Abstract
Histone H2A.Z plays an essential role in regulating transcriptional rates and memory. Interestingly, H2A.Z-bound nucleosomes are located in both transcriptionally active and inactive promotors, with no clear understanding of the mechanisms via which it differentially regulates transcription. We hypothesized that its functions are mediated through recruitment of regulatory proteins to promoters. Using rapid chromatin immunoprecipitation-mass spectrometry, we uncovered the association of H2A.Z-bound chromatin with the metabolic enzymes, oxoglutarate dehydrogenase (OGDH) and acetyl-CoA acyltransferase 2 (ACAA2). Recombinant green florescence fusion proteins, combined with mutations of predicted nuclear localization signals, confirmed their nuclear localization and chromatin binding. Conclusively, chromatin immunoprecipitation-deep sequencing, confirmed the predominant association of OGDH and ACAA2 with H2A.Z-occupied transcription start sites and enhancers, the former of which we confirmed is conserved in both mouse and human tissue. Furthermore, H2A.Z-deficient human HAP1 cells exhibited reduced chromatin-bound metabolic enzymes, accompanied with reduced posttranslational histone modifications, including acetylation and succinylation. Specifically, knockdown of OGDH diminished H4 succinylation. Thus, the data reveal that select metabolic enzymes are assembled at active, H2A.Z-occupied, promoters, for potential site-directed production of metabolic intermediates that are required for histone modifications.
Keywords: H2A.Z, chromatin immunoprecipitation, mitochondria, ACAA2, OGDH, histone acetylation, histone succinylation
Introduction
The highly conserved H2A.Z gene is unique in several aspects that contrast with the genes of core histones; specifically, it is a single copy that is not located within the core histone clusters known in human and mouse genomes, it has introns, and is polyadenylated [1], thus, underscoring its distinct nature. We currently know that it selectively associates with transcriptionally active, as well as, inactive genes. For example, in yeast, Htz1 has been shown to suppress the spread of heterochromatin into transcriptionally active genes near the telomeres [2], while, in contrast, its abundance was shown to negatively correlate with transcriptional rates [3].
Furthermore, changes in growth conditions induced translocation of Htz1 from transcriptionally active to inactive genes [4]. More precisely, Htz1 is found at the transcription start site (TSS) of nearly all genes in euchromatin, in the −1 and +1 nucleosomes flanking a nucleosome free region in the active genes, while present mainly in the −1 nucleosome in inactive genes [5]. Likewise, in Drosophila, H2Av is present at thousands of both transcriptionally active and inactive genes in euchromatin, as well as, in heterochromatic chromocenter of polytene chromosomes [6], whereas its density negatively correlates with that of RNA polymerase II (pol II). Conversely, other studies have shown that H2A.Z vs. H2A at the +1 nucleosome facilitates pol II progression [7]. In murine embryonic stem cells, we also see this bifunctionality, where in the undifferentiated state, H2A.Z, in conjunction with the polycomb subunit Suz12, is present at silenced homeodomain genes involved in differentiation, whereas in committed neuronal progenitor cells, its associates with highly expressed genes [8]. Other than destabilizing nucleosomes at the +1 position, we have little understanding of how H2A.Z selectively regulates transcriptional activation versus deactivation, or if it has any role in metabolism-regulated transcriptional or epigenetics remodeling.
Generally, organisms respond to metabolic cues by exacting a change in gene transcription that influences their development and growth, or homeostasis. These signals include ATP:ADP:AMP and NAD+:NADH ratios, and the availability of metabolites that are involved in histone and DNA modifications – e.g. acetyl-CoA (ac-CoA), α-ketoglutarate (αKG), and succinyl-CoA (suc-CoA), not discounting other acyl-CoAs [9]. In the case of acetyl-CoA, we know that during substrate abundance, citrate is exported from the mitochondria and into the cytosol and nucleus, where ATP citrate lyase (ACLY) converts it into acetyl-CoA, which is a substrate for histone acetylation [10]. Alternatively, during substrate shortage, acetate is imported from the circulation and converted into acetyl-CoA by acyl-CoA synthetase short chain family member 2 (ACSS2) [11]. As for the other CoA-linked metabolites and αKG, the mechanism for nuclear delivery is less well-established. Moreover, the question of how substrates modulate the expression of specific target genes remains a challenge.
The current dogma is that metabolic oxidative enzymes and substrate oxidation are largely confined to the mitochondria. However, recent findings, including our own, challenge this belief. In specific, Sutendra et al, reported the presence of all subunits of the pyruvate dehydrogenase (PDH) complex in human sperm, and normal and cancerous lung epithelial cells [12], while Nagaraj et al reported it in the human 4/8-cell stage zygote [13], and show that it is required for generating acetyl-CoA for histone acetylation. Likewise, Chen et al have recently reported that PDH is present in the nuclei of prostate cancer cells, where it promotes histone acetylation of ‘sterol regulatory element-binding factor’ target genes [14]. On the other hand, oxoglutarate dehydrogenase (OGDH) [15], fumarase [16], and isocitrate dehydrogenase (IDH2) [17] have also been shown to localize to the nucleus, as these findings are further supported by a proteomics study that revealed the presence of all tricarboxylic acid (TCA) cycle enzymes in the nucleus of breast cancer cells [18]. However, except for OGDH in U251 cells [15] and fumarase in U2OS [16], none of the other enzymes have been shown to associate with chromatin. Here we report, that using an unbiased screen for the discovery of proteins that co-localize with H2A.Z-bound chromatin, we uncovered the association of, at least, OGDH and ACAA2 with chromatin in both mouse and human cells. Our recent report shows that H2A.Z is mainly localized to the TSS of most transcribed genes, including housekeeping and inducible genes, where it is at its highest levels, but is relatively low at tissue-specific genes [19]. Therefore, this would position the metabolic enzymes at the TSS of the former groups. In validation, we successfully completed chromatin immunoprecipitation-deep sequencing (ChIP-Seq) using OGDH or ACAA2 antibodies, which revealed the associated of these enzymes with select H2A.Z-occupied TSSs and enhancers. Moreover, the knockout of H2A.Z in HAP1 cell provided evidence that this association is predominantly dependent on H2A.Z. The data also suggest that some histone modifications are reliant on the recruitment of mitochondrial enzymes to chromatin.
Materials and methods
Animal care -
All animal procedures used in this study are in accordance with US National Institute of Health Guidelines for the Care and Use of Laboratory Animals (No. 85-23). All protocols were approved by the Institutional Animal Care and Use Committee at the Rutgers-New Jersey Medical School.
H2A.Z rapid chromatin immunoprecipitation mass spectrometry of endogenous proteins (RIME) -
Male C57/BI, 12 wk-old mice, 10 each, were subjected to a sham or transverse aortic constriction (TAC) procedure. After 1 wk, the hearts were isolated, pooled for each condition, and sent to Active Motif for RIME analysis by anti-H2A.Z (Active Motif, cat # 39943), according to a previously published protocol [20]. The DNA fragments used for ChIP range in size from 200 bp to 1200 bp, with the bulk around 400-500 bp (coverage for ~ 2 nucleosomes).
DATABASE SEARCHING (Active Motif) –
All MS/MS samples were analyzed using X! Tandem (The GPM, thegpm.org; version X! Tandem Alanine (2017.2.1.4)). X! Tandem was set up to search the UP_mouse_CrapE2F1_rev database (unknown version, 106444 entries) assuming the digestion enzyme trypsin. X! Tandem was searched with a fragment ion mass tolerance of 20 PPM and a parent ion tolerance of 20 PPM. Glu->pyro-Glu of the N-terminus, ammonia-loss of the N-terminus, gln- pyro-Glu of the n-terminus deamidated of asparagine and glutamine, oxidation of methionine and tryptophan and dioxidation of methionine and tryptophan were specified in X! Tandem as variable modifications. Each sample was analyzed twice by MS/MS.
CRITERIA FOR PROTEIN IDENTIFICATION (Active Motif) –
Scaffold (version Scaffold_4.8.4, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 50.0 % probability by the Scaffold Local false discovery rates (FDR) algorithm. Peptide identifications were also required to exceed specific database search engine thresholds. X! Tandem identifications required at least. Protein identifications were accepted if they could be established at greater than 5.0% probability to achieve an FDR less than 5.0% and contained at least 1 identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm [21]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
ANALYSIS -
The total spectra counts for all genes in the three samples, were normalized to the corresponding spectra count of the rabbit IgG (control or anti-H2A.Z) used for the immunoprecipitation, which was detected in each sample. Fold enrichment of total spectra for sham/IgG and TAC/IgG, for each gene, was calculated. Seventy-three of these had a ≥ 2-fold enrichment, those are shown in figure 1 and supplementary figure 1S.
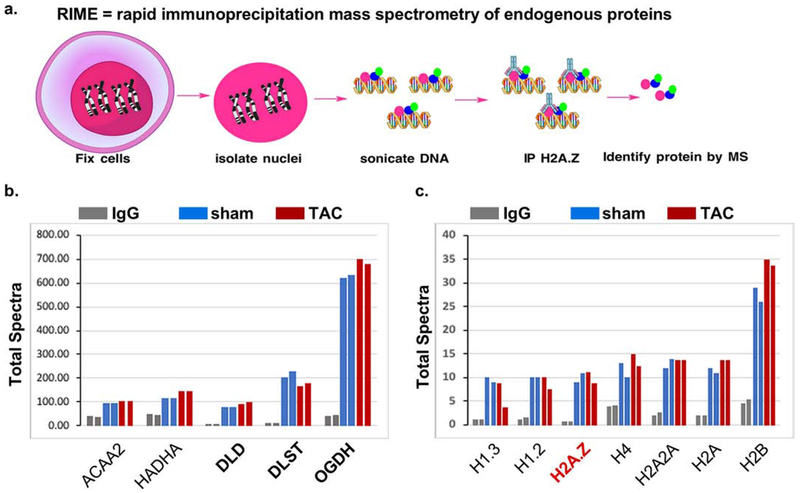
Figure 1. H2A.Z-bound chromatin associates with metabolic enzymes in mouse heart nuclei.
ChIP with anti-H2A.Z or a control IgG was performed on nuclei from a pool of 10 hearts each, from mice subjected to a sham or a transverse aortic constriction operation. The ChIP complex was then subjected to MS/MS, each sample analyzed twice. a. An illustration of the procedure. b-c. Total spectra identified in the IgG, sham, and TAC samples were plotted. These include those with a cutoff of more than 2-fold enrichment versus control IgG, after normalization to the IgG C chain region spectra in each sample, grouped as: b. TCA cycle enzymes and β-oxidation spiral enzymes, and c. histones.
