Abstract
A novel coronavirus (COVID-19) pandemic threatens the world. Here, we first studied the dynamics profile of SARS-CoV-2 from 56 recovered patients with COVID-19. We found viral shedding occurred up to 6 weeks after onset of symptoms. A prolonged observation period is necessary for older patients.
Keywords: COVID-19, SARS-CoV-2, dynamics profile
(See the Editorial Commentary by Tom and Mina on pages 2252–4.)
To date, an outbreak of infectious diseases—coronavirus disease 2019 (COVID-19) associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)—continues in Wuhan, China, and threatens countries such as Korea, Italy, Iraq, and Japan, and others [1, 2]. Over 50 countries are fighting against the disease. As of 4 March 2020, more than 85 000 cases were diagnosed worldwide with a death rate of approximately 2%.
Before this study, some studies reported cases of viral detection by reverse transcription–polymerase chain reaction (RT-PCR) at different timepoints throughout the disease course [3–5]. However, these reports monitored SARS-CoV-2 in the acute phase of infection. Currently, no study has reported the viral dynamics of SARS-CoV-2 infection in a long observation period. Understanding the profile of virus in patients’ respiratory specimens can assist diagnosis and reflect the disease course. Therefore, we collected clinical specimens from 56 patients with COVID-19 and report the results of SARS-CoV-2 detection during the disease course.
METHODS
A total of 56 hospitalized patients (admission date from 21 January to 12 February 2020) with confirmed SARS-CoV-2 infection in 3 branches (Hankou, Sino-French New City, and Optical Valley) of Tongji Hospital at Tongji Hospital of Huazhong University of Science and Technology in Wuhan, China, were included in this study. All enrolled patients had a confirmed diagnosis of COVID-19 according to the diagnosis and treatment guidelines for SARS-CoV-2 from the Chinese National Health Committee (version 5) and the interim guidance from the Centers for Disease Control and Prevention [6, 7]. Throat-swab samples or deep-nasal-cavity-swab samples were collected from patients on different dates after the onset of symptoms. SARS-CoV-2 was detected by real-time RT-PCR assay using a COVID-19 nucleic acid detection kit according to the manufacturer’s protocol (Shanghai Huirui Biotechnology Co, Ltd). Specifically, 2 target genes, including open reading frame 1ab (ORF1ab) and nucleocapsid protein (N), were tested during the real-time RT-PCR assay. If 2 consecutive negative results were achieved, the period between symptom onset and the date of the first negative RT-PCR test result was defined as the viral nucleic acid conversion time. All data (test dates and results of RT-PCR assay) were collected up to the final follow-up date (3 March 2020).
RESULTS
A total of 56 patients diagnosed with COVID-19 were included in this study. According to the guideline, all included patients had mild to moderate disease [7]. No patient was transferred to the intensive care unit (ICU). The median age was 55 years (interquartile range [IQR], 42–68; range, 25–83 years), comprising 34 (60.7%) men and 22 (39.3%) women. At the end of follow-up, all patients recovered and were discharged from the hospital.
The total number of SARS-CoV-2 RT-PCR assays from 56 patients was 299, with 5 tests per patient. The longest duration between RT-PCR test for SARS-CoV-2 was 42 days after the onset of symptoms. The median duration between the onset of symptoms to nucleic acid conversion was 24 days (IQR, 18–31 days). Details on demographic characteristics and RT-PCR test results are shown in Supplementary Table 1.
