Abstract
Bony fish encode multiple multi-gene families of membrane receptors that are comprised of immunoglobulin (Ig) domains and are predicted to function in innate immunity. One of these families, the diverse immunoglobulin (Ig) domain-containing protein (DICP) genes, maps to three chromosomal loci in zebrafish. Most DICPs possess one or two Ig ectodomains and include membrane-bound and secreted forms. Membrane-bound DICPs include putative inhibitory and activating receptors. Recombinant DICP Ig domains bind lipids with varying specificity, a characteristic shared with mammalian CD300 and TREM family members. Numerous DICP transcripts amplified from different lines of zebrafish did not match the zebrafish reference genome sequence suggesting polymorphic and haplotypic variation. The expression of DICPs in three different lines of zebrafish has been characterized employing PCR-based strategies. Certain DICPs exhibit restricted expression in adult tissues whereas others are expressed ubiquitously. Transcripts of a subset of DICPs can be detected during embryonic development suggesting roles in embryonic immunity or other developmental processes. Transcripts representing eleven previously uncharacterized DICP sequences were identified. The assignment of two of these sequences to an unplaced genomic scaffold resulted in the identification of an alternative DICP haplotype that is linked to a MHC class I Z lineage haplotype on zebrafish chromosome 3. The linkage of DICP and MHC Class I genes also is observable in the genomes of the related grass carp (Ctenopharyngodon idellus) and common carp (Cyprinus carpio) suggesting that this is a shared character with the last common Cyprinidae ancestor.
Keywords: innate immunity, immunoglobulin domain, haplotype, polymorphism, immune receptor
Introduction
Zebrafish (Danio rerio) are a well-recognized animal model in developmental biology, immunity and infection, toxicology, as well as cancer (Konantz et al. 2012, Novoa and Figueras 2012, Renshaw and Trede 2012, Sipes et al. 2011, Sullivan and Kim 2008, Veldman and Lin 2008). Investigations in many different zebrafish lines indicate high levels of genetic variation, including copy number variants (CNVs) (Brown et al. 2012). Sequencing of the genome of a single wild-collected zebrafish and comparison to the reference genome revealed 5.2 million single nucleotide polymorphisms and over 1.6 million insertion-deletion variations (Patowary et al. 2013). This extensive genotype variation likely is reflected in phenotypic variation (Loucks and Carvan 2004).
Nearly one third of zebrafish genes shown to be conserved exclusively in the teleost lineage are predicted to encode immune response genes (based on software analyses of protein features) possibly reflecting a likely expansion of immune-related genes in teleost fish (Yang et al. 2013). Immune genes in zebrafish and other fish species are predicted to be under positive selection resulting in high levels of sequence variation (Aparicio et al. 2002, Patowary et al. 2013, Star et al. 2011). In addition to immunoglobulins and T cell antigen receptors, zebrafish possess multiple gene families of immunoglobulin (Ig)-domain containing putative innate immune receptors such as the novel immune-type receptors [NITRs; (Yoder et al. 2001, Yoder et al. 2004, Yoder et al. 2008, Yoder et al. 2010)], novel immunoglobulin-like transcripts [NILTs; (Stet et al. 2005)], leukocyte immune-type receptor [LITRs; (Stafford et al. 2006)], polymeric Ig receptor (pIgR)-like proteins [PIGRLs; (Kortum et al. 2014)] and diverse immunoglobulin domain-containing proteins [DICPs; (Haire et al. 2012)]. These gene families, of which some may be restricted to bony fish, are recently derived, rapidly evolving and are associated with significant polymorphic and haplotypic variation (Haire et al. 2012, Rodriguez-Nunez et al. 2014, Yoder et al. 2010).
The first report of a DICP transcript likely was from an EST project of the common carp (Cyprinus carpio) (Sakai et al. 2005) in which the sequence (GenBank AB098477) erroneously was referred to as a NITR. A subsequent report identified similar sequences on zebrafish chromosome 16 and referred to them as “NITR-WxC” sequences to distinguish them from NITRs (Ohashi et al. 2010); however, only a single zebrafish “NITR-WxC” (DICP) sequence was included in this report (GenBank XM_001345404). Recently twenty-seven DICP genes and pseudogenes were described on zebrafish chromosomes 3, 14 and 16 and were recognized to constitute a single derived multigene family constituting three distinctive groups (Haire et al. 2012). The DICP family possesses two types of Ig ectodomains, D1 and D2, and individual DICPs are predicted to possess one (D1 or D2), two (D1–D2 orientation) or four (D1-D2-D1-D2 orientation) Ig domains. Multiple membrane-bound DICPs possess cytoplasmic immunoreceptor tyrosine-based inhibition motifs (ITIMs) consistent with inhibitory function. A single DICP (dicp2.1) lacks ITIMs but possesses a charged residue within its transmembrane domain indicating that it potentially could partner with an activating adaptor protein (e.g. Dap12, FcRγ, etc). Membrane-bound DICPs lacking these characteristic peptide motifs and secreted DICPs also were identified. Polymorphisms and alternative mRNA processing were shown to contribute to DICP diversity. Recombinant DICP Ig domains bind phospholipids, a property shared with select Ig domains of the mammalian CD300 and TREM receptor families (Cannon et al. 2012, Haire et al. 2012). A specific functional role for DICPs as of yet has not been defined; however, the overall similarities in their structure and ligand recognition to CD300 and TREM proteins suggests that DICPs have a role in mediating innate immunity.
In order to better understand the transcriptional regulation and sequence variation of the DICP family, inter-individual variation of DICP cDNA amplicons within and among three lines of zebrafish (AB, TU and EKW) have been characterized. DICP expression was evaluated from multiple tissues of individual zebrafish, including lymphoid and myeloid cell populations and at different stages of development. DICP amplicons were sequenced to determine polymorphisms and allelic variation between and within lines. Certain DICPs are shown to display restricted tissue expression, whereas others are expressed ubiquitously. Transcripts of several DICPs were detected during embryonic development. Additional DICP genes representing an alternative chromosome 3 haplotype that is linked to a MHC class I Z gene haplotype were described. DICP and MHC Class I sequences were also found to be linked in the genomes of the related grass carp (Ctenopharyngodon idellus) and common carp. Collectively, these findings highlight the sequence complexity and dynamic nature of the DICP family and suggest that DICP and MHC genes may be linked in all cyprinid fishes.
Materials and methods
Zebrafish
All experiments involving live zebrafish were performed in accordance with relevant institutional and national guidelines and regulations and were approved by the North Carolina State University Institutional Animal Care and Use Committee. TU and AB zebrafish were acquired from the Zebrafish International Resource Center (http://zebrafish.org). EKW zebrafish were purchased from EkkWill Waterlife Resources (Ruskin, FL). Adult and embryonic zebrafish were maintained and euthanized as described (Jima et al. 2009).
RNA isolation and Reverse Transcription-PCR for transcript detection
Tissues were dissected from three individual TU, AB and EKW zebrafish. Lymphoid and myeloid cell populations were isolated from pooled kidneys of five EKW zebrafish as described (Traver et al. 2003, Traver 2004). In brief, kidneys from adult zebrafish were dissected and homogenized with a 40-μm nylon-mesh filter in ice-cold PBS supplemented with 5% FBS. Propidium iodide was added to a concentration of 1 μg/ml. Myeloid and lymphoid cells were isolated from this single-cell suspension by sorting based on propidium iodide exclusion, forward scatter and side scatter with a BD FACS Aria II SORP flow cytometer (Beckton Dickinson). Cell populations were sorted twice to optimize cell purity. Zebrafish embryos were collected by natural matings, maintained at 28 °C as described (Westerfield 2007) and ten embryos of each line were pooled at various developmental stages for RNA isolation. Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA). Concentration and purity of RNAs were determined using a Nanodrop.
cDNAs were synthesized from total RNA (1.5 μg for tissues, 0.5 μg for lymphoid and myeloid cells, and 2.0 μg for embryos) using oligo dT primers and SuperScript III Reverse Transcriptase (Life Technologies, Carlsbad, CA). PCR primer pairs were designed by Primer 3 software (Untergasser et al. 2012) to amplify the Ig domains of several related members of the DICP family and to span at least one intron (Fig. 1 and Table 1). Relative gene expression levels were determined by PCR using TITANIUM™ Taq DNA polymerase (Clontech, Mountain View, CA); annealing temperatures, extension times and number of cycles for each primer pair are listed in Table 1. Despite employing various cycling parameters, attempts to detect dicp1.3–4/dicp1.5–6 amplicons (primer pair ATGGCTGATAGGAGTCCTCTGTTTCTGC and GGATGATTCTCTGACCTGAATAGTG) and dicp2.2 (primer pair ATGCTGGGACTGATCATTTTCTGC and ATATCAGCAGCAGCTGGGGTCACTG) with the individual zebrafish in this study were unsuccessful (data not shown). A β-actin primer pair (Yoder et al. 2010) was used as a standard reference while a myeloperoxidase primer pair (MPX) (Yoder et al. 2010) and a T-cell receptor alpha primer pair (TCRa) (Wittamer et al. 2011) served as positive controls for myeloid and lymphoid cells, respectively. Amplicons were cloned into the pGEM®-T-Easy plasmid (Promega) and sequenced.
Fig. 1. Overview of the oligonucleotide primer design employed for amplifying DICP sequences.

Primer pairs are listed on the left. Genes targeted by each primer pair and the overall genomic organization of these genes are listed to the right of each primer pair. Families of DICPs are defined by a number that denotes the DICP cluster (e.g., DICP1 cluster on chromosome 3) and gene names include a second number that denotes the order in which genes were identified (e.g., dicp1.1). Gray rectangles represent exons and black arrowheads approximate the relative location of each primer. Protein domains associated with each exon are indicated above the genomic organization (L: peptide leader sequence, D1: Ig domain, D2: Ig domain, LC: low complexity regions, TM: transmembrane domain, Cyt: cytoplasmic tail). Primer sequences are listed in Table 1.
Table 1.