Culturing rat neonatal cardiac myocytes -
Cardiac myocytes were cultured as described in our previous reports [22]. Briefly, hearts were isolated from 1 day old of Sprague-Dawley rats. After dissociation with collagenase, cells were subjected to Percoll gradient centrifugation followed by differential pre-plating for 30 min to enrich for cardiac myocytes and deplete non-myocyte cells. Myocytes were cultured in Dulbecco’s Modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS). All experiments were initiated after a 24 h culturing period.
Culturing mouse adult cardiac myocytes -
Adult cardiac myocytes were isolated and cultured from C57/BI mice (8-9 wks old), according to the protocol described by Ackers-Johnson et al. [23]. Briefly, mice were anesthesia with Ketamine/Xylazine/Acepromazine (65/13/2 mg/kg) by intraperitoneal injection. The mouse chest cage was then opened, the ascending aorta clamped, and both descending aorta and inferior vena cava cut. First, ethylenediaminetetraacetic acid (EDTA) is injected into the base of the right ventricle, the heart is then transferred into a petri dish, and a second EDTA injection is administered into the left ventricular wall above the apex. Following this, the cells are dissociated using collagenase, and the rod-shaped myocytes are differentially separated by gravity, where calcium is re-introduced. The cells are plated on laminin-coated dishes or glass slides in M199 medium with 5 % FBS for 1 hour, after which the FBS is replaced with 0.1% bovine serum albumin for longer culturing periods.
Human iPSC-derived cardiac myocyte cultures -
Cardiac myocytes derived from human iPSCs were purchased from Cellular Dynamics International and cultured as recommended by the manufacturer.
Colon cancer cell culture -
SW620 were purchased from the American Type Culture Collection (ATCC). Cells were cultured in Leibovitz’s (Gibco) medium with 10% FBS and maintained in a CO2 free incubator.
Human haploid HAP1 and ΔH2A.Z-HAP1 cell cultures -
HAP1 and H2A.Z knockout HAP1 (ΔH2A.Z-HAP1) cell lines were purchased from Horizon Discovery and cultured in Iscove’s Modified Dulbecco’s Medium with 10% FBS, according to the company’s protocol. These cells are fibroblast-like derived from human male chronic myelogenous leukemia (CML) cell line KBM-7. The ΔH2A.Z-HAP1 was generated by CRISPER/Cas, creating a 2bp deletion in exon 3 of H2A.Z. The cells are viable but have a much slower proliferation rate than the parent cell-line.
Construction of GFP fusion proteins -
The plasmids harboring cDNAs of turbo-GFP (tGFP), OGDH (NM_002541), ACAA2 (NM_177470), and IDH2 (NM_002168) were purchased from Origene. The cDNA of the latter three genes were then sub-cloned, in-frame, upstream of tGFP, and the fusion cDNA was subsequently sub-cloned into the pDC316 shuttle plasmid vector (Microbix), which was used to generate recombinant adenoviral vectors via homologous recombination.
Harvesting and immunostaining mouse emobryos -
Embryos were dissected at E10.5 in ice-cold phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde at 4°C overnight, and then washed with ice-cold PBS. Embryos were then embedded into optimal cutting temperature matrix and sectioned in sagittal orientation. For immunofluorescence, sections were incubated with blocking buffer: 5% donkey serum (Sigma, cat # D9663) diluted in PBS containing 0.05% Tween-20 (PBST), for 30 minutes at room temperature. Sections were then incubated with primary antibodies at 4°C overnight: OGDH (Cell Signaling Technology, cat # 26865, 1:100 dilution) and alpha-cardiac actin (Sigma, cat # A9357, 1:300 dilution) diluted in the blocking buffer. Slides were then washed in PBS and incubated with 4′,6-diamidino-2-phenylindole (DAPI) to mark nuclei and with secondary antibodies diluted 1:300 in the blocking buffer for 2 hours at room temperature. 2° antibodies were donkey anti-rabbit-Alexa 488 (Invitrogen, cat # A21206) and donkey anti-mouse-Alexa 555 (Invitrogen, cat # A31570).
Transverse aortic constriction (TAC) in mice -
This was performed as described in our previous reports [24, 25]. Briefly, a 7-0 braided polyester suture was tied around the transverse thoracic aorta, against a 27-gauge needle, between the innominate artery and the left common carotid artery. Control mice were subjected to a sham operation involving the same procedure, minus the aortic constriction.
Echocardiography and doppler -
This was performed as described in our previous reports [24, 25]. Briefly, transthoracic echocardiography was performed using the Vevo 770 imaging system (Visual Sonics, Inc.) with a 707B-30MHz scanhead, encapsulated, transducer. Electrocardiographic electrodes were taped to the four paws, then one dimensional (1D) M-mode and 2D B-mode tracings were recorded from the parasternal short-axis view at the mid papillary muscle level. In addition, pulse-wave Doppler was used to measure blood flow velocity and peak gradient pressure in the aorta. For analysis, we used the Vevo 770 Software (Vevo 770, Version 23), which includes: analytic software package for B-Mode (2D) image capture and analysis; cine loop image capture, display, and review; software analytics for advanced measurements and annotations; and physiological data on-screen trace.
Construction and delivery of recombinant adenovirus (Ad) vector -
Recombinant adenoviral vectors were constructed, propagated, purified, and tittered as described in our previous reports [26–28]. Briefly, short hairpin RNAs (shRNAs) were cloned into pDC311 shuttle (Microbix Biosystems Inc.), downstream of a U6 promoter. These were transfected with the replication-defective Ad5 viral DNA backbone into 293HEK cells, in which a recombination reaction introduces the DNA insert into the viral DNA. Single virus plaques were amplified in 293HEK cells, purified on a CsCl2 gradient, dialyzed, and tittered on 293HEK cells with agarose overlay. Ad vectors were constructed with the following inserts, shRNA targeting OGDH - ccagccactggcaacaagaaTTCAAGAGAAttcttgttgccagtggctggTTTTTT, shRNA targeting H2A.Z - gtcacttgcagcttgctataTTCAAGAGAAtatagcaagctgcaagtgacTTTTTT, or a nonsense control shRNA gaaccgagcccaccagcgagcTTCAAGAGAAgctcgctggtgggctcggttcTTTTTT, the shaded areas are the loop sequence and the terminal 6xT’s is the stop signal for the U6 promoter used for the expression of these shRNAs. Cardiac myocytes were infected with 10-30 multiplicity-of-infection (moi) of the viruses for 24 h or 48 h, as indicated in the figure legends.
Subcellular fractionation and Western blotting -
Proteins were fractionated using the subcellular protein fractionation kit (Thermo Fisher, cat # 78840), according to the manufacturer’s protocols. The cellular fractions were separated on a 4% to 12% gradient SDS-PAGE (Criterion gels, Bio-Rad) and transferred to nitrocellulose membrane. The antibodies used include: anti-turboGFP (Origene, cat # TA150041), -OGDH (E1W8H, Cell Signaling Technology, cat # 26865), -ACAA2 (Origene Technologies, cat # TA506126), -H2A.Z (Active Motif, cat # 39943), -H3 (Active Motif, cat # 61476), -H3 pan-acetyl (Active Motif, cat # 39140), -H3K27 di- and tri-methyl (H3K27me2/3, Active Motif, cat # 39538), -nuclear pore glycoprotein p62 (NUP62, US Biological, cat # USB326547), -transcription factor II B (TFIIB, Cell Signaling Technology, cat # 4169), -H4K12-succinyl (H4K12suc, Epigentek, cat # A70383), -H2A (Active Motif, cat # 35951), -H4 (Upstate, cat # 07-108), -H3K9 mono-, di-, tri-methyl (H3K9me1/2/3, Active Motive, cat # 38241), -AKT1 (Cell Signaling Technology, cat # 9272), -voltage-dependent anion-selective channel 1 (VDAC1, Genscript, cat # A01419), -RNA pol II (Abcam, cat # ab5095), and -complex IV (Thermo Scientific, Cat# 459600). The Western blot signals were detected by the Odyssey imaging system (LI-COR).
Immunocytochemistry -
Cells were seeded on glass chamber slides coated with fibronectin for neonatal myocytes and hiPSC-CM, or with laminin for adult myocytes, fixed with 3% formaldehyde / 0.3% triton x-100, then incubated with antibodies (1:100) or phalloidin (in Tris-buffered saline with 1% bovine serum albumin), washed and mounted using Prolong Gold anti-fade with DAPI (Molecular Probes). The antibodies included: anti-OGDH (Cell Signaling Technology, same as those listed for the Western blotting, above), as well as, anti-OGDH (Sigma, cat # HPA019514), and anti-ACAA2 (Origene Technologies, cat # TA506126). The slides were imaged using Nikon A1R laser scanning confocal microscope with Plan Apo 60x objective.
ChIP-Seq (Active Motif) and data analysis -
Mice were subjected to transverse aortic constriction or a sham operation. After 7 days, cardiac function and structure were assessed by echocardiography, before isolation of the hearts. The hearts were then analyzed by ChIP using the following antibodies: anti-RNA pol II (Abcam, cat # ab5095), -H2A.Z (Active Motif, cat # 39113), -H3K9-acetyl (H3K9ac) (Active Motif, cat # 39918), -TFIIB (Santa Cruz Biotechnology, cat # sc-225), -cyclin-dependent kinase 9 (CDK9, Santa Cruz Biotechnology, cat # sc-8338), -ANP32e (Abeam, cat # ab5993), -ACAA2 (Origene Technologies, cat # TA506126), -OGDH (E1W8H, Cell Signaling Technology, cat # 26865), and -FOXO1 (Santa Cruz Biotechnology, cat # SC-1135), followed by next generation sequencing (Active Motif). We have previously reported the results of our ChIP-Seq for RNA pol II [29], H3K9ac [29], TFIIB [22], H2A.Z [19], and ANP32E [19], and, thus, are not further described here. Briefly, ChIP libraries were sequenced using NextSeq 500, generating 75-nt sequence reads that are mapped to the genome using BWA algorithms. The reads/tags were extended in silico by 150-250 bp at their 3’end (fragments), the density of which is determined along the genome, divided in 32 nt bins, and the results saved in bigWig and BAM (Binary Alignment/Map) files. Fragment peaks were identified using MACS, which identifies local areas of enrichment of Tags, defined as ‘intervals’, while overlapping intervals are grouped into ‘Merged Regions’. The locations and proximities to gene annotations of intervals and active regions are defined and compiled in Excel spreadsheets, which include average and peak fragment densities. Regarding tag normalization and input control, the sample with the lowest number of tags is used for normalization of all samples, while the input is used to identify false positive peaks. The statistics for ACAA2 and OGDH ChIP-Seq results, including total number of reads, peaks, empirical FDR, and peak calling parameters are listed in supplementary Table 1.
