ABSTRACT
The biological basis underlying cognitive dysfunction in women with early-stage breast cancer (BC) remains unclear, but could reflect gene expression changes that arise from the acquisition and long-term retention of soma-wide alterations in DNA methylation in response to chemotherapy. In this longitudinal study, we identified differences in peripheral methylation patterns present in women prior to treatment (T1) and 1 year after receiving chemotherapy (T4) and evaluated relationships among the differential methylation (DM) ratios with changes in cognitive function. A total of 58 paired (T1 and T4) blood specimens were evaluated. Methylation values were determined for DNA isolated from whole blood using a genome-wide array . Cognitive function was measured using the validated, computerized CNS Vital Signs platform. Relationships between methylation patterns and cognitive domain scores were compared using a stepwise linear regression analysis, with demographic variables as covariates. The symptom comparison analysis was restricted to 2,199 CpG positions showing significant methylation ratio changes between T1 and T4. The positions with DM were enriched for genes involved in the modulation of cytokine concentrations. Significant DM ratios were associated with memory domain (56 CpGs). Eight of the ten largest DM ratio changes associated with lack of memory improvement were localized to genes involved in either neural function (ECE2, PPFIBP2) or signalling processes (USP6NL, RIPOR2, KLF5, UBE2V1, DGKA, RPS6KA1). These results suggest that epigenetic changes acquired and retained for at least one year in non-tumour cells following chemotherapy may be associated with a lack of memory improvement following treatment in BC survivors.
KEYWORDS: Breast cancer, chemotherapy, epigenetics, DNA methylation, chemo-brain, cognitive function
Introduction
The five-year survival rate for women with early-stage breast cancer (BC) has risen markedly from 75% in 1976 to 90% in 2016 [1]. Although the survival gains are quite notable, BC survivors continue to report long-term treatment/disease-associated side effects, including perceived deficits in cognition. While the percentage of BC survivors having objective decrements in cognition remains controversial, at least a subset of these women have been shown to have clear decrements in functional status and quality of life [2]. Therefore, the biological basis underlying the development and persistence of cognitive dysfunction in women with early-stage BC remains an important focus of survivorship research. Multiple biological pathways have been explored to identify factors that could provide a logical explanation for the cognitive changes reported over the BC treatment trajectory and in survivorship [3–5] but, to date, no clear causal associations have emerged.
We and others have hypothesized that soma-wide epigenetic alterations acquired following chemotherapy could contribute to cognitive dysfunction [6–8]. While epigenetic alterations in tumour tissue have been studied for over 20 years, with identified alterations showing regions with global/local hypomethylation, as well as regions with hypermethylation [9–12], there is a paucity of studies evaluating ‘bystander’ alterations in methylation patterns in healthy somatic cells following chemotherapy [13–18]. Epigenetic alterations can include post-translational changes involving histones, higher-order chromatin organizational changes, and DNA methylation, with DNA methylation being the most commonly studied epigenetic modification in humans [19]. An emerging body of evidence implicates DNA methylation as a critical mechanism of learning and memory, with gene-specific modifications observed in the development of cognitive dysfunction [20]. For example, in the study of Haberman and colleagues (2012), CpG island DNA methylation in the promotor region of Gabra5 gene, which is highly expressed in the hippocampus, correlates with memory and learning, but its mRNA expression levels decrease with age in a rat model of neurocognitive ageing [20]. Given that DNA methylation is potentially reversible and may be a therapeutic target for some forms of cognitive dysfunction, the primary aim of this longitudinal study was to determine if: (1) changes in DNA methylation patterns were present among women with early-stage BC that were acquired and retained one year post inception of chemotherapy; and (2) differential DNA methylation was associated with cognitive function.
Methods
Study procedures have been reported elsewhere [21]. In the parent study (EPIGEN), women diagnosed with early-stage BC were evaluated across 5 time points: prior to their receipt of chemotherapy (T1), at the time of their 4th chemotherapy treatment (T2), 6 months after the initiation of chemotherapy (T3), one year after the initiation of chemotherapy (T4) and two years after the initiation of chemotherapy (T5). The research team prospectively characterized psychoneurological symptoms (i.e. anxiety, depression, cognitive dysfunction, fatigue, pain, sleep disturbances), inflammatory biomarkers, and epigenetic alterations. For this report, we examined modifications in DNA methylation at two time points (T1 and T4).
