Abstract
Background
Patients with inflammatory bowel disease (IBD) have intestinal inflammation and are treated with immune-modulating medications. In the face of the coronavirus disease-19 pandemic, we do not know whether patients with IBD will be more susceptible to infection or disease. We hypothesized that the viral entry molecules angiotensin I converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) are expressed in the intestine. We further hypothesized that their expression could be affected by inflammation or medication usage.
Methods
We examined the expression of Ace2 and Tmprss2 by quantitative polymerase chain reacion in animal models of IBD. Publicly available data from organoids and mucosal biopsies from patients with IBD were examined for expression of ACE2 and TMPRSS2. We conducted RNA sequencing for CD11b-enriched cells and peripheral and lamina propria T-cells from well-annotated patient samples.
Results
ACE2 and TMPRSS2 were abundantly expressed in the ileum and colon and had high expression in intestinal epithelial cells. In animal models, inflammation led to downregulation of epithelial Ace2. Expression of ACE2 and TMPRSS2 was not increased in samples from patients with compared with those of control patients. In CD11b-enriched cells but not T-cells, the level of expression of ACE2 and TMPRSS2 in the mucosa was comparable to other functional mucosal genes and was not affected by inflammation. Anti-tumor necrosis factor drugs, vedolizumab, ustekinumab, and steroids were linked to significantly lower expression of ACE2 in CD11b-enriched cells.
Conclusions
The viral entry molecules ACE2 and TMPRSS2 are expressed in the ileum and colon. Patients with IBD do not have higher expression during inflammation; medical therapy is associated with lower levels of ACE2. These data provide reassurance for patients with IBD.
Keywords: coronavirus, Crohn disease, ulcerative colitis
INTRODUCTION
The spread of the potentially lethal coronavirus disease 19 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has taken the world health community by surprise. As of April 4, 2020, 3 months after the disease outbreak in Wuhan, China,1-3 COVID-19 was declared a pandemic by the World Health Organization and accounted for more than 973,300 confirmed cases and 55,321 deaths throughout the world.4 COVID-19 typical symptoms include fever, dry cough, fatigue, myalgia, dyspnea, and pneumonia; in most severe cases complications such as acute respiratory distress syndrome can occur, with eventual fatal consequences.5
The structural similarity of SARS-CoV-2 with SARS-CoV, particularly in the spike (S) protein that mediates coronavirus attachment and entry into target cells, has allowed for the rapid identification of the mechanism of viral infection. Like SARS-CoV, SARS-CoV-2 binds to the angiotensin I converting enzyme 2 (ACE2) receptor through the N-terminal S1 subunit,6-8 which is subsequently cleaved by the host transmembrane serine protease 2 (TMPRSS2) to expose the C-terminal S2 subunit that induces virus-cell fusion.8, 9 The ACE2 receptor is found expressed at high levels not only in alveolar type-2 cells in the lung but also in liver cholangiocytes, myocardial cells, esophagus keratinocytes, kidney proximal tubules, bladder urothelial cells, and gastrointestinal epithelial cells.10, 11 Of note, in silico searches in publicly available databases have determined that the small intestine has the highest expression of ACE2 and that TMPRSS2 is also expressed at high levels in both the small intestine and the colon (the Human Protein Atlas12). Furthermore, in vitro assays have shown that the human intestinal epithelial cell (IEC) line Caco-2 expresses both proteins and is easily infected by viral particles bearing the SARS-CoV and SARS-CoV-2 S proteins.8
These findings pose the question of whether the intestine acts as a secondary site for coronavirus tropism and infection. This hypothesis is supported by 2003 and 2020 reports that identified viral replication and nucleocapsid protein staining in small and large intestine samples from 5 SARS patients and 1 COVID-19 patient.13, 14 Moreover, viral RNA can be detected in rectal swabs and stool of SARS and COVID-19 patients long after nasopharyngeal swabs test negative, which indicates that both SARS-CoV and SARS-CoV-2 may actively replicate in the gastrointestinal tract, leading to concerns of fecal-oral transmission.13-17
Similar to SARS, which caused gastrointestinal symptoms in up to 38% individuals of a 138-patient cohort,13 COVID-19 can cause diarrhea, nausea, vomiting, and abdominal pain.18 These symptoms may lead to confusion with respect to flares of inflammatory bowel disease (IBD). In addition, clinicians and patients with IBD are worried about the implications of the SARS-CoV-2 virus and COVID-19 in the context of the underlying disease and the medications to treat it. On the one hand, it is unclear whether gastrointestinal and/or systemic inflammation may alter the expression of ACE2 or TMPRSS2, facilitating viral entry. There is also concern about the medications taken by patients with IBD, which include immunomodulators, biologics, and small molecule inhibitors of Janus kinase1/2, which increase the susceptibility of the reactivation of latent viruses.19 On the other hand, in severe cases of COVID-19, cytokine release syndrome may worsen pulmonary injury, leading to acute respiratory distress syndrome.20 Certain biologics, notably tocilizumab, an anti-interleukin (IL)-6R antibody, may have some efficacy to prevent death. Thus, the balanced effect of medications for IBD to treat the inflammation vs to suppress viral immunity are unclear.
We undertook the current study to understand the regulation of ACE2 and TMPRSS2 in the intestine during inflammatory states and in patients with IBD taking a variety of medications. Our results can edify whether we should be concerned about patients with IBD with active disease and taking medications.
METHODS
Participants and Biospecimen Acquisition
Participants in this study consisted of 65 patients from 2 prospectively collected cohorts within our larger University of Miami Biorepository (3050 participants). We included patients diagnosed with Crohn disease (CD) and ulcerative colitis (UC) with or without active inflammation determined by endoscopy and by histology. Endoscopic tissue was collected from patients who underwent colonoscopy as part of clinical care. Biopsies from the terminal ileum, ascending colon, and sigmoid colon were collected and annotated with respect to inflammation based on endoscopy and pathology. In the first cohort (n = 31, Table 1), biopsy tissue was processed to enrich for lamina propria CD11b+ cells. Our second cohort (n = 34 patients) had baseline tissue biopsies and matched blood samples collected as part of a study to identify markers of response to vedolizumab in which CD4+ memory T-cells and regulatory T-cells were isolated (Tmem/Treg cohort; Table 1). Fourteen to sixteen weeks after starting vedolizumab, blood was collected for effector T-cells and regulatory T-cells, and patients were designated as responders or nonresponders to vedolizumab. This study was performed at the Crohn’s and Colitis Center at the University of Miami and was approved by the University of Miami Institutional Review Board (IRB ID 20150750, 20080011). All clinical and demographic data were collected with written informed consent before enrollment. All authors had access to the analyzed data and approved the final manuscript before submission for publication.
TABLE 1.