In addition, we separately analyzed the fragment densities by gene region, where the average value (Avg Val) of fragment densities at the TSS (−1000 to +1000) and in-gene/gene body (+1000 to 3’ end) regions for all genes were calculated separately. Subsequently, we used these values to sort genes according to TSS-pol II, -ACCA2, or -OGDH, occupancy.
Accessions: the RNA polymerase II and H3K9ac ChIP-Seq data (accession: GSE50637), the TFIIB ChIP-Seq data (accession: GSE56813) and H2A.Z and ANP32E ChIP-Seq data (accession: GSE104702) are available in the Gene Expression Omnibus Datasets.
Integrated genome browser [30] can be downloaded free at: http://bioviz.org/igb/. and EaSeq [31] can be downloaded free at: http://easeq.net/
ChIP-Seq analysis software -
The heatmaps, curves, and histograms shown in figure 4a–c, were generated using EaSeq [31]. Images of sequence alignments of fragments across chromosomal coordinates were generated using the Integrated Genome Browser (Fig. 6–7) [30].
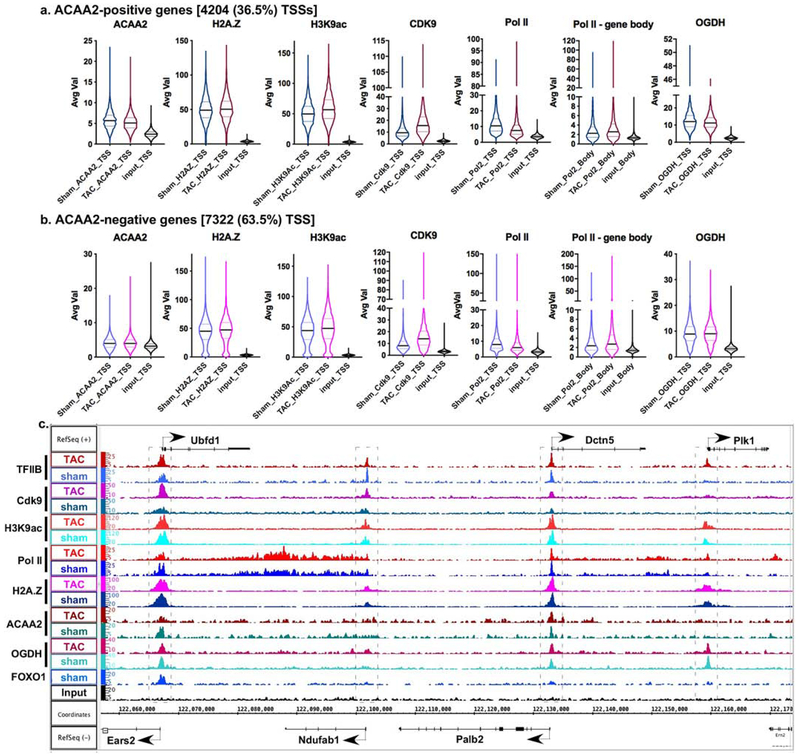
Figure 4. ACAA2 binds selectively to the TSS of genes and is differentially regulated during growth.
Mice were subjected to a sham or TAC operation to induce hypertrophic growth. One-week post-TAC, the hearts were isolated and analyzed by ChIP-Seq for ACAA2. Using the sequence tags, expressed genes (pol II-positive) were sorted into: a. those that bind ACAA2 (ACAA2-positive) and b. ACAA2-negative, at the TSS. They were sorted in parallel with the sequence tags from H2A.Z, H3K9ac, Cdk9, pol II, and OGDH ChIP-Seq, at the TSS (−1000 to + 1000). The results were plotted as violin plots, in which the horizontal solid line represent the median, and the dashed lines the quartiles, and the shape of the violin reflects the tags’ density distribution. c. The alignment of the ChIP-Seq sequence tags for TFIIB, Cdk9, H3K9ac, pol II, H2A.Z, ACAA2, OGDH, and FOXO1 (y-axis) across the genome’s coordinates (x-axis) of a region encompassing Ubfd1, Ndufab, Palb2, Dctn5, and Plk1 genes. The arrow shows the start and direction of transcription.
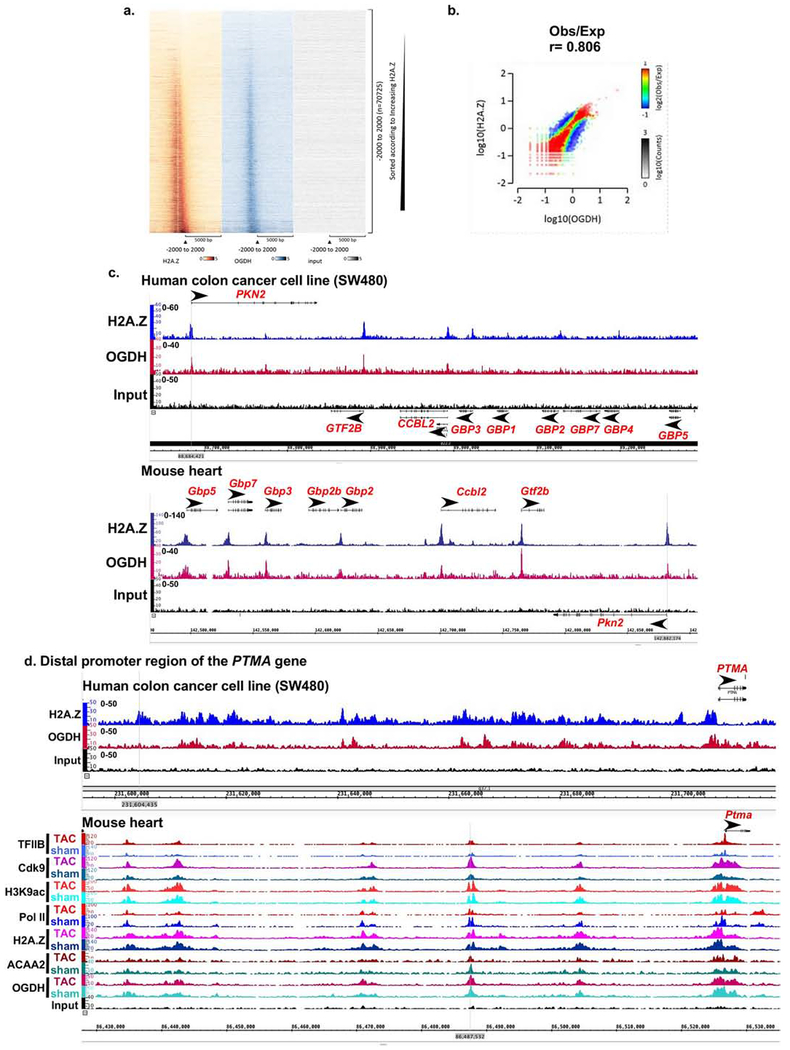
Figure 6. Conserved binding of OGDH to H2A.Z-bound TSSs in human SW480 colon cancer cells.
SW480 cancer cells were cultured, chromatin isolated and subjected to ChIP-Seq using anti-H2A.Z and anti-OGDH. a. Heatmaps of the ChIP-Seq sequence tags H2A.Z (brown), OGDH (blue), and input (grey) aligned at the TSS. The y-axis represents individual positions of bins, and the x-axis represents a region from −5000 to +5000 bp relative to the TSS. b. Histograms showing the distribution of fragments calculated from their overall frequencies in the ChIP-Seq of H2A.Z (y-axis) versus OGDH (x-axis) over the length of the gene and including −2000 bp upstream of the TSS. The x- and y-axes were segmented into 75 bins, and the number of fragments within each bin was counted, color coded, and plotted. The bar to the right of the plot illustrates the relationship between count and coloring. The plots represent pseudo-colored 2D matrices showing observed/expected distribution, calculated from the overall frequencies of fragments on each of the axes. This plot shows the relation between H2A.Z and OGDH relative to what is expected if they occurred by chance. The pseudo-color corresponds to the Obs/Exp ratio, and the color intensity is proportional to the log10 of the number of observed fragments within each bin. These plots suggest that there is a positive correlation between the binding of H2A.Z and OGDH, where the red indicates that this occurs more frequently than expected by chance, as denoted by the correlation coefficient listed above the histogram. c.-d. Both human SW480 colon cancer cell line and mouse heart tissue, were subjected to H2A.Z and OGDH ChIP-Seq using the same antibodies and chromatin concentration. The resulting seqns were aligned across the coordinates for the same genes in the mouse and human genomes, as indicated. c. The region encompassing PKN2, GTF2B, CCBL2, GBP1-5,7 genes that show conserved co-localization of OGDH and H2A.Z at the TSS of the former 3 genes in the mouse and human. d. The region encompassing the distal promoter of the PTMA gene.
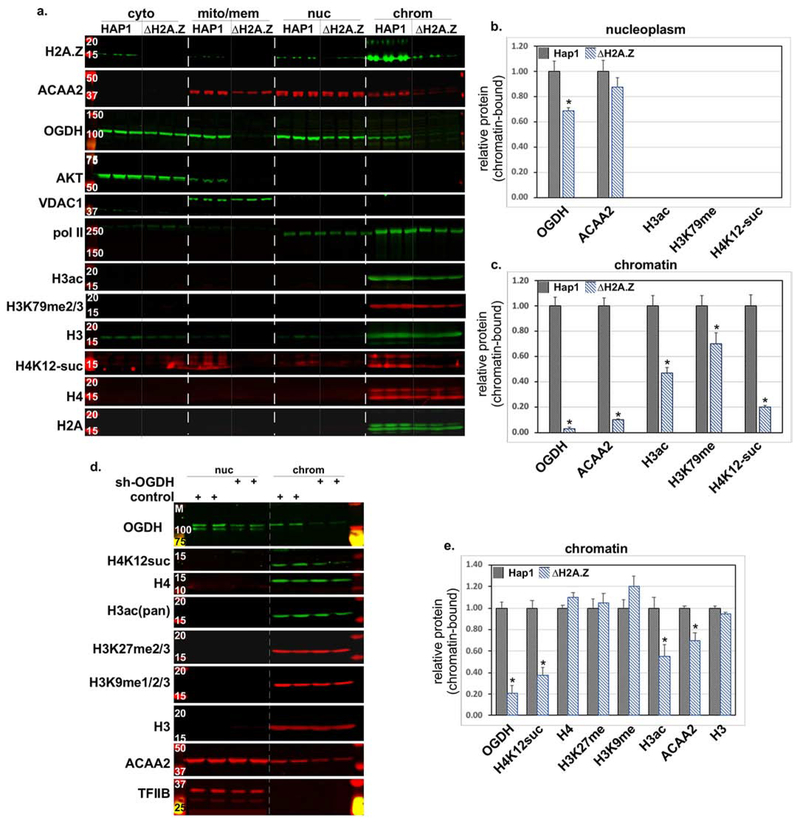
Figure 7. H2A.Z knockout in human HAP1 cells inhibits chromatin binding of ACAA2 and OGDH, and reduces histone modifications.
a. Human HAP1 and H2A.Z-deleted HAP1 (ΔH2A.Z) cells were cultured. Cellular protein was extracted and fractionated into cytosol (cyto), mitochondrial and membrane (mito/mem), nucleoplasm (nuc), and chromatin-bound (chrom), followed by Western blot analysis for the proteins listed on the left of each panel. b.-c. The signals of the bands in the b. nuc and c. chrom fractions were quantified by imageJ, normalized to H2A, or H3 or H4 for the corresponding modified protein, and plotted as the mean ± SEM of the protein in the ΔH2A.Z cells, relative to the parent (HAP1) adjusted to 1. N=3, each, from 3 repeats, *p = 0.001 vs. HAP1. d. shRNA targeting OGDH or a control construct, was delivered to isolated rat neonatal cardiac myocytes using adenoviral vectors (moi 30). After 48 h, protein was extracted, fractionated, and analyzed as described in (a). e. The signals of the bands in the chrom fractions were quantified by imageJ, normalized to H3 or H4, for OGDH or the corresponding modified protein, and plotted as the mean ± SEM of the protein in the control cells, relative to the sh-OGDH-treated adjusted to 1. N=4, each, from 4 repeats, *p = 0.035 versus control.
