Abstract
Data and information technology are key to every aspect of our response to the current coronavirus disease 2019 (COVID-19) pandemic—including the diagnosis of patients and delivery of care, the development of predictive models of disease spread, and the management of personnel and equipment. The increasing engagement of informaticians at the forefront of these efforts has been a fundamental shift, from an academic to an operational role. However, the past history of informatics as a scientific domain and an area of applied practice provides little guidance or prologue for the incredible challenges that we are now tasked with performing. Building on our recent experiences, we present 4 critical lessons learned that have helped shape our scalable, data-driven response to COVID-19. We describe each of these lessons within the context of specific solutions and strategies we applied in addressing the challenges that we faced.
Keywords: COVID-19, applied informatics, lessons learned, analytics
INTRODUCTION
Data and information technology has been key to every aspect of our response to the current coronavirus disease 2019 (COVID-19) pandemic—spanning a spectrum of informing care delivery,1,2 management of personnel and supplies,3 and interaction and communication within and across organizations. Central to these rapid changes has been the increasing engagement of informaticians on the “front lines” of such efforts, a fundamental shift in focus for many from an academic to an operational role. These individuals have become key contributors to a collaborative enterprise involving health system executives, policymakers, public health experts, care providers, and clinical researchers. This reconceptualization of the informaticians’ role is a function of their unique training: with an emphasis on the development and application of methods and tools for translating data into information, and ultimately actionable knowledge, that can inform and improve the practice of medicine and the management of population health.
In contrast to previous pandemics such as severe acute respiratory syndrome (SARS) and Ebola, in which informaticians had a less prominent role in healthcare operations or care delivery,4 the widespread adoption and use of electronic health records (EHRs) has afforded incredible opportunities for developing real-time, data-driven insights that can help in responding to the challenges presented by COVID-19. However, this rapid shift from theory and tool development to applications and operations represents new and uncharted territory for many informaticians. Currently, no roadmap exists for coordinating the development or delivery of informatics solutions in different communities of practice within the necessarily short timelines. This represents a critical gap in knowledge and resources that must be filled as we respond at an unprecedented pace to understand the progression of the pandemic and simultaneously devise data-driven strategies for mitigating its long-term effects from a clinical and a population health standpoint. More importantly, the past history of informatics as a scientific domain and an area of applied practice provides little guidance or prologue for the incredible challenges that we are currently facing.
Building on our recent experiences at Washington University in St. Louis and BJC HealthCare, we have endeavored to apply informatics at the operational, point-of-care, research, and population levels, in order to address the aforementioned challenges facing our healthcare delivery system. In this perspective, we present 4 critical lessons learned from our experience that we believe have helped shape a scalable, data-driven response to COVID-19 and that simultaneously may inform new definitions for the role of informatics practice in such an environment for the foreseeable future.
LESSON 1: WORK TOGETHER, MITIGATE BARRIERS, FILL DATA MANAGEMENT GAPS
This current state of affairs highlights the critical and inexorable relationship between key stakeholders who need to work together while leveraging shared and often fragmented data assets. Central to the ability of these individuals to respond to this crisis is the ability to share ideas, workloads, and leadership at a pace that is not typically experienced in normal operations. Issues we have encountered in this regard include barriers to data access, fragmented data sources, and lack of interoperability of data and systems, affecting our ability to capture the progression of the pandemic to inform timely and critical decisions. These challenges of coordination have been exasperated by “work from home” restrictions.
In particular, in academic medical centers, informatics practice and research are often aligned in parallel functional silos with relatively independent goals. Realigning these efforts across such groups and potential barriers requires both informational and relational coordination (ie, coordinating data, tools, technologies, and people).5 Achieving such coordination can include the selection of “gap filling” technologies (eg, common platforms for messaging, lightweight data management systems, or integrated electronic data capture tools) in the near term, as well as development of plans to build more shared information technology infrastructures that span traditionally separated mission areas in the longer term. It is important that the lessons learned, and solutions selected, during crisis operations inform longer-term data infrastructure planning and should not be considered as ad hoc or otherwise temporary tools or capabilities to be discarded in the future.
