Abstract
Background
The World Health Organization characterizes novel coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), as a pandemic. Here, we investigated the clinical, cytokine levels; T-cell proportion; and related gene expression occurring in patients with COVID-19 on admission and after initial treatment.
Methods
Eleven patients diagnosed with COVID-19 with similar initial treatment regimens were enrolled in the hospital. Plasma cytokine, peripheral T cell proportions, and microfluidic quantitative polymerase chain reaction analyses for gene expression were conducted.
Results
Five patients with mild and 6 with severe disease were included. Cough and fever were the primary symptoms in the 11 COVID-19 cases. Older age, higher neutrophil count, and higher C-reactive protein levels were found in severe cases. IL-10 level significantly varied with disease progression and treatment. Decreased T-cell proportions were observed in patients with COVID-19, especially in severe cases, and all were returned to normal in patients with mild disease after initial treatment, but only CD4+ T cells returned to normal in severe cases. The number of differentially expressed genes (DEGs) increased with the disease progression, and decreased after initial treatment. All downregulated DEGs in severe cases mainly involved Th17-cell differentiation, cytokine-mediated signaling pathways, and T-cell activation. After initial treatment in severe cases, MAP2K7 and SOS1 were upregulated relative to that on admission.
Conclusions
Our findings show that a decreased T-cell proportion with downregulated gene expression related to T-cell activation and differentiation occurred in patients with severe COVID-19, which may help to provide effective treatment strategies for COVID-19.
Keywords: COVID-2019, PBMC, immune response, cytokine, gene expression
Suppressed T-cell immune response and decreased T cells occurred in patients with COVID-19 related to downregulated gene expression involved in T-cell activation and differentiation, especially in severe disease, against SARS-CoV-2 infection and at the early period of treatment.
The rapid outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has grown into a global public health emergency of international concern since November 2019, with reported cases of COVID-19 around the world surpassing 100 000 as of 7 March 2020 [1]. The clinical characteristics of COVID-19 in China and the epidemiologic features of the outbreak in Wuhan and Hubei Province were reported, which helped identify the severity of the disease [2–5].
SARS-CoV-2, a member of Beta-CoV lineage B, was first identified in Wuhan, and can cause clusters of severe respiratory illness [6]. During viral infection, immune responses are triggered by the host against the virus. Many respiratory viruses suppress the innate immune response, providing opportunities for efficient virus replication and the establishment of infections [7]. The interaction of the virus with the cells results in a large number of immune mediators being produced. In response to SARS-CoV-2 infection, infected cells promote the secretion of large amounts of chemokines and cytokines, which have been reported in severe cases of COVID-19 [3]. Similar to severe acute respiratory syndrome (SARS) and Middle East Respiratory syndrome (MERS), the presence of a “cytokine storm” may play a major role in the pathogenesis, and causes inflammatory-induced lung injury and other complications [8].
In Beijing, the capital of China, a total of 428 patients with COVID-19 were confirmed as of 7 March 2020 [9]. Our hospital, a special hospital for infectious diseases, provides the first line of confirmation, diagnosis, and treatment for patients with COVID-19. Despite several studies that reported clinical and plasma cytokine features of COVID-19, only limited information is available on the host innate immune status of SARS-CoV-2–infected patients. The changes in immune-related gene expression levels during SARS-CoV-2 infection are still unclear. Here, we comprehensively evaluated the characteristics of 11 patients with COVID-19 admitted to Beijing YouAn Hospital. We aimed to compare the clinical, cytokine levels and immune-related gene expression characteristics between different clinical stages. Our data provide basic information toward understanding the role of immune responses on the disease process of COVID-19.
METHODS
Patients and Clinical Samples
This study was approved by the Ethics Committee of Beijing YouAn Hospital. Eleven patients diagnosed with COVID-19 with similar initial regimens were enrolled from 31 January to 7 February 2020 in the hospital. All enrolled patients were confirmed to be positive for 2019-nCoV nucleic acid by real-time polymerase chain reaction (PCR). Patients were diagnosed and treated according to the guidance of National Health and Health Commission of China (version 7) and divided into 2 groups, severe or critically severe type (group A) and mild or common type (group B).
All the clinical data on demographics, symptoms, and laboratory results were retrospectively reviewed. Peripheral blood mononuclear cells (PBMCs) and the corresponding serum samples at admission (T1) and after 5 initial treatment days (T2) were collected for CyTOF, microfluidic quantitative PCR (qPCR), and cytokine detection, respectively. The group set as group A1 (group A at admission), group A2 (group A after 5 treatment days), group B1 (group B at admission) and group B2 (group B after 5 treatment days).
Cytokines Detected With Luminex Kits
Each serum sample was analyzed for cytokine levels by a Luminex bead-based MILLIPLEX assay using human cytokine panel kit (Millipore, USA) with a FlexMAP3D (Luminex) platform, and cytokine production data were analyzed using the xPONENT software following the manufacturer’s instructions [10].