Statistical analysis -
The significance of differences between 2 experimental groups was calculated using T-test (equal variance, 2-tailed), where p < 0.05 was considered significant.
Results
Mitochondrial TCA cycle and β-oxidation enzymes localize to the nucleus, in association with H2A.Z-bound chromatin.
In an attempt to discover proteins that associate with H2A.Z-bound nucleosomes, we performed rapid chromatin immunoprecipitation-mass spectrometry of endogenous proteins assay (RIME) [20], which involved chromatin-immunoprecipitation by anti-H2A.Z or a control IgG, followed by mass spectrometry, from the nuclei of mice hearts subjected to transverse aortic constriction (TAC), which imposes work overload on the heart and induces hypertrophic growth of the myocytes, or to a sham operation, as a control (Fig. 1a). This procedure was applied to potentially reveal any cell growth-dependent changes in total protein-chromatin associations and also served as a duplicate sample. The assay resulted in the immunoprecipitation of a total of 73 proteins with ≥ 2x enrichment versus the IgG control, most of which were metabolic enzymes of the tricarboxylic acid cycle (TCA) and beta oxidation spiral (supplementary Fig. 1S–a–g). Figure 1b shows the total spectra of the metabolic enzymes that were most enriched in the RIME assay, including the beta-oxidation spiral enzymes HADHA and ACAA2, and the TCA cycle enzyme, OGDH, and the two subunits that are components of the OGDH enzymatic complex: dihydrolipoyl succinyltransferase (DLST) and dihydrolipoyl dehydrogenase (DLD), plotted versus the IgG control (Fig. 1b). However, there were no striking differences in the total nuclear abundance of these enzymes between the normal and growth-induced hearts. In contrast, other highly expressed mitochondrial proteins, including components of the respiratory complex (e.g. NDUSF7, COX5a, CYC1, and UQCRQ), negative regulators of PDH (e.g. PDK1), and outer mitochondrial membrane proteins (e.g. VDAC1/2), were detected at relatively low and unenriched levels versus IgG; in addition to many others that were undetectable (e.g. PDK4), thus, confirming the selectivity of mitochondrial enzymes that moonlight in the nucleus (supplementary Fig. 1S–h–i). Also, as evidence of the efficacy of the RIME, we identified H2A.Z, in addition to the core histones, in the immunoprecipitated complex (Fig. 1c). Note that OGDH, among other metabolic enzymes identified in the RIME assay, were represented in mass spectrometry data by much higher spectra (~100-700) relative to the histone components (~10-35), however, the fold enrichment was comparable (2-15-fold). One of the reasons for this is the differences in their protein sizes (e.g. OGDH is 116 Kd relative to the 12-14 Kd histones).
Confirming the nuclear localization of metabolic enzymes in rodent and human cells.
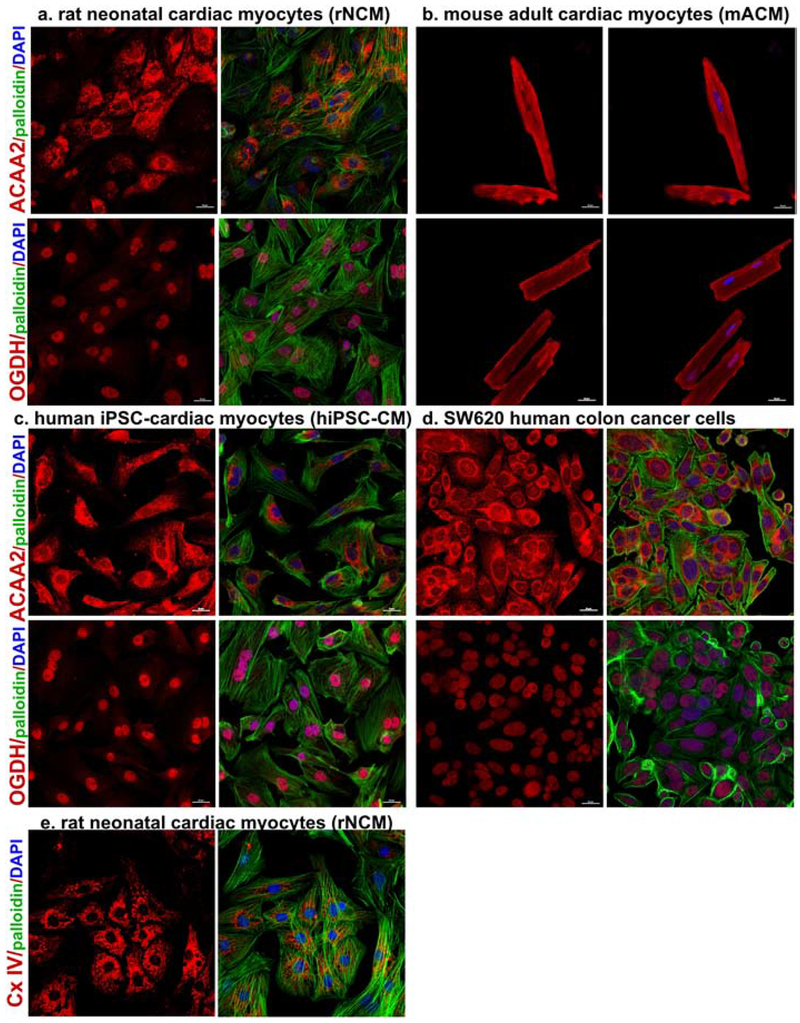
To confirm the results of the RIME regarding the nuclear localization of some of the mitochondrial enzymes, we first used immunocytochemistry (ICC) staining of ACAA2 and OGDH in four cell types, including cultured rat neonatal cardiac myocytes (rNCM), isolated mouse adult cardiac myocytes (mACM), human iPSC-derived cardiac myocytes (hiPSC-CM), and SW620 colon cancer cells (Fig. 2a–d). The results revealed the nuclear localization of OGDH (which had the highest levels of spectra in the RIME assay and 15.8 ± 0.5-fold enrichment / IgG), and ACAA2 (2.5 ± 0.1-fold enrichment / IgG). The ICC with anti-ACAA2 revealed pronounced mitochondrial staining, while the nuclear staining was relatively weaker, albeit distinctively positive (Fig. 2a–d, upper panels) when contrasted with complex IV (Cx IV) staining that was confined to the mitochondria (Fig 2e). On the other hand, immunostaining of OGDH revealed a predominant nuclear versus mitochondrial localization in neonatal myocytes, hiPSC-CM, and SW620 (Fig. 2a, c–d, lower panels), and in the developing embryonic heart (supplementary Fig. 2S). On the other hand, the adult myocytes revealed a more even distribution of OGDH in the mitochondria and nuclei (Fig. 2b, lower panels). Notably, the antibody used in these images targets an epitope near the C-terminal region of the protein (E1W8H, Cell Signaling Technology). Thus, to validate these results, we performed the immunostaining with a second antibody against the N-terminal domain of the protein, which was more favorable for mitochondrial versus nuclear OGDH (Sigma, cat # HPA019514, supplementary Fig. 3S–e). Moreover, images from the Human Protein Atlas [32] show OGDH (also using the latter antibody, Sigma, cat # HPA019514) in the nuclei of cardiac myocytes in normal adult heart tissue and in A-431 squamous carcinoma cells, in addition DLD, a subunit of the OGDH enzyme complex, in the nuclei of U2OS cells, and HADHA in the nuclei of breast cancer tissue (supplementary Fig. 3S–a–d).
Figure 2. Nuclear localization of ACAA2 and OGDH identified by immunocytochemistry.
a. Isolated rat neonatal cardiac myocytes (rNCM), b. mouse adult cardiac myocytes (mACM), c. human iPSC-derived cardiac myocytes (hiPSC-CM), and d. SW620 colon cancer cells, were cultured, fixed, and immune-stained with the anti-ACAA2 or anti-OGDH (red), left panels, in addition to phalloidin (green, except for ACM), and DAPI (blue); the three are overlayed in the right panels. e. rNCM were cultured, fixed, and immune-stained with the anticomplex IV (Cx IV, red), left panel, in addition to phalloidin (green), and DAPI (blue); the three are overlayed in the right panel. The cells were then imaged using confocal microscopy. The scale bars represent 20 μm.
