty.
Gundy et al., 2019 investigated the survival of a human coronavirus (HCoV-229E) and of an animal coronavirus (FIPV, feline infectious peritonitis virus) in tap water (filtered and non-filtered) and wastewater (primary and activated sludge effluents), comparing results with those of Poliovirus-1 (PV-1, Sabin attenuated strain LSc-2ab). In wastewater, the tested CoV died off quite rapidly, with a T99.9 of 2.77–3.54 days at 23 °C. Significantly, the PV-1 lasted 2 to 3 times longer than CoV did, requiring 10.9 days for a comparable reduction in primary wastewater and 5.7 days in secondary effluents. In tap water, CoV reduction was slower than in wastewaters: at 23 °C, the T99.9 was 12.1–12.5 days for HCoV-229E and FIPV, while at 4 °C the same reduction was predicted (by modelling) to be achivable over 100 days. These yields highlight once again that virus survival decreases with increasing temperature. Similarly to the results obtained on wastewater, PV-1 survived six times longer than CoV in both filtered and unfiltered tap water, confirming the observation that non-enveloped viruses display higher resistance in water enviroments compared to enveloped viruses. Another important finding of the study was that CoV inactivation was faster in filtered tap water than unfiltered tap water, suggesting that suspended solids in water can provide protection for viruses adsorbed to these particles.
Finally, a more recent study (Ye et al., 2016) investigated the survival and partitioning of two enveloped viruses, MHV and Pseudomonas phage φ6 and of two non-enveloped viruses, bacteriophages MS2 and T3 in untreated municipal wastewater. Unpasteurized and pasteurized wastewater were spiked with the viral stocks and were then incubated at 25 °C or 10 °C to mimic typical summer and winter wastewater temperatures. Inactivation proceeded faster for the enveloped viruses: in unpasteurized wastewaters at 25 °C, the time to reach a 90% reduction (T90) was 13 h for MHV and 7 h phage φ6, compared to a predicted value of 121 h for the non-enveloped phage MS2. At 10 °C the inactivation kinetics of both MHV and φ6 were, once again, significantly slower than at environmental temperatures, with a T90 of 28–36 h. In pasteurized wastewater, both MHV and phage φ6 lost infectivity at a significantly slower rate compared to unpasteurized wastewater (T90 of 19 h for MHV and 53 h for phage φ6 at 25 °C), possibly due to the reduction of bacterial extracellular enzyme activity and the absence of protozoan and metazoan predation in pasteurized samples. Indeed, it was demonstrated that the presence of an indigenous microbial population has a negative impact on virus survival (Pinon and Vialette, 2018; Rzezutka and, Cook, 2004). Finally, in the same study, Ye et al., reported that up to 26% of the enveloped viruses adsorbed to the solid fraction of wastewater. That means that a reduction of enveloped viruses in wastewaters is provided by solid settling.
3.2. Occurrence of coronavirus in water environments
Two reports specifically addressing detection in water environments of CoV of interest for human health and three metagenomic/virome studies were retrieved through literature search (Table 3).
Sewage discharges from two hospitals in Beijing, China, hosting SARS patients during the 2003 outbreak were analyzed with the aim to investigate whether sewage may be a possible route of transmission for SARS-CoV (Wang et al., 2005b). Both cell culture and RT-PCR were utilized to ascertain viability and detect the virus in sewage. While viral genome was repeatedly detected in hospital sewage before disinfection (10/10 wastewater samples) and, in some cases, after disinfection (3/10 samples), infectious SARS-CoV was never detected in the tested samples. Possible explanation of authors includes viral inactivation by disinfectants (high concentration of disinfectants, were used after a patient had bowel movements), low viral concentration, or loss of infectivity by unknown factors during the concentration process.
