Graphical abstract
Keywords: Multimodal data, Intensive care unit, Medical data mining, Philips IntelliVue patient monitor
Highlights
-
•
Integrated system for ICU multimodal data acquisition, analysis and visualization.
-
•
Multimodal data are acquired and visualized in a user-centered interface.
-
•
Acquired time-series are analyzed for patient state tracking at the bedside.
-
•
Multimodal Human-Computer Interaction assists with clinical decision-making.
Abstract
Modern intensive care units (ICU) are equipped with a variety of different medical devices to monitor the physiological status of patients. These devices can generate large amounts of multimodal data daily that include physiological waveform signals (arterial blood pressure, electrocardiogram, respiration), patient alarm messages, numeric vitals data, etc. In order to provide opportunities for increasingly improved patient care, it is necessary to develop an effective data acquisition and analysis system that can assist clinicians and provide decision support at the patient bedside. Previous research has discussed various data collection methods, but a comprehensive solution for bedside data acquisition to analysis has not been achieved. In this paper, we proposed a multimodal data acquisition and analysis system called INSMA, with the ability to acquire, store, process, and visualize multiple types of data from the Philips IntelliVue patient monitor. We also discuss how the acquired data can be used for patient state tracking. INSMA is being tested in the ICU at University Hospitals Cleveland Medical Center.
1. Introduction
Each year, more than four million acutely ill patients are admitted to intensive care units (ICUs) in the U.S. alone; approximately 500,000 of them do not survive [1], [2], [3]. In extreme situations, like the current COVID-19 pandemic, ICUs are essential for treating critically ill coronavirus patients. Given the high stakes involved, timely and effective care is paramount, and this requires continuous patient surveillance using sophisticated monitoring equipment. As a result, ICUs are complex, data-intensive environments and dozens of systemic parameters are monitored, including heart rate, respiration, arterial blood pressure, oxygen saturation, temperature, end tidal CO2 concentration, etc. Enormous volumes of multimodal physiological data are generated including physiological waveform signals, patient monitoring alarm messages, and numerics and if acquired, synchronized and analyzed, this data can been effectively used to support clinical decision-making at the bedside [10], [17].
Clinical personnel rely on information from these signals provided on the patient monitor display for visual assessment or as numerics in the EMR to understand the current state of the patient, and how it is changing over time. Continuous digital monitoring is intended to allow clinicians to dynamically track changes in patient state more closely than would be possible with more sporadic measurements [4]. The hope has been that this would allow for more accurate diagnosis, earlier anticipation of deterioration, and a clearer understanding of the impact of administered treatments, improving quality of care and lowering costs [5].
Even when data can be viewed in real-time, standard approaches provide little insight into a patient’s actual pathophysiologic state. Understanding the dynamics of critical illness requires precisely time-stamped physiologic data (sampled frequently enough to accurately recreate the detail of physiologic waveforms) integrated with clinical context, but this will produce an overwhelming amount of data— far too much to be routinely reviewed manually. It is thus necessary to develop a data acquisition system that facilitates the access and review of historical data for medical personnel. The acquired data needs to be synchronized across disparate devices, archived, analyzed and presented to clinical personnel in a manner that supports clinical decision-making at the patient bedside. Previous work includes, for example: Matam and Duncan adopt the real-time data recording system used for F1 race cars to acquire and analyze data from bedside monitors in the pediatric intensive care unit [6]. This system supports the storage and review of electrocardiogram (ECG) data retrospectively. Raimond et al. developed a platform called “WaveformECG” that provides interactive analysis, visualization and annotation of ECG signals [7]. Alexander et al. developed an alarm data collection framework to acquire all alarms generated from Philips IntelliVue MP70 patient monitors installed in each ICU room with the objective of reducing false alarms by leveraging annotations provided by clinicians [4]. Hyung and Chul introduced a physiological data acquisition and visualization program “Vital Recorder” with a user-friendly interface similar to that of a video-editing program for anesthesia information management, where Physiological data can be manipulated like editing video clips [8].
Much of the previous work has focused on the acquisition or visualization of certain physiological data, a complete general purpose solution for data collection, analysis and visualization of multimodal ICU data is currently unavailable. ICU clinical personnel need the ability to effectively deal with different data sources on a patient or departmental level, and need advanced analytic methods that transform this data to actionable and clinically meaningful outcomes for each patent. We have been working on building the Integrated Medical Environment (tIME) [10] to address this critical opportunity and in this paper, we discuss an integrated system (INSMA) that supports multimodal data acquisition, parsing, real-time data analysis and visualization in the ICU. In the current implementation, INSMA acquires data from the Philips IntelliVue patient monitor, and has the ability to store and review the multimodal data acquired either in real-time or on request. The system has been tested in the ICU at University Hospitals Cleveland Medical Center, for multimodal data analysis and patient state tracking.
The remainder of the paper is organized as follows. In Section 2, we discuss some related work. In Section 3, we present the INSMA framework, and introduce the details of the data acquisition, parsing and visualization modules. In Section 4, we discuss the applications of our proposed system. We conclude our work and suggest possible future work in Section 5.
2. Related work
ICUs provide treatment to patients with the most severe and life-threatening illnesses and injuries. It requires uninterrupted attentiveness and medical care from various clinical specialists and medical equipment to sustain life and help nurture the patients back to health. Effective and reliable patient monitoring and data analysis are of ultimate importance in the ICU to ensure early diagnosis, timely and informed therapeutic decisions, effective institution of treatment and follow-up [29], [31]. Several clinical information systems have been developed from both industry and academia to meet the demanding needs of the ICU.
General Electric (GE) Co.’s Centricity Critical Care system introduced in 2003 creates actionable insight across the healthcare system and the care pathway in intensive care units, enabling enhanced clinical quality and operational efficiency. The system collects data from monitors and ventilators and displays it in spreadsheets reminiscent of the typical ICU chart. Data are collected from medical devices through device interfaces that connect with GE’s Unity Interface Device network [10]. The Datex-Ohmeda S/5™ Collect program proposed by GE Healthcare can obtain high-resolution data from the Datex-Ohmeda S/5™ series monitors [13]. The program was developed for Windows XP and is not compatible with current Windows operating systems, and the manufacturer does not intend to update it. Philips offers data management solutions that link the Philips ALS monitor/defibrillator and AED and allow quality assurance officers using a direct connection that downloads and forwards every event automatically. Quality assurance officers can then retrieve and review an event summary with confidence [11].