Sample and data collection
A total of 75 women with early stage (I to IIIA) BC initiated data collection following their recruitment through a National Cancer Institute designated cancer centre in Central Virginia and multiple collaborative sites statewide. The study eligibility criteria were: (a) age (at least 21 years old); and (b) a diagnosis of early stage BC with a scheduled clinic appointment to receive chemotherapy. Exclusion criteria were: (a) a history of a previous cancer, or chemotherapy; (b) a diagnosis of dementia; (c) active psychosis; or (d) immune-related diagnoses (e.g., multiple sclerosis, systemic lupus erythematosus). All methods and protocols were approved by the institutional review board for the university health system cancer centre and affiliated institutions. A signed consent form was obtained from each study participant. After obtaining informed consent, participants were asked to complete questionnaires, undergo performance-based cognitive testing via a computerized system and have their blood drawn.
Measures
Covariates
Demographic characteristics, concurrent symptoms, and blood cell compositions (i.e. leukocyte heterogeneity [indirectly inferred using the Houseman et al.’s statistical method [22]]) were included in this analysis as covariates. Comprehensive questionnaires, medical record review and participant interviews were performed at baseline and at subsequent data points for demographic, cancer and cancer treatment-related variables, and concurrent symptoms (perceived stress, anxiety and depressive symptoms, fatigue, and pain) (Table 1). The major cell composition was estimated using the R package ‘minfi’.
Table 1.
Demographics of the study participants undergoing treatment for early stage breast cancer (N = 58).
| Variable | N (%) or mean ± SD (range) |
|---|---|
| Age | 51.48 ± 10.52 (23, 69) |
| Race | |
| African-American | 19 (32.8%) |
| Caucasian | 39 (67.2%) |
| Others | 0 (0.0%) |
| BMI (kg/m2) | 30.27 ± 7.31 (19.11, 54.34) |
| Current smoking status | |
| Yes | 11 (19.0%) |
| No | 47 (81.0%) |
| Stage | |
| I | 16 (27.6%) |
| IIA | 26 (44.8%) |
| IIB | 11 (19.0%) |
| IIIA | 5 (8.6%) |
| Triple negative | |
| Yes | 19 (32.8%) |
| No | 39 (67.2%) |
| Neoadjuvant | |
| Yes | 6 (10.3%) |
| No | 52 (89.7%) |
| Radiation treatment | |
| Yes | 44 (75.9%) |
| No | 14 (24.1%) |
| Chemotherapy regimen | |
| AC | 1 (1.7%) |
| CMF | 2 (3.4%) |
| TAC | 25 (43.1%) |
| TC | 19 (32.8%) |
| TCH | 11 (19.0%) |
| BFI (Total) | 2.25 ± 2.71 (0, 9.5) |
| PSS | 2.15 ± 0.35 (1.5, 3) |
| HADS | 5.67 ± 2.69 (0, 12) |
| BPI (Pain severity) | 1.69 ± 2.27 (0, 7) |
Abbreviations: AC, doxorubicin (Adriamycin), cyclophosphamide (Cytoxan); BFI, brief fatigue inventory; BMI, body mass index; BPI, brief pain inventory; CMF, cyclophosphamide, methotrexate and fluorouracil; HADS, hospital anxiety and depression scale; PSS, perceived stress scale; SD, standard deviation; TAC, docetaxel (Taxotere), doxorubicin (Adriamycin), cyclophosphamide (Cytoxan); TC, docetaxel (Taxotere) and cyclophosphamide (Cytoxan); TCH, docetaxel (Taxotere), carboplatin (Paraplatin), trastuzumab (Herceptin).
Cognitive function
A performance-based computerized neurocognitive testing system, CNS Vital SignsTM (CNSVS, https://www.cnsvs.com), was used to evaluate multiple cognitive domains [23]. Test results were obtained in subject (raw) scores, age-matched standard scores, and percentile ranks. The CNSVS scores individual tests and calculates a report of the clinical domains of neurocognitive functioning. In this analysis, memory, psychomotor speed (e.g., finger tapping, symbol digit coding), reaction time (i.e. how quickly subject responds), complex attention (i.e. continuous performance, shifting attention), and cognitive flexibility (i.e. shifting attention) domains were assessed. Descriptions of these domains and their relevance are given in Supplementary Table 1. The subscales of the CNSVS have good test-retest reliability: correlation coefficients for attention, memory, psychomotor speed, cognitive flexibility, and reaction time were 0.65, 0.66, 0.88, 0.71, and 0.75, respectively [23]. This test has been commonly used in women with BC [24,25].