In the second study specifically addressing CoV detection in water, Blanco et al. (2019) investigated the occurrence of these viruses in surface waters of Wadi Hanifa, Riyadh, using a broad-range RT-PCR for the detection of Alpha- and Betacoronavirus. Of the 21 tested samples, only one sample was positive for CoV. Upon sequence analysis, the positive sample was found to be closely related to a novel rodent/shrew-specific clade within lineage A of Alphacoronavirus, reported in Asia and Europe.
Three metagenomic studies have detected CoV in water matrices: two focused on class B biosolids from wastewater treatment facility (Bibby et al., 2011; Bibby& Peccia, 2013), and one on different type of water (river, lake, reservoir) (Alexyuk et al., 2017). The study of Bibby and coworkers identified a large variety of both enveloped and non-enveloped viruses in biosolids, including coronavirus, Herpesvirus, Torque Teno virus and Parechovirus. Interestingly, all these groups of viruses were highly represented in compared to Adenovirus, which have been for long time considered the most abundant viral genus in biosolids (Bibby et al., 2011). In detail, 10 CoV sequences were identified, nine of which related to HCoV-229E and one to HCoV-HKU1.
Two years later, another paper from the same authors described the diversity of viruses in sewage sludge samples (influents and effluents) with comparable results: emerging viruses such as coronavirus, Klassevirus, and cosavirus were detected in abundance in the sample (Bibby& Peccia, 2013). Coronaviruses were detected in 83% of samples and coronavirus HKU1 was the second most prevalent RNA virus. Interestingly, coronavirus showed a higher relative abundance in influent samples compared to effluent ones.
Finally, Alexyuk et al. (2017) studied the viromes sampled in surface water (river, lake and water reservoir). While the majority of the sequences were related to autochthonous viruses, typical for aquatic ecosystems, allochthonous viruses, such families as Coronaviridae, Reoviridae and Herpesviridae were also detected, suggesting anthropogenic pollution of the three selected water environments. In detail, Coronaviridae were detected in all of the three environments, ranging from 0.002% to 0.009% of the total sequences depending on sample.
To complete the picture on the occurrence of coronavirus in water environments, after the initial submission of this scoping review, while the paper was under review, novel papers, some of which published as preprint, have demonstrated the occurrence of SARS-CoV-2 in municipal wastewaters worldwide, and, specifically, in the Netherlands (Medema et al., 2020), in Massachusetts (Wu et al., 2020), in Australia (Ahmed et al., 2020), France (Wurtzer et al., 2020), and Italy (La Rosa et al. 2020; submitted).
3.3. Methods for concentration of enveloped viruses from water matrices
Five studies (Table 4) investigated concentration methods for CoV in waters and the associated recovery efficiency.
The first study investigating CoV recovery from waters was published more than 30 years ago (Collomb et al., 1986) using, for the spiking experiments, a bovine enteric coronavirus, and assessing a concentration procedure based on viral adsorption on glass-powder at acid pH followed by alkaline-pH elution. Since CoV is sensitive to acid (pH 3) and alkaline pH (pH ≥ 10), adsorption was optimal at pH 3.3 and elution at pH 9. Under such conditions, the overall efficiency of the concentration method appeared to be between 24% and 28%. Unfortunately, since it was not possible to retrieve the full text of this publication, no further information, beside those included in the abstract, could be reported.
Following the SARS outbreak of 2003, Wang et al. (2005c) performed a study to evaluate the recovery from spiked sewage of SARS-CoV and of a surrogate virus, bacteriophage f2. The concentration procedure foresaw the use of positively charged electropositive filter media particle (silica gel plus Al(OH)3), packed in a glass column according to a protocol previously described by Li et al. (1998) for enteric viruses. Hospital sewage and domestic sewage (100 ml) were spiked with SARS-CoV and phage f2, passed through the glass column, eluted from the filter media with 3 × nutrient broth (pH 7.2), and then polyethylene glycol (PEG) precipitated. The procedure gave recoveries of SARS-CoV ranging from 0% (sewage from a housing estate) to 21.4% (sewage from the hospital), with an average of 1.02%.The recovery of phage f2 under the same conditions were significantly higher (from 33.6% to more than 100%). This method therefore seemed more suitable for the concentration of enveloped viruses, in agreement with the initial study proposing its use, that showed recoveries of enterovirus and hepatitis A virus from tap water ranging from 88.7% to 96.0% (Li et al., 1998).