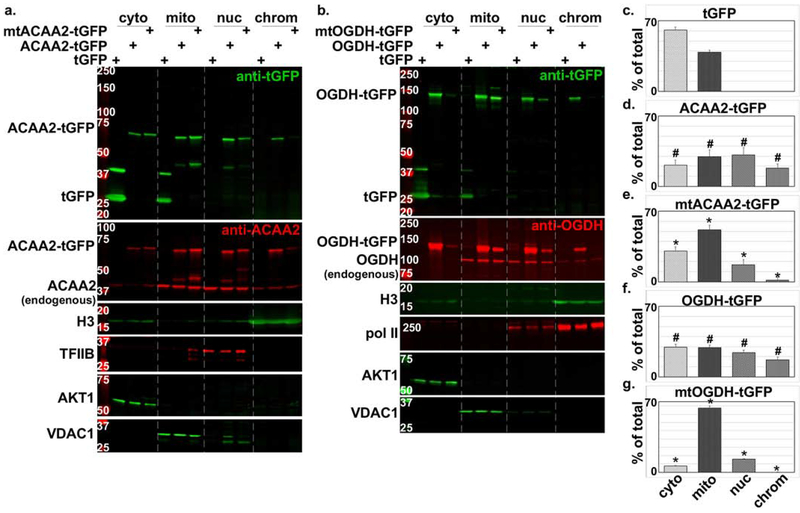
To validate these results, we generated C-terminal tGFP-fusion proteins with ACAA2 and OGDH. In addition, we identified putative nuclear localization signals (NLS) in these enzymes that we mutated to generate NLS-defective tGFP fusion proteins (supplementary Fig. 4S). The constructs were delivered to cultured cardiac cells via adenoviral vectors. Figure 3 shows that while the unfused tGFP was predominantly located in the cytosol and to a lesser extent in the mitochondrial/membrane (mito/mem) fraction, its fusion with ACAA2 or OGDH, resulted in its import into the nucleus and detection in the chromatin-bound protein fractions (Fig. 3a–b, upper 2 panels). Likewise, the endogenous enzymes were detected in the mito/mem fraction (VDAC1-positive fraction), in addition to the nuclear and chromatin fractions (TFIIB- and histone H3-positive fractions, respectively, Fig. 3a–b, second panels). Ultimately, substitutions of key lysine residues with glutamine in the putative NLS of ACCA2 (mtACAA2) or OGDH (mtOGDH) significantly reduced their nuclear import and chromatin associations, proving that this localization is specific and requires an NLS (Fig. 3a–b, 3c–g). Note, the chromatin bound proteins were not subjected to crosslinking, thus, only the directly- or tightly-associated proteins are retained in this fraction, such as the histones and RNA polymerase II. A comparison between the distributions of ACAA2-tGFP vs. NLS mtACAA2-tGFP proteins in the different fractions show that the % of total of the latter in the cytosol and mitochondrial fractions is significantly higher than that observed with the wild type protein, as the mutant is excluded from the nucleoplasm and chromatin fractions (Fig. 3a, 3d–e). Similarly, the mtOGDH-tGFP protein is predominantly in the mitochondrial, while significantly excluded from the nucleus, relative to the wild type protein (Fig. 3b, 3f–g). Similar results were observed in the human colon cancer SW480 cells (supplementary Fig. 5S). These data confirm that ACAA2 and OGDH reside in the nucleus in significant concentrations and that they harbor an NLS that mediates their nuclear import.
Figure 3. Nuclear localization of ACAA2 and OGDH confirmed by tGFP-fusion proteins and NLS mutations.
a-b. Cardiac myocytes were infected with a 10-20 moi of recombinant adenoviruses harboring turbo-GFP (tGFP) or a. wt ACAA-tGFP or an NLS mutant (mtACAA2-tGFP), or b. wt OGDH or an NLS mutant (mtOGDH-tGFP). After 18 h, the cellular proteins were fractionated into cytosol (cyto), mitochondrial and membrane (mito), nucleoplasm (nuc), and chromatin-bound (chrom) protein fractions that were then analyzed by Western blotting for the proteins listed on the left of each panel. The fusion proteins were detected by anti-GFP (upper panels, a-b) and anti-ACAA2 or anti-OGDH (second from the top panels, a-b), which also detect the endogenous proteins. AKT1, VDAC1, TFIIB or Pol II, and H3, were immunodetected for use as internal controls for the corresponding cell fractions. The signals for the c. tGFP, and tGFP-fusion proteins d. ACAA2-tGFP, e. mtACAA2-tGFP, f. OGDH-tGFP, g. mtOGDH-tGFP, were quantified using imageJ, normalized to internal controls, and plotted as the mean ± SEM of % total protein in all 4 fractions. Error bars represent SEM, N=3 from 3 repeats. #p ≤ 0.05 vs. tGFP, *p = 0.0095 vs. wt tGFP-ACAA2 or -OGDH fusions, in corresponding fractions.
ACAA2 and OGDH associate with H2A.Z-bound transcription start sites -
The co-precipitation of mitochondrial enzymes with chromatin-bound H2A.Z indicates that these enzymes associate with chromatin and co-localize with H2A.Z at TSSs aand/or enhancers. To confirm this, we performed a ChIP-Seq assay using anti-ACAA2 or anti-OGDH on chromatin extracted from normal vs. growth-induced hearts. The ChIP-Seq statistics, including the number of tags, peaks, and empirical false discovery rates, are reported in supplementary Table 1. The results show that ACAA2 and OGDH, similar to H2A.Z [19], predominantly associate with TSSs, as demonstrated by the heatmaps of the sequence tags and the curves of the average peak density signals aligned to a region encompassing −2000 to +2000 bp from the TSS (supplementary Fig. 6S). Additionally, the binding of the ACAA2 and OGDH coincided with that of H2A.Z at values higher than expected for a random event (r=0.813, 0.803, 0.888, 0.894), in normal or growth-induced hearts for each enzyme, respectively (supplementary Fig. 7S–a–d). Moreover, ACAA2’s and OGDH’s chromatin binding sites extensively overlap (r= 0.86, supplementary Fig. 7S–e–f).
ACAA2 ChIP-Seq -
The average fragment densities of the sequence tags from the ACAA2 ChIP-Seq analysis were sorted into ACAA2-positive and -negative TSSs (−1000 to +1000 bp from TSS) for transcriptionally active genes [RNA polymerase II (pol II)-positive], and aligned with those of H2A.Z, OGDH, H3K9ac (a histone mark that is associated with active promoters), TFIIB (which demarcates the TSS), RNA pol II (which reflects transcriptional activity), and CDK9 (which reflects transcriptional elongation). The data were graphed as violin plots representing the median, quartiles, and probability density of the tags (Fig. 4a–b). This revealed that ACAA2 associates with the TSS of 4204 genes (36.5% of genes expressed in the heart) that are all H2A.Z-bound (approx. 90% of expressed genes are H2A.Z-bound). Conversely, not all H2A.Z-bound genes are associated with ACAA2 (Fig. 4b), suggesting that this a selective versus constitutive association. Notably, ACAA2-positive TSSs exhibits a higher median of bound H2A.Z (8% and 6% higher for normal and growth-induced hearts, respectively) and the H3K9ac mark (13.6% and 12% higher for normal and growth-induced hearts, respectively) relative to those negative for ACAA2, as reflected in the graphs (Fig. 4a–b). Indeed, the means of these differences are significant (supplementary Fig. 8S–a–b). Interestingly, both H2A.Z and H3K9ac show similar patterns of tag probability density, in the absence vs. presence of ACAA2, demonstrating a positive association between the two marks. ACAA2-positive TSSs also exhibit significantly higher levels of pol II and Cdk9 peaks, denoting higher transcriptional activity. While, cumulatively, ACAA2 preferentially associates with transcriptionally active genes, changes in its abundance does not always correlate with changes in transcriptional activity when observed on a gene-by-gene basis. Of the 4204 ACAA2-positive genes, 697 exhibited ≥ 1.25-fold significant upregulation of ACAA2, while 1203 genes exhibited ≤ 0.75-fold significant downregulation, and 1900 genes with minimal or no change of ACAA2, in growth-induced versus normal hearts (supplementary Fig. 8S–e–f and 8S–i–j). In general, functional pathway analysis shows that these three categories of ACAA2-bound genes encompass pathways involved in endoplasmic protein processing and proteolysis, metabolism, and RNA transport and protein synthesis, respectively (supplementary Tables 2–4).
For a gene-specific view, the ACAA2 ChIP-Seq tags were aligned across the chromosomal coordinates, along with those of H2A.Z, OGDH, H3K9ac, TFIIB, RNA pol II, and CDK9 (Fig. 4c). In addition, we also aligned these with FOXO1 ChIP-Seq tags to demonstrate the selectivity of the bindings of the various factors to the TSSs. The image shows the location and densities of their bindings across a genomic region encompassing the Ubfdl, Ndufab1, Palb2, Dctn5, and Plk1 genes. This revealed the binding of ACAA2 to the TSSs of Ubfd1 and Dctn5, overlapping with H2A.Z, OGDH, TFIIB, pol II and H3K9ac peaks. Those genes are regulated by a pause-release of pol II, as evidence by the reduction in the pol II peak at the TSS that is associated with an increase in Cdk9 and pol II downstream of the genes’ 3’ end. In contrast, ACAA2 does not bind the TSSs of Ndufab1 or Plk1, which reveals the selectivity of its binding. Moreover, FOXO1 was only detected at the TSS of Ubdfd1, underscoring the selectivity of binding to TSSs. Conversely, the cardiac-specific genes (Ryr2 and Actn2), which have no visible H2A.Z binding, coincide with undetectable ACAA2 or OGDH (Fig. 5d), suggesting an H2A.Z-dependent mechanism for recruiting these enzymes to the TSS.
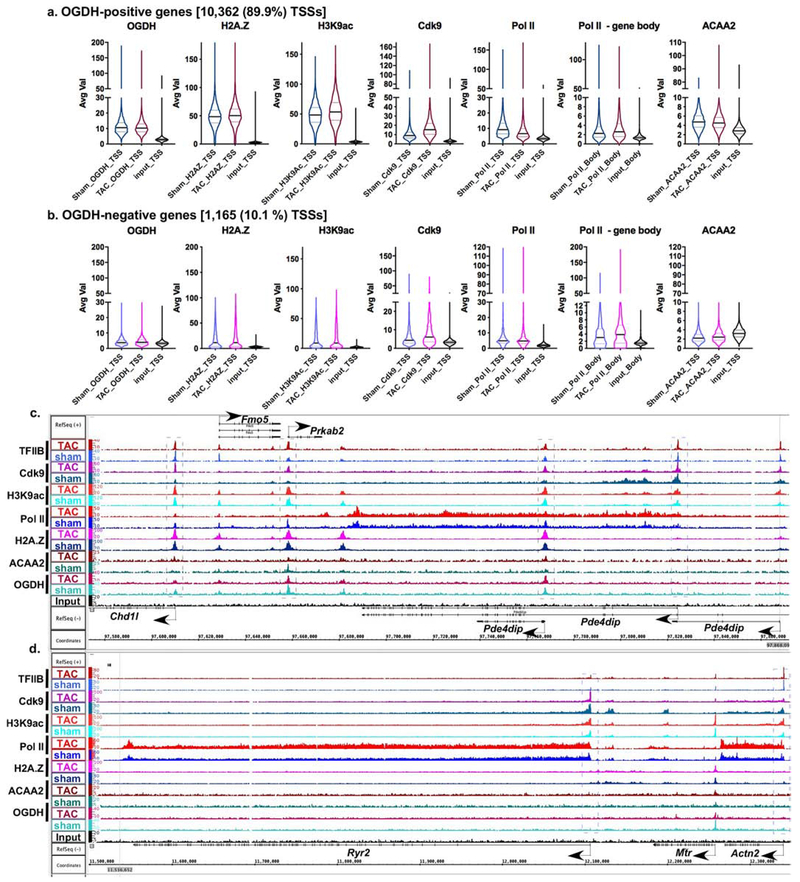
Figure 5. OGDH binds to the TSSs of 89.9% of expressed genes in consistent overlap with H2A.Z.