Primer sequences and PCR cycling parameters for detection and recovery of DICP sequences
| Titanium Taq DNA polymerase | KAPA HiFi DNA polymerase | |||||
|---|---|---|---|---|---|---|
| Primer Pair | Forward primer Reverse primer | Annealing Temp (°C) | Extension time (s) | Annealing Temp (°C)a | Extension time (s)a | No. of cycles |
| DICP1.1/2/9/11/16/19 | ATGGCTGATAGGAGTCCTCTGTTTCTGC GCCAAAGCAGCAGCAGAAAGCAGAACTG |
70 | 30 | NA | NA | 35 |
| DICP1.7/8/17/22 | ATGGCTGATAGGAGTCCTCTGTTTCTGC ACACCACAGACGTCAGCAGAGAAGAGTC |
68 | 30 | 68 | 30 | 35 |
| DICP1.22 | CTGCAGACCATCAGAAACTGAC AGTGGACAGTAAACGTGTCTGAG |
70 | 30 | NA | NA | 35 |
| DICP2.1 | ATGCTGGGACTGATCATTTTCTGC GCAGTAGGAAAACTTAGTGCTGCCATCTCA |
68 | 30 | 68 | 30 | 35 |
| DICP3.1 | AAAGGAACCAKYGGCGCAGAAGACA GTACTTGACAGTAAATGAGGCTGAGG |
68 | 30 | 60 | 30 | 35 |
| DICP3.2/3 | ACAGGTACCARTGGAGCAGAAGACA GTACTTGACAGTAAATGAGGCTGAGG |
68 | 30 | NA | NA | 35 |
| DICP3.6 | ACTGGTACCAGTGGAGTAGAAGAGA GTACTTGACAGTAAATGAGGCTGAGG |
TD 70–60b | 30 | NA | NA | 35 |
| DICP1.1 (genomic only) | GATCAGTACTGCAGCACATCAGGA TTGTTGTTGTTGTTGTTGTTGG |
67 | 30 | NA | NA | 40 |
| DICP1.22_nested-RACE | GATGGTTTGTTAAGTTCTCAGC | 65 | 60 | NA | NA | 25 |
| DICP1.23_nested-RACE | GCAGTTCACAAACCAAACACAGCAGCG | 65 | 60 | NA | NA | 25 |
| DICP1.24_nested-RACE | GGAGACCAGAAACAAGGCAATGAT | 65 | 60 | NA | NA | 25 |
| DICP1.25_nested-RACE | CAGCTGAACTGGTTGTTTTAGGGTT | 65 | 60 | NA | NA | 25 |
| DICP1.26_nested-RACE | TCCAGATCTCACAGCTAAAGCAG | 65 | 60 | NA | NA | 25 |
| DICP1.30_nested-RACE | AGGAGGGAATCAGAAACAAGGCAATGAC | 65 | 60 | NA | NA | 25 |
NA = not applicable
Touch-down PCR during which the annealing temperature is lowered from 70°C to 60°C by 0.5°C per cycle for 20 cycles and then an additional 15 cycles are completed with a 60°C annealing temperature
Rapid amplification of cDNA ends (RACE) for amplification of DICP transcripts
A rapid amplification of cDNA ends (RACE) strategy was used to amplify and clone the 3′ ends of new DICP transcripts identified in this study. Total RNA from the kidney of TU zebrafish 1 and EKW zebrafish 2, the liver of EKW zebrafish 1, and AB zebrafish embryos at 36 hours post fertilization (hpf) were reverse transcribed using the GeneRacer™ oligo dT primer and Superscript™ III Reverse Transcriptase supplied with the GeneRacer™ kit and amplification strategies were conducted as recommended by the manufacturer (Invitrogen). An initial “touchdown” PCR [denaturing at 95 °C for 30 sec, touchdown annealing at 70 to 68 °C for 30 sec (during which the annealing temperature is lowered by 0.5 °C per cycle), and extension at 72 °C for 90 sec (5 cycles); immediately followed by denaturing at 95 °C for 30 sec, annealing/extension at 68 °C for 90 sec (25 cycles)] was performed with the DICP1.1/2/9/11/16/19 and DICP1.7/8/17/22 Forward primers (Table 1) in combination with the GeneRacer™ 3′ primer (Invitrogen). Subsequently, nested PCRs were performed with the PCR products from the touchdown PCR along with gene specific nested primers (Table 1) and the GeneRacer™ 3′ nested primer (Invitrogen) with the cycling parameters listed in Table 1. PCR products were cloned into pGEM®-T Easy (Promega) and sequenced. Attempts to amplify the 3′ ends of dicp1.26, dicp1.27, dicp1.28 and dicp1.29 transcripts were unsuccessful (data not shown).
Reverse Transcription-PCR amplification for sequence analyses
In order to define DICP sequence variation between individual zebrafish, RT-PCR was performed from zebrafish kidney cDNA using the high fidelity proofreading KAPA HiFi DNA Polymerase (Kapa Biosystems) and the DICP1.7/8/17/22, DICP2.1 and DICP3.1 primer pairs. Primer sequences and cycling parameters are listed in Table 1 and Fig. 1. Amplicons were cloned into the pGEM®-T Easy plasmid and sequenced.
Haplotype analyses
To investigate a predicted alternative DICP haplotype, PCR was performed using genomic DNA from adult zebrafish, the DICP1.1 and DICP1.22 primer pairs, Titanium Taq DNA polymerase and the cycling parameters described in Table 1. Genomic DNA was obtained from fin clips of the adult zebrafish described above using a modified HotSHOT protocol (Meeker et al. 2007). The linkage of MHC class I Z lineage genes with this DICP haplotype was confirmed by genomic PCR using the MHC class I primers and cycling conditions described previously (Dirscherl and Yoder 2014). Genomic DNA from zebrafish with defined MHC class I Z gene haplotypes were kindly provided by Hayley Dirscherl (Dirscherl and Yoder 2014). Amplicons were cloned into the pGEM®-T-Easy plasmid and sequenced.
Sequence analyses
The sequences obtained from the DICP transcripts were translated in silico and predicted protein domains identified by SMART software (Letunic et al. 2012). The nucleotide and amino acid (aa) sequences encoded by the DICP transcripts were used as queries for BLAST searches of the zebrafish reference genome (Howe et al. 2013), the nucleotide collection, the high throughput genomic sequences (HTGS) and the non-redundant protein sequences from the NCBI.
Sequence alignments were generated using ClustalW2 (Larkin et al. 2007). Phylogenetic trees were constructed with the Neighbor-Joining method (Saitou and Nei 1987) and 1000 bootstrap replicates using MEGA5 (Tamura et al. 2011).
Data access
All new DICP sequences reported here are provided in Online Resource 1 and have been deposited in the GenBank database under accession numbers KT585285 - KT585478.
Results
DICP transcript detection and nomenclature
Twenty-seven DICP genes have been identified from the zebrafish reference genome (version Zv8) as well as from individual genomic (BAC) clones and can be placed into three groups, (DICP1, DICP2 and DICP3) based on sequence similarity and chromosomal location (chromosomes 3, 14 and 16, respectively) (Haire et al. 2012). In order to define the normal expression of various DICP genes, seven primer pairs (DICP1.1/2/9/11/16/19, DICP1.7/8/17/22, DICP1.22, DICP2.1, DICP3.1, DICP3.2/3 and DICP3.6) were employed to amplify a range of different DICP transcripts (Fig. 1 and Table 1). For example, the primer pair DICP1.1/2/9/11/16/19 was designed to amplify transcripts encoded by the dicp1.1, dicp1.2, dicp1.9, dicp1.11, dicp1.16 and dicp1.19 genes. Although the DICP1.7/8/17/22 primer pair was designed to amplify transcripts of the previously described dicp1.7, dicp1.8 and dicp1.17 genes, it also amplified transcripts of dicp1.22, a previously uncharacterized DICP gene (see below). Subsequently, a DICP1.22 primer pair was designed to specifically amplify a 554 base pair (bp) amplicon of dicp1.22 (Fig. 1 and Table 1). RT-PCR was employed to evaluate DICP expression in various immune-related tissues from nine individual zebrafish from the TU, AB and EKW lines (Fig. 2), lymphoid and myeloid cells from the EKW line (Fig. 3) and from various embryonic stages of development from the TU, AB and EKW lines (Fig. 4). Variable expression patterns were observed between zebrafish lines as well as individuals of the same line. Amplicons were cloned and sequenced to verify that they represent DICPs. In order to distinguish among transcript variants of the same gene, sequence identity numbers are included as superscripts after each gene symbol (e.g. transcript variant 5571 for dicp1.1 is shown as dicp1.15571). Details of all new DICP sequences identified in this study are provided in Online Resource 1.
Fig. 2. DICP gene expression in immune-related tissues.
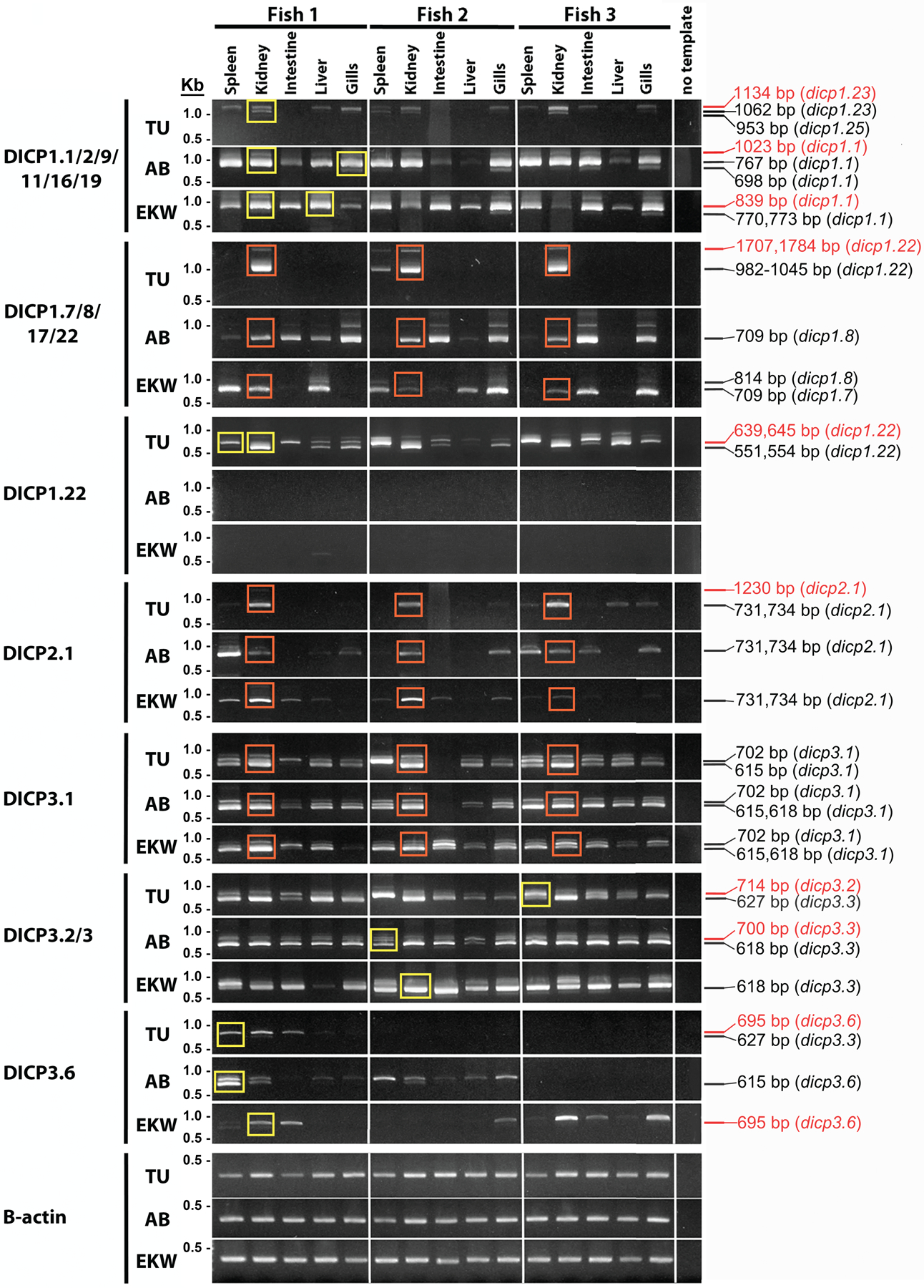
DICP expression was evaluated using primer pairs listed in Table 1 and tissues from nine individual adult zebrafish of the TU, AB and EKW lines. RT-PCR amplicons shown were generated with Titanium Taq DNA polymerase and yellow rectangles indicate those products that were cloned and sequenced to confirm their identity. Orange rectangles indicate amplicons that subsequently were generated with a proofreading DNA polymerase (KAPA HiFi) for evaluation of sequence variation. The size and identity of recovered amplicons is listed on the right of the gel image with red text indicating nonfunctional transcripts. β-actin expression was used as a reference for cDNA quantity and quality.