Ye et al. (2016) evaluated three methods for separating and concentrating viruses from the liquid fraction of municipal wastewater: i) PEG precipitation, ii) ultracentrifugation, iii) ultrafiltration with centrifugal devices (Ye et al., 2016). Wastewater (250 ml for PEG precipitation and ultrafiltration and 60 ml for ultracentrifugation) was spiked with the rodent coronavirus Murine Hepatitis Virus (MHV) and with the non-enveloped phage MS2. Low mean recoveries (∼5%) were achieved for both MHV and MS2 with the ultracentrifugation method. This result was suggested to be related to virus inactivation by the high g-force of the ultracentrifugation. Recovery of MHV was low (∼5%) also with the PEG precipitation method, whose performance for MS2 concentration was instead significantly higher (43.1%). Finally, the optimized ultrafiltration protocol adopted in the study provided the highest recoveries for both viruses: 25.1% for MHV and 55.6% for phage MS2. Results of Ye and colleagues suggested that the PEG precipitation method, which is effective at recovering non-enveloped viruses from water samples, may be not optimal for recovering infective enveloped viruses, while ultrafiltration could be successfully applied for recovering CoV. However, in this study, only small volumes of wastewater were tested using centrifugal ultrafilters. Since viruses in water matrices may occur in very low numbers, there is a need for analytical methods suitable to process large volumes of water. It was therefore concluded by the authors that further progress could be made through the optimization of hollow fiber ultrafilters and tangential flow ultrafiltration to allow concentration of CoV in waters from larger volumes of water.
Abd-Elmaksoud and coworkers (2014) measured the effectiveness of glass wool filtration to simultaneously concentrate a variety of waterborne viral and bacterial pathogens typically found in runoff from agricultural fields using dairy manure as fertilizer. Viruses of bovine origin were used to spike 20 L of dechlorinated tap water, including Bovine Coronavirus (BoCoV), Bovine Viral Diarrhea Virus types 1 and 2 (BVDV1 and BVDV2), Bovine Rotavirus group A (BoRV), and Poliovirus 3 (Sabin). Moreover, E. coli O157:NM, and Campylobacter jejuni were selected as bacterial pathogen. Three water turbidity levels were evaluated, prepared by mixing dried agricultural soil into the 20 L of water. After glass wool filtration, elution was performed with 3% beef extract-glycine buffer (pH 9.5), followed by flocculation with PEG 8000. Recovered organisms were enumerated by qPCR. Results showed that glass wool filtration is a cost-effective method for the concentration of several waterborne pathogens simultaneously. In details, the average recoveries (across the different turbidity levels tested) were: 18.1% for BoCoV, 22.1% for BoRV, 15.6% and 19.7% for BVDV1 and BVDV2 respectively. Higher recovery efficiencies were obtained for the non-enveloped virus Poliovirus 3 (57.9%), and for the bacterial microorganisms E. coli O157:NM (54.8%) and C. jejuni (32.7%). However, the authors advised to use caution in the interpretation of these comparative results since the quantity of pathogens used for spiking varied for the different microorganisms, precluding a clear differentiation of the effects on recovery efficiency of seeding quantities and pathogen type.