RNA Extraction and cDNA Synthesis
Total RNA was extracted from PBMCs using the RNeasy Plus Micro Kit (Qiagen, Germany) according to the manufacturer’s instructions. The quantity of total RNA was measured by Qubit (Thermo Fisher, USA). cDNAs were synthesized from and prepared using the SuperScript II first-strand cDNA synthesis kit (Thermo Fisher, USA) according to the manufacturer’s protocol.
Microfluidic qPCR Preparation and Gene Expression Analyses
cDNA purification and dilution according to the instructions as described by Fluidigm (Fluidigm, USA) using probe-based TaqMan assays use primers to detect sequence-specific cDNA probes. A total of 108 specific target genes are shown in Supplementary Table 1. Briefly, the gene expression assay was performed with the GE 96.96 integrated fluidic circuits (IFCs) using the Juno system for IFC preparation and Biomark HD for qPCR thermal cycling and data acquisition. Each value obtained is an average of 2 independent biological replicates, and the experiment was repeated 2 times for some genes as a batch effect correction. As described before, the GAPDH gene expression levels were used for data standardization, and the average gene expression levels of 7 normal PBMCs (NC group) were used as the sample reference. The fold-change (FC) was calculated using the 2−ΔΔCt method.
Bioinformatics Analysis
Differentially expressed genes (DEGs) were obtained using the MeV tool, to compare the FC between the NC group and group A or group B using a Mann-Whitney test with statistical significance considered at a 2-sided P value less than .05. Further DEGs were hierarchically clustered in the MeV tool. Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of differentially expressed genes were implemented on the metascape [11]. Identified GO terms and KEGG pathways with a corrected P < .05 were considered significantly enriched. Protein–protein interaction (PPI) analysis of DEGs was based on tissue-specific PPI data from the DifferentialNet database, and the images were examined using the NetworkAnalyst platform [12].
Mass Cytometry Antibody Staining of the CD45 Barcode and Data Acquisition in Helios
All samples were incubated with cisplatin (195-Pt) and then quenched with cell staining buffer (Fluidigm) for viability evaluation by mass cytometry. A CD45 barcode was applied to minimize intersample staining variation, and stained with CD45 100 antibodies labeled with different metals before being pooled together. The original flow cytometry standard data were normalized, and the results of each run were collected, then analyzed with R and PhenoGraph.
Statistical Analysis
Comparisons of differences between group A and group B were calculated using Mann-Whitney, chi-square, or Fisher’s exact test. Statistical significance was set at P < .05 for 2-sided tests. Correlation analyses were performed using a Spearman’s rank test. All statistical analyses were performed using the SPSS software package (version 17.0; SPSS Inc, USA).
RESULTS
Patient Demographics and Baseline Characteristics
Eleven patients with COVID-2019 were included in this study: 6 diagnosed as having severe or critically severe disease (group A) and 5 as having mild or common disease (group B) on admission. Demographic and clinical characteristics of the patients with COVID-2019 on admission are shown in Table 1. With regard to basic characteristics, the median age in group A (67 years) was significantly older than that in group B (45 years) (P = .043). Gender, occupation, source of infection, symptoms, and comorbidities were not significantly different between group A and group B. Overall, the primary symptoms in the 11 patients with COVID-2019 were cough (9/11, 81.82%) and fever (8/11, 72.73%). In the blood laboratory findings, we noticed that most of the results were not significantly different between the 2 groups, except for neutrophil count and albumin and C-reactive protein levels. A higher neutrophil count (median, 6.135 group A vs 2.34 group B; P = .045) and higher C-reactive protein level (median, 78.3 group A vs 13.7 group B; P = .006) were found in group A than in group B, while a relative lower level of albumin was more prone to be observed in group A (median, 30.35 group A vs 39.1 group B; P = .018). The blood laboratory results of all the patients except for 2 patients from group B were compared within groups, and no significant differences were found between groups A1 and A2 or groups B1 and B2 after 5 days of initial treatment (Supplementary Table 2).
Table 1.