Study Demographics
| Total Patients Enrolled | Study Total | CD11b | Tmem/Treg |
|---|---|---|---|
| (n = 65) | (n = 31) | (n = 34) | |
| Disease | |||
| UC | 31 (47.69%) | 15 (48.3%) | 16 (47.1%) |
| CD | 34 (52.3%) | 16 (51.6%) | 18 (52.9%) |
| Age at time of collection, y (mean) | 43.81 (18-79) | 45 (18-72) | 42.7% (20-79) |
| Years of disease (mean) | 23.01 (1-65) | 30 (10-61) | 16.2% (1-65) |
| Gender | |||
| Male | 31 (47.6%) | 15 (48.3%) | 16 (47.1%) |
| Female | 34 (52.3%) | 16 (51.6%) | 18 (52.9%) |
| Race | |||
| White | 61 (93.8%) | 29 (93.5%) | 32 (94%) |
| African American | 2 (3%) | 0 (0%) | 2 (5.88%) |
| Asian | 1 (1.5%) | 4 (0%) | 0 (0%) |
| Mixed | 1 (1.5%) | 3 (9.67%) | 0 (0%) |
| Ethnicity | |||
| Not Hispanic | 39 (60%) | 18 (58%) | 21 (61.7%) |
| Hispanic | 26 (40%) | 13 (41.9%) | 13 (38.2%) |
| Jewish | |||
| Yes | 21 (32.3%) | 11 (35%) | 10 (29%) |
| No | 42 (64.6%) | 20 (67.7%) | 22 (64.7%) |
| Unknown | 2 (3%) | 0 (0%) | 2 (5.88%) |
| Medication | |||
| Anti-TNF | 24 (36.9%) | 9 (29%) | 15 (44%) |
| Vedolizumab | 7 (10.76%) | 6 (19.3%) | 1 (2.9%) |
| Ustekinumab | 3 (4.61%) | 4 (12.9%) | 1 (2.9%) |
| Thiopurines | 12 (18.46%) | 2 (6.45%) | 10 (29.4%) |
| Mesalamine | 14 (21.5%) | 8 (25.8%) | 6 (17%) |
| Steroids | 15 (23 %) | 7 (22.5%) | 8 (23.5%) |
| No medications | 10 (15.38%) | 5 (16%) | 5 (14.7%) |
Anti-TNF indicates anti-tumor necrosis factor.
Cell Isolation From Biopsies and Blood
Tissue biopsies collected during colonoscopy were placed in Hypothermosol solution (MilliporeSigma, St. Louis, MO) for preservation during transport. To obtain single-cell suspensions, samples were first depleted of IECs by incubation in 10 mM dithiothreitol followed by 0.5 mM EDTA solutions in Dulbecco's Modified Eagle Medium media containing penicillin/streptomycin. Dissociation was achieved by digesting tissues in a mixture containing 250 µg/mL Liberase (MilliporeSigma) and 10 µg/mL DNase I (Lucigen Corporation, Middleton, WI) in Dulbecco's Modified Eagle Medium at 37°C for 20 minutes. Digested tissue was mechanically dissociated by pipetting, and single cells were rinsed and filtered using a 70 µm strainer.
In the CD11b cohort, single-cell suspensions were counted and conditioned for magnetic sorting by labeling with 20 µL of CD11b MicroBeads per 107 cells (Miltenyi Biotec GmbH, Gaithersburg, MD). Magnetic separation was performed by positive selection using LS columns (Miltenyi Biotec GmbH). For the purposes of this study, we defined lamina propria phagocytes as CD11b+ cells, which include macrophages, dendritic cells, granulocytes, and natural killer cells. Purity assessment of the CD11b+ target cells was analyzed using flow cytometry analysis from 12 inflamed and 12 noninflamed biopsies. After the enrichment protocol by magnetic separation, we obtained an average purity of 70.7%.
In the Tmem/Treg cohort, effector T-cells (CD3+ CD4+ CD25- CD45RO+ CCR7-) and regulatory T-cells (CD3+ CD4+ CD25+ CD45RO+ CCR7-) were isolated from tissue biopsies and peripheral blood mononuclear cells by flow cytometry and cell sorting. Single cells were counted and labeled using fluorescent-tagged antibodies targeting specific antigens on the cell surface to identify and segregate these T-cell subpopulations (Supplementary Table 1). Rhodamine 123 fluorescent dye (MilliporeSigma) was also included as a functional reporter for permeability glycoprotein 1 expression. Peripheral blood mononuclear cells were isolated by means of a Histopaque-1077 gradient (MilliporeSigma) following manufacturer recommendations, quantified, and labeled following the same procedure described for tissue biopsies. All flow cytometry analysis and cell sorting were performed using BD FACSAria IIu (BD Bioscience, San Jose, CA).
RNA Extraction and Next-Generation Sequencing
The RNA from the CD11b-enriched cell population was isolated by means of the RNeasy Mini Kit (Qiagen, Germantown, MD), quantified by nanodrop, and sequenced at the University of Miami Onco-Genomics Shared Resource. Libraries were prepared using the KAPA RNA HyperPrep Kit with RiboErase (HMR; Roche Life Science, Wilmington, MA), quality-checked, normalized to concentration, pooled, and sequenced on a 200 bp paired-end run using both lanes of the NovaSeq 6000 S1 flow cell (100 cycles; 1.6 billion flow cell). Sixty barcoded libraries were sequenced across 2 lanes, 4.18 billion pass filter paired-end reads (2.09 billion clusters) were generated, and 472 million paired-end reads (236 million clusters) were unindexed.
Total RNA from the Tmem/Treg subsets from blood and tissue was isolated by using the miRNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions and quality-checked by means of the high-sensitivity Agilent Bioanalyzer RNA Pico LabChip analysis (Agilent, Santa Clara, CA). Genome-wide transcriptional profiles (RNA sequencing) to characterize T-cell parameters were assessed using the SMART-Seq HT Kit (Takara Bio USA, Mountain View, CA). Libraries were individually indexed and pooled in groups of 20. Each pool was run on a NextSeq500 flow cell (9 flow cells total) generating 75-base single reads with an average of 20 million reads per sample (3.5 billion reads total).
Animal Model, Collection of Samples, and Epithelial Cell Isolation
Seven-week-old C57Bl/6 mice purchased from Jackson Laboratory were housed in specific pathogen-free conditions with a controlled temperature of 20 ± 2°C and free access to food and water. One week later, mice of both sexes were randomly assigned cages and given 5 days to adapt before the beginning of experiments. Mice undergoing the chemically induced model of colitis received 3% dextran sodium sulfate (DSS; 40-50 kDa; Affymetrix/USB, ThermoFisher Scientific, Waltham, MA) in drinking water for either 2, 4, or 6 consecutive days. The DSS was replaced every other day. To minimize variability in our determinations, all mice were euthanized on the same day (for a diagram of the study design, please refer to Burgueño et al21). The weight loss for each mouse was assessed daily during DSS treatment; mice losing more than 30% of initial body weight or displaying severe bloody diarrhea with lack of exploratory behavior were immediately euthanized. No animals died or met the endpoint criteria before the end of the study. All experiments were performed with the approval of the Institutional Animal Care and Use Committee at the University of Miami (Protocols 17–196 and 18–169). The University of Miami is internationally accredited by the Association for Assessment and Accreditation of Laboratory Animal Care.