Blanco et al. (2019) used adsorption to glass wool, followed by elution with alkaline buffer and subsequent secondary concentration through PEG 6000 precipitation. The viruses used for the experimental procedures were Hepatitis A virus (HAV, non-enveloped) and the porcine coronavirus Transmissible Gastroenteritis Virus (TGEV). Large volumes of water (5 L and 50 L) were used for method optimization and performance characterization. Several steps of the elution procedure were modified compared to other published glass wool protocols to improve the recovery of TGEV and virus recoveries were ascertained by real-time qPCR. Recovery of the initial experiments (5 L of water, adsorption to the positively charged glass wool matrix, elution with glycine/beef extract buffer at pH 9.5 with 10 min of contact) showed that TGEV efficiently adsorbed onto the glass wool (attachment of 57.1%) but it was poorly eluted from it, with an overall recovery of 2.6%. The increase of buffer pH to 11.0 provided an improvement of elution efficiency, and a final recovery of 28.8%; further performance improvements could be obtained by changing the length of the elution incubation. Subsequent experiments for the concentration of HAV and TGEV from 50 L of spiked water samples were therefore all performed using an elution buffer pH 11.0. Results showed that addition of Tween 80 hampered the recovery of TGEV, possibly by damaging the lipid-containing envelope of viruses. Recirculation of the eluent at pH 11.0 for 20 min was instead beneficial to the elution, and provided recoveries of 18.0% and 23.9% for TGEV and HAV, respectively. Similarly, increasing PEG concentration from 10 to 20% in the secondary concentration, showed a significant improvement of the recovery (51.3% and 47.2% for TGEV and HAV, respectively). Following optimization of the method, the procedure provided a recovery efficiency of 5.1% for TGEV and 4.5% for HAV in spiked surface water. Overall, the study by Blanco et al. (2019) clearly demonstrated that the concentration procedures commonly used for non-enveloped viruses need adaptation to yield satisfactory performances on enveloped viruses like CoV.
To summarize, this scoping review has highlighted several aspects of coronavirus research that need to be explored in depth.
-
1)
The evidence of the presence of CoV in waters is currently very scarce and there is no evidence that human CoV are present in surface or groundwater sources or transmitted through contaminated drinking-water.
-
2)
Environmental factors, such as temperature, seem to affect the ability of CoV to persist in water. Further studies are needed to investigate CoV persistence in water in relation to climatic and seasonal conditions.
-
3)
Although different studies showed different viral inactivation rates for CoV in water, based on the type of virus and the type of water, generally, there is evidence that CoV is generally considered unstable in the environment and is more susceptible to oxidants, such as chlorine than non-enveloped viruses.
-
4)
Based on the few available data, methods commonly used to concentrate and recover non-enveloped enteric viruses from wastewater and other water matrices may not be appropriate to recover CoV. Therefore, future research should focus on the development of robust methods for concentrating CoV and other enveloped viruses from large volumes of waters and from different types of water.
4. Conclusion
The evidence-based knowledge here reported can be a key support for risk analysis in natural water resources and integrated water cycle, according to the water and sanitation safety planning approaches, as well as for the management and control of water-related risks during the pandemic COVID-19 caused by SARS-CoV2.