Often, the commercial off-the-shelf products do not support the acquisition, archiving, or annotations of high-resolution physiologic data with bedside observations for clinical applications. Systems have also been developed in academic settings primarily to support clinical research. Tsui et al. developed a system to acquire, model, and predict ICP in the ICU using wavelet analysis for feature extraction [12]. Goldstein et al. proposed and developed a physiologic data acquisition system that could capture and archive parametric data, but the annotation of important clinical events such as changes in a patient’s condition or timing of drug administration, was limited [21]. Kool et al. reported that they collected numerical data at five-second intervals from the Datex Ohmeda S/5™ monitoring system using their own information management system [14]. Liu et al. [15] also reported the collection of vital signal data from 32 surgical patients, from Philips IntelliVue MP series monitors, using a self-developed program that was not disclosed. Lee and Jung developed an anaesthesia information management system (AIMS) for the acquisition of high-quality vital signal data (Vital Recorder) to support research [8]. Physiological data of surgical patients were collected from 10 operating rooms by Vital Recorder through the patient monitor, anaesthesia machine and bispectral index monitor. Winslow et al. proposed a platform called WaveformECG for visualizing, annotating, and analyzing ECG data [7]. As discussed in the first section, these systems only focus on acquiring and analyzing one or two types of physiological data, and that is not sufficient for ICU applications.
Matam and Duncan used real-time data recording software, ATLAS from McLaren Electronics Systems that continuously monitor and analyze data from F1 racing cars to implement a similar real-time data recording platform system adapted with real time analytics to suit the requirements of the intensive care environment [6]. The parameter data recorded by Philips MP30 bedside monitors can be transferred to the server in real-time. However, such a third-party data acquisition tool is not flexible enough to customize the functions according to the clinician’s requirements, and the compatibility of the data format is another issue.
To address the issues described above, our research proposed the INSMA with the aim of obtaining clinical physiological data including electroencephalography (EEG), electrocardiography (ECG), photoplethysmogram (PPG), peripheral capillary oxygen saturation (SpO2), blood pressure (BP) and other signals to be acquired and stored for data sharing, mining, analysis and visualization. The primary data source in our first implantation comes from the IntelliVue MP (Philips, Germany) series of monitors.
3. INSMA
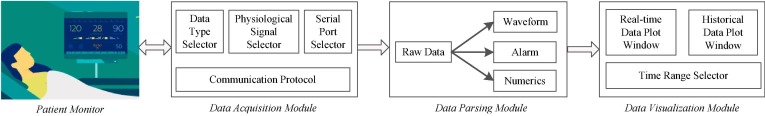
INSMA contains three independent but data related modules: data acquisition module, parsing module and visualization module. Fig. 1 shows the INSMA architecture and its data flow. The Data Acquisition Module establishes the connection with the patient monitor and requests the physiological measurements that are acquired. The raw multimodal data obtained from the monitor includes physiological waveforms, alarm messages and numeric (vitals) data. The specific types of data to be acquired can be chosen by the users according to their needs through the “Data Type Selector” and “Physiological Signal Selector”. The data transport rate can also be set by the “Serial Port Selector”. Once the raw data has been acquired from the monitor, the Data Parsing Module will process, parse and transform the data into a time-series using the physiological identifiers or codes provided by the monitor. The Data Visualization Module will display the graphs for the parsed time-series results. It can plot both real-time signals and historical data given a time range. All three modules are developed using MFC and C/C++, so that they all run in the same operating environment and use compatible data formats, and therefore provide a complete solution for data acquisition, parsing and visualization. The details of these modules are discussed in next sections.
Fig. 1.
INSMA Architecture: Data Acquistion, Parsing and Visualization modules. INSMA acquires data from patient monitors in real-time through a variety of different types of network communication.
3.1. Patient monitor
The bedside patient monitor is the most common long-term monitoring medical device used in an ICU. It is used to continuously monitor the physiological parameters of an patient through specially designed sensors, signal acquisition modules, and invasive or noninvasive interfaces: cardiac activity including ECG and heart rate, circulation including blood pressure & cardiac output indices, SPO2, respiratory function including respiration rate, oxygenation, capnography, and Brain through EEG waveforms and derived indicators, temperature and metabolic rate, etc.
The Philips IntelliVue MP70 is a bedside patient monitoring device that displays various physiological waves (e.g. ECG and Blood Pressure) and provides important functions such as displaying numeric vitals data (e.g. heartrate, oxygen saturation) and performing alarm functions based on minimum and maximum limits set by the clinical staff in the monitor. A variety of sensors and associated clinical measurement modules can be connected to the monitor, and these modules are generally interchangeable with other monitors provided by Philips [9]. One of these modules is the Philips VueLink module that provides an interface to more than 100 third-party specialty measurement devices like mechanical ventilators, gas analyzers, anesthesia machines, etc. The Philips IntelliVue MP70 is the monitor INSMA uses to collect high-resolution physiological signals from patients in ICU.
Using a communication interface protocol, data from the Philips IntelliVue Patient Monitor MP70 can be transferred via the Local Area Network (LAN) Interface or Medical Information Bus (MIB/RS232) Interface to an external computer. For the LAN interface, the transport protocol is the standard UDP/IP protocol. For the MIB/RS232 interface, two transport protocols are supported: a fixed baudrate protocol at 19200 or 115200 baud and a protocol with baudrate negotiation (Auto Speed). INSMA can connect and acquire data using either the UDP/IP/LAN protocol/interface or the MIB/RS232 protocol/interface. The Fixed Baudrate Protocol provides a transport protocol with minimal overhead and complexity, so we describe how INSMA uses the Fixed Baudrate Protocol through the MIB/RS232 interface. The Fixed Baudrate Protocol is a connection-oriented, message-based request/respond protocol, based on an object-oriented model concept. All information is stored as attributes within a set of defined object types. The following objects are defined in the protocol: Medical Device System (MDS), Alert Monitor, Numeric, and Patient Demographics. In order for a client application to access the attributes of instantiated objects, it first has to poll the MDS object. Then, the client gets the information of the instantiated object via queries that return the attribute values of these objects. After building the association, the following data can be accessed from the IntelliVue monitor: all measurement numerics and alarm data (real-time update rates up to 1024 ms), wave data, and patient demographic data entered by the user in the IntelliVue monitor.