Fig. 3. DICP gene expression in myeloid and lymphoid cells.
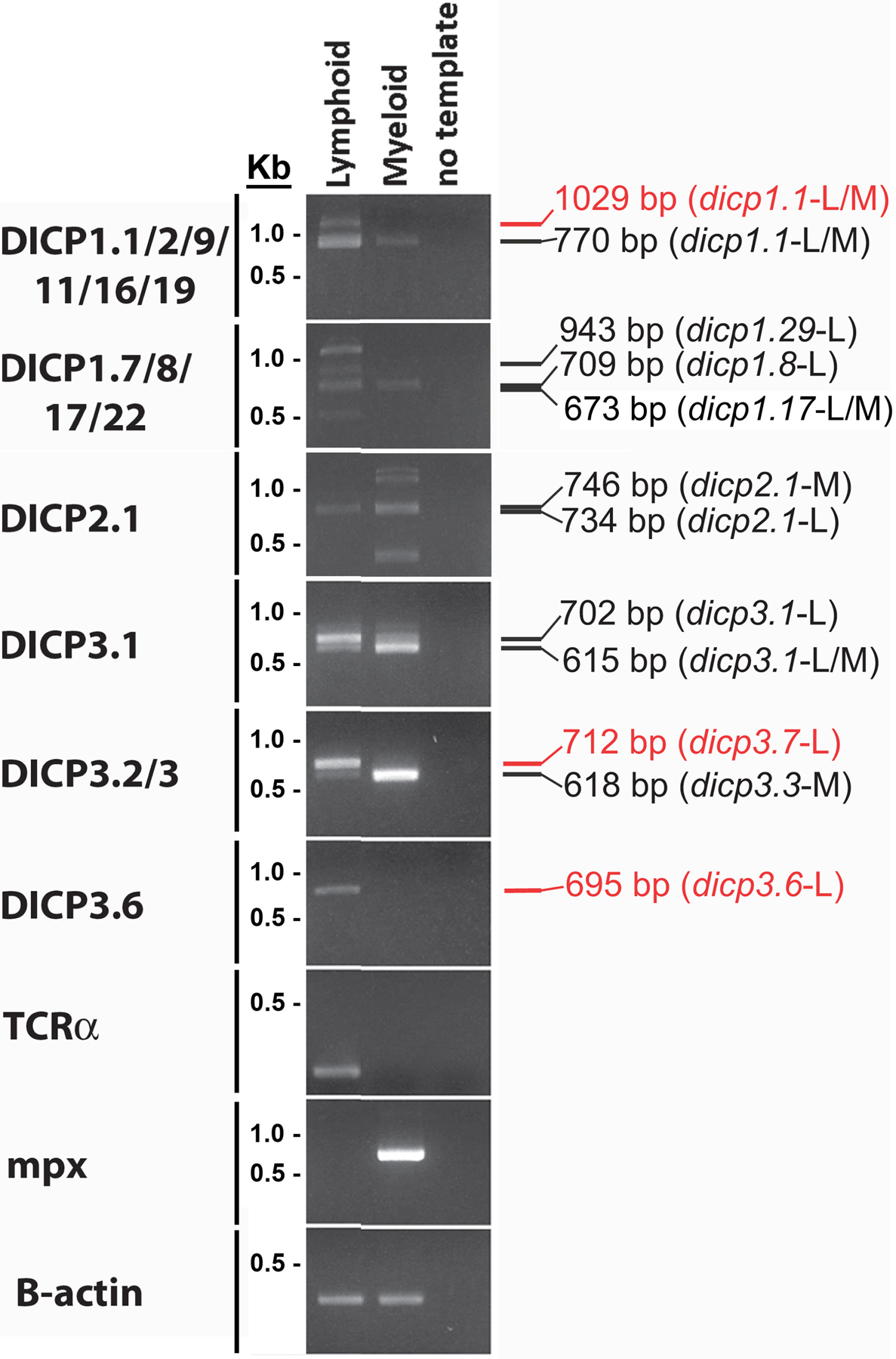
Kidney cells from five adult EKW zebrafish were pooled and lymphoid and myeloid cells sorted by flow cytometry. DICP transcripts were amplified by RT-PCR with Titanium Taq DNA polymerase and all amplicons detected were cloned and sequenced to confirm their identity. The size and identity of recovered amplicons is listed on the right of the gel image; red text indicates nonfunctional transcripts. RT-PCR of myeloperoxidase (mpx) provides a positive control for myeloid cells and TCR-α provides a positive control for T lymphocytes. β-actin expression was used as a reference for cDNA quantity and quality.
Fig. 4. DICP gene expression during zebrafish development.

RT-PCR was employed to detect DICP transcripts at different developmental stages from TU, AB and EKW zebrafish lines. Ten embryos were pooled for each cDNA template. RT-PCR was employed with Titanium Taq DNA polymerase. Yellow rectangles indicate amplicons that were cloned and sequenced to confirm their identity. The size and identity of recovered amplicons is listed on the right of the gel image; red text indicates nonfunctional transcripts. β-actin expression was used as a reference for cDNA quantity and quality.
DICP expression in adult zebrafish tissues
Although the sequence of several DICP1 amplicons recovered from adult zebrafish tissues (Fig. 2) correspond with DICP1 sequences that were described previously (Haire et al. 2012), some primer pairs produced different size amplicons in a zebrafish line-dependent manner. For example, the DICP1.1/2/9/11/16/19 primer pair generated amplicons of ~700–800 bp from tissues from AB and EKW zebrafish, but TU zebrafish gave rise to amplicons of ~950–1050 bp. Sequencing of the smaller amplicons from AB and EKW fish revealed different dicp1.1 transcripts from AB fish (dicp1.15571 and dicp1.15574) and EKW fish (dicp1.15544, dicp1.15577 and dicp1.15578). Sequencing of the larger amplicons from the TU individuals revealed new DICP sequences, dicp1.23, dicp1.24, dicp1.25 and dicp1.30, which are discussed below. Similar observations were made for amplicons generated by the DICP1.7/8/17/22 primer pair: amplicons from AB and EKW fish were ~700–800 bp and amplicons from TU fish were ~1000 bp. Sequencing of the smaller AB and EKW amplicons revealed dicp1.7 and dicp1.8 transcripts (dicp1.73986, dicp1.83864 and dicp1.83994). Sequencing of the larger TU amplicons revealed a new DICP, dicp1.22 (dicp1.223796, dicp1.223797, dicp1.223800, dicp1.223803, dicp1.223806 and dicp1.223816). A primer pair designed to amplify only dicp1.22 subsequently generated amplicons from TU tissues, but failed to amplify detectable amplicons from the AB and EKW individuals in this study (Fig. 2). The DICP1.7/8/17/22 primer pair also generated multiple dicp1.17 amplicons from the AB, EKW and TU individuals, which were not visualized but could be identified by cloning and sequencing [dicp1.173810, dicp1.173725, dicp1.173735, dicp1.173860, dicp1.173863, dicp1.173872, dicp1.173875, dicp1.173979, and dicp1.173980, as well as amplicons reflecting two new genes, dicp1.27 and dicp1.28 (discussed below)].
Comparable overall expression of the putative activating DICP, dicp2.1 (Haire et al. 2012), is observed in multiple tissues from adult TU, AB and EKW individuals. Although some minor variation in patterns of dicp2.1 expression was observed between individual zebrafish, expression was consistently highest in kidney and spleen (Fig. 2).
The DICP3.1 primer pair generated multiple D1–D2 dicp3.1 amplicons from nearly all examined tissues from all zebrafish lines (Fig. 2). Some of the recovered sequences were more similar to the dicp3.134H11 allele previously identified in BAC CH73–34H11 (GenBank: FP929011), whereas the remaining dicp3.1 sequences were more similar to the dicp3.11952 allele from genomic scaffold 1952 (GenBank: NW_001877662.2) of the zebrafish Zv8 reference genome (Haire et al. 2012). Although the DICP3.2/3 primer pair generated amplicons from all tissues and fish that were examined, the only functional transcripts were recovered were from dicp3.3. The transcript recovered from the TU fish (dicp3.35639) was most similar to dicp3.31952 encoded in genomic scaffold 1952 (Haire et al. 2012). In contrast, the dicp3.3 sequences recovered from the AB and EKW fish (dicp3.35641, dicp3.35644 and dicp3.35647) were most similar to the dicp3.3322B17 allele identified in BAC CH73–322B17 (GenBank: FP015862). The DICP3.6 primer pair produced amplicons from only six of the nine individuals examined: one of three TU fish, two of three AB fish and three of three EKW fish (Fig. 2). Sequencing of the amplicons revealed only one functional transcript, which was from an AB fish (dicp3.65618). The DICP3.6 primer pair also amplified a dicp3.3 sequence from the spleen of a TU fish (dicp3.35617). Sequencing of additional amplicons from multiple DICP3 primer pairs revealed additional new, but non-functional, DICP gene sequences (dicp3.7 and dicp3.8), which are discussed below.