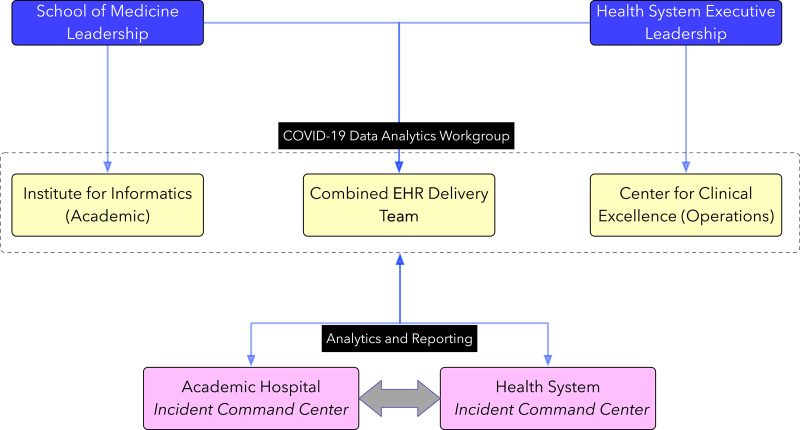
Toward this end, we have created a cross-institutional data analytics leadership team, spanning our operational analytics, EHR delivery, and academic informatics units, that (1) coordinates access to data and data products and, further, (2) acts as a gatekeeper for clinical and research data requests to ensure economies of scale, while (3) providing guidance for epidemiologic modeling efforts. Further, this combined unit is acting as a source of data analytic “intelligence” for the COVID-19 “incident command centers” at both our academic hospital and broader system-wide levels, thus ensuring consistency of data and analytics capabilities being provided to all key decision makers (see Figure 1). This approach is collectively serving to drive applied informatics and data analytics across our 2 organizations while leveraging all relevant resources and personnel in a structured manner.
Figure 1.
Structure of the integrated team for cross-institutional management of resources and analytics. COVID-19: coronavirus disease 2019; EHR: electronic health record.
LESSON 2: DEVELOP LOCALLY, SHARE REGIONALLY AND NATIONALLY
Given the scale of the current pandemic, the needs of healthcare systems across the country are very similar: understand disease incidence and progression in the community, quantify resource capabilities, predict a proximal future state (in days and weeks), and develop adaptive strategies to respond to the changing demands. With the primary source of data for many of these efforts being derived from the institution’s EHR, it is critical to build, optimize, and share ensuing tools across institutions. In addition, it is important to create persistent data models that can reduce the duplication of efforts. This requires the development of reliable and reproducible data queries, analytics, and dashboards that can be broadly shared across health systems using similar EHRs or equivalently interoperable platforms. Further, ensuring that these capabilities are reliable and reproducible can and should rely on close and regular interaction between informatics professionals and the healthcare information technology “operators” responsible for the delivery of the EHR, all working as a seamless team.
At an even more granular level, we have learned that building and delivering such persistent data models and analytical products requires critical thinking regarding the elements that are most relevant for the needs of the intended audience(s). During a pandemic, it is vital that local, regional, and national partners that share common analytical needs receive the necessary information in an integrated manner. We have found that an efficient approach is to identify a finite list of variables that fall in the need to have, not nice to have category to share with collaborators (Table 1). This can be achieved through the generation of a minimum viable dataset and expanding it with the changing organizational needs in an iterative manner (eg, starting with a small and concise set of data, rather than letting “perfection be the enemy of the good”). Further, if those same data elements—and potentially their associated data queries—can be shared across healthcare systems and stakeholder entities, local analyses can be replicated on a more comprehensive regional or national level, informing action at a larger scale.
Table 1.
Data elements that were initially considered in a minimum viable dataset for research and operational purposes
| ID | Demographics | Influenza-like-illness symptoms | Respiratory tests | Care trajectory (with dates) | Clinical characteristics |
|---|---|---|---|---|---|
| Patient ID, facility ID | Age, race, gender, zip code | Fever, cough, sore throat | RPP/RVP Flu RSV by PCR, rapid influenza test, COVID-19 RNA test | ED, hospital, ICU, death (if relevant) | Need for ventilation, intubation, clinical comorbidities |
COVID-19: coronavirus disease 2019; ED: emergency department; ICU: intensive care unit; PCR: polymerase chain reaction; RPP: respiratory pathogen profile; RSV: respiratory syncytial virus.
Importantly, and extending beyond local impact, such coordinated efforts can help to harness and harmonize data collected across healthcare systems with persistent and shareable data models, leading to consistent messaging and policy decisions, as well as the ability to share critical data-driven insights across traditional organizational boundaries, thus meeting population health needs. Data harmonization efforts serve to coordinate institutions to, for example, adequately estimate the trajectory of the current pandemic and track how it can affect both care management and capacity management across healthcare systems. Finally, creating shared resources can also enable smaller health centers to track and share their progress and benefit from the activities of larger and more well-resourced organizations, with an ensuing benefit at the population level. Finally, EHR vendor organizations can also play a key role in the wide dissemination successful data models, dashboards, or analytics.