3.2. Data acquistion module
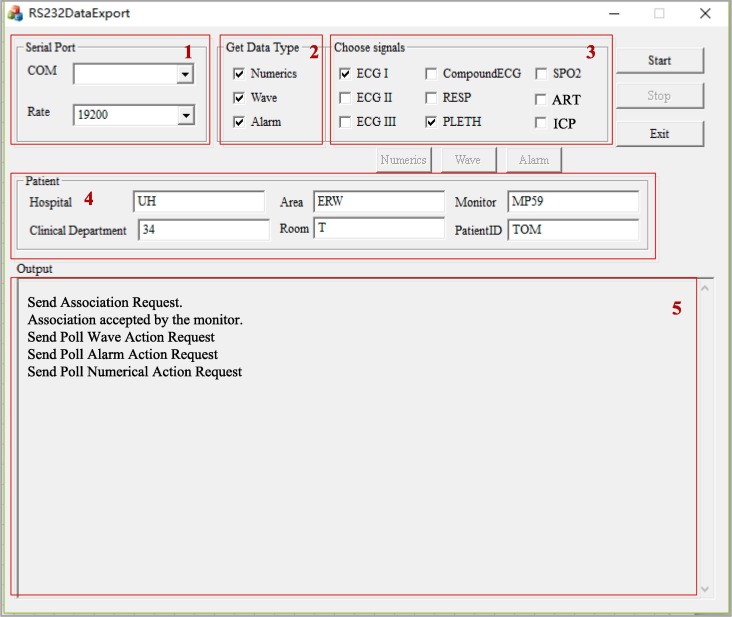
The Data Acquisition Module collects and stores real-time data from patient monitors in intensive care units for further data analytics that supports clinical decision-making. We developed an interface using MFC, to make the data acquisition process easy to be controlled by users. Fig. 2 shows the interface with function areas 1–5.
Fig. 2.
Data Acquisition Interface with special features labeled 1–5.
Functionality includes: Serial Port Selection, Data Type Selection, Signal Selection, Start-Stop, Patient Demographic Display and Output Display. The (1) Serial Port Selector lets the user set the initialization parameters of the communication port and the baudrate rate for serial port communication. The MSComm Control handler will transfer the parameters to messages to initialize serial communication. The (2) Data Type Selector allows users to decide the type of data that will be polled in the Poll Data Request. The backend will be automatically enabled or disabled when the checkbox is checked or unchecked. The (3) Signal Selector allows users to choose the expected signals to observe. The (4) Patient Information area shows the patient demographic information. All the commands are displayed in the (5) Output area, including the requests that are sent and the results that are received. It works like a printout log, to show whether the messages are sent correctly, or what types of messages are received.
The data acquisition and preprocessing tools can perform high-resolution recording and processing tasks, such as simultaneously recording of 1–3 ECG (at 500 samples/s) channels, and additionally up to 8 non-ECG (at 125 or 62.5 samples/s) waves, along with other signal types such as all available numeric values and alert messages. After the program is started, a text file including the monitoring results of the selected data type and signals will be generated for further analysis. The file is named by the date time and patient’s demographic information.
3.3. Data parsing module
The Data Parsing Module runs synchronously with the Data Acquisition Module to continuously parse real-time data being streamed from the monitor to increase the efficiency of the data collection and archiving process and to also make the system suitable for real-time applications in the clinical environment. For the fixed baudrate communication protocol, each transmitted byte consists of one start bit, 8 data bits (no parity) and one stop bit. The monitor will process up to 4 frames within 128 ms, and the framing structure is shown in Fig. 3 .
Fig. 3.
The framing structure.
A frame starts with the Beginning Of Frame (BOF) and Header Information (Hdr), and then followed by the Association Control or Data Export Command message as the User Data, and a 16 bit Frame Check Sequence using the CRC-CCITT algorithm (FCS). The frame ends with the End Of Frame (EOF).
When parsing the data, we first identify each frame by locating its BOF (0xC0) and EOF (0xC1), and get the type and length of the message from Hdr. Next, we interpret the time stamp from the User Data. In the Data Export Protocol defined by Philips IntelliVue monitor, the time stamps contain two types of data: absolute time and relative time.
The absolute time stamp is the time stamp generated and sent only once at the beginning of the communication according to the system date and time set of the monitor, with 1 s resolution. All the following messages sent from monitor will only contain relative time stamp information with resolution in milliseconds. With this relation, all sampled data and alarm events can be mapped to a time axis accordingly. We then interpret the multimodal data in the User Data field.
For the numeric data, we need to interpret the physiological parameters of Numerics Observed Value and Compound Numerics Observed Value. The Numerics Observed Value attribute represents the measured value, along with state and identification data. Each observed value of numeric data has the same data structure. The Compound Numerics Observed Value attribute represents multiple measured values contained in one Numeric object, along with state and identification data. For example, a blood pressure measurement can include systolic, diastolic and mean values in a single Numeric object.
The Alarm object represents the overall device alert condition and contains the global status and a list of active patient alarms. There are two groups of alarms available in the IntelliVue monitor: the active Technical Alarm (T-Alarm) and the active Patient Alarm (P-Alarm). T-Alarm messages indicate that the monitor cannot measure or detect alarm conditions reliably. P-Alarm indicates a relevant deviation from a normal clinical state of patient. It can be a high priority alarm signal such as a potentially life-threatening situation (for example, asystole), or a lower priority alarm signal like a respiration alarm limit violation. T-Alarm and P-Alarm both have the same alarm data structure, including the alarm source, code, type, state, etc.
For the waveform signals, the IntelliVue patient monitor supports the wave types (Table 1 ) that are defined by sample period, sample size, array size, update period and bandwidth requirement.
Table 1.