Biological samples
Genomic DNA was isolated from whole blood using the Puregene DNA isolation kit (Qiagen, Valencia, CA). An aliquot of 500 μg of DNA per specimen was bisulfite converted using an EZ DNA Methylation Kit (Zymo Research, Irvine, CA) and then hybridized to a genome-wide Infinium HumanMethylation450K BeadChip (HM450K; Illumina, San Diego, CA), with both of these procedures being performed according to the manufacturer’s protocols (at HudsonAlpha Institute for Biotechnology).
Statistical analysis
Statistical analyses were performed in R statistical software v3.5.2. Descriptive statistics were presented for demographic data and outcomes, with means, standard deviations (SDs) and ranges. If CNSVS scores were smaller than 40, they were replaced with 40 [23]. As higher scores on reaction time and complex attention domains indicate lower cognitive function, they were reversed scored for the analysis. To determine the changes over time in each cognitive domain, linear mixed effects models were fit to model each domain separately. Raw methylation sequencing data were processed using the R programming environment ‘minfi’ Bioconductor package. There were 61 subjects with methylation data available for both T1 and T4. Following a quality control workflow with R package ‘MethylAid’ [26], three subjects were excluded, to yield a total sample size of 58 paired specimens. These subjects were excluded because they had CpG positions with probe detection call rates of less than 95% (pre-established quality control criteria). For the 485,512 CpG positions which remained eligible for analysis, methylation ratio intensities (β-values) were calculated using the following formula: M divided by U + M + α, where M = methylated; U = unmethylated; and α = 100. Wilcoxon signed rank tests were used to compare paired methylation ratios at each CpG position, followed by a Bonferroni adjustment for 485,512 separate statistical tests on P-values. Significant differentially methylated positions (DMPs) with an adjusted P-value of less than 0.05 were selected. Next, a multiple linear regression model was used to examine potential association between the change of each cognitive domain from T1 to T4 as the outcome variable and the change of each of the selected methylation CpG positions as the predictor variable of interest while controlling for effects of demographic covariates as well as change of major cell type proportions. Due to the relatively small sample size, the model size was reduced by excluding irrelevant covariates via stepwise model selection. Covariates selected for inclusion in the model were based on reports of potential relationships from the extant literature, and included age, race, current smoking status, body mass index (BMI), tumour stage, hormone receptor status, neoadjuvant, radiation therapy, fatigue, perceived stress, anxiety, depressive symptoms, and pain. In addition, to account for the impact of cell composition, we added the changes of six major cell proportions between T1 and T4, to the covariate list, including CD8 + T cells, CD4 + T cells, Natural Killer Cells, B Cells, Monocyte, and Granulocyte, with the values for these cell types being estimated using the R package ‘minfi’. The Akaike information criterion (AIC) was used for model selection procedures [27]. Potential relationships between DMPs and multiple cognitive function domains were identified using a false discovery rate (FDR) control at q < 0.1. The DMPs were annotated and mapped to the nearest genes using R package ‘IlluminaHumanMethylation450kanno.ilmn12.hg19’ and ‘FDb.InfiniumMethylation.hg19’ from Bioconductor. In addition, gene set enrichment analysis for biological functions was conducted using the genomic regions enrichment of annotations tool (GREAT) online software (http://great.stanford.edu/great/public/html/). The background was set to the total Illumina 450K array. Only CpG positions with methylation Δβ value > 5% at a FDR rate < 0.05 were included in the annotation analysis. Lastly, we predicted transcription factors located near 10 top-ranked DMPs through in silico analysis. The DNA sequences around the methylation locations were obtained using ENSENBL, and the promotor binding sites located near the methylated points were determined using Alibaba 2.1 [28].