To create such a common ground, both locally and regionally, our focus has been on creating initial minimum viable products (data, analytics, dashboards, epidemiologic modeling) that formed the basis for discussions with local and regional partners. These minimum viable products were then used as the foundation for developing larger data sharing agreements across institutions in the region, state, and country. This iterative approach benefited from a structured approach toward meeting the informatics needs of multiple local and regional stakeholders.
LESSON 3: ADAPT, NOT BUILD ANEW, AND DELIVER KNOWLEDGE AT THE RIGHT TIME, PLACE, AND FORMAT
In order to address the rapidly changing analytic needs for capacity planning and epidemic modeling, it is important to rely on data infrastructures that are currently available, not those we wish we had or what we may be able to build in the future. This is because the time scales for response during a pandemic are in the order of hours and days, not weeks or months. Further, we must consider how data-driven insights will be delivered “at scale,” which in the light of limited technical resources during an ongoing pandemic, must utilize existing technology infrastructure. For example, analytic or visualization efforts should rely on existing, open-source platforms that are flexible and allow for wide-scale use across platforms and devices.6 These flexible platforms also will allow for rapid integration of new data (eg, from a health information exchange or a state-wide hospital association) and help in delivering multiscaled visualizations with limited customization.
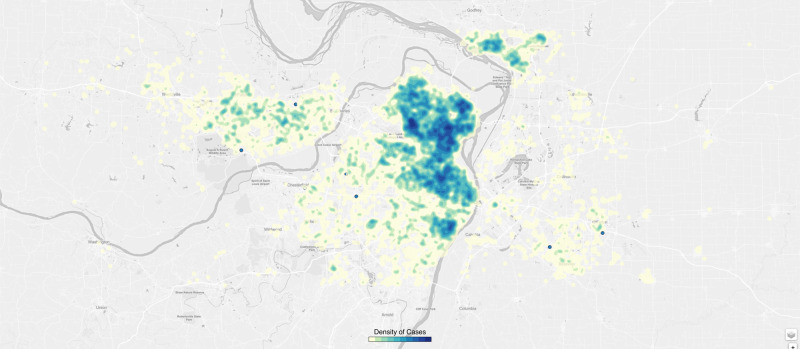
End users find data presented as time trends or as geospatial analyses helpful for informing care delivery and for anticipating patient surges. Our efforts have been focused on the use of open source platforms such as D36 to provide actionable information for internal and local public health authorities. We have developed multiple visualizations to identify “hot spots” of infections relying on zip code–level maps and more granular street-level data (with appropriate security to protect privacy). Such visualizations will be augmented in the future to identify communities that are at risk for increased infection and those that would benefit from intensive contact tracing or public health messaging around social distancing (see Figure 2).
Figure 2.
A 2-dimensional density plot of influenza-like illness and coronavirus disease–positive patients presenting to clinics (with the darker blue dots showing higher-density areas).
LESSON 4: SUPPORTING AN EVOLVING RESEARCH ENTERPRISE
In a recently published editorial, McDermott and Newman7 highlighted the importance of developing alternative approaches to continuing research, especially randomized controlled clinical trials, during the current pandemic. Their suggestions focused on utilizing remote modes for continuing data collection for trials, which included telehealth or video visits for patient follow-up, electronic consent, mobile health applications for patient monitoring, and the use of electronic tools for remote collection of data.7,8 Such recommendations are consistent with the fact that while operational needs must be addressed first and foremost during a pandemic, we cannot abandon the role of research in driving our response to COVID-19. In this context, it is also important to guard against duplicative data collection efforts for clinical care and research projects so as not to increase the clinical workload during a pandemic.
Critical research questions can and should be answered, including the diagnosis of patients with greater sensitivity and specificity with respect to specific symptomatic presentations, the modeling of individual and population trajectories, and the evaluation of novel therapeutics through both traditional and pragmatic clinical trials. In all of these cases, and in many others with equal importance, the key question is how we pursue such research in a timely manner while facing the realities of both resource constraints and safety issues when seeking to interact with COVID-19 patients in order to conduct such studies.