Different Types Of Waveform Signals Supported By The Monitor.
| Wave Type | Sample rate | Sample Period | Sample Size | Array Size | Update Period | Bandwidth Requirement |
|---|---|---|---|---|---|---|
| ECG | 500 samples/s | 2 ms | 16 bits | 128 samples | 256 ms | 1064 bytes/s |
| Compound ECG | 250 samples/s | 4 ms | 16 bits | 3*64 samples | 256 ms | 1640 bytes/s |
| Non-ECG waves | 125/62.5 samples/s | 8/16 ms | 16 bits | 32/16 samples | 256 ms | 296/168 byte/s |
The data acquisition module can poll up to three ECG waves simultaneously with sample rate at 500 samples/s. The ECG compound wave can contain up to three channels with 250 samples/s for each. Up to eight non-ECG waves (125 or 62.5 samples/s) can be polled simultaneously by selecting the appropriate labels. In the data parsing module, all these polled wave signals can be identified by their physiological identifier, and the values will be interpreted in a binary data format.
3.4. Data visualization module
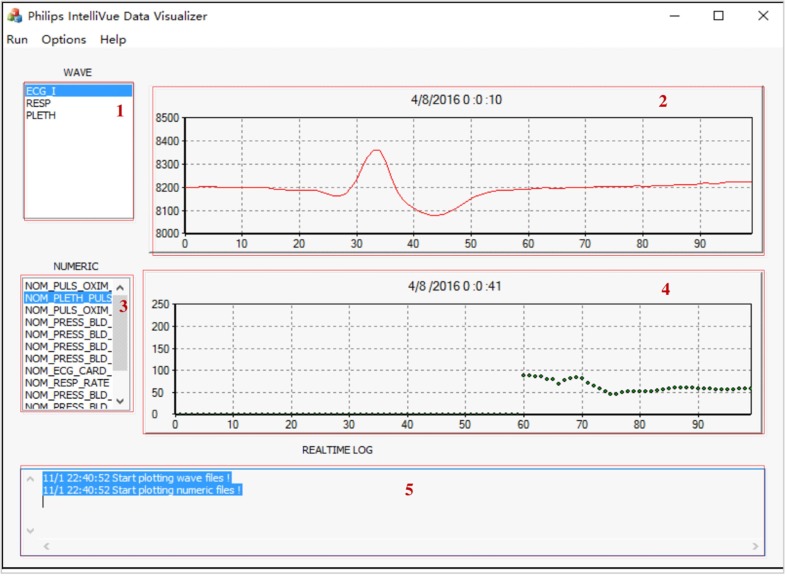
The Data Visualization Module can display the multimodal data including the wave signals and numeric data of patients. Fig. 4 illustrates the main interface.
Fig. 4.
The main interface of the Data Visualization Module. The data is from a patient with three measurements: ECG I, RESP and PLETH.
After picking up the type of wave signals in the selector (1), the corresponded real-time flow of data will display on the charting control panel (2) in a continuous way. The charting control can plot continuous data and is able to display the data quickly by plotting only the new-coming points of data, not the complete series. When a data point is added to a series, the chart control does not refresh the display totally, only the last point (or last line section) will be drawn, which is quite efficient. The charting control module was based on the open-source project provided by Cedric Moonen.1 The timestamp is displayed above the chart. The selector (3) lists all the numeric data types obtained from the parsed results. When a numeric data type is chosen, its value will be displayed in the panel (4) in real-time. The list in (3) will update automatically when the program finds “new” data type in the parsed results of that numeric data. The numeric data is displayed as markers (dots) instead of a continuous curve, and each dot represents one data value at that time point. Different types of waveforms or numeric values are displayed in different colors, so it is easier for users to distinguish them. All the control commands are displayed in panel (5) in a log style. The patient’s ID, wave and numeric data type list can be set in the “Option” menu.
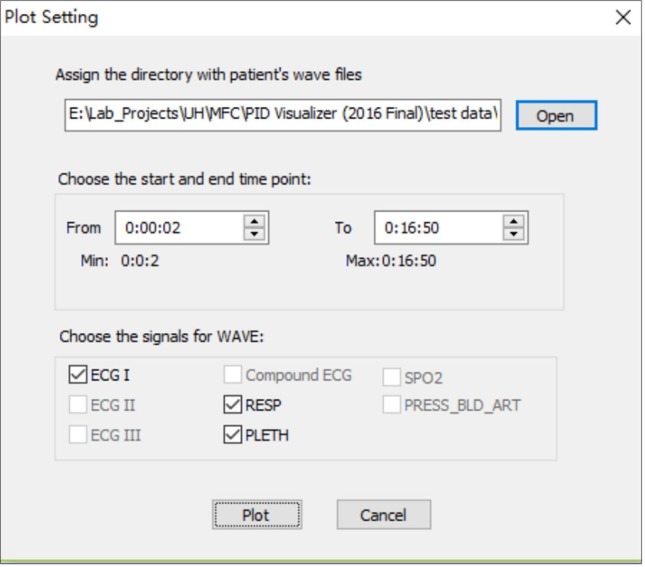
In addition to providing the patient’s physiological status in a real-time mode, the Data Visualization Module can also display waveforms from archived data. Users first need to choose the patient and types of waves that are to be visualized, and then set the time range in the “Plot Setting” window, as shown in Fig. 5 .
Fig. 5.
Plot Setting dialogue window.
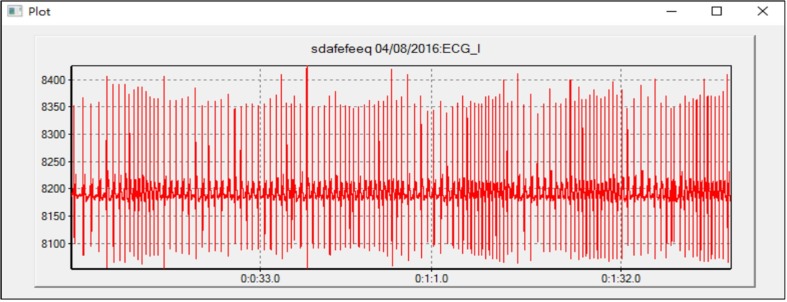
The Teechart charting library in MS VC++ is used to draw the waveforms. Fig. 6 shows an example of displaying the ECG I waveform for 2 min from 0:0:2 to 0:2:2. The x-axis shows the time line, with the scale of milliseconds and format “HH:MM:SS.SSS. The figure is scalar, so it is available to focus on one given segment of data and zoom in to check more details of the signal.
Fig. 6.