Results
Sample characteristics
All 58 participants received chemotherapy. Nearly half of the chemotherapy participants (N = 25, 43.1%) were treated with a docetaxel (Taxotere), doxorubicin (Adriamycin) and cyclophosphamide (Cytoxan) (TAC) regimen. Nineteen (32.8%) received a docetaxel and cyclophosphamide (TC) regimen. Eleven (19.0%) were treated with docetaxel, carboplatin (Paraplatin) and trastuzumab (Herceptin) (TCH) regimen. The remaining participants (N = 3) were treated with doxorubicin and cyclophosphamide (AC) regimen or cyclophosphamide, methotrexate and fluorouracil (CMF) regimen. The study sample consisted of a majority of Caucasian non-Hispanic women (67.2%), with an average age of 51.48 ± 10.52 years. Cancer stages of participants were Stage I (27.6%), Stage IIA (44.8%), Stage IIB (19.0%), and Stage IIIA (8.6%). A majority of participants were diagnosed with non-triple negative BC (67.2%), did not receive neoadjuvant therapy (89.7%), but did receive radiation treatment (75.9%), and were non-smokers (81.0%). The average BMI of the participants was 30.27 ± 7.31 kg/m2, which falls within the obese range (Table 1).
Cognitive domain outcomes
As shown in Table 2, five cognitive domain outcomes, i.e. memory, psychomotor speed, reaction time, complex attention, and cognitive flexibility, were assessed at baseline, 6-months, and 1-year after the initiation of chemotherapy. Except for psychomotor speed, mean scores of all cognitive domains were below 100 at T1. Over one year, significant improvements from baseline were observed for psychomotor speed (F = 7.180, P = .001), reaction time (F = 18.193, P < .001), complex attention (F = 6.775, P = .002), and cognitive flexibility (F = 21.673, P < .001). The greatest difference occurred in cognitive flexibility (improved). Moderate increases in scores were observed in psychomotor speed, complex attention, and reaction time, suggesting that they showed improved perception, attention, response and coordination, and increased complex attention over time, as well as quicker reaction at T4 compared to T1. However, the mean score for the memory domain was not significantly different across timepoints (F = 0.102, P = .903), indicating a lack of improvement/change for this component.
Table 2.
Cognitive domain outcomes (N = 58).
| Cognitive domain | Baseline (prior to their receipt of chemotherapy) | 6-month after the initiation of chemotherapy | 1-year after the initiation of chemotherapy | F (P-value) |
|---|---|---|---|---|
| Memory | 97.4 (13.9) [67, 127] |
98.2 (19.6) [41, 140] |
98.5 (17.6) [51, 126] |
0.102 (.903) |
| Psychomotor speed | 114.0 (23.5) [62, 172] |
117.4 (27.1) [39, 186] |
120.8 (26.4) [39, 190] |
7.180 (.001) |
| Reaction time | 99.3 (17.1) [39, 126] |
103.0 (15.6) [36, 130] |
105.9 (12.9) [54, 133] |
18.193 (<.001) |
| Complex attention | 99.8 (13.8) [64, 121] |
101.9 (19.1) [38, 121] |
106.0 (12.5) [39, 121] |
6.775 (.002) |
| Cognitive flexibility | 98.4 (16.4) [58, 127] |
104.2 (20.3) [27, 126] |
108.2 (14.8) [40, 131] |
21.673 (< .001) |
Values are standardized scores and presented as mean and standard deviation (SD). The minimum and maximum values (range) were presented under the mean scores. As higher scores on reaction time and complex attention domains indicate lower cognitive function, they were reversed scored for the analysis.
P < .05 denotes significant differences between three time points.
Association between CpG methylation levels and cognitive function
DNA methylation ratios at 485,512 CpG dinucleotides were evaluated and identified 2,199 DMPs in the T1 (baseline) compared to T4 (one year after chemotherapy initiation) specimens (P = 1.03 × 10−7), as shown in Figure 1a. The mean value of methylation ratio changes for the 2,199 CpG positions was −0.0367, suggesting that the overall methylation ratios decreased by 3.67% over one year (consistent with hypomethylation changes). The majority of CpG positions with significant changes showed a decrease in DNA methylation ratios (N = 2,113 [96%]). Biological functions for the 2,199 significant DMPs were investigated using the GREAT online tool [29]. These regions showing significant changes corresponded with a total of five clusters related to: (a) negative regulation of cytoplasmic mRNA processing body assembly (P = 9.22 × 10−7); (b) regulation of interleukin-8 (IL-8) production (P = 2.31 × 10−5); (c) dimethylallyl diphosphate biosynthetic process (P = 3.98 × 10−5); (d) positive regulation of chemokine biosynthetic process (P = 5.21 × 10−5); and (e) regulation of cellular senescence (P = 5.36 × 10−5). The biological process with the highest degree of enrichment was ‘negative regulation of cytoplasmic mRNA processing body assembly’, and the biological function with the highest number of total regions and genes involved was ‘regulation of interleukin-8 production’. A list of significant genes that emerged from this assessment for each of these biological function clusters is provided in Table 3.