Demographic and Clinical Characteristics of Patients With COVID-19 on Admission
| Group A (n = 6) | Group B (n = 5) | P | All Patients (n = 11) | |
|---|---|---|---|---|
| Characteristics | ||||
| Age, median (range),a,b years | 67 (46–78) | 45(29–65) | .043 | 63 (29–78) |
| Male,c n (%) | 3 (50) | 2 (40) | 1.00 | 5 (45.45) |
| Occupation, retired and farmer,c n (%) | 5 (83.33) | 1 (20) | .080 | 6 (54.55) |
| Close-contact exposure,c n (%) | .545 | |||
| Native | 3 (50) | 1 (20) | 4 (36.36) | |
| Foreign provinces | 3 (50) | 4 (80) | 7 (63.64) | |
| Symptoms,c n (%) | ||||
| Fever | 5 (83.33) | 3 (60) | .545 | 8 (72.73) |
| Cough | 5 (83.33) | 4 (80) | 1.00 | 9 (81.82) |
| Shortness of breath | 4 (66.67) | 0 (0) | .061 | 4 (36.36) |
| Sore throat | 1 (16.67) | 1 (20) | 1.00 | 2 (18.18) |
| Nausea | 2 (33.33) | 0 (0) | .455 | 2 (18.18) |
| Headache | 0 (0) | 1 (20) | .455 | 1 (9.09) |
| With chronic medical illness,c,d n (%) | 3 (50) | 0 (0) | .182 | 3 (27.27) |
| Incubation period,a,e days | 5 (3-7) | 4 (2-7) | .514 | 5 (2-7) |
| Initial treatment,c n (%) | ||||
| Oxygen therapy | 5 (83.33) | 2 (40) | .242 | 7 (63.64) |
| Antiviral treatment | 5 (83.33) | 2 (40) | .242 | 7 (63.64) |
| Glucocorticoids | 2 (50) | 0 (0) | .455 | 2 (18.18) |
| Traditional Chinese medicine | 3 (50) | 4 (80) | .545 | 7 (63.64) |
| Blood laboratory findingsa,b | ||||
| Leucocytes, ×109/L | 7.45 (2.2–10.6) | 3.93 (2.8–4.4) | .068 | 4.350 (2.2–10.6) |
| Neutrophils, ×109/L | 6.135 (1.80–9.24) | 2.34 (1.2–2.81) | .045 | 2.81 (1.20–9.24) |
| Lymphocytes, ×109/L | 0.38 (0.20–1.61) | 1.2 (0.95–1.41) | .273 | 0.96 (0.20–1.61) |
| Platelets, ×109/L | 255 (160–501) | 204 (120–257) | .273 | 212 (120–501) |
| Hemoglobin, g/L | 122 (96–163) | 140 (134–145) | .313 | 134 (96–163) |
| Prothrombin time, seconds | 12.55 (11–13) | 12.8 (11–14) | .521 | 12.70 (11–14) |
| Albumin, g/L | 30.35 (28.3–38.6) | 39.1 (36.3–42.2) | .018 | 36.3 (28.3–42.2) |
| Alanine aminotransferase, U/L | 34 (6–159) | 25 (5–114) | .361 | 30 (5–159) |
| Aspartate aminotransferase, U/L | 37.5 (17–97) | 35 (28–64) | .465 | 35 (17–97) |
| Creatine kinase, U/L | 78.5 (40–513) | 98 (39–362) | .583 | 98 (39–513) |
| Lactic acid, mmol/L | 1.98 (0.91–4.01) | 1.39 (0.94–1.79) | .068 | 1.79 (0.91–4.01) |
| Creatinine, μmol/L | 65 (43–121) | 74 (46–81) | .584 | 72 (43–121) |
| Procalcitonin, ng/mL | 0.145 (0.1–0.47) | 0.12 (0.11–0.12) | .111 | 0.12 (0.1–0.47) |
| C-reactive protein, mg/L | 78.3 (27.5–150.3) | 13.7 (1.1–18.4) | .006 | 27.50 (1.1–150.3) |
Abbreviation: COVID-19, coronavirus disease 2019.
aMann-Whitney test.
bMedian (minimum–maximum).
cFisher exact test.
dChronic medical illness, including cardiovascular and cerebrovascular diseases and endocrine system disease.
eIncubation period results indicate the potential earliest date of symptom onset (such as cough and fever).