Plot one type of wave data: ECG I from 0:0:2 to 0:2:2.
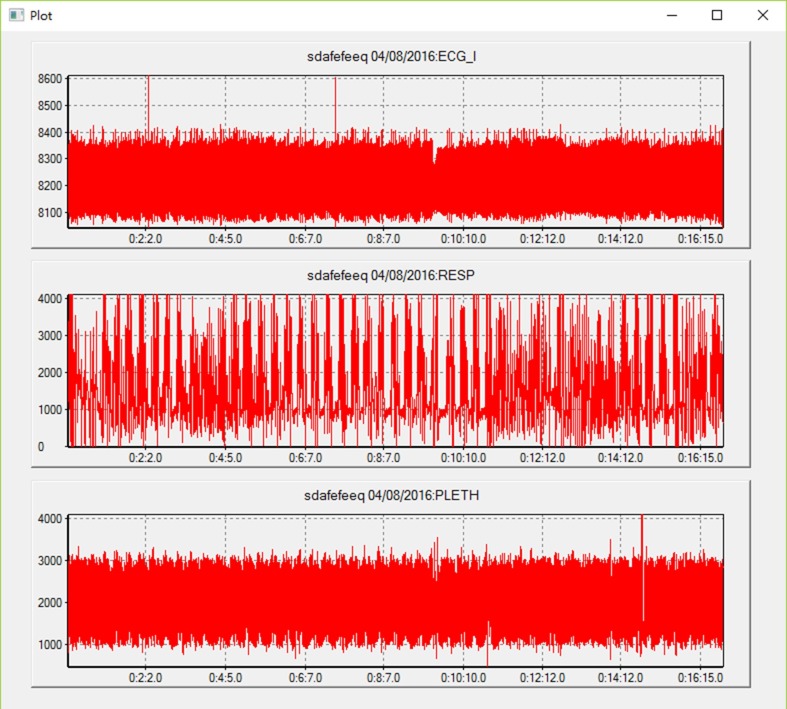
If multiple types of waveform signals are chosen, they will be displayed in one panel with the same but independent horizontal time axes because the signals are simultaneously acquired from the monitor, but they may have different vertical axes because their value scales or ranges may be different. Clinicians can check multiple physiological measurements of a patient at one time and find the correlation among them. Fig. 7 shows an example of displaying three types of waveforms: ECG I, RESP and PLETH from the time 0:0:2 to 0:16:50.
Fig. 7.
Display of three types of wave data: ECG I, RESP and PLETH from 0:0:2 to 0:16:50 in one panel.
4. Discussion and application
4.1. Multimodal data analysis
In order to evaluate the performance of INSMA, the system was deployed in the Neurological Surgery ICU at University Hospitals Cleveland Medical Center using a DELL minicomputer with an Intel(R) Dual Core Celeron processor, 2 GB of RAM, and 240 GB hard disk storage. Results to date indicate that it is reliable for collecting data from the patient monitor. The waveform signals (e.g. ECG, respiration, PLETH/CO2), numeric signals and alarm event signals when streamed continuously from the monitor over a 24-hour period generate an approximately 400 MB data file for each patient. The parsed results will be larger in size because the absolute time stamp information is added to each sample point in the file.
Table 2 shows the parsed results of a patient monitored on March 20, 2019 in the ICU. ECG I, Arterial Blood Pressure (ART) and Intracranial Pressure (ICP) were continuously monitored. Clinical personnel are able to check on how the patient is progressing by observing the ART and ICP waveforms, and how they are temporally correlated. We also collected alarm messages and numeric measurements in the parsed results.
Table 2.
The Parsed Result of a monitored patient after craniotomy.
| Data Type | Size (MB) | Sample Points | |
|---|---|---|---|
| Alarm message | 15.1 | 81,995 | |
| Numerical measurement | 9.36 | 81,850 | |
| Waveform signals | ECG I | 633 | 14,489,216 |
| ART | 135 | 2,738,912 | |
| ICP | 136 | 3,738,912 | |
The analysis of ICU data from patients in the clinical setting is generally limited to visualizing waveform and numeric data and computing simple values such as average heartrate, average respiratory rate, average blood oxygen saturation, etc. In INSMA, each waveform can be analyzed independently or in conjunction with other waveforms to extract more information, as shown in Fig. 7. It is possible to zoom in to provide additional waveform details for visual inspection or apply different analytical analysis techniques to single or multiple waveform signals to better understand the status of the patient and support clinical decision-making.
4.2. Patient state tracking
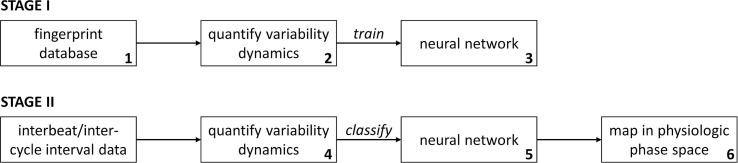
It has been well established that feature extraction for quantifying the complexity and/or variability in physiological time-series data can provide important information related to health and disease [28]. Specifically, even though temporal patterns of variability can be leveraged as powerful diagnostic and/or prognostic indicators, the current use of beat-to-beat and cycle-to-cycle variability dynamics at the bedside is hampered by: (1) lack of high-resolution real-time multimodal clinical data, (2) non-trivial interpretation and integration of these variability metrics into clinical workflows, and (3) lack of a unified framework for classifying variability dynamics into meaningful clinical categories. Algorithms that quantify variability dynamics over multiple temporal scales, such as multiscale entropy (MSE) and multifractal detrended fluctuation analysis (MFDFA) have shown a lot of promise as diagnostic tools in clinical research settings, but the difficulty in interpreting these measures by non-specialists prevents their routine implementation and use in the ICU. The acquired data from patient monitors can be used to develop novel and generalizable methods for quantifying and tracking patient state in real-time [19]. We are developing a patient state tracking system based on the analysis of physiologic time-series dynamics as shown in Fig. 8 .
Fig. 8.
Patient state tracking using acquired multimodal data. Numbers in bottom-right hand corner correspond to numbering below-this stage is performed once for every variability dynamics algorithm of interest.