Further researches are needed to study the potential presence and fate of coronavirus and other enveloped viruses in municipal wastewater and drinking water and to develop robust methods for water analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Amer H.M. Bovine-like coronaviruses in domestic and wild ruminants. Anim. Health Res. Rev. 2018;19(2):113–124. doi: 10.1017/S1466252318000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abd-Elmaksoud S., Spencer S.K., Gerba C.P., Tamimi A.H., Jokela W.E., Borchardt M.A. Simultaneous concentration of bovine viruses and agricultural zoonotic bacteria from water using sodocalcic glass wool filters. Food and Environmental Virology. 2014;6:253–259. doi: 10.1007/s12560-014-9159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J. Science of the Total Environment; 2020. First Confirmed Detection of SARS-CoV-2 in Untreated Wastewater in Australia: A Proof of Concept for the Wastewater Surveillance of COVID-19 in the Community. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexyuk M.S., Turmagambetova A.S., Alexyuk P.G. Comparative study of viromes from freshwater samples of the Ile-Balkhash region of Kazakhstan captured through metagenomic analysis. VirusDis. 2017;28:18–25. doi: 10.1007/s13337-016-0353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environmental Science & Technology. 2013;47:1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Viau E., Peccia J. Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Lett. Appl. Microbiol. 2011;52:386–392. doi: 10.1111/j.1472-765X.2011.03014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco A., Abid I., Al-Otaibi N., Perez-Rodriguez F.J., Fuentes C., Guix S., Pinto R.M., Bosch A. Glass wool concentration optimization for the detection of enveloped and non-enveloped waterborne viruses. Food and Environmental Virology. 2019;11:184–192. doi: 10.1007/s12560-019-09378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonadonna L., La Rosa G. A review and update on waterborne viral diseases associated with swimming pools. Int J Environ Res Public Health. 2019;16(2):166. doi: 10.3390/ijerph16020166. Published 2019 Jan 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43:1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collomb J., Laporte J., Vautherot J.F., Schwartzbrod L. Recherche des coronavirus dans l’eau. Note I. Adsorption et élution des coronavirus sur poudre de verre [Research on coronaviruses in water. I. Adsorption and elution of the coronavirus on glass powder]. Virologie. 1986;37(2):95–105. [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper F., Ou Z., Huang Y.T. Human coronaviruses are uncommon in patients with gastrointestinal illness. J. Clin. Virol. 2010;48(2):131–133. doi: 10.1016/j.jcv.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A.M., Mariñas B.J., Lu Y., Shisler J.L. Waterborne viruses: a barrier to safe drinking water. PLoS Pathog. 2015;11(6) doi: 10.1371/journal.ppat.1004867. Published 2015 Jun 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020 doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy P., Gerba C., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ Virol. 2019;1(1):10. 2009. [Google Scholar]
- Hamza I.A., Bibby K. Critical issues in application of molecular methods to environmental virology. J Virol Methods. 2019;266:11–24. doi: 10.1016/j.jviromet.2019.01.008. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakbaeva E.T., Khetsuriani N., Beard R.S., Peck A., Erdman D., Monroe S.S., Tong S., Ksiazek T.G., Lowther S., Pandya-Smith I., Anderson L.J., Lingappa J., Widdowson M.A. SARS-associated coronavirus transmission, United States. Emerg. Infect. Dis. 2004;10:225–231. doi: 10.3201/eid1002.030734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevšnik M., Steyer A., Zrim T. Detection of human coronaviruses in simultaneously collected stool samples and nasopharyngeal swabs from hospitalized children with acute gastroenteritis. Virol. J. 2013;10:46. doi: 10.1186/1743-422X-10-46. Published 2013 Feb 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Fratini M., della Libera S., Iaconelli M., Muscillo M. Emerging and potentially emerging viruses in water environments. Ann. Ist. Super Sanita. 2012;48(4):397–406. doi: 10.4415/ANN_12_04_07. [DOI] [PubMed] [Google Scholar]
- G. La Rosa, M. Iaconelli, P. Mancini, G. Bonanno Ferraro, C. Veneri, L. Bonadonna, L. Lucentini, E. Suffredini First detection of SARS-CoV-2 in untreated wastewaters in Italy. 10.1101/2020.04.25.20079830. [DOI] [PMC free article] [PubMed]
- Li J.W., Wang X.W., Rui Q.Y., Song N., Zhang F.G., Ou Y.C., Chao F.H. A new and simple method for concentration of enteric viruses from water. J. Virol. Methods. 1998;74:99–108. doi: 10.1016/s0166-0934(98)00078-0. [DOI] [PubMed] [Google Scholar]