Interleukin-10 Was Significantly Different Between Group A and Group B on Admission and During Early Treatment
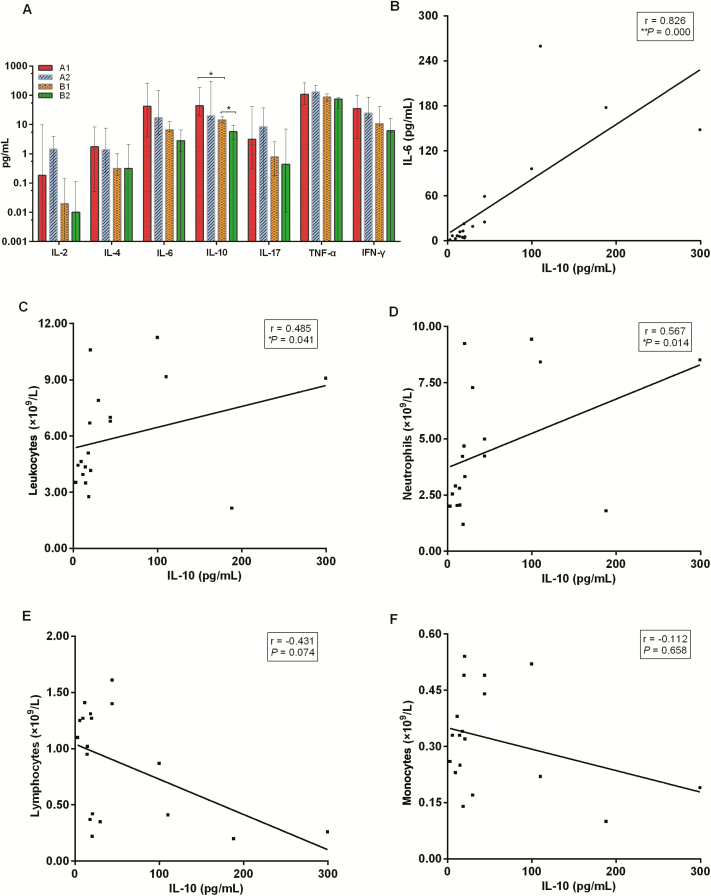
Plasma cytokines including interleukin (IL)-2, IL-4, IL-6, IL-10, IL-17, tumor necrosis factor α (TNF-α), and interferon-γ (IFN-γ) were evaluated in relation to disease severity (A1 vs B1; A2 vs B2) and disease progression over time (A1 vs A2; B1 vs B2) (Figure 1A). IL-10 levels were only significantly different between group A1 versus group B1 (P = .020) and group A1 and group A2 (P = .050). Specifically, IL-10 levels were higher in group A1 (median, 41.91 pg/mL; min-max 20.23–187.98 pg/mL) than in group B1 (median, 14.40 pg/mL; min-max 11.56–18.47 pg/mL) on admission. After 5 days of initial treatment, no significant differences were observed between groups A1 and A2 (median, 20.155 pg/mL; min-max 14.71–299.43 pg/mL) groups, while the levels of IL-10 level in group B2 decreased (median, 5.70 pg/mL; min-max 3.00–9.41 pg/mL) compared with those in group B1 (median, 14.40 pg/mL; min-max 11.56–18.47 pg/mL). The potential correlations between the IL-10 level of all COVID-19 cases and other cytokines or the corresponding immune cell counts were analyzed with the Spearman rank order correlation test (Figure 1 and Supplementary Figure 1). We found that the IL-10 level positively correlated with IL-6 level (Figure 1B) (r = 0.826, P = .000), leukocyte counts (Figure 1C) (r = 0.485, P = .041), or neutrophil counts (Figure 2D) (r = 0.567, P = .014). No significant correlation was observed between the IL-10 levels and lymphocyte counts (Figure 1E) (r = −0.431, P = .074), monocyte counts (Figure 1F) (r = −0.112, P = .658), and the remaining cytokines (Supplementary Figure 1).
Figure 1.
A, Plasma cytokine levels of patients with COVID-19 on admission and after initial treatment. Each bar and error bar represents median value and range. B–F, correlation between IL-10 level of all COVID-19 cases and IL-6 levels (B), the corresponding leukocyte counts (C), neutrophil counts (D), lymphocyte counts (E), and monocyte counts (F). P values (2-sided) and r values are based on Spearman rank test. *P < .05; **P < .01. Abbreviations: COVID-19, coronavirus disease 2019; IFN-γ, interferon-γ; IL, interleukin; TNF-α, tumor necrosis factor α.
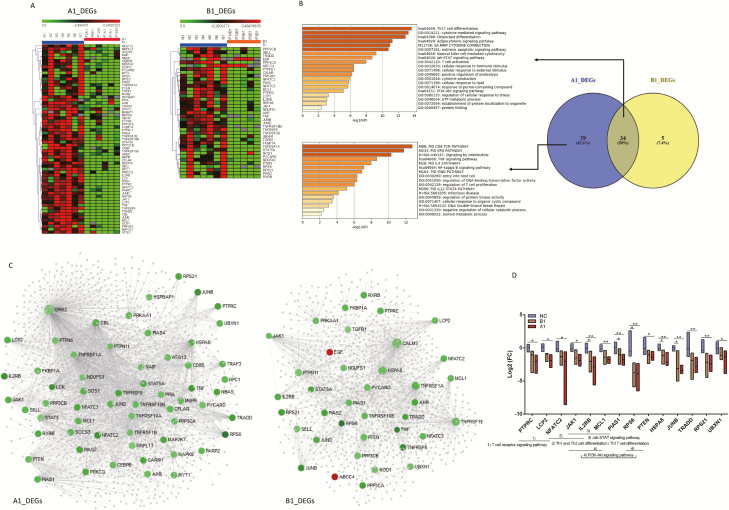
Figure 2.