In STAGE I, synthetic non-stationary time-series exhibiting multifractal dynamics are generated and validated to mimic the beat-to-beat dynamics expected in an individual exhibiting long-term stable dynamics in the ICU. Progressive amounts of white noise are added to the time-series and then the time-series are integrated to generate their brown noise equivalent and to generate a fingerprint database of synthetic time-series that span multifractal time-series dynamics from white noise and brown noise. The fingerprint database is then processed using any algorithm that quantifies multiscale variability dynamics (e.g. Multi-Scale Entropy in our application), and the curated database is divided into classes based on how generated profiles clustered. Next, a feedforward artificial neural network (ANN) is trained on the curated database to classify the MSE profiles according to the specified classes. A 2-dimensional physiologic phase space is created for the ANN classes and based on the data, the classes are rearranged so that actual transitions are more likely to occur between adjacent classes.
In STAGE II, the data is analyzed using the beat-to-beat or cycle-to-cycle time-series data that is of interest. A new dataset is analyzed (Step 4) with the same algorithm used in Step 2, the ANN classifies the output of the algorithm in Step 5, and the result of the ANN classification is then mapped into the physiological phase space in Step 6.
This methodology reduces the dimensionality of multiscale variability dynamics in a clinically relevant manner, thereby facilitating the development of clinician-centric visualization tools that can be implemented in a bedside display, and easily integrated in the ICU workflow as a generalized early warning system for clinical decompensation in ICU patients [18]. Any algorithm that quantifies multiscale variability dynamics [16], [22] can be used to process the waveform data in order to classify the information extracted from the raw data in an intuitive and physiologically relevant manner [23], [24], and thus to facilitate the incorporation of subtle and dynamic fluctuations in physiological waveform data. By assessing the current status of a patient in the ICU, the system will provide a wealth of information on future trajectories for extracting related clinical information [25], [26], [27].
5. Conclusions and future work
The amount of data that is available for clinicians to use in support of real-time patient care at the bedside is growing rapidly as a result of advances in medical monitoring and imaging technology. Advances in informatics, whether through data acquisition, physiologic alarm detection, or signal analysis and visualization for decision support have the potential to markedly improve patient treatment in ICUs. Clinical monitors have the ability to collect and visualize important numerics or waveforms, but more work is needed to interface to the monitors and acquire and synchronize multimodal physiological data across a diverse set of clinical devices. Patient monitors offer the opportunity to acquire a number of different physiological signals in a single device, but in certain cases there are other monitors and devices whose data is critical to patient care, but do not interface to the patient monitor. The tIME framework that we are developing is directly addressing this unmet clinical need. An integrated solution for multimodal data acquisition, parsing and visualization in the ICU (INSMA) presented in this paper is an important first step in achieving this overall vision [19]. Particularly in the Neuro-Intensive Care Unit, there are a variety of different devices that provide valuable information for patient care that do not interface directly to a patient monitor including EEG signal data, real-time tissue blood flow (perfusion) data, and advanced hemodynamic data monitoring (e.g. continuous cardiac output) that are cornerstones in the management of critically ill patients. There are options, for example, with Nihon-Kohden EEG acquisition systems to collect patient vitals (similar to a bedside patient monitor) as well as interface to specialized devices such as for hemodynamic monitoring. Simultaneous acquisition of data from Philips patient monitors and Nihon-Kohden EEG systems in the ICU was done to augment data provided in the MIMIC study [20]. The objective of data acquisition was to stream real-time data from both monitors for archiving in a single biorepository. This provides valuable data for research, but the intent of tIME is to stream data for real-time patient care at the bedside. INSMA is an important first step, and we have also developed data techniques for synchronizing data acquisition from a variety of different ICU devices as a core technology for future implementations of tIME in the ICU. We have also demonstrated that patient data acquired from the patient monitor can be used for patient state tracking. The prototype system we developed was optimized to identify the type of dynamics observed in cardiac (ECG or blood pressure) beat-to-beat time-series data collected from ICU patients. The prototype system has been tested using ICU patient data from ECG to understand how variability in the heartbeat time-series can be used to dynamically track patient state [18].
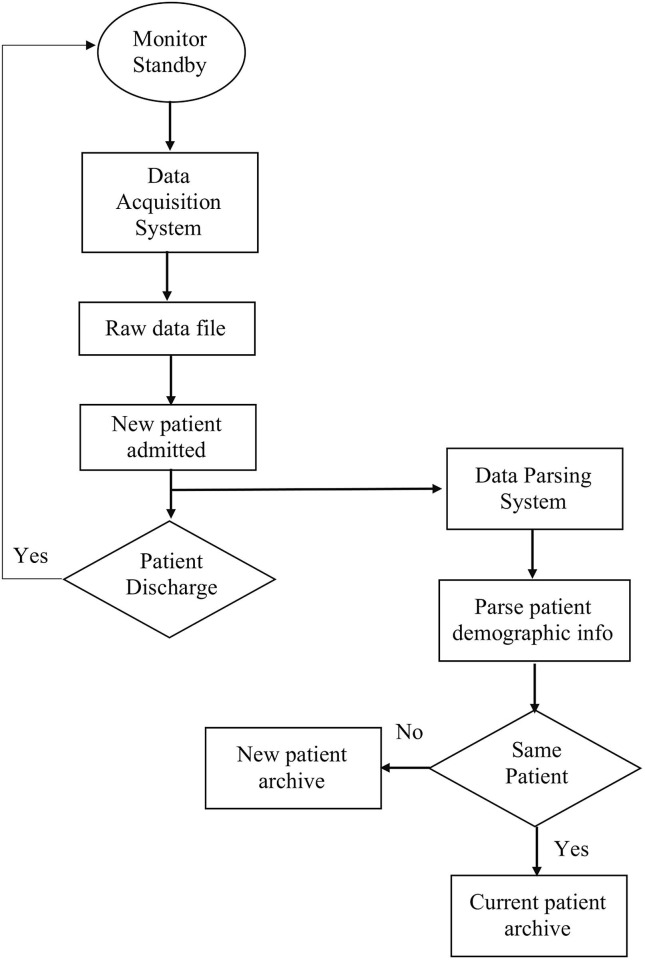
In the current development of INSMA and tIME, we have implemented the INSMA software on Lenovo ThinkCentre M600 computers, and currently have one system connected to the Philips patient monitor in the Neurosurgery ICU at University Hospitals Cleveland Medical Center under the direction of Dr. DeGeorgia to continuously collect patient data. We are completing the development of 21 additional INSMA units that will be connected to each of the Neurosurgery ICU beds. The INSMA system operates in background and once setup for data collection at the bedside and does not require attention from any clinical ICU personnel. INSMA allows the client to send messages to request for patient demographic information, and we implemented a patient demographic request function that monitors any modification of patient information. New patient demographic information will be entered when patients admitted into the ICU are connected to the monitor, and this information will be requested and stored in the raw data file. When the patient is discharged and a new patient is admitted and connected to the monitor, the parsing algorithm will capture the change of patient demographic information (e.g. First Name, Last Name, Age, Weight) and a new patient archive corresponding to the new patient identifier will be created, as shown in Fig. 9 .