DICP expression in zebrafish lymphoid and myeloid cells
DICP transcripts were recovered in varying relative abundance from lymphoid and myeloid cells isolated from adult EKW zebrafish (Fig. 3). The DICP1.1/2/9/11/16/19 primer pair amplified functional transcripts of DICPs that were identified previously (Haire et al. 2012) such as dicp1.1 transcripts that were recovered from both myeloid and lymphoid cells (dicp1.15544, dicp1.15475 and dicp1.15605) but were expressed at higher levels in the lymphocyte population (Fig. 3). This same primer pair revealed a dicp1.16 functional transcript from myeloid cells (dicp1.165478). Using the DICP1.7/8/17/22 primer pair, multiple amplicons of different sizes were obtained from lymphoid and myeloid cDNA (Fig. 3), including transcripts of dicp1.8 and dicp1.17 from lymphoid cells (dicp1.85502 dicp1.175503 and dicp1.175505) and dicp1.17 transcripts from myeloid cells (dicp1.173863, dicp1.175506 and dicp1.175508). Sequencing of the leukocyte amplicons generated by the DICP1.7/8/17/22 primer pair revealed one new DICP gene sequence (dicp1.29), which is discussed below. The DICP1.22 primer pair produced no detectable amplicons from leukocytes of EKW individuals (data not shown), which may reflect the absence of this gene from the genomes of the fish from which leukocytes were isolated (discussed below).
The expression of the putative activating DICP, dicp2.1, was detected in both leukocyte lineages. The DICP2.1 primer pair generated one predominant band of ~750 bp from lymphoid cell cDNA and several bands that range from 400 to 1500 bp from the myeloid cell cDNA (Fig. 3). These amplicons include functional dicp2.1 transcripts from lymphoid (dicp2.15599 and dicp2.15601) and myeloid cells (dicp2.15602 and dicp2.15603); however, both lymphocyte transcripts possess a deletion within the D1 or D2 domain. Although additional amplicons were not identified from the myeloid cell cDNA, the ~400 bp amplicon might correspond to a functional dicp2.1 transcript encoding a single D2 Ig domain that was identified from AB zebrafish kidneys (dicp2.14150 amplicon of 434 bp). Similarly, the ~1200 bp amplicon might correspond to non-functional dicp2.1 transcripts (described below).
The DICP3.1 primer pair generated amplicons from both myeloid (dicp3.14445 and dicp3.14446) and lymphoid (dicp3.14484) cells (Fig. 3) that also had been recovered from zebrafish kidneys, as well as amplicons representing new dicp3.1 transcripts from myeloid cells (dicp3.15270 and dicp3.15272) and lymphoid cells (dicp3.15275, dicp3.15280 and dicp3.15282). Transcripts from myeloid cells were more similar to the dicp3.134H11 allele, except for dicp3.14445, which was more similar to the dicp3.11952 allele (Haire et al. 2012). All lymphoid dicp3.1 transcripts were more similar to the dicp3.11952 allele. The DICP3.2/3 primer pair recovered functional dicp3.3 transcripts (dicp3.35283, dicp3.35284, dicp3.35286, dicp3.35288, dicp3.35289 and dicp3.35290) from myeloid cells and one functional dicp3.3 transcript from lymphoid cells that included a deletion within the D1 domain (dicp3.35297). The DICP3.6 primer pair produced only non-functional dicp3.6 transcripts from lymphocytes. Sequencing of the leukocyte amplicons generated by the DICP3 primer pairs revealed a functional dicp3.7 transcript and additional non-functional dicp3.7 and dicp3.8 transcripts (see below).
Variable DICP expression during zebrafish embryo development
Diverse expression patterns of DICP1 genes are revealed at different stages of embryonic development (Fig. 4). The DICP1.1/2/9/11/16/19 primer pair generated amplicons exhibiting the same relative pattern of expression from TU, AB and EKW embryos; however, differences in amplicon length are evident between different developmental stages and genetic backgrounds. The earliest developmental stage at which transcripts can be detected is 12 hpf, with amplicon lengths ranging from 407 to 1020 bp. By 36 to 48 hpf, some of these amplicons were not detectable but ~1000 bp amplicons were detected in the three zebrafish lines. Sequence analysis of these amplicons detected previously described DICP transcripts (Haire et al. 2012), including dicp1.9 and dicp1.19 (dicp1.95432, dicp1.195446, dicp1.195468 and dicp1.195582), as well as transcripts of new DICP genes (dicp1.24, dicp1.25 and dicp1.26; see below). Amplicons generated with the DICP1.7/8/17/22 primer pair displayed similar relative lengths and patterns between TU and EKW embryos (Fig. 4); functional dicp1.8 transcripts can be detected at 6 dpf (dicp1.85479, dicp1.85488 and dicp1.85625). Functional transcripts of dicp1.17 also were recovered from 6 dpf TU (dicp1.175480 and dicp1.175482) and EKW embryos (dicp1.175489). The most abundant functional transcripts from AB embryos were shown to be dicp1.17 at 24 hpf (dicp1.175484 and dicp1.175485), including one dicp1.17 transcript that lacks a transmembrane domain (dicp1.175483). The DICP1.22 primer pair generated two amplicons from embryos of all three zebrafish lines (Fig. 4). The shorter amplicon corresponds to a functional transcript of dicp1.22 (dicp1.225517, dicp1.225518 and dicp1.225519); the larger amplicon, which likely corresponds to a non-functional dicp1.22 transcript, was not sequenced (see below). The dicp1.22 transcripts in embryos of the three zebrafish lines contrast with the expression observed in the adult tissues where transcripts were detected only in the TU line, negating the possibility that dicp1.22 or a specific allele of this gene is present only in the TU line. The only DICP with definitive maternal transcripts in the 1-cell embryo stage (0–1 hpf) was dicp1.22 from the AB line.
Expression of the putative activating receptor, dicp2.1, appears to be absent or at very low levels during embryonic development with amplicons first being identifiable from 6 dpf TU and AB embryos (Fig. 4). Sequencing of these amplicons revealed several functional dicp2.1 transcripts (dicp2.15537, dicp2.15538, dicp2.15540, dicp2.14073, dicp2.15667 and dicp2.15670). Although dicp2.1 amplicons were not visible in the gel from EKW embryos, multiple functional dicp2.1 amplicons were recovered from 6 dpf EKW embryos (dicp2.15549, dicp2.15550, dicp2.15551, dicp2.15552, dicp2.15593, dicp2.15595 and dicp2.15598), including two transcripts with a deletion in the D2 domain (dicp2.15594 and dicp2.15597).
Transcripts of DICP3 genes could be detected by 6 hpf with expression maintained throughout embryonic development (with the exception of dicp3.6, which was not detected in AB and EKW embryos). The DICP3.1 primer pair detected multiple transcripts from the three genetic backgrounds (dicp3.15610, dicp3.15611 and dicp3.15614); larger transcripts with additional coding sequence between the D1 and D2 domains (dicp3.14484 and dicp3.15613) also were identified. Amplicons displaying highly similar lengths and patterns were recovered from the AB and EKW embryos with the DICP3.2/3 primer pair, including functional dicp3.3 transcripts (dicp3.35652 and dicp3.35653). A dicp3.3 transcript also was recovered from TU embryos that lacked the D1 domain (dicp3.35648). The DICP3.6 primer pair revealed only non-functional dicp3.6 transcripts from TU embryos.
New DICP sequences
DICP transcripts possessing D1 domains that were <90% identical to any previously described DICP were designated as a new gene, such as the dicp1.22 transcripts that were recovered from kidney cDNA of TU zebrafish (Fig. 2) and described above. In addition, the DICP1.1/2/9/11/16/19 primer pair generated amplicons from the TU zebrafish that were larger than the amplicons detected in the AB and EKW fish (Fig. 2). Although the shorter AB and EKW amplicons represent dicp1.1 transcripts (see above), sequence of the larger amplicons from a TU zebrafish reveal two new DICPs, dicp1.23 and dicp1.25 (dicp1.235657, dicp1.235658 and dicp1.255664). Four additional DICP amplicon sequences were identified, although they cannot be observed in Fig. 2. The DICP1.1/2/9/11/16/19 primer pair yielded dicp1.24 and dicp1.30 amplicons (dicp1.245542 and dicp1.305543) from EKW zebrafish liver and the DICP1.7/8/17/22 primer pair yielded dicp1.27 and dicp1.28 amplicons (dicp1.273985 and dicp1.283991) from EKW zebrafish kidney.
New DICPs also were recovered from zebrafish leukocytes (Fig. 3) and embryos (Fig. 4). The DICP1.7/8/17/22 primer pair from lymphocyte cDNA yielded dicp1.29, which encodes a single D1 Ig domain transcript (dicp1.295504). The DICP3.1 and DICP3.2/3 primer pairs amplified non-functional transcripts of two new genes, dicp3.7 and dicp3.8 from kidney cDNA (Online Resource 1); a functional dicp3.7 transcript was recovered from myeloid cell cDNA that encodes two Ig (D1–D2) domains (dicp3.75271). Functional transcripts of dicp1.24 and dicp1.25 as well as an additional gene dicp1.26 (dicp1.245436, dicp1.245437, dicp1.245438, dicp1.245440, dicp1.245442, dicp1.245447 dicp1.255427, dicp1.255580, dicp1.265443) were recovered from embryo cDNA with the DICP1.1/2/9/11/16/19 primer pair. The dicp1.26 amplicon encodes two Ig domains (D1–D2). Phylogenetic analyses of all predicted DICP Ig domains are consistent with the classification of these sequences as new DICP genes (Fig. 5).
Fig. 5. Phylogenetic comparison of newly identified DICP Ig domains with previously described sequences.