A, DEGs in patients with COVID-19 on admission. The DEG expression profiles of mRNAs are shown by a heatmap, and the colors represent the FC values. B, Venn diagram for DEGs of A1 and B1 groups and their corresponding enrichment analysis using metascape. All statistically enriched terms were identified, and cumulative hypergeometric P values and enrichment factors were calculated. C, PPI network of DEGs of A1 and B1 groups. The PPI network was drawn using NetworkAnalyst platform based on tissue-specific PPI data from the DifferentialNet database. The red circles represent the upregulated genes, the green circles downregulated genes, and gray circles (relatively small circles) indicate not included in DEGs. D, Kruskal-Wallis 1-way ANOVA was used to screen the 14 significantly different genes among the Normal, A1, and B1 groups. Then the multiple comparisons with all pairwise were performed. Statistical significance was set at a 2-sided P < .05 and adjusted P < .05. *P < .05, **P < .01. Abbreviations: ANOVA, analysis of variance; COVID-19, coronavirus disease 2019; DEG, differentially expressed gene; FC, fold-change; PPI, protein–protein interaction; Th, T-helper.
Numbers of A1_DEGs Were Significantly Higher Than B1_DEGs in COVID-19
We performed microfluidic qPCR to investigate the possible immune-related gene expression change in the 108 target genes in the 11 patients with COVID-19. The DEGs of group A1 and group B1 relative to the normal group at admission were analyzed. Comparisons between the 2 groups were made with the Kruskal–Wallis test. The results showed that A1_DEG numbers (63/108, 58.33%) were significantly greater than B1_DEG numbers (39/108, 36.11%) (P = .001). A total of 63 downregulated mRNAs were differentially expressed in the A1 group relative to the NC group, while B1 group there were 37 downregulated and 2 upregulated mRNA. These differentially expressed mRNAs were used for subsequent analysis. Cluster analysis of the above DEGs was conducted using heatmaps (Figure 2A).
Enrichment Analysis of DEGs Caused by COVID-19
We performed an intersection analysis between the A1_DEGs and B1_DEGs. Thirty-four genes overlapped between the 2 groups, and 29 genes were specific to the A1 group (Figure 2B). Enrichment analyses were respectively performed on significantly differently expressed mRNAs in group A1 versus NC and group B1 versus NC. We found that both group A and group B DEGs enriched involved in T-helper 17 (Th17) cell differentiation, cytokine-mediated signaling pathways, sa matrix metalloproteinases (mmp) cytokine connection, natural killer cell–mediated cytotoxicity, and T-cell activation (Figure 2B), while several enriched terms, such as the CD8 TCR pathway, EPO pathway, signaling by interleukins, the TNF signaling pathway, entry into host cells, and regulation of T-cell proliferation, were closely specific to group A (Figure 2B). The summaries of genes that were involved in the significant enriched pathways are listed in Supplementary Tables 3 and 4.
Protein–Protein Interaction Analysis Revealed the Key Genes Related to Immune Response in COVID-19
The tissue-specific PPI data were collected from the DifferentiaNet database using the NetworkAnalyst platform to screen the key genes involved in immune-related genes of COVID-19. According to the DEGs from the above 2 groups, the corresponding significant genes are mapped to the corresponding molecular interaction database (Figure 2C). The key genes with the top degree (>20) and significant FC (|log2 FC| >1) involved in the pathways interaction network were screened, which may play an important role in COVID-19 (Supplementary Tables 4 and 5). The FCs of 14 key genes were significantly different among NC, A1, and B1 (all P < .05; rank order: NC > B1 > A1); then further multiple comparisons were conducted that showed interleukin 2B (IL-2B), protein inhibitor of activated STAT 1 (PIAS1), ribosomal protein S6 (RPS6), heat shock protein family A member 8 (HSPA8), and JunB proto-oncogene (JUNB) (adjusted P < .05) (Figure 2D).
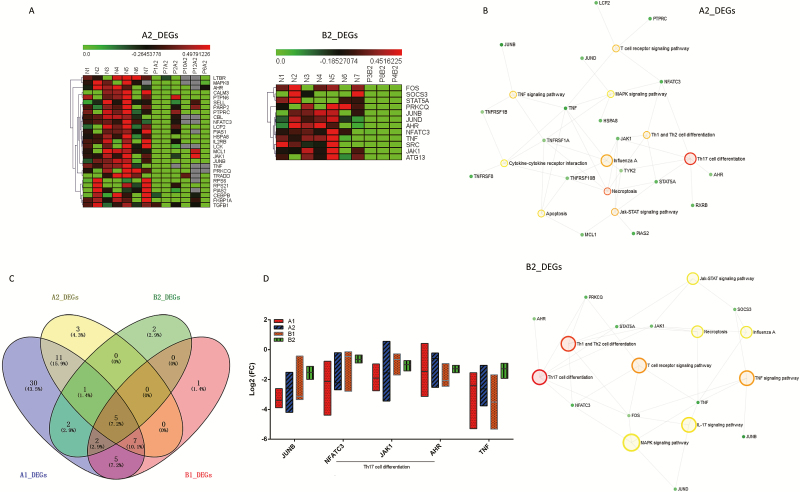
DEG Numbers Decreased in COVID-19 After Initial Treatment
We then focused on the DEG numbers in the 2 groups after initial treatment in the hospital. The DEG numbers in group A2 (27/108, 25%) were significantly decreased compared with the patients in group A1 (P = .000). And a similar trend was found in group B2 (12/108, 11.11%; P = .000). A heatmap shows the cluster analysis for the 27 and 12 DEGs for the A2 and B2 groups (Figure 3A). Then the above 27 and 12 DEGs were conducted KEGG enrichment analysis, respectively. Th17-cell differentiation, Th1- and Th2-cell differentiation, necroptosis, the Jak-STAT signaling pathway, the T-cell receptor signaling pathway, the TNF signaling pathway, and the MAPK signaling pathway were common significantly enriched terms both in the A2 and B2 groups (Figures 3B). Then we compared the differences and similarities between the 2 groups of patients at 2 time points. Specifically, 5 DEGs were common to the A1, B1, A2, and B2 groups (Figure 3C), including JUNB, NFATC3, JAK1, AHR, and TNF, which were variously downregulated relative to the NC group, respectively, although there were no significant differences between the 4 groups (Figure 3D) (all P > .05).