Mice were euthanized by cervical dislocation under isoflurane (Piramal Critical Care, Bethlehem, PA) anesthesia. Subsequently, the duodenum, terminal ileum, and colon were removed, flushed, cut down the center, and pinned flat on a Sylgard-coated petri dish. One longitudinal section of each segment of the gut was placed in Trizol (ThermoFisher Scientific) and used for RNA isolation. The rest of these longitudinal sections were incubated in either a 5 mM (duodenum and ileum) or a 20 mM (colon) EDTA chelation buffer prepared in Hank’s balanced salt solution for 60 minutes at 80 rpm in an orbital shaker. Next, EDTA was washed with Hank’s balanced salt solution and the crypts were released by gentle agitation. Collected crypts were spun and lysed in Trizol. The remaining longitudinal tissue was snap-frozen for myeloperoxidase (MPO) determinations. 21
Quantitative Polymerase Chain Reaction
Whole tissues or IECs lysed in Trizol were homogenized using the BeadBlasterTM24 (Benchmark Scientific, Sayreville, NJ), and RNA was isolated using phenol-chloroform extraction. We retro-transcribed 500 ng of RNA using the PrimeScript RT reagent kit (Takara Bio USA) and amplified the resulting cDNA on a LightCycler 480 II instrument (Roche Life Science) in the presence of selected primers (Supplementary Table 2) using the SYBR Premix Ex Taq (Takara Bio USA). Relative mRNA expression was calculated by means of the change in cycle threshold (ΔΔCt) method and normalized to the geometric mean of the housekeeping genes βActin and Hprt. Basal mRNA levels of Ace2 and Tmprss2 were compared with those of the housekeeping gene βActin by calculating the difference between their Ct as described elsewhere.22 According to their ΔCt to βActin, genes were classified as highly expressed (ΔCt < 5 cycles), intermediate (5 < ΔCt < 15 cycles), low (ΔCt > 15 cycles), and undetectable (ΔCt > 40 cycles).
Bioinformatics and Statistical Analysis
Raw CEL files from 4 publicly available microarray studies were downloaded from the Gene Expression Omnibus (GEO) database: GSE107810,23 GSE87317,24 GSE59071,25 and GSE75916.26 We tested for differential gene expression using the Transcriptome Analysis Console (Ver. 4.0.2, ThermoFisher Scientific). Reads from CD11b-enriched and effector or regulatory T-cell RNA sequencing were mapped to the human genome (GRCh38) using the STAR (Ver. 2.5.0) aligner.27 Raw counts were generated based on Ensembl genes (GENCODE Ver. 26 or Ensembl release 91) with featureCounts (Ver. 1.5.0)28 or HTSeq (Ver. 0.11).29 To normalize different numbers of reads in the samples, we used the DESeq2 median of ratios method.30 We calculated transcripts per million (TPM) values with a custom python script by using the longest transcript for each gene extracted from the gtf annotation file. To assess the association between different clinical characteristics with gene expression levels, we used the Spearman rank correlation for continuous variables, the Wilcoxon rank sum test for binary variables, and an analysis of variance for categorical variables with more than 2 levels. To assess the effects of medications on gene expression levels, we used mixed-effects models with normalized and log-transformed gene expression levels as the outcome variable, medication group as the main independent variable, and tissue type as a covariate variable. In the analysis of the TMPRSS2 gene, we also included disease duration. Moreover, patient random effects were included in the mixed models to account for correlations in gene expression levels resulting from multiple samples within the same subject. Statistical analyses were performed using the R software (https://www.r-project.org/).
Data from animal experiments are presented as mean values ± SD. Data analysis and plots were performed using Prism 8.0 (GraphPad Software, San Diego, CA) and compared using a 1-way or 2-way analysis of variance, as indicated in figure captions. A P value of <0.05 was considered significant.
Ethical Considerations
The studies involving human participants were reviewed and approved by the University of Miami Leonard Miller M. School of Medicine Institutional Review Board. The patients/participants provided their written informed consent to participate in this study. The animal studies were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Miami.
RESULTS
Ace2 and Tmprss2 Are Highly Expressed in Mouse Gut Epithelial Cells
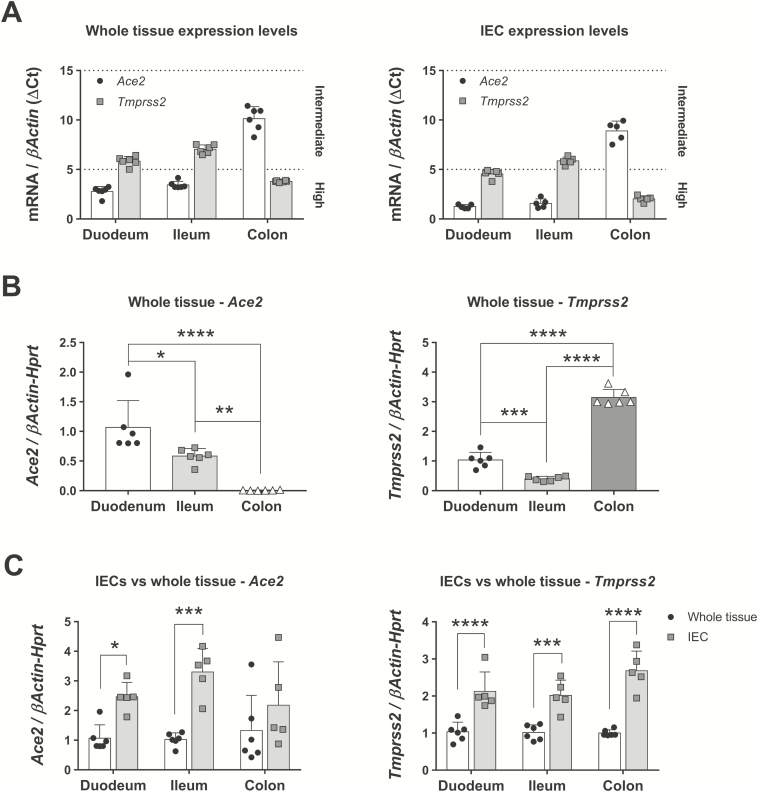
We first sought to characterize the expression of the 2 proteins involved in viral infection, ACE2 and TMPRSS2, throughout the gut. Whole tissues and IECs were collected from the duodenum, terminal ileum, and mid-colon of C57Bl/6 mice and analyzed for the mRNA expression of both proteins by means of quantitative polymerase chain reaction. We identified Ace2 as a gene with high expression levels in whole tissue and IECs from the duodenum and the terminal ileum (Fig. 1A). Conversely, Tmprss2 was highly expressed in the whole colon and IECs from the duodenum and the colon (Fig. 1A). We then compared the expression of both genes in these 3 segments. Expression of Ace2 was highest in the duodenum and progressively reduced toward the ileum (Fig. 1B, left panel). Notably, the level of Ace2 transcripts was markedly reduced in the colon (duodenum = 1.07 ± 0.45 vs ileum = 0.58 ± 0.13 vs colon = 0.006 ± 0.005 fold, n = 6), suggesting that this location could be less prone to infection by SARS-CoV-2. In contrast, the expression of Tmprss2 was highest in the colon, followed by the duodenum and the terminal ileum (duodenum = 1.03 ± 0.26 vs ileum = 0.4 ± 0.08 vs colon = 3.14 ± 0.27 fold, n = 6; Fig. 1B, right panel). From these results, we speculate that the duodenum would be the most accessible area for the virus to infect the gut.