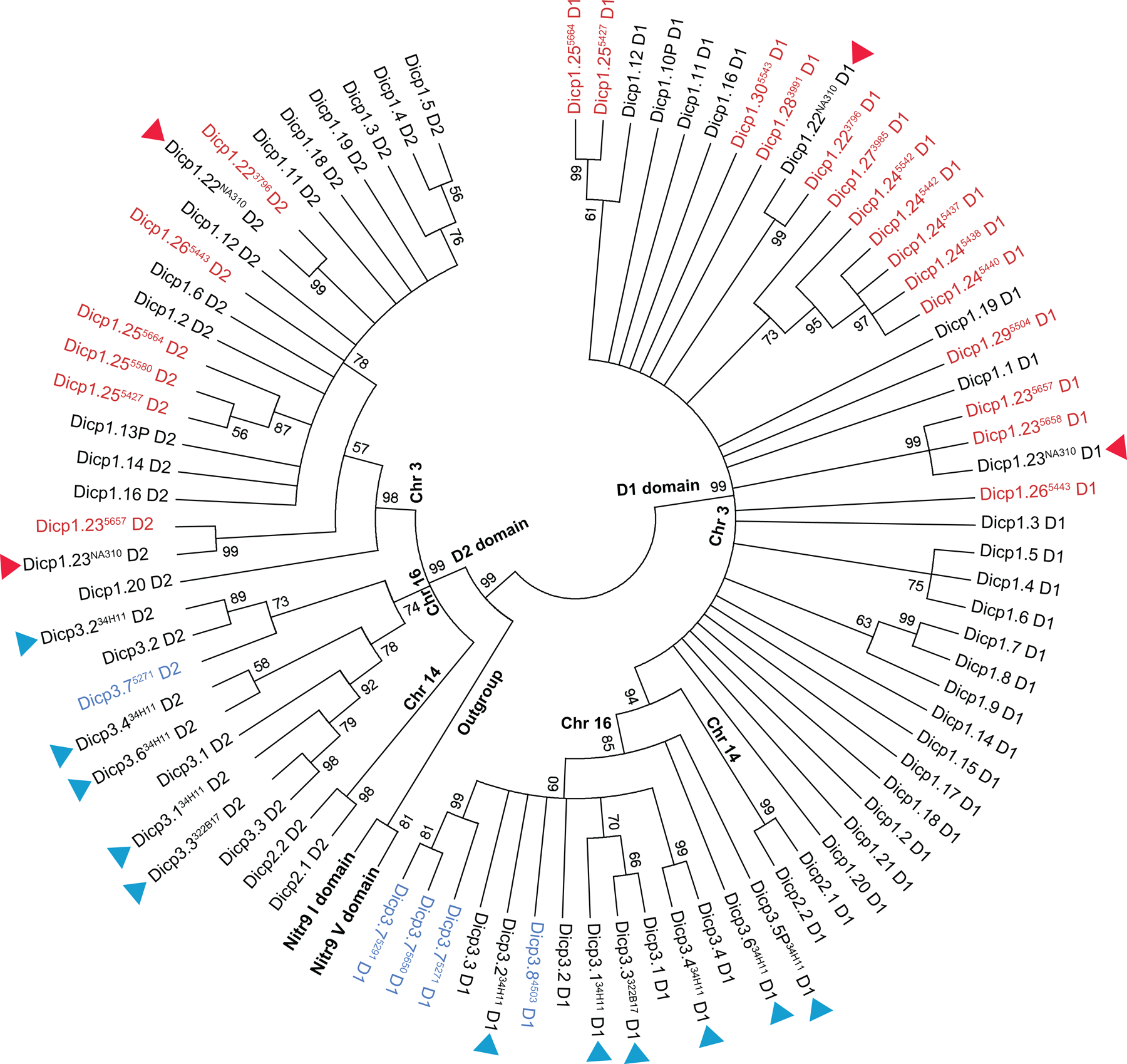
New DICP sequences that group with the DICP1 genes on chromosome 3 are in red text. New DICP sequences that group with the DICP3 genes on chromosome 16 are in blue text. DICP sequences encoded by the unplaced genomic scaffold NA310 (GRCz10 reference genome) are indicated by red triangles. All additional sequences were reported previously (Haire et al. 2012), including those predicted from genomic clones (BAC CH73–34H11 and BAC CH73–322B17), which are indicated by blue triangles. Nitr9 Ig V and I domains (Wei et al 2007; Yoder 2009) were used as an outgroup (bold characters). The percentage of replicate trees in which the associated taxa cluster together (bootstrap values) are shown next to the branches; values less than 50 are not shown.
DICP sequence analyses
In order to investigate the inter-individual sequence variation of select DICP genes, a high fidelity DNA polymerase was used for RT-PCR with the same individual zebrafish evaluated in Fig. 2. The cloning and sequencing of multiple DICP1.7/8/17/22, DICP2.1 and DICP3.1 amplicons from each individual revealed differences in sequences and splicing (Figs. 6–7; Online Resource 2 – Table S2; Online Resource 3 – Sequence Variation and – Figs. S1–S6). In silico translation of the amplicons revealed that all of the amplified transcripts encode one or two Ig domains. A phylogenetic comparison of these DICP Ig domains with all other DICP Ig domains demonstrated that the majority of these Ig domains grouped with the D1 and D2 Ig domains of Dicp1.7, Dicp1.8, Dicp1.17, Dicp1.22, Dicp2.1 and Dicp3.1, as expected (Online Resource 3 – Fig. S7).
Fig. 6. Exon-intron architecture of previously identified DICPs.

The exon-intron organization of transcript variants encoding previously described DICPs were compared with the DICP genes present in the reference genome. Sequence identifier numbers or GenBank accession numbers are listed to the right of each transcript. Red text indicates predicted non-functional transcripts. Details are provided in Online Resource 3 – DICP exon-intron architecture.
Fig. 7. Exon-intron architecture of newly identified DICPs.

The exon-intron organization of transcript variants encoding newly identified DICPs were predicted by comparison to DICP genes present in the reference genome and/or to similar DICP transcripts previously identified. Sequence identifier numbers or GenBank accession numbers are listed to the right of each transcript. Red text indicates predicted non-functional transcripts. Sequence identifiers shown in parentheses represent RACE clones. Details are provided in Online Resource 3 – DICP exon-intron architecture.
Additional sequencing of several DICP transcripts with a non-high fidelity DNA polymerase also revealed evidence of alternative splicing and allelic variation (Figs. 6–7 and Online Resource 3). The exon-intron organization of these DICPs was predicted based on the DICP genes described in the Zv9 zebrafish reference genome. Numerous transcripts likely reflect alternative mRNA splicing events, including exon skipping, intron retention, and alternative 3′ and 5′ splice sites, although these sequences may also reflect haplotypic variants that have gained or lost sequences (as compared to the reference genome). Insertions or deletions inside of an exon also were identified that might indicate either an alternative splicing inside of the exon or these insertions/deletions already were part of the gene. A possible instance of exon shuffling or recombination was identified in the dicp1.83994 transcript, which possesses two exons from the dicp1.17 gene (Fig. 6 and Online Resource 3 – DICP exon-intron architecture). Transcripts from multiple DICP genes contain a predicted premature termination codon (PTC). These transcripts likely correspond either to pre-messenger RNAs (pre-mRNA) that possess a PTC by intron retention or mRNAs reflecting alternative splicing that results in a frameshift and a PTC. A detailed description of observed polymorphisms, alternative splicing and PTCs are presented in Online Resource 3.
Structural features of new DICPs
By overlapping cDNA sequences obtained by RT-PCR and 3′ RACE, full-length sequences were predicted for Dicp1.22, Dicp1.23, Dicp1.24, Dicp1.25 and Dicp1.30 (Online Resource 3 – Figs. S8–S9). Dicp1.22 possesses two Ig domains, a transmembrane domain and a cytoplasmic tail that lacks any known signaling motifs. Dicp1.23 is predicted to be a secreted protein. Although the dicp1.23 transcripts possess an exon that could encode a transmembrane domain, a frameshift 5′ of this exon results in the use of an alternative reading frame and no transmembrane domain (Online Resource 3 – DICP exon-intron architecture). Dicp1.24 possesses a single D1 domain and three cytoplasmic ITIMs and an ITIM-like sequence (itim). Dicp1.25 possesses D1 and D2 ectodomains as well as three cytoplasmic ITIMs. Dicp1.26 possesses two Ig domains and likely a transmembrane domain; however 3′ sequence were not recovered. Dicp1.27, Dicp1.28, Dicp1.29 and Dicp1.30 each possess a D1 domain and a transmembrane domain; however, 3′ sequences were recovered only for dicp1.30 which encodes two ITIMs (Online Resource 3 – Figs. S8–S9).
Linkage of DICP and MHC class I Z haplotypes on zebrafish chromosome 3
In order to identify genomic sequences that encode the newly identified DICPs, the Ig domains of the eleven new DICPs were used as queries for BLAST searches of the zebrafish reference genome (GRCz10), the nucleotide collection (nt), the unfinished high throughput genomic sequence (HTGS) and the non-redundant protein sequence (nr) databases. Only dicp1.22 and dicp1.23 produced significant matches. Specifically, genomic sequences matching dicp1.22 and dicp1.23 transcripts were identified in unplaced genomic scaffold NA310 (GenBank: NW_003336703.1) (Fig. 8a). Scaffold NA310 also encodes the mhc1zka (GenBank NM_001302245), ccdc134l (coiled-coil domain-containing protein 134-like; GenBank XM_003201673) and gimap4l (GTPase IMAP family member 4-like; GenBank XM_001920324) genes which are predicted to represent an alternate haplotype for the mhc1zja (GenBank NM_001109718), ccdc134 (coiled-coil domain-containing protein 134; GenBank XM_003198004) and gimap8 (GTPase IMAP family member 8; GenBank XM_001919280) genes on chromosome 3 (Online Resource 3 – Fig. S10) (Dirscherl and Yoder 2014). Based on the linkage of dicp1.22 and dicp1.23 to mhc1zka/ccdc134l/gimap4l and the linkage of dicp1.1 – dicp1.21 to mhc1zja/ccdc134/gimap8 (Fig. 8a and Online Resource 3 – Fig. S10), these sequences were predicted to represent alternate haplotypes with differing gene content for both the DICP gene cluster and the MHC class I Z gene cluster on chromosome 3.
Fig. 8. Alternative haplotypes for the chromosome 3 DICP gene cluster.