- Wang X.W.1, Li J.S., Guo T.K., Zhen B., Kong Q.X., Yi B., Li Z., Song N., Jin M., Wu X.M., Xiao W.J., Zhu X.M., Gu C.Q., Yin J., Wei W., Yao W., Liu C., Li J.F., Ou G.R., Wang M.N., Fang T.Y., Wang G.J., Qiu Y.H., Wu H.H., Chao F.H., Li J.W. Excretion and detection of SARS coronavirus and its nucleic acid from digestive system. World J. Gastroenterol. 2005 Jul 28;11(28):4390–4395. doi: 10.3748/wjg.v11.i28.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney K.R., Gong Y.Y., Lewis T.G. Environmental transmission of SARS at Amoy gardens. J. Environ. Health. 2006;68:26–30. [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 in sewage. 2020. [DOI] [PubMed]
- Moreira N.A., Bondelind M. Safe drinking water and waterborne outbreaks. J. Water Health. 2017;15(1):83–96. doi: 10.2166/wh.2016.103. [DOI] [PubMed] [Google Scholar]
- Pinon A., Vialette M. Survival of viruses in water. Intervirology. 2018;61(5):214–222. doi: 10.1159/000484899. [DOI] [PubMed] [Google Scholar]
- Risku M., Lappalainen S., Räsänen S., Vesikari T. Detection of human coronaviruses in children with acute gastroenteritis. J. Clin. Virol. 2010;48(1):27–30. doi: 10.1016/j.jcv.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinol M., Girones R. Summary of excreted and waterborne viruses. In: Rose J.B., Jiménez-Cisneros B., editors. Global Water Pathogen Project. UNESCO; MI: 2017. http://www.waterpathogens.Orghttp://www.waterpathogens.Org/book/summary-Of-Excreted-And-Waterborne-Viruses Michigan State University R. Girones (Eds) Part 3 Viruses), E. Lansing. [DOI] [Google Scholar]
- Rzezutka A., Cook N. Survival of human enteric viruses in the environment and food. FEMS Microbiol. Rev. 2004;28(4):441–453. doi: 10.1016/j.femsre.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Tekes G., Thiel H.J. Feline coronaviruses: pathogenesis of feline infectious peritonitis. Adv. Virus Res. 2016;96:193–218. doi: 10.1016/bs.aivir.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A., Dina J., Gouarin S., Petitjean J., Corbet S., Freymuth F. Detection of the new human coronavirus HKU1: a report of 6 cases. Clin. Infect. Dis. 2006;42(5):634–639. doi: 10.1086/500136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J.S., Jin M., Zhen B., Kong Q.X., Song N., Xiao W.J., Yin J., Wei W., Wang G.J., By Si, Guo B.Z., Liu C., Ou G.R., Wang M.N., Fang T.Y., Chao F.H., Li J.W. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J Virol Methods. 2005 Jun;126(1–2):171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J.S., Guo T.K. Concentration and detection of SARS coronavirus in sewage from Xiao tang Shan hospital and the 309th hospital [published correction appears in J virol methods. 2005 dec;130(1-2):210] J Virol Methods. 2005;128(1–2):156–161. doi: 10.1016/j.jviromet.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Vlasova A.N., Kenney S.P., Saif L.J. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol. 2019;34:39–49. doi: 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . fourth ed. 2017. Guidelines for Drinking-Water Quality.https://www.who.int/water_sanitation_health/publications/drinking-water-quality-guidelines-4-including-1st-addendum/en/ incorporating the 1st addendum. [Google Scholar]
- Wigginton K.R., Ye Y., Ellenberg R.M. Emerging investigators series: the source and fate of pandemic viruses in the urban water cycle. Environ. Sci.: Water Res. Technol. 2015;1:735. [Google Scholar]
- Wong A.C.P., Li X., Lau S.K.P., Woo P.C.Y. Global epidemiology of bat coronaviruses. Viruses. 2019;11(2):174. doi: 10.3390/v11020174. Published 2019 Feb 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Moniz K., Erickson T., Chai P., Thompson J., Alm E. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. 2020. medRxiv preprint. [DOI] [PMC free article] [PubMed]
- Wurtzer S., Marechal V., Mouchel J.M. 2020. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases. medRxiv preprint. [DOI] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for Gastrointestinal Infection of SARS-CoV-2 [published online ahead of print, 2020 Mar 3] Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]