FIGURE 1.
Ace2 and Tmprss2 expression in mouse gut and IECs in steady-state conditions. mRNA expression of Ace2 and Tmprss2 was analyzed by qPCR in different segments of the gut and in IECs isolated from these segments. A, Whole tissue expression levels of both genes in whole tissue or IECs as related to normal βActin expression. Note that lower ΔCt values mean higher expression levels, closer to those of βActin. N = 6 mice. B, Ace2 and Tmprss2 expression in whole tissue from selected gut locations. N = 6 mice; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 as determined by 1-way ANOVA followed by Tukey posthoc test. C, Ace2 and Tmprss2 in regional IECs normalized to their matched whole tissue expression. N = 5-6 mice; *P < 0.05, ***P < 0.001, and ****P < 0.0001 as determined by 2-way ANOVA followed by Sidak posthoc test. ANOVA indicates analysis of variance; qPCR, quantitative polymerase chain reaction.
Given that both genes are expressed at high levels in alveolar type-2 cells in the lung,31 we next sought to determine whether IECs are the main cells expressing these receptors. For that purpose, we normalized the expression of Ace2 and Tmprss2 in IECs to that of whole tissues. We found that Ace2 was significantly increased in IECs isolated from the duodenum and the ileum (Fig. 1C, left panel), whereas the expression of transcripts for Tmprss2 was significantly increased in the IECs of all the gut segments (Fig. 1C, right panel). These results show that IECs have high production of ACE2 and TMPRSS2 in the gut and may be targets during viral infection.
Inflammation Correlates With a Significant Downregulation of Epithelial Ace2
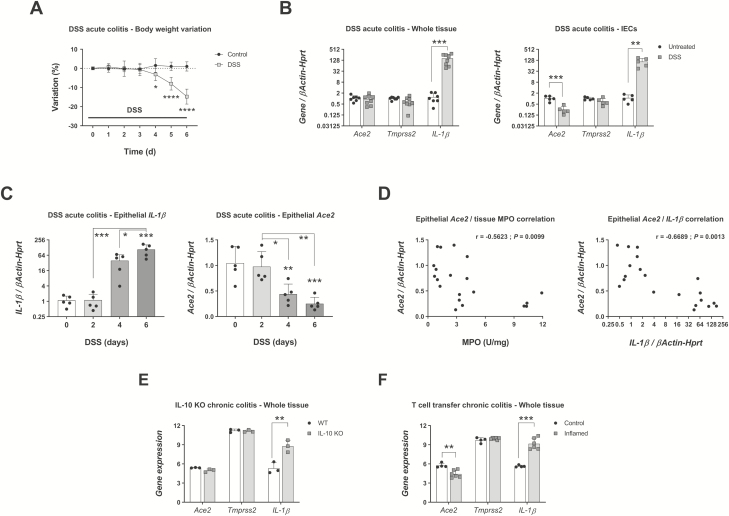
To determine the alterations in the expression of Ace2 and Tmprss2 during inflammation, we analyzed the levels of these transcripts in whole tissues vs isolated colonic IECs collected from C57Bl/6 mice undergoing a model of acute colitis. Administration of the colitogenic agent DSS for 6 days induced inflammation, as shown by marked body weight loss (Fig. 2A) and a significant upregulation of IL-1β in colon tissue (untreated colon = 1.2 ± 0.71 vs inflamed colon = 162.9 ± 99.13 fold, n = 7-8; Fig. 2B, left panel). Histologic characterization of inflammation and quantitative neutrophil infiltration in this experiment have been reported elsewhere.21 In contrast to IL-1β, no changes were seen in the expression of Ace2 and Tmprss2 in the whole colon tissue.
FIGURE 2.
Ace2 and Tmprss2 expression in mouse colon and IECs during inflammation. C57Bl/6 mice treated with 3% DSS for indicated times and expression of Ace2, Tmprss2, and IL-1β transcripts in the colon determined by qPCR. A, Body weight variation. N = 7-8 mice; *P < 0.05, ****P < 0.0001 as determined by 2-way ANOVA followed by Sidak posthoc test. B, Gene expression on day 6 of inflammation in whole colon and IECs isolated from colon. N = 7-8 (whole tissue) or n = 5 (colon IECs); **P < 0.01, ***P < 0.001 as determined by unpaired 2-tailed Student t test. C, Time course expression of IL-1β and Ace2 in colonic IECs isolated on days 0, 2, 4, and 6 of DSS treatment. N = 5 mice; IL-1β on day 6 vs day 0, ***P < 0.001; Ace2 on day 4 vs day 0, **P < 0.01; Ace2 on day 6 vs day 0, ***P < 0.001 as determined by 1-way ANOVA followed by Tukey posthoc test. D, Correlation between epithelial Ace2 expression levels and matched colon determinations of MPO (left panel) or epithelial IL-1β (right panel) on days 0, 2, 4, and 6 of DSS. N = 20 mice; Ace2/MPO, r = –0.5623, **P = 0.0099; Ace2/IL-1β, r = –0.6689, **P = 0.0013. E, Expression of Ace2, Tmprss2, and IL-1β in whole colon tissues of wild type and IL-10 KO mice euthanized after development of spontaneous colitis (GSE107810). N = 3 mice; **FDR = 0.0016. F, Expression of Ace2, Tmprss2, and IL-1β in whole colon tissues of recombination-activating gene 1 KO mice injected with regulatory T-cells (control) or naïve T-cells (inflamed). N = 4-6 mice; **FDR = 0.002, ***FDR = 1.95E-5. ANOVA indicates analysis of variance; qPCR, quantitative polymerase chain reaction.