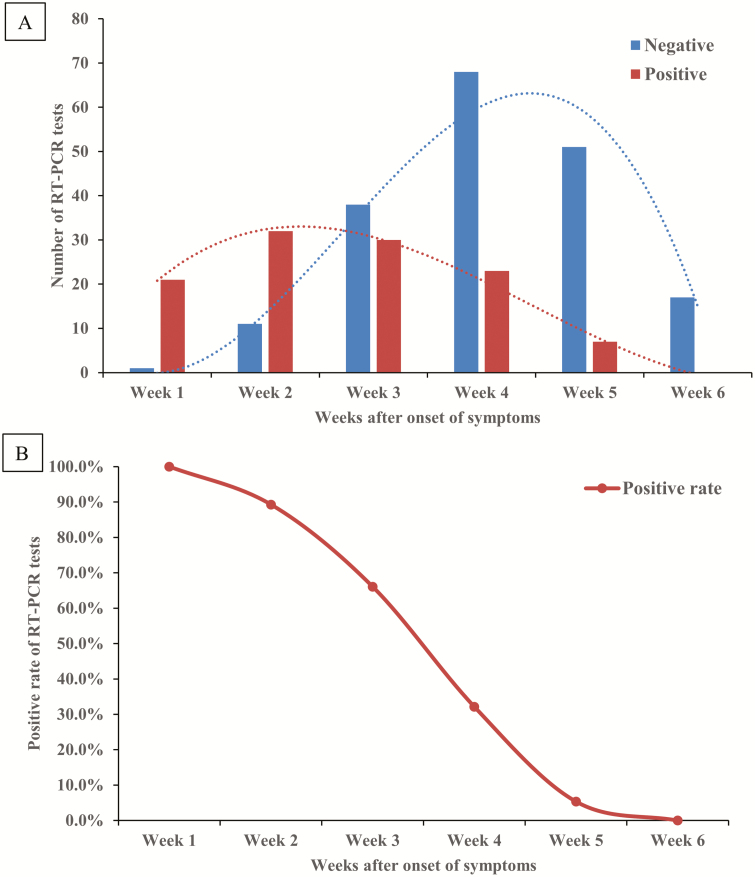
The number of positive and negative results of RT-PCR tests are shown in Figure 1A. In first 3 weeks after symptoms onset, majority results of RT-PCR for SARS-CoV-2 were positive. From week 3 after symptom onset, the number of negative RT-PCR results increased. All RT-PCR test results were negative at week 6 after onset (Supplementary Table 2). The positive rate of RT-PCR test results was highest at week 1 (100%), followed by 89.3%, 66.1%, 32.1%, 5.4%, and 0% at weeks 2, 3, 4, 5, and 6, respectively (Figure 1B).
Figure 1.
Dynamic profile of RT-PCR for SARS-CoV-2. A, Dynamic profile of SARS-CoV-2 detected by RT-PCR from 56 patients with COVID-19 (n = 299). Numbers of the positive (red bars) and negative (blue bars) results of SARS-CoV-2 RT-PCR tests were summed on weeks after the onset of symptoms. B, Positive rate of SARS-CoV-2 detected by RT-PCR from 56 patients with COVID-19 (n = 299). Percentages of positive results of SARS-CoV-2 RT-PCR test were calculated on weeks after the onset of symptoms. Abbreviations: COVID-19, coronavirus disease 2019; RT-PCR, reverse transcription–polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
We divided patients into nonprolonged and prolonged shedding groups according to nucleic acid conversion time (≤24 days or >24 days). As shown in Supplementary Table 3, patients with prolonged viral shedding tended to be older (P = .011) and were more likely to have comorbidities such as diabetes (P = .016) and hypertension (P = .006).
DISCUSSION
This study is the first case series from 56 patients with COVID-19 with 299 samples of RT-PCR tests for SARS-CoV-2 detection. Our preliminary results are notable for providing evidence of an SARS-CoV-2 dynamic profile in infected patients.
Genomic studies have shown that SARS-CoV-2 shares approximately 80% identity sequencing with SARS-CoV, which caused a global epidemic with 8096 confirmed cases worldwide in 2002–2003 [8]. Presumed person-to-person transmission of SARS-CoV-2 was suggested based on epidemiologic and clinical evidence [9, 10]. Although SARS-CoV-2 shares similar sequencing characteristics with SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), study of case series suggested that the viral nucleic acid shedding pattern of patients infected with SARS-CoV-2 is different from that of SARS-CoV, which had modest viral loads in the early stage and peaked approximately 10 days after symptom onset [11].
Our study collected serial RT-PCR test results from 56 patients who had recovered from COVID-19 and investigated the dynamic profile during the disease course. We showed that the majority of patients had positive results on RT-PCR tests for SARS-CoV-2 within 3 weeks after the onset of symptoms. The negative results on RT-PCR tests for SARS-CoV-2 began to be dominant from week 4 after the onset of symptoms, and by the end of follow-up (6 weeks) all results of RT-PCR tests were negative. The positive rate of RT-PCR test results kept declining during the observation period (6 weeks) (Figure 1). The above findings suggest that SARS-CoV-2 viral replication has a relatively long period in infected patients.
Our study attempted to explore the correlation between clinical characteristics and viral shedding in patients with COVID-19. We found that patients with prolonged viral nucleic acid conversion tended to be older and have more comorbidities. Previous studies suggested that coronavirus is more likely to infect older individuals, in whom the immunopathogenesis and induction of a proinflammatory cytokine storm might be responsible [12]. Older patients with impaired immune function might have a prolonged period of viral elimination.