Figure 3.
A, DEGs in patients with COVID-19 after initial treatment. The DEG expression profiles of mRNAs are shown by a heatmap, and the colors represent the FC values. B, All DEGs from groups A2 and B2 were conducted by using KEGG enrichment analysis, respectively. Large hollow circles indicate the pathway name and small solid circles represent the gene. C, Venn diagram for DEGs of the A1, B1, A2, and B2 groups, and 5 DEGs were common to the 4 groups. D, FC of JUNB, NFATC3, JAK1, AHR, and TNF relative to the NC group, respectively, were compared within the 4 groups using Kruskal-Wallis test (all P > .05). Statistical significance was set at a 2-sided P value < .05. Abbreviations: COVID-19, coronavirus disease 2019; DEG, differentially expressed gene; FC, fold-change; Th, T-helper.
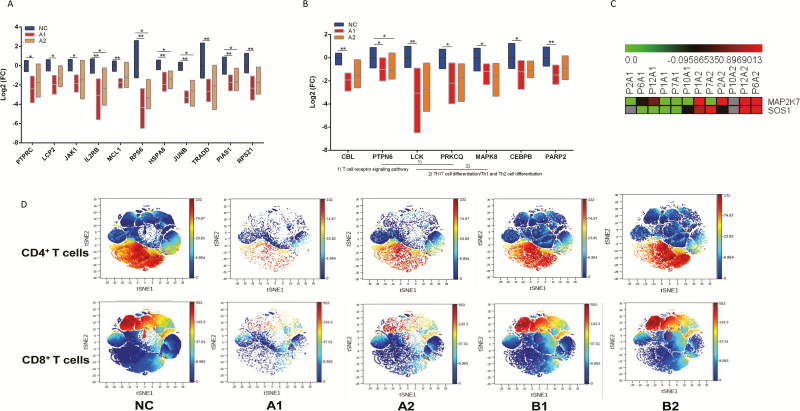
Potential Key Genes Related to COVID-19 Immune Response and T-Cell Responses in Different Clinical Types
Finally, we focused on the DEGs that changed during groups A1 and A2. The 14 key genes from Figure 2D were observed in A2 group, which showed FC of 11 in 14 genes were significantly different among NC, A1, and A2 groups. IL2B, RPS6, HSPA8, JUNB, and PIAS1 were all downregulated in groups A1 and A2 based on the following multiple comparisons (Figure 4A) (adjusted P < .05). The group A–specific DEGs were defined according to get intersection DEGs of A1 and A2 but excluding B1, which showed 7 mRNAs including CBL, PTPN6, LCK, PRKCQ, CEBPB, MAPK8, and PARP2 were considered as severe COVID-19–specific DEGs (Figure 4B) (all P < .05 among NC, A1, and A2 groups), and PTPN6 was downregulated in both groups A1 and A2 that relative to the NC group (adjusted P < .05). After initial treatment, MAP2K7 and SOS1 of group A2 were the only 2 significant DEGs that were upregulated relative to group A1, which are involved in the T-cell-receptor signaling pathway and Fc epsilon RI signaling pathway, which play important roles in disease progression through immune-related signaling pathways (Figure 4C). Using CyTOF mass cytometry (Figure 4D), we found that group A1 showed significantly decreased proportions of T cells (median %, 35 vs 64), CD4+ T cells (median %, 24 vs 35), and CD8+ T cells (median, 11 vs 26) compared with NC, respectively. After initial treatment (A2), the proportion of CD4+ T cells (median %, 34 vs 35, P = 1.000) was similar to that in NC but excluded T cells (median %, 49 vs 64, P = .027) and CD8+ T cells (median %, 14 vs 26, P = .007). For group B1, there were significantly reduced T cells (median %, 46 vs 64, P = .036) and CD8+T cells (median %, 12 vs 26, P = .036) relative to NC, but all returned to levels similar to NC after initial treatment.