Next, we analyzed isolated colonic IECs and observed a significant upregulation of IL-1β (P < 0.01) that was accompanied by a downregulation of Ace2 (P < 0.001; Fig. 2B, right panel). To improve our understanding of the regulation of Ace2 during inflammation, we analyzed the expression of this gene in colonic IECs isolated on days 0, 2, 4, and 6 of DSS-induced colitis. We previously reported that on day 4 after DSS commencement, mice showed crypt destruction, increased immune cell recruitment, and a trend of increased neutrophil infiltration as determined by MPO activity.21 Subsequently, MPO levels were significantly increased on day 6 after DSS. Here we also report a marked although not significant increase in the expression of epithelial IL-1β on day 4 that became significant by day 6 of DSS administration (day 0 = 1.1 ± 0.5 vs day 4 = 39.38 ± 27.3 vs day 6 = 108.2 ± 54.25 fold, n = 5; Fig. 2C, left panel). In these conditions, Ace2 transcripts were significantly downregulated on days 4 and 6 (day 0 = 1.05 ± 0.33 vs day 4 = 0.44 ± 0.2 vs day 6 = 0.25 ± 0.13 fold, n = 5; Fig. 2C, right panel), suggesting that the onset of inflammation leads to a reduction in the expression of this gene in colonic IECs. To corroborate these findings, we analyzed the correlation between the epithelial expression of Ace2 and the MPO values of matched whole colon tissue. We observed a significant negative correlation (r = –0.5623, P = 0.0099) between both parameters (Fig. 2D, left panel), indicating that higher amounts of neutrophil infiltration are associated with reduced expression of Ace2. Furthermore, the correlation between Ace2 and IL-1β expressed in IECs was more significantly negative (r = –0.6689, P = 0.0013; Fig. 2D, right panel), suggesting that the inflammation status in the IECs determines the transcriptomic profile of Ace2. These results suggest that active inflammation reduces the epithelial expression of Ace2 although whole tissue levels remain similar.
To investigate whether these findings are shared with chronic mouse models of IBD, we analyzed the expression of Ace2, Tmprss2, and IL-1β in publicly available microarray datasets. In the IL-10 knockout (KO) spontaneous model of colitis, the whole colon tissue of IL-10 KO mice euthanized after development of colitis23 showed a significant increase in the expression of IL-1β [false discovery rate (FDR) = 0.0016], whereas Ace2 and Tmprss2 remained stable (Fig. 2E). In the T-cell transfer model of chronic colitis,24 inflammation was accompanied by upregulation of IL-1β (FDR = 1.95E-5) and downregulation of Ace2 (control = 5.75 ± 0.35 vs inflamed = 4.43 ± 0.57 arbitrary units, n = 4-6; Fig. 2F) in whole colon tissue. Taken together, our results in 3 different mouse models of IBD indicate that both acute and chronic inflammation are associated with a stable or decreased expression of Ace2 and Tmprss2 in colonic tissue.
Expression of ACE2 and TMPRSS2 Is Not Increased in Colonic Mucosa of Patients With IBD
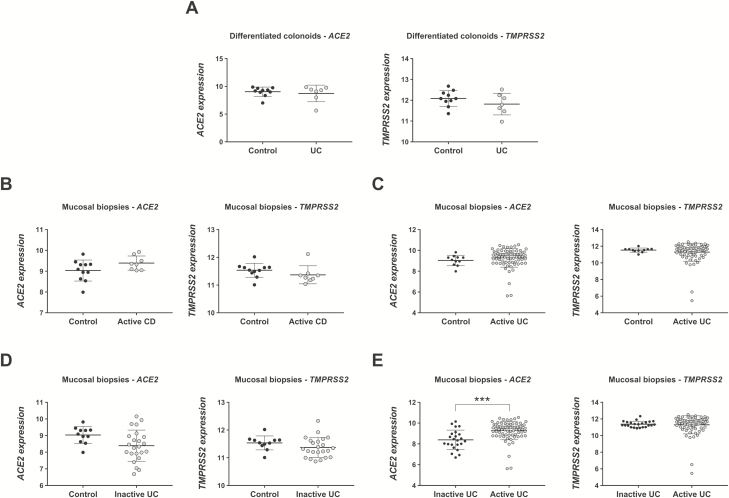
We then sought to determine whether patients with IBD show alterations in the expression of SARS-CoV-2 entry genes. Given that intestinal organoids retain not only genetic32, 33 but also functional,34 epigenetic,35 and transcriptomic26 features of their original tissue, we reasoned that their transcriptional profiles can reliably represent those of IECs in patients with IBD. Therefore, we analyzed a gene expression dataset consisting of a microarray performed on differentiated colon organoids developed from 8 UC patients and 11 control patients without IBD (GSE7591626). Previous analyses in this dataset showed that differentiated colonoids from patients with UC have a transcriptomic signature that is different from that of non-IBD colonoids even in steady-state conditions.26 However, the expression of ACE2 and TMPRSS2 was similar between differentiated colonoids of patients with UC and control patients here (Fig. 3A), suggesting that colon IECs from patients with UC in baseline conditions are not at higher risk of inducing SARS-CoV-2 tropism.
FIGURE 3.
ACE2 and TMPRSS2 expression in human IECs and mucosal biopsies. The expression of both SARS-CoV-2 entry genes was analyzed in 2 gene datasets. A, Expression of ACE2 and TMPRSS2 in differentiated human colonoids from control patients (n = 10) and patients with UC (n = 7) (GSE75916). B-E, Expression of ACE2 and TMPRSS2 in mucosal biopsies from control patients (n = 11), patients with active CD (n = 8), patients with active UC (n = 74), and patients with inactive UC (n = 23) (GSE59071); ***FDR = 8.63E-5.
To characterize the expression of ACE2 and TMPRSS2 during inflammation, we analyzed a second microarray dataset consisting of colonic biopsies taken from the edge of inflamed ulcers in a cohort of 97 patients with UC (74 with active disease, 23 with inactive disease), 8 patients with CD, and 11 control patients screened for polyps (GSE5907125). Similar to our results in whole colon tissue from inflamed mice, the expression of ACE2 and TMPRSS2 in mucosal biopsies was similar between control patients and patients with active UC or CD (Figs. 3B-D). Nevertheless, we observed a modestly higher expression of ACE2 in active UC when compared to inactive UC (active UC = 9.46 ± 0.89 vs inactive UC = 8.35 ± 0.94 arbitrary units, FDR = 8.63E-5; Fig. 3E, left panel). Taken together with our mouse model observations, these data show that expression of both genes is stable in colon tissue during inflammation.