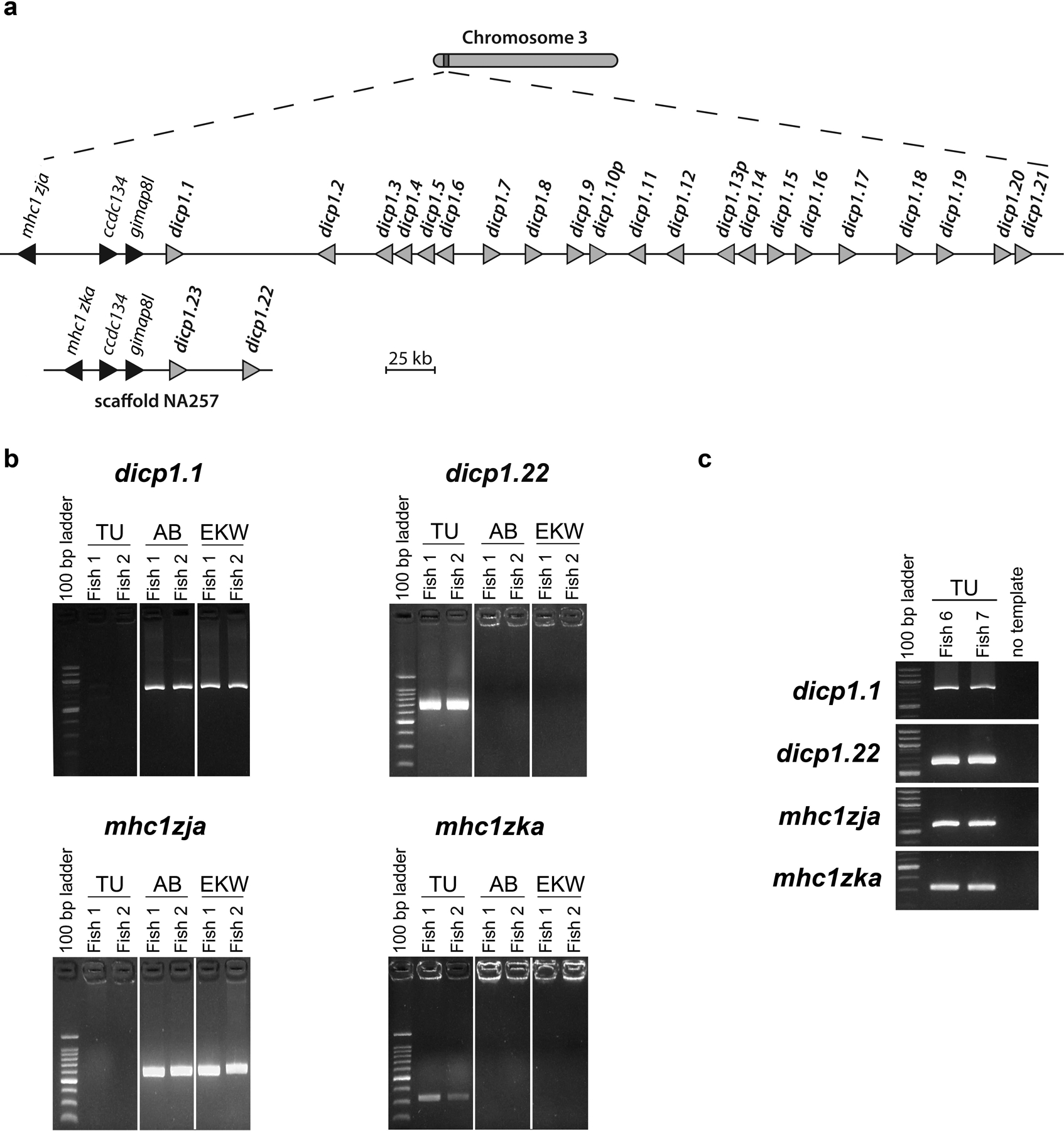
(a) Relative chromosomal positions of the DICP1 genes on chromosome 3 (scaffold CTG10218) compared to the relative positions of the DICP1 genes found on unplaced scaffold NA310. Gray triangles represent a single DICP gene, except for dicp1.3–4 and dicp1.5–6 that are predicted to be encoded in single genes. Predicted pseudogenes are indicated with a “p” at the end of the gene symbol. Black triangles represent linked non-DICP genes. The ccdc134 or ccdc134l and the gimap8l or gimpa4l genes along with a member of the MHC class I Z lineage are present in both regions. A detailed sequence identity comparison is provided in Online Resource 3 – Fig. S10. (b) Genomic PCRs for dicp1.1, dicp1.22, mhc1zja and mhc1zka using genomic DNA from individual TU, AB and EKW zebrafish analyzed in Fig. 2 and predicted to be homozygous for one of the two haplotypes depicted in panel a. (c) Genomic PCRs for dicp1.1, dicp1.22, mhc1zja and mhc1zka using gDNA from individual TU zebrafish previously shown to be heterozygous for the two MHC class I Z gene haplotypes depicted in panel a (Dirscherl and Yoder 2014).
Genomic PCR analyses were employed with individual zebrafish predicted to be homozygous for the different haplotypes (dicp1.1 and dicp1.22) depicted in Fig. 8a, and shown to express either dicp1.1 or dicp1.22 transcripts (Fig. 2). Genomic amplicons for dicp1.1 were identified in zebrafish that express dicp1.1 and genomic amplicons for dicp1.22 were identified in zebrafish that express dicp1.22 (Figs. 2 and 8b). Individuals encoding dicp1.1 (and not dicp1.22) also encode mhc1zja (and not mhc1zka) and individuals encoding dicp1.22 (and not dicp1.1) also encode mhc1zka (and not mhc1zja) (Fig. 8b). These observations support the hypothesis that dicp1.1 and dicp1.22 represent different DICP haplotypes that are tightly linked to different MHC class I Z haplotypes and that these individual zebrafish are homozygous for a single haplotype. Genomic PCR analyses with DNA from zebrafish that are heterozygous for the two MHC class I Z gene haplotypes depicted in Fig. 8a (Dirscherl and Yoder 2014) indicate that both dicp1.1 and dicp1.22 haplotypes are present (Fig. 8c).
Linkage of DICP and MHC Class I genes in cyprinid fishes
Data-mining available genomic sequence databases in 2012 identified definitive DICP sequences in zebrafish and common carp (Haire et al 2012) which are both members of the Cyprinidae family. Although DICP-like sequences were identified in Atlantic salmon (Salmo salar), Japanese pufferfish (Takifugu rubripes), green spotted pufferfish (Tetraodon nigroviridis) and Nile tilapia (Oreochromis niloticus) which belong to the Salmonidae, Tetraodontidae and Cichlidae families (Haire et al 2012) it is unknown if they share an evolutionary origin.
In order to investigate if DICP genes could be identified in other species of fish, DICP Ig domains from zebrafish and common carp were used as queries for tBLASTn searches of the teleost genome sequence databases currently available in Ensembl (v82). Definitive DICP sequences were not identifiable in any current genome assemblies of Amazon molly (Poecilia formosa), cave fish (Astyanax mexicanus), cod (Gadus morhua), Japanese pufferfish, medaka (Oryzias latipes), platyfish (Xiphophorus maculatus), stickleback (Gasterosteus aculeatus), green spotted pufferfish or Nile tilapia. As none of these fish are in the Cyprinidae family, it is possible that definitive DICPs may be restricted to cyprinid species. In order to determine if DICP and MHC genes are linked in other cyprinids, the recently reported genomes of the grass carp (Wang et al 2015) and common carp (Xu et al 2014) were searched and definitive evidence for their linkage was established. Conserved synteny was observed between zebrafish chromosome 3, grass carp scaffold CI01000243 and common carp Linkage Group 38 (Online Resource 3 – Fig. S11) demonstrating that DICPs are present and linked to MHC Class I Z sequences in three different subfamilies of cyprinids. Assignment of carp sequences to the DICP family and the MHC Class I Z lineage is supported by phylogenetic analyses (Online Resource 3 – Fig. S12). As annotation has not been performed on the carp gene models, the predicted protein sequences referred to in Online Resource 3 – Figs. S11 and S12 are provided in Online Resource 3 – Fig. S13 and the BLAST results from using each carp sequence as a query to search the NCBI database of non-redundant (nr) zebrafish proteins (queried Nov 2015) are provided in Online Resource 3 – Table S4.
Discussion
The zebrafish DICPs exhibit similarities in structure and ligand recognition to members of the mammalian CD300, TREM and FcR-like (FCRL) families (Haire et al. 2012). Certain CD300 and TREM receptors bind specific subsets of phospholipids which may reflect roles in differentiating pathogens, mediating phagocytosis of apoptotic cells and/or recognizing activated lymphocytes (Borrego 2013, Cannon et al. 2012, Pelham and Agrawal 2014). Although recombinant forms of certain DICPs also bind specific subsets of phospholipids, the overall role of DICPs in immune function remains unknown. In order to better characterize the DICPs we provide a detailed examination of the expression patterns and sequence diversity of the DICP gene family between individual zebrafish from different genetic backgrounds. The comparison of nearly 200 new DICP transcript sequences identified eleven new DICP gene sequences, revealed extensive polymorphic and haplotypic variation between DICPs of individual zebrafish, and documented transcripts that likely reflect alternative mRNA splicing that, in many cases, resulted in presumably non-functional transcripts.
Each primer pair employed to detect DICP expression yielded distinctly different patterns from adult tissues and leukocytes. Although DICPs are differentially expressed in immune tissues, a nearly universal feature of the DICP genes is their expression in the hematopoietic kidney (Fig. 2). The expression of DICPs in several immune-related tissues and their heterogeneous expression within these tissues is reminiscent of the expression patterns of CD300 in mammals (Clark et al. 2009). DICPs are expressed in both myeloid and lymphoid cell lineages with certain DICPs being expressed at higher levels in one lineage (Fig. 3). By way of comparison, expression of mammalian CD300 genes is most abundant in monocytes, but certain CD300 transcripts also are expressed at high levels in T or NK cells (Clark et al. 2009).
Expression of DICPs was evaluated at different developmental stages and transcripts of multiple DICPs (e.g. dicp1.22, dicp3.1, dicp3.3) were detected as early as 6 hpf, whereas transcripts of other DICPs (e.g. dicp2.1) were not detected until well after embryogenesis (Fig. 4). The embryonic expression of certain DICPs raises the possibilities that they may play important and early roles in innate immunity within the developing embryo or that they may play specific roles in embryonic development.