Figure 4.
A, FCs of 11 of 14 genes from Figure 2D that were compared between the A1 and A2 groups using Kruskal-Wallis test (all P < .05). Statistical significance was set at a 2-sided P value < .05. B, The FCs of group A–specific DEGs including CBL, PTPN6, LCK, PRKCQ, CEBPB, MAPK8, and PARP2 were compared among NC, A1, and A2 group using Kruskal-Wallis test (all P < .05). Statistical significance was set at a 2-sided P value < .05. C, The DEG expression profiles of mRNAs are shown by a heatmap, and the colors represent the FC values. DEGs of group A2 relative to A1 are shown by a heatmap, including MAP2K7 and SOS1, which were upregulated in group A2 after initial treatment. D, The Mass Cytometry (CyTOF)-based analysis discovered T-cell signatures in peripheral blood of patients with COVID-19. Expression patterns of T cells, CD4+ T cells, and CD8+ T cells in PhenoGraph analysis. Populations identified by the relative expression of CyTOF markers are indicated by differences in expression patterns in the NC, A1, A2, B1, and B2 groups. *P < .05, **P < .01. Abbreviations: COVID-19, coronavirus disease 2019; DEG, differentially expressed gene; FC, fold-change; Th, T-helper.
DISCUSSION
The current outbreak of COVID-19 was first reported from Wuhan and declared a Public Health Emergency of International Concern on 30 January 2020 by WHO. Based on the data of 1099 patients with laboratory-confirmed COVID-19 from 552 hospitals in 30 provinces of China, the most common symptoms of COVID-19 are fever and cough, followed by other respiratory symptoms [2]. Similarly, in the present study, the primary symptoms in the 11 patients with COVID-19 were cough (9/11, 81.82%) and fever (8/11, 72.73%). Older individuals (group A) were prone to develop the severe stage of disease. In terms of laboratory findings, higher neutrophil counts and higher C-reactive protein levels were found in severe (group A) cases, which is consistent with other COVID-19 reports [4, 13]. After 5 days of initial treatment, no significant differences were found in blood laboratory results in either type of disease.
The immune system plays an important role in resisting respiratory viruses, and incomplete, delayed, weakened, or strong immune responses of the host may cause tissue damage [7]. It has been reported that IL-2R, IL-6, IL-10, IP-10, MCP-1, MIP-1A, or TNF-α increased in the majority of severe cases [3, 13]. We also found that IL-10 levels fluctuated in different COVID-19 progressions, which was the only difference between the A1 and B1 groups and A2 and B2 groups, respectively. Interestingly, after 5 days of initial treatment, IL-10 in the mild group (group B) was significantly decreased, although it maintained a similar level in the severe group (group A). The above results suggest the early high level of serum proinflammatory cytokines, indicating a potential cytokine storm–mediated disease severity, which is similar to SARS-CoV and MERS-CoV infections [14, 15]. Dysregulated immune responses occur in patients with COVID-19 [16]. IL-10 can be produced by most cells of the immune system, and suppresses the immune response by preventing various cell types from inhibiting the synthesis of a number of cytokines, including IFN-γ, IL-2, IL-3, IL-5, IL-6, IL-8, IL-12, TNF, and GM-CSF [17]. The anti-inflammatory properties of IL-10 play a key role in determining the outcome of the infection, which shows that the absence of IL-10 leads to better pathogen clearance [18].
The results of microfluidic qPCR and subsequent bioinformatics analysis demonstrated that SARS-CoV-2 infection could affect the mRNA expression level related to immune function in COVID-19 cases. Our study revealed the number of misregulated genes increased as the disease progressed when compared with NC group, as more DEGs were found in severe cases. DEGs of both patients with severe and mild disease participated in pathway such as Th17-cell differentiation, the cytokine-mediated signaling pathway, sa mmp cytokine connection, natural killer cell–mediated cytotoxicity, and T-cell activation. While DEGs specific to severe cases were mainly responsible for the CD8 TCR pathway, the EPO pathway, signaling by interleukins, the TNF signaling pathway, entry into host cells, and regulation of T-cell proliferation enrichment terms, these results indicated that SARS-CoV-2 infection may result in specific Th1/Th17 inactivation and impaired inflammatory responses, which may be related to the decreased proportion of T cells, especially in patients with severe COVID-19. Fourteen key genes related to immune functions in COVID-19 cases were screened, which revealed that FC downregulated more in group A1 than in group B1, both relative to the NC group. Like influenza viruses, coronaviruses (CoVs) such as SARS-CoV and MERS-CoV also use a combination of ways to achieve host halting cellular protein expression, both at the transcriptional and the translational levels [7]. Innate and adaptive immune are response to viral infection during infection. Coronaviruses infect macrophages and present CoV antigens to T cells, a process that could result in T-cell activation and differentiation including the production of cytokines associated with different T-cell subpopulations, and a subsequent large release of cytokines for immune response expansion [19]. However, in the patients with severe COVID-19 before treatment we observed the opposite phenomenon, with decreased T-cell numbers and suppressed cytokine levels except for IL-10.