ACE2 and TMPRSS2 Expression Is Stable During Inflammation in CD11b-Enriched Cells
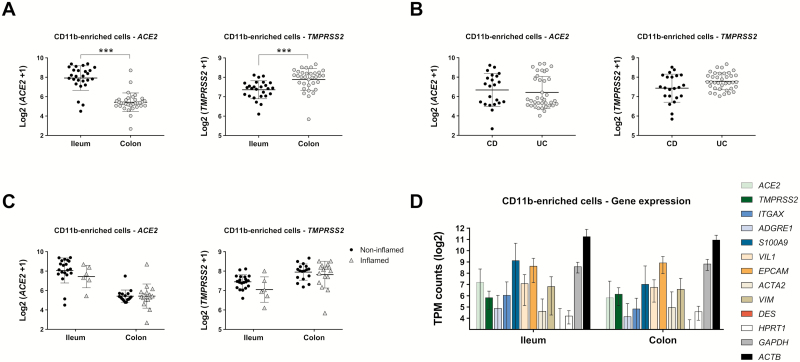
Our expression data in the mouse model and human microarray data indicated that ACE2 and TMPRSS2 are expressed in the gut and may be regulated by inflammation. Given those findings, we examined isolated CD11b+ phagocytic cells from biopsy specimens taken from patients with well-characterized CD and UC. The CD11b+ cells contain a population of neutrophils, macrophages, and dendritic cells in the gut and form part of the first line of defense against pathogens crossing the epithelial barrier. These cells were isolated using magnetic sorting and had a purity of about 70%, with other cell types, including IECs, making up the rest.36 The RNA of CD11b-enriched cells was sequenced and analyzed for the expression of SARS-CoV-2 entry genes. We found that ACE2 expression was higher in the ileum (ileum = 7.92 ± 1.27 vs colon = 5.42 ± 0.96 log2 [ACE2 + 1], n = 26-33; Fig. 4A, left panel) whereas TMPRSS2 was significantly more expressed in the colon (ileum = 7.36 ± 0.46 vs colon = 7.89 ± 0.56 log2 (TMPRSS + 1), n = 26-33; Fig. 4A, right panel), confirming our results in mice. Additional comparisons across all samples from CD vs UC patients showed no significant differences (Fig. 4B). We also looked at the effect of inflammation in the ileum and colon regardless of disease type. We used the strictest definition of inflammation based on the microscopic appearance as evaluated by dedicated gastrointestinal pathologists. We did not find significant differences in the expression of these 2 genes in the presence or absence of inflammation in the ileum or the colon (Fig. 4C). These results support the notion that our CD11b-enriched population of cells express ACE2 and TMPRSS2.
FIGURE 4.
ACE2 and TMPRSS2 expression in CD11b-enriched cell isolates from biopsies of patients with IBD. Cells isolated from mucosal biopsies were analyzed via RNA sequencing and expression of SARS-CoV-2 entry genes was determined. Expression levels shown were analyzed using mixed-effects models to properly account for correlations within patients; these models correct the effect of multiple samples collected from the same patient. A, Differential expression of genes in ileum (n = 26) and colon (n = 33) samples from n = 31 patients; ***P < 0.001. B, Differential expression of genes in CD (n = 22) and UC (n = 37) samples from n = 31 patients. C, Differential expression of genes in different segments of the gut segregated by inflamed and noninflamed pathology. D, TPM of selected genes expressed in CD11b-enriched cell isolates and segregated by gut location; n = 26 (ileum) and n = 33 (colon) samples.
We next wanted to determine the level of expression of ACE2 and TMPRSS2 compared with that of genes with known mucosal functions. To understand the relevance of the gene expression values in the RNA sequencing data, we calculated the TPM for genes of interest. In general, TPM considers both the depth of sequencing and the length of the transcript. We analyzed the TPM for genes expressed in immune populations such as ITGAX in dendritic cells, ADGRE1 in macrophages, and S100A9 in neutrophils; VIL1 and EPCAM in IECs; ACTA2 and VIM in fibroblasts and mesenchymal cells; and DES in muscle cells. We also determined the TPM of structural and enzymatic housekeeping genes such as ACTB, GAPDH, and HPRT1 for comparison. We found that the expression of ACE2 and TMPRSS2 was comparable to the expression of genes involved in immune, epithelial, and mesenchymal processes and of housekeeping genes such as HPRT1 (Fig. 4D). These data suggest that the expression of ACE2 and TMPRSS2 in CD11b-enriched cells is comparable to that of other functional mucosal-expressed genes.
Medication Regimen and Duration of Disease Can Alter the Expression of ACE2 and TMPRSS2 in CD11b-Enriched Cells
Thus far, older male patients have been more likely to succumb to COVID-19. We analyzed the expression of ACE2 and TMPRSS2 stratified by age and gender and saw no significant differences (Supplementary Fig. 1). We also found no alterations in gene expression based on race/ethnicity, although our patients were primarily non-Hispanic whites and Hispanic whites.
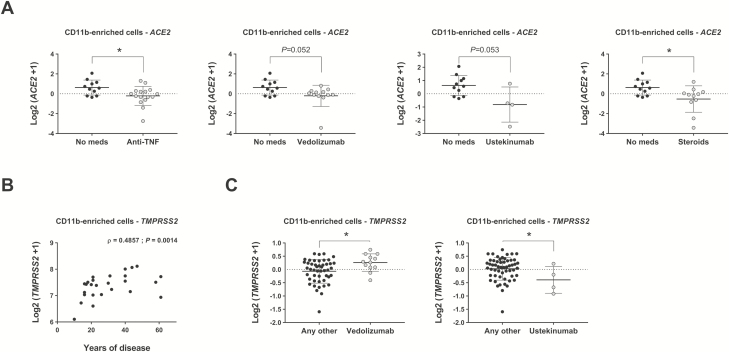
Patients with IBD are on a variety of immune-modulating medications, which may be linked to alterations in ACE2 and TMPRSS2 expression. Therefore, we next compared the expression of ACE2 and TMPRSS2 in CD11b-enriched isolates stratified by patient medication. Using a mixed-effects model considering individual correlations, we found that patients on anti-tumor necrosis factor drugs, vedolizumab, ustekinumab, and steroids had lower levels of ACE2 compared with patients on no medications (Fig. 5A). Furthermore, this model identified a significant positive correlation between the expression of TMPRSS2 and the years with disease, suggesting that this gene increases with IBD over time (Fig. 5B). Comparison of residuals also showed an increased expression of TMPRSS2 in patients treated with vedolizumab when compared with patients taking any other drugs (Fig. 5C, left panel). Finally, we also found a downregulation of TMPRSS2 in patients treated with ustekinumab when compared with those taking any other medications (Fig. 5C, right panel). Taken together, we can conclude that medications used to treat IBD do not adversely affect expression of the main viral target, ACE2, but years of IBD diagnosis can affect gene expression in the intestine and should be accounted for.
FIGURE 5.
Effect of medication and years of disease in expression of ACE2 and TMPRSS2. RNA sequencing of CD11b-enriched cells isolated from patients with IBD analyzed for the expression of ACE2 and TMPRSS2 using mixed-effects models (see “Methods” section for details). All medications numbered in Table 1 were analyzed; only significant comparisons are represented. A, Expression of ACE2 in patients taking anti-TNF drugs, vedolizumab, ustekinumab, or steroids vs no medication; *P < 0.05. B, Expression of TMPRSS2 correlated to duration of disease using Spearman correlation; ρ = 0.4857, **P = 0.0014. C, Expression of TMPRSS2 in patients taking vedolizumab and ustekinumab compared with patients receiving any other medication; *P < 0.05.