As a result of errors in sampling and testing, false-negative results on RT-PCR for SARS-CoV-2 are very common in clinical settings. Meanwhile, it is recommended by the current diagnosis and treatment guidelines for SARS-CoV-2 from the Chinese National Health Committee that the criteria for discharging a patient include the relief of symptoms, improvement in radiography, and 2 consecutive negative RT-PCR results for SARS-CoV-2 [7]. In our study we found 2 consecutive negative RT-PCR test results followed by a positive result in 4 patients (patient numbers 20, 24, 37, and 56) (Supplementary Table 1). A recent report by Lan et al [13] found positive RT-PCR test results in cases of patients who had recovered from COVID-19. These infected patients could be the source of transmission. The above findings question the current criteria of discharge.
Evidence suggested that the outbreaks of COVID-19 may be correlated to its rapid person-to-person transmission ability [2]. Since specific treatment has not been validated for COVID-19, traditional public health tactics—isolation, quarantine, and community containment—are critical to control the spread [14–16]. This preliminary study has found evidence of the dynamic profile of SARS-CoV-2 in non-ICU patients with COVID-19 during the disease course. According to the results in our study, we suggest prolonged observation and repeat confirmation of RT-PCR tests from respiratory specimens for safe discharge and discontinuation of quarantine.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. All authors participated in the study design. A. T. X. and S. Z. conceived the study, analyzed the data, and drafted the manuscript. Y. X. T. helped critically revise the manuscript and collected data. All authors have agreed on the final version and meet the major criteria recommended by the ICMJE (http://www.icmje.org/)
Acknowledgments. The authors thank Ms Cheng Chen for English-grammar correction of this manuscript. This study was approved by the Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. All procedures followed in this study were in accordance with the 1964 Helsinki Declaration and later versions. Oral consent was obtained from patients involved before enrollment when data were collected retrospectively. The database used and/or analyzed during the current study is not publicly available (to maintain privacy) but can be available from the corresponding author on reasonable request.
Financial support. No funding resources were declared for this study.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. J Med Virol. Published online January 16, 2020. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zunyou W, Jennifer MG. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA. Published online February 24, 2020. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 3. Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. Published online February 19, 2020. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pan Y, Zhang DT, Yang P, et al. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. Published online February 24, 2020. doi. 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. Published online March 11, 2020. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Interim infection prevention and control recommendations for patients with confirmed coronavirus disease 2019 (COVID-19) or persons under investigation for COVID-19 in healthcare settings. Updated: February 21, 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/infection-control.html. Accessed 1 March 2020.
- 7. China National Health Commission. Diagnosis and treatment of 2019-nCoV pneumonia in China. Version 5. In Chinese. Published February 8, 2020. Available at: http://www.nhc.gov.cn/yzygj/s7653p/202002/d4b895337e19445f8d728fcaf1e3e13a/files/ab6bec7f93e64e7f998d802991203cd6.pdf. Accessed 3 March 2020.
- 8.World Health Organization. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. Geneva, Switzerland: World Health Organization, 2004. Available at: https://www.who.int/csr/sars/country/table2004_04_21/en. Accessed 1 March 2020. [Google Scholar]
- 9. Ping Y, Jiang Z, Zhengdong Z, et al. A familial cluster of infection associated with the 2019 novel coronavirus indicating potential person-to-person transmission during the incubation period. J Infect Dis. Published February 18, 2020. doi: 10.1093/infdis/jiaa077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan JF, Yuan S, Kok K, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. Published online January 24, 2020. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peiris JS, Chu CM, Cheng VC, et al. ; HKU/UCH SARS Study Group . Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 2003; 361:1767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. Published online February 28, 2020. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lan L, Dan X, Guangming Y, et al. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. Published online February 27, 2020. doi: 10.1001/jama.2020.2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. Published on January 29, 2020. N Engl J Med. 2020. doi: 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McCloskey B, Heymann DL. SARS to novel coronavirus—old lessons and new lessons. Epidemiol Infect 2020; 148:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Du Z, Wang L, Cauchemez S, et al. Risk for transportation of 2019 novel coronavirus disease from Wuhan to other cities in China. Emerg Infect Dis 2020; 26:1049–52. doi: 10.3201/eid2605.200146 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.