Mice were subjected to a sham or TAC operation. One-week post-TAC, the hearts were isolated and analyzed by ChIP-Seq for OGDH. a. Using the sequence tags, expressed genes (pol II-positive) were sorted into: a. those that bind OGDH (OGDH-positive) and b. OGDH-negative, at the TSS. They were sorted in parallel with the sequence tags from H2A.Z, H3K9ac, Cdk9, pol II, and ACAA2 ChIP-Seq, at the TSS (−1000 to + 1000). The results were plotted as violin plots, in which the horizontal solid line represent the median, and the dashed lines the quartiles, and the shape of the violin reflects the tags’ density distribution. c-d. The alignment of the ChIP-Seq sequence tags for TFIIB, Cdk9, H3K9ac, pol II, H2A.Z, ACAA2, and OGDH (y-axis) across the genome’s coordinates (x-axis) of a region encompassing c. Chd1l, Fmo5, Prkab2, and Pde4dip, and d. Ryr2 and Actn2. The arrow shows the start and direction of transcription.
OGDH ChIP-Seq -
Next, the average fragment densities of the sequence tags from the OGDH ChIP-Seq analysis were sorted into OGDH-positive and -negative TSSs (−1000 to +1000 bp from TSS) of transcriptionally active genes (pol II-positive), in parallel with those of H2A.Z, H3K9ac, TFIIB, Cdk9, and RNA pol II. The data were graphed as violin plots representing the median, quartiles, and probability density of the tags. This analysis revealed that OGDH preferentially associates with H2A.Z-bound TSSs with substantially higher H2A.Z densities (4.3- and 4.5-fold higher medians in sham and TAC hearts, respectively, vs. OGDH-negative genes), which included 89.9% (10,362) of expressed genes (Fig. 5a–b). This also coincides with substantially higher levels of H3K9ac (5.3- and 6.5-fold higher medians for sham and TAC hearts, respectively, vs. OGDH-negative genes) and Cdk9 (2.25- and 2.5-fold higher medians for sham and TAC hearts, respectively, vs. OGDH-negative genes) at the TSSs. Indeed, the means of these differences are significant (supplementary Fig. 8S–c–d). The reduction in TSS-pol II, associated with an increase in TSS-Cdk9 and incremental increase in gene body-pol II, in growth-induced vs. normal hearts, is characteristic of a pause-release in transcription (Fig. 5a–b). Of the 10,362 OGDH-positive genes, 992 exhibited ≥1.25-fold significant increase of OGDH, while 993 exhibited ≤ 0.75-fold significant downregulation in OGDH at the TSSs, in growth-induced versus normal hearts (supplementary Fig. 8S–g–h and Fig. 8S–k–l). Similar to ACCA2, there is no correlation of the changes in OGDH abundance with the those observed in Cdk9, H3K9ac, or pol II in normal versus growth-induced hearts, and, thus, changes in transcriptional activity, when investigated on a gene-by-gene basis. In general, functional pathway analyses show that genes that exhibit upregulation of OGDH during cardiac growth include a preponderance of metabolic genes, while those that show downregulation include pathways in cancer and endocytosis (supplementary Tables 5–6). In contrast to OGDH-positive genes, a more uniform increase in pol II across the gene was observed in the OGDH-negative genes, in growth-induced versus normal hearts, characteristic of de novo recruitment of pol II versus pause-release (Fig. 5b, d). Gene ontology analysis of these genes included the terms sarcomere, Z disc, myofibril,..etc. that characterize cardiac muscle-specific genes (supplementary Table 7). These genes have relatively very low or undetectable H2A.Z (Fig. 5b), as seen in figure 5d in the cardiac-specific genes, Ryr2 and Actn2, and as we have previously reported [19].
For a genomic, gene-specific view, the ChIP-sequence tags for OGDH were aligned with those of H3K9ac, TFIIB, RNA pol II, Cdk9, and ACAA2 across the chromosomal coordinates (Fig. 5c). The image shows the binding sites and densities of the genes’ in the normal versus growth-induced hearts, across a region encompassing the Chd1l, Fmo5, Prkab2 and Pde4dip genes. Shown, is the 3 TSSs of the Pde4dip gene, which differentially bind OGDH; the first and second TSSs have no H2A.Z, and, thus, no OGDH, while the third TSS has both, in addition to ACAA2, in the growth-induced heart. Chd1 and Fmo5, also have OGDH peaks overlapping with H2A.Z, albeit not as prominent as that seen with Prkab2. Consistently, cardiac-specific genes - the majority of which have little to no detectable H2A.Z - are devoid of OGDH, as seen in the Ryr2 and Actn2 genes (Fig. 5d). Thus, OGDH-positive TSSs coincide with H2A.Z-positive TSSs, whereas, H2A.Z-positive TSSs are not necessarily OGDH-positive, revealing the selective nature of this association and suggesting that it is a regulated process.
OGDH and H2A.Z ChIP-Seq in human colon cancer cells -
The strict overlap of OGDH with H2A.Z-bound TSS, reflects the selectivity of this association. To validate this finding and determine its conservation in human cells, we performed OGDH and H2A.Z ChIP-Seq in human colon cancer SW480 cells (Fig. 6a). The number of peaks in the cancer cells were lower than those observed in the mouse heart (4,924 versus 15,653, respectively, see supplementary Table 1a and 1c), however, it adhered to the same pattern, as it always coincided with H2A.Z-bound TSS (Fig. 6a–c). For example, GBP1,4-5,7 genes, which had little or no H2A.Z were devoid of OGDH (Fig. 6c). In contrast, the TSSs of these genes, which were H2A.Z-bound in the mouse genome, were occupied by OGDH. However, not all H2A.Z-bound TSSs were occupied by OGDH in the human genome, as seen with GBP3 gene (Fig. 6c). Specifically, the ChIP-Seq results reveal that the human genome has 24,126 H2A.Z peaks versus 4,924 OGDH peaks (supplementary Table 1c), suggesting a selective versus constitutive association. In addition, to the colocalization of these molecules at TSSs, they also selectively overlap at specific upstream enhancer sites, as seen in the PTMA (Fig. 6d) and the FOS distal promoter regions (supplementary Fig. 9S–a), of both the mouse and human genes.
While all the of OGDH peaks at the TSSs overlapped with H2A.Z, intriguingly, we identified 53 peaks in the mouse genome that were specifically located in the terminal exon of zinc finger protein (ZFP) genes, which did not overlap with H2A.Z peaks (supplementary Fig. 9S–b). Notably, this appeared to be a conserved phenomenon in the human genome as well (supplementary Fig. 9S–c). Overall, the results provide evidence of the specificity and conserved nature of the association of OGDH with H2A.Z-bound TSSs, as well as, its H2A.Z-independent binding to the terminal exons of some ZFP’s. These results suggest that the binding of OGDH to H2A.Z-occupied chromatin is not necessarily a direct association with H2A.Z. Alternatively, OGDH may also bind to additional chromatin-bound factors.
Knockout of H2A.Z in human HAP1 cells reduces chromatin association of metabolic enzymes and posttranslational histone modifications.
To validate the above data and investigate the extent of OGDH’s and ACAA2’s dependence on H2A.Z for association with the genome, we analyzed human haploid (HAP1) cells with a 2 bp deletion in exon 3 of H2A.Z (ΔH2A.Z). These cells are viable, however, they proliferate at ~1/4 of the rate of the parent cells (supplementary Fig. 10S). Fractionating the cellular proteins and analyzing it by Western blotting, we confirmed that H2A.Z is deficient in the ΔH2A.Z cells (Fig. 7a). We did, however, detect a residual band that is equivalent in size to H2A.Z, in the ΔH2A.Z cells, which could either be non-specific binding of the antibody to other histones or binding to H2A.Z.2 that differs from H2A.Z (H2A.Z.1) by only 3 amino acids.
The results of this knockout show that the nucleoplasm levels of ACAA2 were not significantly reduced, while those of OGDH were down by ~30%. Otherwise, the chromatin-bound fractions for both were reduced by > 90% (Fig. 7a–c), suggesting specific chromatin exclusion of both enzymes in the absence of H2A.Z. Notably, loss of H2A.Z reduced the overall expression of these enzymes, as observed in the mito/mem fractions, relative to other proteins including Vdac1, H2A, or H4 (Fig. 7a). Intriguingly, we also observed that mitochondrial, but not cytosolic OGDH, was eliminated by ΔH2A.Z, the cause of which remains to be investigated. These findings coincided with reductions in the histone marks, including H3-ac and more robustly H4K12-succinyl, paralleled by slight reduction of chromatin-associated pol II (Fig. 7a–b). Consistently, knockdown of H2A.Z using shRNA in cultured cardiac myocytes reduced chromatin-bound ACAA2, in parallel with histone modifications (supplementary Fig. 11S). In contrast, the mitochondrial proteins VDAC1 and PDK1 exhibited no nuclear presence (see supplementary Fig. 1S) and were, thus, used as mitochondrial-specific loading markers. The data suggest that H2A.Z is largely necessary for the association of ACAA2 and OGDH with chromatin, and for histone modifications.
Knockdown of OGDH inhibits H4 succinylation.
OGDH has been reported to bind to chromatin in U251 glioblastoma cells where it mediates succinylation of H3K79 [15]. To test its function, we knocked it down using shRNA (sh-OGDH) in cultured myocytes. This treatment induced a significant reduction of chromatin-bound OGDH (79 ± 7 %), and H4K12-suc (62 ± 7 %) versus control levels, but not of H3K27me2/3, H3K9me1/2/3, H3, or H4 (Fig. 7c–d). We have to caution, though, that due to the limited anti-H4K12-suc antibodies that are currently commercially available, we were unable to validate these results with a second similar antibody, especially that we observe a non-specific signal in the mito/mem fraction. We also observed a slight reduction in chromatin-bound ACAA2, suggesting its codependence on OGDH or histone modifications. We conclude that OGDH is required for histone succinylation, plausibly through conversion of alpha-ketoglutarate into succinyl-CoA at TSSs. On the other hand, we predict the reduction in H3 acetylation is secondary to the reduction in chromatin-bound ACAA2.