ACE2 and TMPRSS2 Expression Is Minimal in Effector and Regulatory T-Cells in the Intestine and in the Periphery of Patients With IBD
Peripheral blood T-cells in patients infected with COVID-19 show functional alterations that suggest active participation and even exhaustion during the antiviral immune response.37 We had the opportunity to assess whether T-cells can potentially mediate viral entry via ACE2 and TMPRSS2. Blood samples and matched tissue samples were obtained in patients with CD and UC. Effector T-cells (CD3+ CD4+ CD25- CD45RO+ CCR7-) and regulatory T-cells (CD3+ CD4+ CD25+ CD45RO+ CCR7-) were isolated from tissue biopsies and from blood. In general, ACE2 and TMPRSS2 were expressed at undetectable levels in effector T-cells and regulatory T-cells in blood. We found that ACE2 was detectable in blood-derived effector T-cells and regulatory T cells in 2/62 and 5/41 samples, respectively (Supplementary Table 3). In biopsy samples, the expression of ACE2 was seen in memory T-cells and regulatory T-cells in 5/47 and 3/20 samples, respectively. All expression levels were low. By contrast, TMPRSS2 was more highly expressed in lamina propria effector T-cells and regulatory T-cells (23/47 and 13/20, respectively; Supplementary Table 3). These data led us to conclude that the T-cell compartment in the intestine is less susceptible as a target for viral entry.
Vedolizumab works by inhibiting the trafficking of α4β7+ T cells to the gut. Therefore, we wanted to understand whether vedolizumab was associated with an increase in peripheral T-cells expressing ACE2 and TMPRSS2. To that end, we examined the effect of vedolizumab treatment on peripheral blood T-cells 14-16 weeks after initiating vedolizumab treatment. We found no increase in the expression of ACE2 and TMPRSS2 in peripheral T-cells (Supplementary Table 4). These data provide further reassurance that biologic therapy is likely to be safe in patients with IBD in the face of the COVID-19 epidemic.
DISCUSSION
At this time, patients with IBD and their physicians are unsure of what to do in the face of the COVID-19 pandemic. In particular, they are juggling the questions of which medications are safe for patients with IBD to continue, when to start new medications, and whether or not it is better to treat active inflammation or wait until after the pandemic is over. Furthermore, SARS-CoV-2 RNA has been identified in the stool of 53% of patients hospitalized with COVID-19 and viral nucleocapsid positive staining has been found in biopsies of the stomach, duodenum, and rectum,14 suggesting that the virus can infect the gut. Consequently, this information raises additional concerns about the risks of performing endoscopy, leading to the cancellation of elective endoscopic procedures during this time. The real patient data to answer these questions will only be available with time—but that is time we do not have.
The receptor ACE2 and the serine protease TMPRSS2 are essential for the interaction with the viral spike protein S and for membrane fusion resulting in viral entry to host cells.8, 9, 38 Whereas little is known about the biological function and regulation of TMPRSS2, the expression of ACE2 in kidney epithelial cells is downregulated in vitro by cytokines such as IL-4 and interferon-γ 39. These findings suggest that inflammation may modulate the expression of these proteins. Therefore, we undertook this study to determine their expression levels in the gut of patients with IBD as a surrogate for whether patients with IBD are at greater risk of viral infection— because of either local inflammation or medications. We have addressed these questions using a combination of publicly available datasets from IBD samples, our own well-annotated samples from patients with IBD who have undergone RNA sequencing, and 3 animal models of IBD. We can say based on both the animal and human data that ACE2 and TMPRSS2 are expressed in the ileum and the colon. Although we do not have the corresponding human data, the duodenum seems to have the highest expression of Ace2 and Tmprss2 in mice, suggesting that it may be the most accessible location for viral entry in the event of fecal-oral transmission.
Because ACE2 and TMPRSS2 are expressed in lung epithelial cells,31 we sought to further explore their expression in IECs in homeostatic (noninflamed) conditions and during inflammation. We found that overall, ACE2 was more highly expressed in the ileum than in the colon and that TMPRSS2 was the mirror image and more highly expressed in the colon than in the ileum. Under homeostatic conditions, mouse IECs had a high expression of Ace2 and Tmprss2 when compared to whole tissue. In the DSS acute colitis model, we found that isolated IECs very clearly decreased the expression of Ace2 in the face of inflammation. However, the whole intestinal mucosa in both patients and mice had similar expression of ACE2 and TMPRSS2 in the presence or absence of inflammation. The only exception was UC; there was a slightly higher level of ACE2 expression in active UC than in inactive UC. In innate immune cells isolated from biopsies of patients with CD and UC, we also did not see a change with inflammation in the expression of these genes. These data should offer reassurance that our patients are not simply more susceptible to COVID-19 infection with respect to the gastrointestinal tract.
Stratifying patients by medication showed that the medications used to treat IBD did not increase expression of these genes, and there was a trend for a decrease in the expression of ACE2 in patients on anti-tumor necrosis factor drugs, vedolizumab, ustekinumab, and steroids compared to those taking no medications. Obviously, these findings do not take into account the systemic effects of these medications in the face of COVID-19, but at least they do not suggest that they lead to global changes in the gene expression of these critical players.
We wanted to have a sense of how the expression of ACE2 and TMPRSS2 compares with that of other relevant genes in the intestinal epithelium and in the lamina propria to gain insight into whether the level of expression can be interpreted as biologically relevant. Indeed, we found that the expression level based on transcripts per million was in the midrange of the expression of some typical epithelial and immune genes that would otherwise be expressed in the gut. Our study cannot conclude whether ACE2 and TMPRSS2 protein expression tracks with its transcriptional level or whether functionally the expression permits viral entry. We also had a mixture of CD11b+ cells (approximately 70.7%) and IECs and fibroblasts. In situ hybridization staining of ACE2 mRNA in the Human Protein Atlas12 database has shown a clear epithelial pattern, which suggests that the expression levels we report were likely a hybrid of innate immune cells and IECs, with a higher contribution of IECs. Nevertheless, CD11b+ cells and IECs are on the front lines of protection against pathogens and may be coopted to permit viral entry.
We had the opportunity to look at T-cell expression of these 2 viral entry proteins and found that in the peripheral T-cells, neither effector T-cells nor regulatory T-cells expressed ACE2 or TMPRSS2. In the intestine, there was some expression of TMPRSS2 in both effector T-cells and regulatory T-cells, but again very little ACE2, and the levels of expression of these genes were much lower than what we saw in the CD11b-enriched population. This is consistent with a recent report that found low ACE2 expression in lymphocyte populations in the oral mucosa.40 Note that we studied peripheral blood T-cells from patients after induction of vedolizumab treatment. Vedolizumab results in an increase in α4β7+ T-cells in the periphery after therapy. We did not see a shift in the peripheral T-cells expressing these genes. Taken together, our findings suggest that it would be difficult for T-cells to mediate viral entry and replication in IBD.