In-depth sequence analyses of the DICP amplicons generated from different tissues, leukocytes and embryos and from different zebrafish lines reveal a large number of transcript variants that reflect the loss of exons or gain of predicted introns which likely represent alternative splice variants, allelic variants or undefined DICP genes. A number of transcript variants differ in the length and sequence of the stalk domains, which are located either between the Ig domains or between an Ig domain and the transmembrane domain. In contrast, the Ig domains of transcript variants display high sequence conservation, perhaps reflecting a role for different stalk domains to act as flexible spacers for ligand binding. Stalk domains in some immune receptors have been implicated in ligand binding and intracellular signaling suggesting that the observed differences may directly influence function (Berry et al. 2013, Hartmann et al. 2012, Moody et al. 2001, Xu et al. 2006). Alternative splicing, which can lead to the loss of an Ig domain (e.g. dicp2.1 and dicp3.1), likely contributes to the complexity of the immune receptor repertoire (Maisey and Imarai 2011).
Multiple DICP transcript variants introduce PTCs (Online Resource 1) that may: 1) correspond to pre-mRNAs which have not been fully processed, 2) encode truncated, secreted DICPs or 3) be targeted for elimination through the nonsense-mediated mRNA decay (NMD) pathway (Kervestin and Jacobson 2012). In addition to RNA surveillance, the NMD pathway employs mRNA containing PTCs to control other cellular functions (Hamid and Makeyev 2014), including the regulation of gene expression during hematopoiesis (Frischmeyer-Guerrerio et al. 2011, Pimentel et al. 2014, Wong et al. 2013) as well as physiological responses to bacterial infection (Gloggnitzer et al. 2014, Kalyna et al. 2012). Certain DICPs exhibit a higher number of transcripts containing PTCs than functional transcripts, such as dicp1.23, dicp3.2, dicp3.6 and dicp3.7, in contrast to other DICPs, such as dicp1.8, dicp1.17, dicp2.1 and dicp3.1, which present higher numbers of apparently functional transcripts (Online Resource 3 – Fig. S14). Most DICP transcripts encoding PTCs were identified from lymphoid cells although a few were recovered from myeloid cells. The source of these differences is unknown and it is recognized that the efficiency of each PCR may influence these observations.
Sequence analyses also identified several DICP transcripts encoding D1 domains that were less than 90% identical to previously described DICP D1 domains (Haire et al. 2012). Included in this group are eleven new DICP genes, dicp1.22 – dicp1.30, dicp3.7 and dicp3.8. These new genes encode inhibitory receptors (dicp1.24, dicp1.25 and dicp1.30), secreted proteins (dicp1.23 and a dicp1.17 splicing isoform) and proteins with ambiguous function (dicp1.22). Only two of these genes, dicp1.22 and dicp1.23, could be mapped to the reference genome (GRCz10), where they are encoded in scaffold NA310, which also encodes one MHC class I Z gene cluster haplotype (Dirscherl and Yoder 2014). Scaffold NA310 likely represents an alternative haplotype for both MHC class I Z genes and DICP genes on chromosome 3. The identification of transcripts for dicp1.24-dicp1.30 without representation in the genomic databases suggests that they may be present on the haplotype which encodes dicp1.22 and dicp1.23 or that additional DICP haplotypes remain to be identified for this locus on chromosome 3.
Definitive DICPs have been identified in three subfamilies of the Cyprinidae family: Rasborinae (zebrafish), Leuciscinae (grass carp) and Cyprininae (common carp). The relationship of DICP sequences to DICP-like sequences in non-cyprinid teleosts remains to be elucidated. Although it is possible that DICPs may be restricted to cyprinids, DICPs more likley share an ancient origin with DICP-like sequences from other teleost lineages. The identification of DICPs and their linkage to MHC class I sequences in multiple cyprinid lineages indicates that the DICPs and their linkage to MHC class I has been conserved for at least 70 million years which is the estimated divergence time of the Cyprinidae family (Broughton et al 2013). The linkage of Ig domain-containing innate immune receptor genes (such as those encoding DICPs) to MHC genes supports the model that similar genes were linked in the ancient “proto-MHC” (or “Ur-MHC”) (Kasahara 1999; Abi Rached et al. 1999). This model places the primordial MHC genes in the same chromosomal region as Ig domain- containing receptors, which would be precursors to modern mammalian receptors encoded at the leukocyte receptor complex (LRC) (Flajnik and Kasahara 2010). The proposal that the precursors of the mammalian LRC and MHC were once genetically linked is supported by their demonstrated functional relationship—many receptors encoded at the LRC bind MHC class I proteins as markers of “self” (Vivier et al. 2008). The linkage of DICP and MHC genes in cyprinids may have originated from the proto-MHC; however, it is not known if they are linked functionally. In support of an ancient linkage, DICP-like sequences from medaka (reported as “NITR- WxC” sequences) are linked to MHC class II sequences (Ohashi et al. 2010). The functional significance of this linkage remains to be elucidated and further studies are required to investigate the DICP-like sequences outside of cyprinid fishes.
It is likely that additional haplotypes of the DICP3 cluster will be identified on chromosome 16 as: 1) genes encoding dicp3.7 and dicp3.8 are absent from the current reference genome, 2) the dicp3.6 gene is absent from earlier versions of the reference genome (Zv8) but has been identified in two BAC clones (Haire et al. 2012) and 3) dicp3.6 amplicons only have been detected in certain individual zebrafish (Fig. 2) and only in embryos of the TU line (Fig. 4). The possibility of multiple DICP haplotypes is supported by the high frequency of CNVs observed at all three DICP gene clusters (Rodriguez-Nunez et al. 2014). CNVs provide an important evolutionary strategy for chromosomal rearrangement as well as the creation of novel loci and commonly occur in immune-related gene clusters, such as the mammalian MHC and KIR loci (Dirscherl and Yoder 2014, Traherne et al. 2010). As different haplotypes of immune-related genes have been correlated with resistance or susceptibility to infections and autoimmunity (Jackson et al. 2007, Olsson and Holmdahl 2012, Pelak et al. 2011), it will be of interest to determine if different DICP haplotypes influence individual fitness of zebrafish.
Supplementary Material
ESM_1. Table S1. Complete DICP amplicon sequence data including: cDNA source, DNA polymerase, nucleotide and amino acid sequence and GenBank accession numbers.
ESM_2. Table S2. Percent (nucleotide) identity between DICP amplicons generated with a high fidelity DNA polymerase and previously described DICP sequences.
ESM_3.DICP sequence variation; DICP exon-intron architecture Figures S1 – S14; Tables S3 – S4.
Acknowledgements
We thank Hayley Dirscherl (NC State University) for helpful discussions and zebrafish genomic DNA samples, John Whitesides and Patti McDermott (Duke Human Vaccine Institute, Duke University School of Medicine) for assistance with cell sorting, Diana Skapura (All Children’s Hospital) for assistance with DNA sequencing and Barbara Pryor for editorial assistance. The authors are supported by the National Institutes of Health (R01 AI057559 to G.W.L. and J.A.Y. and R01 AI23337 to G.W.L.) and the National Evolutionary Synthesis Center (NESCent), NSF #EF‐0905606 (D.J.W.).
Footnotes
Electronic Supplementary Material. The online version of this article contains supplementary material, which is available to authorized users.
References
- Abi Rached L, McDermott MF, Pontarotti P (1999) The MHC big bang. Immunol Rev 167:33–44. doi: 10.1111/j.1600-065x.1999.tb01380.x [DOI] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E et al. (2002) Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297:1301–1310. doi: 10.1126/science.1072104 [DOI] [PubMed] [Google Scholar]
- Berry R, Ng N, Saunders PM et al. (2013) Targeting of a natural killer cell receptor family by a viral immunoevasin. Nat Immunol 14:699–705. doi: 10.1038/ni.2605 [DOI] [PubMed] [Google Scholar]
- Borrego F (2013) The CD300 molecules: an emerging family of regulators of the immune system. Blood 121:1951–1960. doi: 10.1182/blood-2012-09-435057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton RE, Betancur-R R, Li C, Arratia G, Ortí G (2013) Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution. PLoS Curr. April 16;5. doi: 10.1371/currents.tol.2ca8041495ffafd0c92756e75247483e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KH, Dobrinski KP, Lee AS et al. (2012) Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proceedings of the National Academy of Sciences 109:529–534. doi: 10.1073/pnas.1112163109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon JP, O’Driscoll M, Litman GW (2012) Specific lipid recognition is a general feature of CD300 and TREM molecules. Immunogenetics 64:39–47. doi: 10.1007/s00251-011-0562-4 [DOI] [PubMed] [Google Scholar]
- Clark GJ, Ju X, Tate C, Hart DN (2009) The CD300 family of molecules are evolutionarily significant regulators of leukocyte functions. Trends Immunol 30:209–217. doi: 10.1016/j.it.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Dirscherl H, Yoder JA (2014) Characterization of the Z lineage Major histocompatability complex class I genes in zebrafish. Immunogenetics 66:185–198. doi: 10.1007/s00251-013-0748-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M (2010) Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet 11(1):47–59. doi: 10.1038/nrg2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer-Guerrerio PA, Montgomery RA, Warren DS et al. (2011) Perturbation of thymocyte development in nonsense-mediated decay (NMD)-deficient mice. Proc Natl Acad Sci U S A 108:10638–10643. doi: 10.1073/pnas.1019352108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloggnitzer J, Akimcheva S, Srinivasan A et al. (2014) Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe 16:376–390. doi: 10.1016/j.chom.2014.08.010 [DOI] [PubMed] [Google Scholar]
- Haire RN, Cannon JP, O’Driscoll ML et al. (2012) Genomic and functional characterization of the diverse immunoglobulin domain-containing protein (DICP) family. Genomics 99:282–291. doi: 10.1016/j.ygeno.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid FM, Makeyev EV (2014) Emerging functions of alternative splicing coupled with nonsense-mediated decay. Biochem Soc Trans 42:1168–1173. doi: 10.1042/BST20140066 [DOI] [PubMed] [Google Scholar]