Within the applied clinical informatics realm opportunities exist for leveraging extensions to current EHR platforms for more efficient engagement with participants who are already enrolled in trials. For example, the use of PROMIS (Patient-Reported Outcomes Management Information Systems) applications9 as a standardized mechanism to capture physical, behavioral, and mental health outcomes affords both clinical and research benefits; its EHR integration allows for longitudinal tracking with features for automated administering, scoring, reviewing, and diagnostic interpretation.10 Similarly, we have independently used electronic data capture tools such as REDCap (Research Electronic Data Capture) for research data capture and also to import data from the EHR to streamline data collection, leading to considerable time savings (eg, using REDCap Fast Health Interoperability Resources).11 Finally, we have developed centralized data marts with comprehensive patient data on COVID-19 from testing to admission and discharge (where relevant) with longitudinal clinical information such as procedures and medications. This data mart has been designed to be extensible with potential connections to biospecimen banks and sequencing information, as they become available.
CONCLUSION
The COVID-19 pandemic elucidates the essence of a true informatics problem, one that is first and foremost concerned with translating data into actionable knowledge, in this case at a speed that we have not previously needed. The stress on the system is unprecedented, and we must act using data-driven solutions. Informaticians can serve to link healthcare, research, and public health stakeholders and enable systems-level responses to the current crisis. As we have noted, achieving this potential impact is not easy, given that issues such as barriers to access to data and data integration, lack of interoperability of communication channels and file sharing tools, and an inability to capture the “big picture” of the pandemic given a limited, often “local,” slice of data are pervasive. While we have not yet emerged from the current and high-intensity phase of this pandemic, we have sought to share key lessons learned at Washington University in St. Louis and BJC HealthCare, which seek to address and overcome the barriers in both the near and longer term. Our hope in doing so is to advance the state of informatics knowledge and practice as our community seeks to contribute to addressing this unprecedented public health crisis.
AUTHOR CONTRIBUTIONS
TGK, REF, and PROP developed the initial framework with input from AML and KFW. All authors contributed to the writing of the manuscript and gave approval for the final manuscript.
ACKNOWLEDGMENTS
We thank Dr. Clay Dunagan, Dr. Dennis Goldfarb, and Rachel Komeshak for their input and contributions.
CONFLICT OF INTEREST STATEMENT
The authors have no conflicts of interest to disclose.
REFERENCES
- 1. Reeves JJ, Hollandsworth HM, Torriani FJ, et al. Rapid response to COVID-19: Health informatics support for outbreak management in an Academic Health System. J Am Med Inform Assoc 2020. Apr 27 [E-pub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Judson TJ, Odisho AY, Neinstein AB, et al. Rapid design and implementation of an integrated patient self-triage and self-scheduling tool for COVID-19. J Am Med Inform Assoc 2020. Apr 8 [E-pub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Turer RW, Jones I, Rosenbloom ST, Slovis C, Ward MJ.. Electronic personal protective equipment: a strategy to protect emergency department providers in the age of COVID-19. J Am Med Inform Assoc 2020. Apr 2 [E-pub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fidler DP, Gostin LO.. The WHO pandemic influenza preparedness framework: a milestone in global governance for health. JAMA 2011; 306 (2): 200–1. [DOI] [PubMed] [Google Scholar]
- 5. Malone TW, Crowston K.. The interdisciplinary study of coordination. ACM Comput Surv 1994; 26 (1): 87–119. [Google Scholar]
- 6.D3 (Data-Driven Documents). https://d3js.org/ Accessed April 15, 2020.
- 7. McDermott MM, Newman AB.. Preserving clinical trial integrity during the coronavirus pandemic. JAMA 2020. Mar 25 [E-pub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 8. Nicol GE, Piccirillo JF, Mulsant BH, Lenze EJ.. Action at a distance: geriatric research during a pandemic. J Am Geriatr Soc 2020. Mar 24 [E-pub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cella D, Yount S, Rothrock N, et al. The Patient-Reported Outcomes Measurement Information System (PROMIS): progress of an NIH Roadmap cooperative group during its first two years. Med Care 2007; 45 (Suppl 1): S3–S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grossman LV, Mitchell EG. Visualizing the patient-reported outcomes measurement information system (PROMIS) measures for clinicians and patients. In: AMIA Annual Symposium Proceedings, 2017; Washington, DC: American Medical Informatics Association. [PMC free article] [PubMed]
- 11. Metke-Jimenez A, Hansen D.. FHIRCap: Transforming REDCap forms into FHIR resources. AMIA Jt Summits on Transl Sci Proc 2019; 2019: 54–63. [PMC free article] [PubMed] [Google Scholar]