In summary, we show that ACE2 and TMPRSS2 are indeed expressed in the mucosa of the intestine, making the gut a plausible target for viral infection, and that the expression of these genes is not altered in patients with IBD. Our study has implications for the management of IBD and even for procedures such as fecal microbial transplantation for Clostridium difficile and other diseases. It highlights that the IECs and perhaps the innate immune cells in the ileum and colon can permit viral entry. However, there was no untoward effect of inflammation or of the medications used to treat patients with IBD, which should offer reassurance that continuing the medications is still best to prevent flares of disease and the requirement for potential hospitalization.
CONCLUSIONS
Our data in animal models and samples of patients with IBD indicate that neither inflammation nor IBD medications increase the expression of the viral entry molecules ACE2 and TMPRSS2 in the gut, supporting the notion that patients with IBD are not at increased risk of gastrointestinal infection by SARS-CoV-2.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the Flow Cytometry Shared Resource staff at the University of Miami for participating in the design of the flow cytometry panel used for T-cell characterization and for performing the cell sorting. We also acknowledge the Onco-Genomics Shared Resource staff at the University of Miami and The Scripps Research Institute Genomics staff at Scripps Research for performing the RNA sequencing of CD11b-enriched and T-cell samples, respectively.
Supported by: This work was supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (grant number R01DK09907), Takeda Pharmaceutical, U.S.A. (grant number IISR-2014-1000892), Inc., Pfizer (ASPIRE award number WI227247), The Micky & Madeleine Arison Family Foundation Crohn’s & Colitis Discovery Laboratory, and the Martin Kalser Chair (to coauthor MTA).
Conflicts of interest: AR has served as a consultant to GenapSys Inc. AD has served as a scientific advisory board member for GI Health Foundation, has a funded grant from Takeda, and received honoraria from AMBIM. DHK has served as a scientific advisory board member for AbbVie and as a consultant for Cleveland Clinic and currently serves as a trainer or lecturer for PRIME Continuing Medical Education and The Academy for Continued Healthcare Learning. MTA has served as a scientific advisory board member for Boehringer Ingelheim Pharmaceuticals, Gilead, AbbVie, Seres Therapeutics, Shire, and Landos Biopharma; serves as a trainer or lecturer for Imedex, Focus Medical Communications, and Cornerstones Health, Inc.; has served as a consultant for Ferring Pharmaceuticals, Allergan, Amgen, Celltrion Healthcare CO, Millennium Pharmaceuticals, Theravance Biopharma Inc., and UCB Biopharma SRL; and has funded projects by Pfizer, Prometheus Laboratories, and Takeda Pharmaceuticals. OMD received honoraria from Pfizer and has a funded grant from Pfizer. This does not alter the authors’ adherence to the journal’s policies on sharing data and materials. All other authors declare no conflict of interest.
REFERENCES
- 1. Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu N, Zhang D, Wang W, et al. ; China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020. doi: 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. WHO. Coronavirus disease 2019 (COVID-19) Situation Report- 74, 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200403-sitrep-74-covid-19-mp.pdf?sfvrsn=4e043d03_14. [Google Scholar]
- 5. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li W, Moore MJ, Vasilieva N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glowacka I, Bertram S, Müller MA, et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zou X, Chen K, Zou J, et al. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qi F, Qian S, Zhang S, et al. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. 2020. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 13. Leung WK, To KF, Chan PK, et al. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xiao F, Tang M, Zheng X, et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hindson J. COVID-19: faecal-oral transmission? Nat Rev Gastroenterol Hepatol 2020. doi: 10.1038/s41575-020-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu Y, Li X, Zhu B, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nature Med. 2020; 26:502–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wong SH, Lui RN, Sung JJ. Covid-19 and the digestive system. J Gastroenterol Hepatol. 2020. doi: 10.1111/jgh.15047. [DOI] [PubMed] [Google Scholar]
- 19. Fernández-Ruiz M. Assessment of latent infections in patients receiving biological therapies. Rev Esp Quimioter. 2019;32 Suppl 2:63–68. [PMC free article] [PubMed] [Google Scholar]
- 20. Wang W, He J, Lie P, et al. The definition and risks of cytokine release syndrome-like in 11 COVID-19-infected pneumonia critically ill patients: disease characteristics and retrospective analysis. medRxiv 2020. doi: 10.1101/2020.02.26.20026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burgueño JF, Fritsch J, Santander AM, et al. Intestinal epithelial cells respond to chronic inflammation and dysbiosis by synthesizing H2O2. Front Physiol. 2019;10:1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burgueño JF, Barba A, Eyre E, et al. TLR2 and TLR9 modulate enteric nervous system inflammatory responses to lipopolysaccharide. J Neuroinflammation. 2016;13:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ahn J, Son S, Oliveira SC, Barber GN. STING-dependent signaling underlies IL-10 controlled inflammatory colitis. Cell Rep. 2017;21:3873–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lyons J, Ghazi PC, Starchenko A, et al. The colonic epithelium plays an active role in promoting colitis by shaping the tissue cytokine profile. PLoS Biol. 2018;16:e2002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vanhove W, Peeters PM, Staelens D, et al. Strong upregulation of AIM2 and IFI16 inflammasomes in the mucosa of patients with active inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:2673–2682. [DOI] [PubMed] [Google Scholar]
- 26. Dotti I, Mora-Buch R, Ferrer-Picón E, et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 2017;66:2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. [DOI] [PubMed] [Google Scholar]
- 29. Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hamming I, Timens W, Bulthuis ML, et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fujii M, Shimokawa M, Date S, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18:827–838. [DOI] [PubMed] [Google Scholar]
- 33. Suzuki K, Murano T, Shimizu H, et al. Single cell analysis of Crohn’s disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties. J Gastroenterol. 2018;53:1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Middendorp S, Schneeberger K, Wiegerinck CL, et al. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells. 2014;32:1083–1091. [DOI] [PubMed] [Google Scholar]
- 35. Kraiczy J, Nayak KM, Howell KJ, et al. DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development. Gut. 2019;68:49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dheer R, Davies JM, Quintero MA, et al. Microbial signatures and innate immune gene expression in lamina propria phagocytes of inflammatory bowel disease patients. Cell Mol Gastroenterol Hepatol. 2020;9:387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng HY, Zhang M, Yang CX, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith DM, Waite M. Phosphatidylinositol hydrolysis by phospholipase A2 and C activities in human peripheral blood neutrophils. J Leukoc Biol. 1992;52:670–678. [DOI] [PubMed] [Google Scholar]
- 39. de Lang A, Osterhaus AD, Haagmans BL. Interferon-gamma and interleukin-4 downregulate expression of the SARS coronavirus receptor ACE2 in Vero E6 cells. Virology. 2006;353:474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.