Discussion
In this study, we have identified the association of ACAA2 and OGDH with TSSs and enhancers of transcriptionally active genes in a H2A.Z-dependent fashion. These were discovered by an unbiased screen using anti-H2A.Z chromatin immunoprecipitation-mass spectrometry. This approach provides the unique advantage of identifying proteins that associate with nucleosome-packaged H2A.Z vs. free H2A.Z. Therefore, we predict that these associations might not all be direct interactions with H2A.Z, but, alternatively, indirect association with other H2A.Z-dependent chromatin-binding factors. For example, Wang et al, reported that OGDH interacts with KAT2A in the nucleus [15], whereas, Jiang et al reported that fumarase interacts with H2A.Z in U2OS cells after exposure of the cells to ionizing radiation [16]. On the other hand, using direct pull-down of H2A.Z or using it as bait in a yeast-two-hybrid screen, we identified its direct interaction with ANP32e, but not OGDH or ACAA2 [19]. Similarly, several unbiased screens for identifying proteins that directly interact with H2A.Z, have not revealed its interaction with any of these enzymes [33]. This suggests that, at least, the association of OGDH and ACAA2 with chromatin may be dependent on nucleosome-packaged vs. free H2A.Z. Furthermore, the absence of these enzymes from intergenic or intragenic regions, or inactive genes that are packaged with H2A and devoid of H2A.Z, supports the conclusion these enzymes co-localize with H2A.Z versus H2A.
We confirmed our RIME findings for the interaction of OGDH and ACAA2 with nucleosomal H2A.Z by immunocytochemistry staining, fractionation of cellular proteins and Western blot analysis, GFP-fusion proteins combined with putative NLS mutations, ChIP-Seq, and H2A.Z knockout. These approaches were performed in both rodent and human cell lines, including isolated mouse and rat myocytes, human iPSC-derived cardiac myocytes, human colon cancer cells (SW480), and human near-haploid cells (Hap1) with or without a H2A.Z deletion. All findings were consistent with selective H2A.Z-dependent TSS association of ACAA2 and OGDH.
H2A.Z is a highly conserved histone variant that plays an essential role in sensing and responding to metabolic and environmental cues, however, the underlying mechanisms remain elusive. It has, though, been shown to interact with 93 proteins, mostly identified by unbiased screens using co-fractionation, affinity capture-MS, and affinity capture-Western (compiled in BioGRID [34]), some of which may mediate its functions. These proteins include core histones [35, 36], proteins that regulate H2A.Z’s chromatin deposition (e.g. INO80 complex subunit C [37], vacuolar protein sorting 72 [38], ANP32E [19, 39]), epigenetic regulators (e.g. E1A binding protein p400, lysine acetyltransferase 5, and histone deacetylase 1 and 2 [36]), in addition to its interaction with the metabolic enzymes ACAA2 and fumarase, observed in an interactome identified by co-fractionation [40], although only the latter has been validated [16]. Otherwise, most of these interactions were not identified or validated in the context of nucleosomal-bound H2A.Z. Using the rapid immunoprecipitation-mass spectrometry of endogenous proteins assay approach [20] with anti-H2A.Z we identified its interaction with core histones, as well as, ACAA2 and OGDH, among other enzymes that we have not validated as of yet (supplementary Fig. 1S). The fact that these enzymes are associated with chromatin at the TSS may explain how metabolites can be directly delivered to target genes where they are used as substrates for histone modifications (e.g. acetyl-CoA, succinyl-CoA, among other short acyl-CoA metabolites) or as co-factors (αKG) for histone modifying enzymes, thereby, allowing promoters to sense and respond to metabolic cues through modulation of their activity or transcriptional memory. Consistent with an H2A.Z-dependent recruitment, genes that are devoid of H2A.Z, also lack ACAA2 or OGDH at their TSS, whereas, knockdown or knockout of H2A.Z abrogates their chromatin association. On the other hand, we speculate that the absence of H2A.Z and metabolic enzymes at the promoters of constitutively expressed, tissue-restricted genes (e.g. sarcomeric proteins), which distinguish an organ’s unique functionality, ensures these are not impacted by metabolic fluctuations, as one would expect. This contrasts with housekeeping and inducible genes that would be immediately modulated in response to changes in oxygen and/or metabolic substrate availabilities, as a mechanism of cellular adaptation, and as candidates for transcriptional memory. Our results are also consistent with a proteomics study that identified all TCA cycle enzymes in the nucleus of normal and cancer cells [18].
H2A.Z is required for transcriptional memory in yeast where it is incorporated in newly deactivated INO1 at the nuclear periphery [41, 42]; in hippocampal memory in mice, where it negatively controls fear memory through suppressing the expression of specific memory-activating genes [43]; and is necessary for neurogenesis and normal behavioral traits in mice [44]. As well established, memory is a function of epigenetics, wherein histone acetylation is an essential regulator, as demonstrated by Mews et al [45]. In that study, the authors reported that ACSS2 (a cytosolic and nuclear enzyme that converts acetate into acetyl-CoA) binds near the TSS of hippocampal neuronal genes, where its knockdown diminishes long-term spatial memory and an acetylation-dependent cognitive process. These reports suggest a potential link between H2A.Z and the intricate and precise regulation of core histone modifications that underlie memory processes. Concordantly, we show that knockdown or knockout of H2A.Z reduces the association of metabolic enzymes with chromatin, which is paralleled with a significant reduction in histone H3 acetylation and succinylation, whereas OGDH knockdown induces a robust decline in H4K12 succinylation. The association of metabolic enzymes with the TSS of genes could potentially explain the mechanism of targeted histone modifications, and, therefore, the direct regulation of transcriptional activity and memory via glucose and fatty acid metabolism. Furthermore, identifying other metabolic enzymes that are recruited to the genome could help explain how other short chain acyl-CoAs (butyryl-, hydroxyisobutyryl, crotonyl-, and proprionyl-CoA), which cannot be exported from the mitochondria, may be delivered to genes in a targeted fashion.
While acetylation and methylation of histones are key players in transcriptional regulation and memory, succinylation is another modification that has received less attention. The alpha-ketoglutarate dehydrogenase complex catalyzes the conversion of αKG into succinyl-CoA as a source of cellular succinyl-CoA. However, there is no known route via which this metabolic intermediate is delivered to the nucleus. Interestingly, the interaction between OGDH and lysine acetyltransferase 2A in the nucleus mediates the succinylation of H3K79 [15]. The study showed that ChIP-Seq of OGDH, in U251 glioblastoma cells, identified 249 peaks, mainly enriched within 2 Kb of the TSS. This agrees with what we observed in our study, where OGDH is enriched at −2000 to +2000 surrounding the TSSs. The differences, though, are that we identified 4,924 peaks in human SW480 cancer cells, and 16,790 peaks in the mouse heart, at the TSSs, only in the presence of H2A.Z. We predict that the difference in the number of peaks is likely due to differences in the cell types and/or the antibodies used for ChIP-Seq. Other support of nuclear localization of mitochondrial enzymes was reported by Jiang et al, showing that phosphorylated fumarase interacts with H2A.Z in response to ionizing radiation-induced activation of DNA-dependent protein kinase in U20S cells, a function that regulates DNA repair [16].
The presence of nucleosomal H2A.Z does not always correlate with transcriptional activity, as noted by others and us. Not only do we show that constitutively-expressed, tissue-restricted genes, which have the highest level of cellular expression, have little to no H2A.Z at the TSS or in the gene body, but conversely, we found that developmental suppressed/unexpressed genes (e.g. Wnt1, Noggin, Tbx1) have substantial amounts of H2A.Z at the TSS and in gene body (supplementary Fig. 12S and Table 8). On the other hand, moderately-expressed housekeeping genes that are amenable to incremental modulation by external stimuli have the highest levels of H2A.Z at their TSS, whereas, inducible genes have relatively lower levels of H2A.Z at the TSS that extends into the gene body [19]. The latter pattern enhances the responsiveness of inducible genes to stimuli, as previously reported in Arabidopsis thaliana [46]. These results support a role for H2A.Z in positive and negative transcriptional regulation, and transcriptional memory of developmental genes. In contrast, the data demonstrate that neither H2A.Z, nor the metabolic enzymes it associates with, are required for constitutive transcription of tissue-restricted genes. Thus, we conclude that our findings add a new level of understanding to the intricacies of the transcriptional machinery and its regulation by metabolism.
Supplementary Material
Highlights.
H2A.Z chromatin immunoprecipitation – mass spectrometry reveals the association of H2A.Z-occupied chromatin with a wide range of metabolic enzymes including oxoglutarate dehydrogenase (OGDH) and acetyl-CoA acyltransferase 2 (ACAA2).
ACAA2 and OGDH chromatin immunoprecipitation – sequencing (ChIP-Seq) reveals the selective association of these enzymes with H2A.Z-occupied transcription start sites and enhancers.
Both human cancer cells and mouse heart reveal the selective co-localization of OGDH and H2A.Z in promoters.
Knockout of H2A.Z in Hap1 cells reduces the expression of these enzyme and their chromatin association.
Knockout of H2A.Z reduces histone modifications including acetylation and succinylation, whereas knockdown of OGDH reduces succinylation of H4.
Acknowledgement
We thank Dr. Junichi Sadoshima, Chairman of the Department of Cell Biology and Molecular Medicine, Rutgers University, for his support. This work was supported by National Institute of Health funding to [1R01HL119726 to M.A.]
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data access
All newly generated raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE119391, in private status with a secure token for reviewers (mjitowoyhnstpyd).