We also found that the DEG numbers in the 2 groups after initial treatment in the hospital were decreased compared with those on admission. The DEGs after initial treatment in COVID-19 cases both mainly enriched on Th1-, Th2-, and Th17-cell differentiation; the Jak-STAT signaling pathway; the T-cell-receptor signaling pathway; and the TNF signaling pathway. Potential key genes related to COVID-19 immune response that involved in T cell activation and differentiation were screened. After initial treatment in severe cases, MAP2K7 and SOS1 were upregulated relative to that on admission, which are involved in the T-cell-receptor signaling pathway and the Fc epsilon RI signaling pathway, which play important roles in disease progression. MAP2K7 activates the c-Jun kinases JNK1 and JNK2 in response to T-cell activation signals and mediates T-cell gene expression programs and cytokine expression [20]. SOS1 significantly contributes to ERK activation during sustained TCR signaling and T-cell activation [21]. We also found that, after initial treatment, the CD4+ T cells of patients with severe disease returned to normal proportions, and CD8+ T cells improved slightly, although were still less than normal.
Our study has some notable limitations. First, the limited sample size with the similar initial regimens in the same period may limit the robustness of the conclusions. Second, only 108 mRNAs gene expression level were preliminary observed in this study, which related to the reduced T cells proportions. A larger cohort with a change in immune response after infection with SARS-CoV-2 should be addressed in the key stages including admission, hospitalization, discharge, and follow-up after cure in the near future. Despite that, our study demonstrated the change in immune response in patients with COVID-19 through downregulated genes at the transcriptional levels involved in T-cell activation and differentiation, especially in patients with severe disease. Although clinical blood laboratory findings did not show significant differences after initial treatment, IL-10 level, T-cell proportions, and related mRNA expression levels changed with the disease progression. These findings will help us extend our understanding of the SARS-CoV-2 infection mechanism, and may help elucidate the COVID-19 infection and provide a basis for future novel immune therapeutic strategies.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was supported by the National Natural Science Foundation of China (grant numbers 81672026 and 81601796), the COVID-19 Key Technology Research and Development Funding of Beijing Hospital Authority, National Science and Technology Major Special Program of the 13th Five-Year Plan (grant number 2018ZX10302205-005), and a Capital Health Research and Development special grant (grant number 2018-1-1151).
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. World Health Organization. Coronavirus disease (Covid-2019) situation reports–47. 2020. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports [Google Scholar]
- 2. Guan WJ, Ni ZY, Hu Y, et al. Clinical c haracteristics of coronavirus disease 2019 in China. N Engl J Med 2020. doi:10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020; 395:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-cov-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 2020. doi:10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu N, Zhang D, Wang W, et al. ; China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kikkert M. innate immune evasion by human respiratory RNA viruses. J Innate Immun 2020; 12:4–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol 2020; 38:1–9. [DOI] [PubMed] [Google Scholar]
- 9.The People’s Government of Beijing Municipality. Coronavirus disease (Covid-2019) situation reports Beijing, China 2020. Available at: http://www.beijing.gov.cn/ywdt/zwzt/yqfk/yqbb/202003/t20200308_1838807.html
- 10. Faresjö M. A useful guide for analysis of immune markers by fluorochrome (Luminex) technique. Methods Mol Biol 2014; 1172:87–96. [DOI] [PubMed] [Google Scholar]
- 11. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019; 10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 2019; 47:W234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020; 580:E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mahallawi WH, Khabour OF, Zhang Q, Makhdoum HM, Suliman BA. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine 2018; 104:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wong CK, Lam CW, Wu AK, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol 2004; 136:95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with covid-19 in Wuhan, China. Clin Infect Dis 2020. doi:10.1093/cid/ciaa248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ding Y, Chen D, Tarcsafalvi A, Su R, Qin L, Bromberg JS. Suppressor of cytokine signaling 1 inhibits IL-10-mediated immune responses. J Immunol 2003; 170:1383–91. [DOI] [PubMed] [Google Scholar]
- 18. Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol 2010; 10:170–81. [DOI] [PubMed] [Google Scholar]
- 19. Li G, Fan Y, Lai Y, et al. Coronavirus infections and immune responses. J Med Virol 2020; 92:424–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martinez NM, Agosto L, Qiu J, et al. Widespread JNK-dependent alternative splicing induces a positive feedback loop through CELF2-mediated regulation of MKK7 during T-cell activation. Genes Dev 2015; 29:2054–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Poltorak M, Meinert I, Stone JC, Schraven B, Simeoni L. Sos1 regulates sustained TCR-mediated Erk activation. Eur J Immunol 2014; 44:1535–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.