Fig. 9.
Flow chart of patient data management.
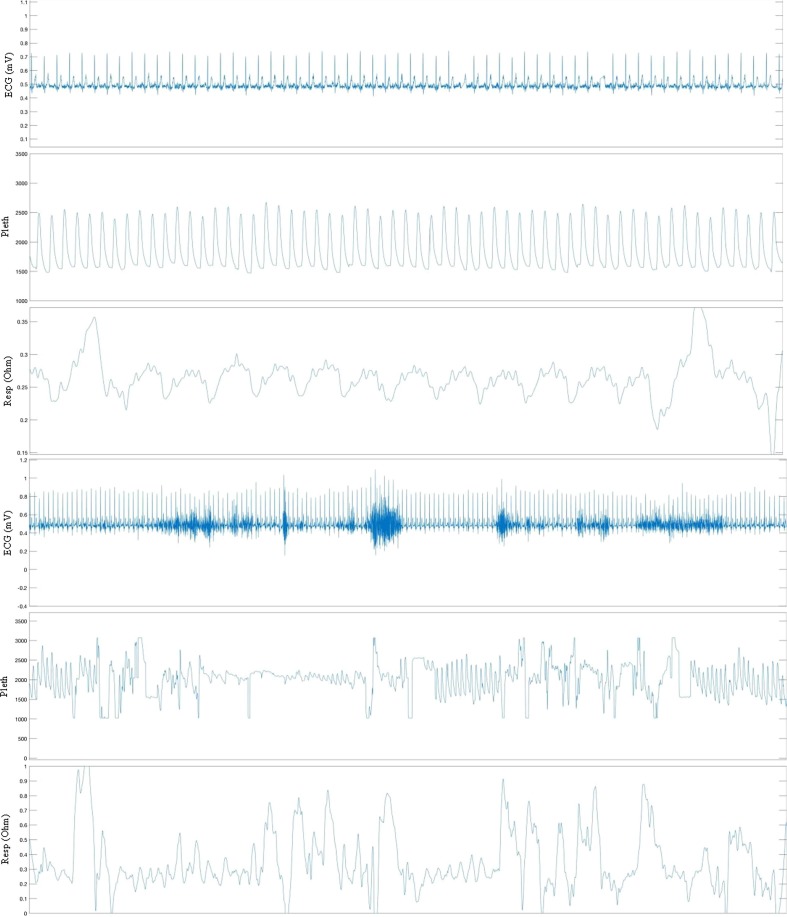
Each unit is equipped with a wireless communication link that supports remotely monitoring the INSMA units only from within the hospital firewall to protect the privacy and security of the data, and then also moves the data to a permanent secure data storage unit. Example data from patient monitoring is shown in Fig. 10 .
Fig. 10.
Parsed data from INSMA. ECG, Pleth and Respiration waveforms (top and bottom panels) selected from a recording of a patient in the Neurosurgery ICU.
The future of critical care will require “information management”, that includes the real-time collection, integration, and interpretation of various types of physiological data from multiple sources. The possible research work will focus on (1) the integration and analysis of massive heterogenous medical data to provide scientific decision-making with machine learning methods [9], [30], and (2) the acquisition and processing of vast amount of multi-channel high-density and real-time streaming data using multivariate and nonlinear time series analysis methods to facilitate rapid diagnosis and treatment [31]. Patient care in the ICU can be significantly improved through the application of complex system analysis and information management methods.
CRediT authorship contribution statement
Yingcheng Sun: Conceptualization, Methodology, Software, Visualization, Writing - original draft. Fei Guo: Conceptualization, Software, Methodology, Visualization. Farhad Kaffashi: Conceptualization, Methodology, Formal analysis. Frank J. Jacono: Resources, Data curation, Validation. Michael DeGeorgia: Resources, Data curation, Validation. Kenneth A. Loparo: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported in part by AHRQ Grant 1R18HS022860-01A1 (PI: Dr. Leo Kobayashi).
Footnotes
References
- 1.Dasta J.F., McLaughlin T.P., Mody S.H., Piech C.T. Daily cost of an intensive care unit day: the contribution of mechanical ventilation. Crit. Care Med. 2005;33:1266–1271. doi: 10.1097/01.ccm.0000164543.14619.00. [DOI] [PubMed] [Google Scholar]
- 2.Zimmerman J.E., Wagner D.P., Draper E.A., Wright L., Alzola C., Knaus W.A. Evaluation of acute physiology and chronic health evaluation III predictions of hospital mortality in an independent database. Crit. Care Med. 1998;26:1317–1326. doi: 10.1097/00003246-199808000-00012. [DOI] [PubMed] [Google Scholar]
- 3.Lwin A.K., Shepard D.S. The Leapfrog group; Washington, D.C.: 2008. Estimating Lives and Dollars Saved from Universal Adoption of the Leapfrog Safety and Quality Standards: 2008 Update. [Google Scholar]
- 4.Roederer A., Dimartino J., Gutsche J., Mullen-Fortino M., Shah S., Hanson C.W., Lee I. 2016 IEEE First International Conference on Connected Health: Applications, Systems and Engineering Technologies (CHASE) IEEE; 2016. Clinician-in-the-Loop Annotation of ICU Bedside Alarm Data; pp. 229–237. [Google Scholar]
- 5.Raghupathi W., Raghupathi V. Big data analytics in healthcare: promise and potential. Health Inform. Sci. Syst. 2014;2(1):3. doi: 10.1186/2047-2501-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matam B.R., Duncan H. Technical challenges related to implementation of a formula one real time data acquisition and analysis system in a paediatric intensive care unit. J. Clin. Monit. Comput. 2018;32(3):559–569. doi: 10.1007/s10877-017-0047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winslow R.L., Granite S., Jurado C. WaveformECG: a platform for visualizing, annotating, and analyzing ECG Data. Comput. Sci. Eng. 2016;18(5):369. doi: 10.1109/MCSE.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee H.C., Jung C.W. Vital recorder—a free research tool for automatic recording of high-resolution time-synchronised physiological data from multiple anaesthesia devices. Sci. Rep. 2018;8(1):1527. doi: 10.1038/s41598-018-20062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gjermundrod H., Papa M., Zeinalipour-Yazti D., Dikaiakos M.D., Panayi G., Kyprianou T. Twentieth IEEE International Symposium on Computer-Based Medical Systems (CBMS'07) IEEE; 2007. Intensive care window: A multi-modal monitoring tool for intensive care research and practice; pp. 471–476. [Google Scholar]
- 10.De Georgia M.A., Kaffashi F., Jacono .J., Loparo K.A. Information technology in critical care: review of monitoring and data acquisition systems for patient care and research. Sci World J. 2015 doi: 10.1155/2015/727694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philips, Streamlining Data Management Workflow, 2010. http://incenter.medical.philips.com.