Disclosure declaration
The authors declare that they have no conflicts of interest with the contents of this article.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- [1].Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ, The human and mouse replication-dependent histone genes, Genomics., 80 (2002) 487–498. [PubMed] [Google Scholar]
- [2].Meneghini MD, Wu M, Madhani HD, Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin, Cell, 112 (2003) 725–736. [DOI] [PubMed] [Google Scholar]
- [3].Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL, Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling, Proc Natl Acad Sci U S A., 102 (2005) 18385–18390. Epub 12005 Dec 18312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang H, Roberts DN, Cairns BR, Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss, Cell., 123 (2005) 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD, Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin, Cell., 123 (2005) 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Leach TJ, Mazzeo M, Chotkowski HL, Madigan JP, Wotring MG, Glaser RL, Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster, J Biol Chem, 275 (2000) 23267–23272. [DOI] [PubMed] [Google Scholar]
- [7].Weber CM, Ramachandran S, Henikoff S, Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase, Mol Cell., 53 (2014) 819–830. doi: 810.1016/j.molcel.2014.1002.1014. [DOI] [PubMed] [Google Scholar]
- [8].Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA, H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment, Cell., 135 (2008) 649–661. doi: 610.1016/j.cell.2008.1009.1056. Epub 2008 Nov 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].van der Knaap JA, Verrijzer CP, Undercover: gene control by metabolites and metabolic enzymes, Genes Dev., 30 (2016) 2345–2369. doi: 2310.1101/gad.289140.289116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB, ATP-citrate lyase links cellular metabolism to histone acetylation, Science., 324 (2009) 1076–1080. doi: 1010.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yoshii Y, Furukawa T, Yoshii H, Mori T, Kiyono Y, Waki A, Kobayashi M, Tsujikawa T, Kudo T, Okazawa H, Yonekura Y, Fujibayashi Y, Cytosolic acetyl-CoA synthetase affected tumor cell survival under hypoxia: the possible function in tumor acetyl-CoA/acetate metabolism, Cancer Sci., 100 (2009) 821–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E, Michelakis ED, A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation, Cell., 158 (2014) 84–97. doi: 10.1016/j.cell.2014.1004.1046. [DOI] [PubMed] [Google Scholar]
- [13].Nagaraj R, Sharpley MS, Chi F, Braas D, Zhou Y, Kim R, Clark AT, Banerjee U, Nuclear Localization of Mitochondrial TCA Cycle Enzymes as a Critical Step in Mammalian Zygotic Genome Activation, Cell., 168 (2017) 210–223.e211. doi: 210.1016/j.cell.2016.1012.1026. Epub 2017 Jan 1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen J, Guccini I, Di Mitri D, Brina D, Revandkar A, Sarti M, Pasquini E, Alajati A, Pinton S, Losa M, Civenni G, Catapano CV, Sgrignani J, Cavalli A, D’Antuono R, Asara JM, Morandi A, Chiarugi P, Crotti S, Agostini M, Montopoli M, Masgras I, Rasola A, Garcia-Escudero R, Delaleu N, Rinaldi A, Bertoni F, Bono J, Carracedo A, Alimonti A, Compartmentalized activities of the pyruvate dehydrogenase complx sustain lipogenesis in prostate cancer, Nat Genet., 50 (2018) 219–228. doi: 210.1038/s41588-41017-4002-41583. Epub 42018 Jan 41515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang Y, Guo YR, Liu K, Yin Z, Liu R, Xia Y, Tan L, Yang P, Lee JH, Li XJ, Hawke D, Zhen Y, Qian X, Lyu J, He J, Xing D, Tao YJ, Lu Z, KAT2A coupled with the alpha-KGDH complex acts as histone H3 succinyltransferase, Nature., 552 (2017) 273–277. doi: 210.1038/nature25003. Epub 22017 Dec 25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jiang Y, Qian X, Shen J, Wang Y, Li X, Liu R, Xia Y, Chen Q, Peng G, Lin SY, Lu Z, Local generation of fumarate promotes DNA repair through inhibition of histone H3 demethylation, Nat Cell Biol., 17 (2015) 1158–1168. doi: 1110.1038/ncb3209. Epub 2015 Aug 1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Haraguchi CM, Mabuchi T, Yokota S, Localization of a mitochondrial type of NADP-dependent isocitrate dehydrogenase in kidney and heart of rat: an immunocytochemical and biochemical study, J Histochem Cytochem., 51 (2003) 215–226. doi: 210.1177/002215540305100210. [DOI] [PubMed] [Google Scholar]
- [18].Qattan AT, Radulovic M, Crawford M, Godovac-Zimmermann J, Spatial distribution of cellular function: the partitioning of proteins between mitochondria and the nucleus in MCF7 breast cancer cells, J Proteome Res., 11 (2012) 6080–6101. doi: 6010.1021/pr300736v. Epub 302012 Nov 300738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shin H, He M, Yang Z, Jeon YH, Pfleger J, Sayed D, Abdellatif M, Transcriptional regulation mediated by H2A.Z via ANP32e-dependent inhibition of protein phosphatase 2A, Biochim Biophys Acta., 1861 (2018) 481–496. doi: 410.1016/j.bbagrm.2018.1003.1002. Epub 2018 Mar 1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mohammed H, Taylor C, Brown GD, Papachristou EK, Carroll JS, D’Santos CS, Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes, Nat Protoc., 11 (2016) 316–326. doi: 310.1038/nprot.2016.1020. Epub 2016 Jan 1021. [DOI] [PubMed] [Google Scholar]
- [21].Nesvizhskii AI, Keller A, Kolker E, Aebersold R, A statistical model for identifying proteins by tandem mass spectrometry, Anal Chem., 75 (2003) 4646–4658. [DOI] [PubMed] [Google Scholar]
- [22].Sayed D, Yang Z, He M, Pfleger J, Abdellatif M, Acute Targeting of General Transcription Factor IIB Restricts Cardiac Hypertrophy via Selective Inhibition of Gene Transcription, Circ HF, 8 (2015) 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ackers-Johnson M, Li PY, Holmes AP, O’Brien SM, Pavlovic D, Foo RS, A Simplified, Langendorff-Free Method for Concomitant Isolation of Viable Cardiac Myocytes and Nonmyocytes From the Adult Mouse Heart, Circ Res., 119 (2016) 909–920. doi: 910.1161/CIRCRESAHA.1116.309202. Epub 302016 Aug 309208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Han M, Yang Z, Sayed D, He M, Gao S, Lin L, Yoon SH, Abdellatif M, GATA4 Expression is Primarily Regulated via a miR-26b-Dependent Posttranscriptional Mechanism During Cardiac Hypertrophy, Cardiovasc Res, 93 (2012) 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M, MicroRNAs play an essential role in the development of cardiac hypertrophy, Circ Res, 100 (2007) 416–424. [DOI] [PubMed] [Google Scholar]
- [26].Chen IY, Lypowy J, Pain J, Sayed D, Grinberg S, Alcendor RR, Sadoshima J, Abdellatif M, Histone H2A.z is essential for cardiac myocyte hypertrophy but opposed by silent information regulator 2alpha, J Biol Chem, 281 (2006) 19369–19377. [DOI] [PubMed] [Google Scholar]
- [27].Lypowy J, Chen IY, Abdellatif M, An alliance between Ras GTPase-activating protein, filamin C, and Ras GTPase-activating protein SH3 domain-binding protein regulates myocyte growth, J Biol Chem, 280 (2005) 25717–25728. [DOI] [PubMed] [Google Scholar]
- [28].Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M, Downregulation of MiR-199a Derepresses Hypoxia-Inducible Factor-1{alpha} and Sirtuin 1 and Recapitulates Hypoxia Preconditioning in Cardiac Myocytes, Circ Res, 104 (2009) 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sayed D, He M, Yang Z, Lin L, Abdellatif M, Transcriptional regulation patterns revealed by high resolution chromatin immunoprecipitation during cardiac hypertrophy, J Biol Chem., 288 (2013) 2546–2558. doi: 2510.1074/jbc.M2112.429449. Epub 422012 Dec 429410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nicol JW, Helt GA, Blanchard SG Jr., Raja A, Loraine AE, The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets, Bioinformatics, 25 (2009) 2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K, An interactive environment for agile analysis and visualization of ChIP-sequencing data, Nat Struct Mol Biol., 23 (2016) 349–357. doi: 310.1038/nsmb.3180. Epub 2016 Feb 1029. [DOI] [PubMed] [Google Scholar]
- [32].Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J,Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F, Proteomics. Tissue-based map of the human proteome, Science., 347 (2015) 1260419. doi: 1260410.1261126/science.1260419. [DOI] [PubMed] [Google Scholar]
- [33].Zhang Y, Ku WL, Liu S, Cui K, Jin W, Tang Q, Lu W, Ni B, Zhao K, Genome-wide identification of histone H2A and histone variant H2A.Z-interacting proteins by bPPI-seq, Cell Res., 27 (2017) 1258–1274. doi: 1210.1038/cr.2017.1112. Epub 2017 Sep 1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M, BioGRID: a general repository for interaction datasets, Nucleic Acids Res., 34 (2006) D535–539. doi: 510.1093/nar/gkj1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wratting D, Thistlethwaite A, Harris M, Zeef LA, Millar CB, A conserved function for the H2A.Z C terminus, J Biol Chem., 287 (2012) 19148–19157. doi: 19110.11074/jbc.M19111.317990. Epub 312012 Apr 317999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wuhr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar RA, Harris T, Artavanis-Tsakonas S, Sowa ME, De Camilli P, Paulo JA, Harper JW, Gygi SP, The BioPlex Network: A Systematic Exploration of the Human Interactome, Cell., 162 (2015) 425–440. doi: 410.1016/j.cell.2015.1006.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, Conaway JW, YY1 functions with INO80 to activate transcription, Nat Struct Mol Biol., 14 (2007) 872–874. doi: 810.1038/nsmb1276. Epub 2007 Aug 1026. [DOI] [PubMed] [Google Scholar]
- [38].Latrick CM, Marek M, Ouararhni K, Papin C, Stoll I, Ignatyeva M, Obri A, Ennifar E, Dimitrov S, Romier C, Hamiche A, Molecular basis and specificity of H2A.Z-H2B recognition and deposition by the histone chaperone YL1, Nat Struct Mol Biol., 23 (2016) 309–316. doi: 310.1038/nsmb.3189. Epub 2016 Mar 1014. [DOI] [PubMed] [Google Scholar]
- [39].Mao Z, Pan L, Wang W, Sun J, Shan S, Dong Q, Liang X, Dai L, Ding X, Chen S, Zhang Z, Zhu B, Zhou Z, Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z, Cell Res., 24 (2014) 389–399. doi: 310.1038/cr.2014.1030. Epub 2014 Mar 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kristensen AR, Gsponer J, Foster LJ, A high-throughput approach for measuring temporal changes in the interactome, Nat Methods., 9 (2012) 907–909. doi: 910.1038/nmeth.2131. Epub 2012 Aug 1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH, H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state, PLoS Biol., 5 (2007) e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Light WH, Brickner DG, Brand VR, Brickner JH, Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory, Mol Cell., 40 (2010) 112–125. doi: 110.1016/j.molcel.2010.1009.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zovkic IB, Paulukaitis BS, Day JJ, Etikala DM, Sweatt JD, Histone H2A.Z subunit exchange controls consolidation of recent and remote memory, Nature., 515 (2014) 582–586. doi: 510.1038/nature13707. Epub 12014 Sep 13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Shen T, Ji F, Wang Y, Lei X, Zhang D, Jiao J, Brain-specific deletion of histone variant H2A.z results in cortical neurogenesis defects and neurodevelopmental disorder, Nucleic Acids Res., 46 (2018) 2290–2307. doi: 2210.1093/nar/gkx1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mews P, Donahue G, Drake AM, Luczak V, Abel T, Berger SL, Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory, Nature., 546 (2017) 381–386. doi: 310.1038/nature22405. Epub 22017 May 22431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Coleman-Derr D, Zilberman D, Deposition of histone variant H2A.Z within gene bodies regulates responsive genes, PLoS Genet, 8 (2012) e1002988. doi: 1002910.1001371/journal.pgen.1002988. Epub 1002012 Oct 1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.