- Hartmann J, Tran TV, Kaudeer J et al. (2012) The stalk domain and the glycosylation status of the activating natural killer cell receptor NKp30 are important for ligand binding. J Biol Chem 287:31527–31539. doi: 10.1074/jbc.M111.304238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF et al. (2013) The zebrafish reference genome sequence and its relationship to the human genome. Nature 496:498–503. doi: 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AN, McLure CA, Dawkins RL, Keating PJ (2007) Mannose binding lectin (MBL) copy number polymorphism in Zebrafish (D. rerio) and identification of haplotypes resistant to L. anguillarum. Immunogenetics 59:861–872. doi: 10.1007/s00251-007-0251-5 [DOI] [PubMed] [Google Scholar]
- Jima DD, Shah RN, Orcutt TM et al. (2009) Enhanced transcription of complement and coagulation genes in the absence of adaptive immunity. Mol Immunol 46:1505–1516. doi: 10.1016/j.molimm.2008.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M, Simpson CG, Syed NH et al. (2012) Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res 40:2454–2469. doi: 10.1093/nar/gkr932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M (1999) The chromosomal duplication model of the major histocompatibility complex. Immunol Rev 167:17–32. doi: 10.1111/j.1600-065x.1999.tb01379.x [DOI] [PubMed] [Google Scholar]
- Kervestin S, Jacobson A (2012) NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 13:700–712. doi: 10.1038/nrm3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konantz M, Balci TB, Hartwig UF et al. (2012) Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann N Y Acad Sci 1266:124–137. doi: 10.1111/j.1749-6632.2012.06575.x [DOI] [PubMed] [Google Scholar]
- Kortum AN, Rodriguez-Nunez I, Yang J et al. (2014) Differential expression and ligand binding indicate alternative functions for zebrafish polymeric immunoglobulin receptor (pIgR) and a family of pIgR-like (PIGRL) proteins. Immunogenetics 66:267–279. doi: 10.1007/s00251-014-0759-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40:D302–5. doi: 10.1093/nar/gkr931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loucks E, Carvan MJ 3rd (2004) Strain-dependent effects of developmental ethanol exposure in zebrafish. Neurotoxicol Teratol 26:745–755. doi: 10.1016/j.ntt.2004.06.017 [DOI] [PubMed] [Google Scholar]
- Maisey K, Imarai M (2011) Diversity of teleost leukocyte molecules: role of alternative splicing. Fish Shellfish Immunol 31:663–672. doi: 10.1016/j.fsi.2010.08.001 [DOI] [PubMed] [Google Scholar]
- Meeker ND, Hutchinson SA, Ho L, Trede NS (2007) Method for isolation of PCR-ready genomic DNA from zebrafish tissues. BioTechniques 43:610, 612, 614. doi: 000112619 [DOI] [PubMed] [Google Scholar]
- Moody AM, Chui D, Reche PA, Priatel JJ, Marth JD, Reinherz EL (2001) Developmentally regulated glycosylation of the CD8alphabeta coreceptor stalk modulates ligand binding. Cell 107:501–512. doi: S0092-8674(01)00577-3 [DOI] [PubMed] [Google Scholar]
- Novoa B, Figueras A (2012) Zebrafish: model for the study of inflammation and the innate immune response to infectious diseases. Adv Exp Med Biol 946:253–275. doi: 10.1007/978-1-4614-0106-3_15 [DOI] [PubMed] [Google Scholar]
- Ohashi K, Takizawa F, Tokumaru N et al. (2010) A molecule in teleost fish, related with human MHC-encoded G6F, has a cytoplasmic tail with ITAM and marks the surface of thrombocytes and in some fishes also of erythrocytes. Immunogenetics 62:543–559. doi: 10.1007/s00251-010-0460-1 [DOI] [PubMed] [Google Scholar]
- Olsson LM, Holmdahl R (2012) Copy number variation in autoimmunity--importance hidden in complexity?. Eur J Immunol 42:1969–1976. doi: 10.1002/eji.201242601 [DOI] [PubMed] [Google Scholar]
- Patowary A, Purkanti R, Singh M et al. (2013) A sequence-based variation map of zebrafish. Zebrafish 10:15–20. doi: 10.1089/zeb.2012.0848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelak K, Need AC, Fellay J et al. (2011) Copy number variation of KIR genes influences HIV-1 control. PLoS Biol 9:e1001208. doi: 10.1371/journal.pbio.1001208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham CJ, Agrawal DK (2014) Emerging roles for triggering receptor expressed on myeloid cells receptor family signaling in inflammatory diseases. Expert Rev Clin Immunol 10:243–256. doi: 10.1586/1744666X.2014.866519 [DOI] [PubMed] [Google Scholar]
- Pimentel H, Parra M, Gee S et al. (2014) A dynamic alternative splicing program regulates gene expression during terminal erythropoiesis. Nucleic Acids Res 42:4031–4042. doi: 10.1093/nar/gkt1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw SA, Trede NS (2012) A model 450 million years in the making: zebrafish and vertebrate immunity. Dis Model Mech 5:38–47. doi: 10.1242/dmm.007138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Nunez I, Wcisel DJ, Litman GW, Yoder JA (2014) Multigene families of immunoglobulin domain-containing innate immune receptors in zebrafish: Deciphering the differences. Dev Comp Immunol 46:24–34. doi: S0145-305X(14)00037-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425 [DOI] [PubMed] [Google Scholar]
- Sakai M, Kono T, Savan R (2005) Identification of expressed genes in carp (Cyprinus carpio) head kidney cells after in vitro treatment with immunostimulants. Dev Biol (Basel) 121:45–51 [PubMed] [Google Scholar]
- Sipes NS, Padilla S, Knudsen TB (2011) Zebrafish?As an integrative model for twenty-first century toxicity testing. Birth Defects Research Part C: Embryo Today: Reviews 93:256–267. doi: 10.1002/bdrc.20214 [DOI] [PubMed] [Google Scholar]
- Stafford JL, Bengten E, Du Pasquier L et al. (2006) A novel family of diversified immunoregulatory receptors in teleosts is homologous to both mammalian Fc receptors and molecules encoded within the leukocyte receptor complex. Immunogenetics 58:758–773. doi: 10.1007/s00251-006-0134-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star B, Nederbragt AJ, Jentoft S et al. (2011) The genome sequence of Atlantic cod reveals a unique immune system. Nature 477:207–210. doi: 10.1038/nature10342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stet RJ, Hermsen T, Westphal AH et al. (2005) Novel immunoglobulin-like transcripts in teleost fish encode polymorphic receptors with cytoplasmic ITAM or ITIM and a new structural Ig domain similar to the natural cytotoxicity receptor NKp44. Immunogenetics 57:77–89. doi: 10.1007/s00251-005-0771-9 [DOI] [PubMed] [Google Scholar]
- Sullivan C, Kim CH (2008) Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol 25:341–350. doi: 10.1016/j.fsi.2008.05.005 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traherne JA, Martin M, Ward R et al. (2010) Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum Mol Genet 19:737–751. doi: 10.1093/hmg/ddp538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D (2004) Cellular dissection of zebrafish hematopoiesis. Methods Cell Biol 76:127–149 [DOI] [PubMed] [Google Scholar]
- Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI (2003) Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol 4:1238–1246. doi: 10.1038/ni1007 [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T et al. (2012) Primer3--new capabilities and interfaces. Nucleic Acids Res 40:e115. doi: gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman MB, Lin S (2008) Zebrafish as a developmental model organism for pediatric research. Pediatr Res 64:470–476. doi: 10.1203/PDR.0b013e318186e609 [DOI] [PubMed] [Google Scholar]
- Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S (2008) Functions of natural killer cells. Nat Immunol 9(5):503–10. doi: 10.1038/ni1582 [DOI] [PubMed] [Google Scholar]
- Wang Y, Lu Y, Zhang Y et al. (2015) The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat Genet. 47(6):625–31. doi: 10.1038/ng.3280. [DOI] [PubMed] [Google Scholar]
- Wei S, Zhou JM, Chen X et al. (2007) The zebrafish activating immune receptor Nitr9 signals via Dap12. Immunogenetics. 59(10):813–21. doi: 10.1007/s00251-007-0250-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M (2007) The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio). Univ. of Oregon Press, Eugene, OR [Google Scholar]
- Wittamer V, Bertrand JY, Gutschow PW, Traver D (2011) Characterization of the mononuclear phagocyte system in zebrafish. Blood 117:7126–7135. doi: 10.1182/blood-2010-11-321448 [DOI] [PubMed] [Google Scholar]
- Wong JJ, Ritchie W, Ebner OA et al. (2013) Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154:583–595. doi: 10.1016/j.cell.2013.06.052 [DOI] [PubMed] [Google Scholar]
- Xu C, Call ME, Wucherpfennig KW (2006) A membrane-proximal tetracysteine motif contributes to assembly of CD3deltaepsilon and CD3gammaepsilon dimers with the T cell receptor. J Biol Chem 281:36977–36984. doi: 10.1074/jbc.M607164200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Zhang X, Wang X et al. (2014) Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat Genet. 46(11):1212–9. doi: 10.1038/ng.3098. [DOI] [PubMed] [Google Scholar]
- Yang L, Zou M, Fu B, He S (2013) Genome-wide identification, characterization, and expression analysis of lineage-specific genes within zebrafish. BMC Genomics 14:65-2164-14-65. doi: 10.1186/1471-2164-14-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Turner PM, Wright PD et al. (2010) Developmental and tissue-specific expression of NITRs. Immunogenetics 62:117–122. doi: 10.1007/s00251-009-0416-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA (2009) Form, function and phylogenetics of NITRs in bony fish. Dev Comp Immunol 33:135–144. doi: 10.1016/j.dci.2008.09.004 [DOI] [PubMed] [Google Scholar]
- Yoder JA, Cannon JP, Litman RT, Murphy C, Freeman JL, Litman GW (2008) Evidence for a transposition event in a second NITR gene cluster in zebrafish. Immunogenetics 60:257–265. doi: 10.1007/s00251-008-0285-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Litman RT, Mueller MG et al. (2004) Resolution of the novel immune-type receptor gene cluster in zebrafish. Proc Natl Acad Sci U S A 101:15706–15711. doi: 0405242101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Mueller MG, Wei S et al. (2001) Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian leukocyte receptor cluster. Proc Natl Acad Sci U S A 98:6771–6776. doi: 10.1073/pnas.121101598 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ESM_1. Table S1. Complete DICP amplicon sequence data including: cDNA source, DNA polymerase, nucleotide and amino acid sequence and GenBank accession numbers.
ESM_2. Table S2. Percent (nucleotide) identity between DICP amplicons generated with a high fidelity DNA polymerase and previously described DICP sequences.
ESM_3.DICP sequence variation; DICP exon-intron architecture Figures S1 – S14; Tables S3 – S4.