- 12.Tsui F., Ching-Chung L., Sun M. Acquiring, modeling, and predicting intracranial pressure in the intensive care unit. Biomed. Eng. 1998;8:96–108. [Google Scholar]
- 13.Schmidt G.N., Bischoff P., Standl T., Hellstern A., Teuber O., Esch J.S. Comparative evaluation of the Datex-Ohmeda S/5 Entropy Module and the Bispectral Index® monitor during propofol–remifentanil anesthesia. Anesthesiology: J. Am. Soc. Anesthesiologists. 2004;101(6):1283–1290. doi: 10.1097/00000542-200412000-00007. [DOI] [PubMed] [Google Scholar]
- 14.Kool N.P., van Waes J.A., Bijker J.B., Peelen L.M., van Wolfswinkel L., de Graaff J.C., van Klei W.A. Artifacts in research data obtained from an anesthesia information and management system. Can. J. Anesthesia/Journal canadien d'anesthésie. 2012;59(9):833–841. doi: 10.1007/s12630-012-9754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu D., Görges M., Jenkins S.A. University of Queensland vital signs dataset: Development of an accessible repository of anesthesia patient monitoring data for research. Anesth. Analg. 2012;114(3):584–589. doi: 10.1213/ANE.0b013e318241f7c0. [DOI] [PubMed] [Google Scholar]
- 16.Y. Sun, K. Loparo, Context aware image annotation in active learning, in: 2019 19th Industrial Conference on Data Mining, vol. 1, Springer, 2019, pp. 251–262.
- 17.Jacono F., DeGeorgia M.A., Wilson C., Dick T.E., Loparo K.A. Data acquisition and complex systems analysis in critical care: developing the intensive care unit of the future. J. Healthcare Eng. 2010;1(3):337–355. [Google Scholar]
- 18.Vandendriessche et al., A framework for patient state tracking by classifying multiscalar wavform features, IEEE Trans. Biomed. Eng. 64(12) (2017). [DOI] [PMC free article] [PubMed]
- 19.DeGeorgia M., Loparo K., editors. Neurocritical Care Inforamtics: Translating Raw Data into Bedside Action. Springer; 2019. [Google Scholar]
- 20.Yoon D., Lee S., Kim T.Y. System for collecting biosignal data from multiple patient monitoring systems. Healthcare Inform. Res. 2017;23(4):333–337. doi: 10.4258/hir.2017.23.4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein B., McNames J., McDonald B.A. Physiologic data acquisition system and database for the study of disease dynamics in the intensive care unit. Crit. CareMed. 2003;31(2):433–441. doi: 10.1097/01.CCM.0000050285.93097.52. [DOI] [PubMed] [Google Scholar]
- 22.Y. Sun, X. Cai, K. Loparo, Learning-based adaptation framework for elastic software systems, in: 2019 IEEE 31st International Conference on Software Engineering & Knowledge Engineering (SEKE), vol. 1, pp. 281–286.
- 23.Y. Sun, K. Loparo, Context aware image annotation in active learning with batch mode, in: 2019 IEEE 43rd Annual Computer Software and Applications Conference (COMPSAC), vol. 1. IEEE, 2019, pp. 952–953.
- 24.Sun Y., Kolacinski R., Loparo K. February. Eliminating search intent bias in learning to rank. IEEE; 2020. pp. 108–115. [Google Scholar]
- 25.Y. Sun, K. Loparo, Information extraction from free text in clinical trials with knowledge-based distant supervision, in: 2019 IEEE 43rd Annual Computer Software and Applications Conference (COMPSAC), vol. 1, IEEE, 2019, pp. 954–955.
- 26.Y. Sun, K. Loparo, Knowledge-guided text structuring in clinical trials, in: 2019 19th Industrial Conference on Data Mining (ICDM), vol. 1, Springer, 2019, pp. 211–219.
- 27.Q. Li, Y. Sun, B. Xue, Complex query recognition based on dynamic learning mechanism, J. Comput. Inform. Syst. 8(20) (2012) 1–8.
- 28.Sun Y., Liang X., Loparo K. Vol. 1. Springer; 2019. A common gene expression signature analysis method for multiple types of cancer; pp. 185–196. (2019 19th Industrial Conference on Data Mining (ICDM)). [Google Scholar]
- 29.Yue L., Tian D., Chen W., Han X., Yin M. Deep learning for heterogeneous medical data analysis. World Wide Web. 2020:1–23. [Google Scholar]
- 30.Sun Y., Loparo K. Opinion spam detection based on heterogeneous information network. 2019 IEEE 31st International Conference on Tools with Artificial Intelligence (ICTAI) 2019:1156–1163. [Google Scholar]
- 31.Xu Y., Biswal S., Deshpande S.R., Maher K.O., Sun J. Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. 2018. July. Raim: Recurrent attentive and intensive model of multimodal patient monitoring data; pp. 2565–2573. [Google Scholar]