Figure 1.
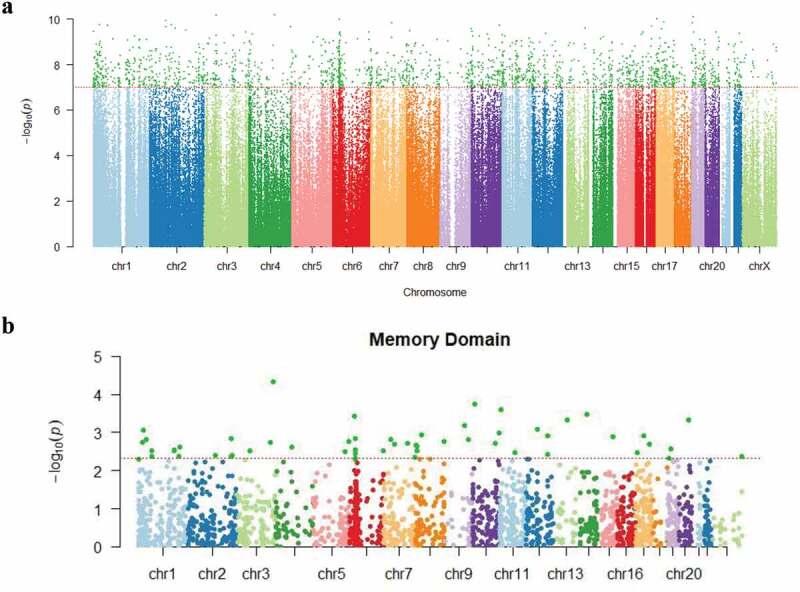
(a). Manhattan plot showing P values for changes in methylation at the individual CpG positions from baseline to one year after the initiation of chemotherapy. The Y axis shows log transformed P values. The X axis shows the chromosomal location of the 485,512 CpG positions evaluated. The dotted horizontal line indicates the threshold for significance (P = 1.03 × 10−7). (b). Manhattan plot for P values for associations between CpG methylation ratio differences and memory domain score. The Y axis shows log transformed P values. The X axis shows the chromosomal location of the 2,199 positions with significant differences in methylation ratios from baseline to 1 year post chemotherapy induction. The dotted horizontal line indicates the threshold for significance (P = 4.91 × 10−3) in associations between memory domain scores and methylation ratio differences, with these 56 CpG sites being highlighted in dark green.
Table 3.
Lists of significant biological functions among the genomic regions that include 2,199 differentially methylated CpG positions between baseline (prior to their receipt of chemotherapy [T1]) and one year after the initiation of chemotherapy (T4).
| Biological Process | Hyper rank | Hyper raw P-value | Hyper FDR Q-value |
Hyper fold enrichment |
Hyper total regions |
Hyper foreground gene hits | Total genes annotated | Genes |
|---|---|---|---|---|---|---|---|---|
| Negative regulation of cytoplasmic mRNA processing body assembly | 1 | 9.2158 × 10−7 | 9.6213 × 10−3 | 132.4726 | 5 | 1 | 1 | PATL2 |
| Regulation of interleukin-8 production | 6 | 2.3100 × 10−5 | 4.0193 × 10−2 | 2.8012 | 1734 | 11 | 43 | TLR5, BCL10, TNF, AFAP1L2, WNT5A, F2RL1, TLR4, GDF2, NLRP10, ARRB1 |
| Dimethylallyl diphosphate biosynthetic process | 9 | 3.9826 × 10−5 | 4.6198 × 10−2 | 4.5804 | 530 | 1 | 2 | IDI1 |
| Positive regulation of chemokine biosynthetic process | 13 | 5.2148 × 10−5 | 4.1879 × 10−2 | 4.8847 | 452 | 5 | 10 | TNF, MYD88, WNT5A, IL4, IFNG |
| Regulation of cellular senescence | 14 | 5.3600 × 10−5 | 3.9970 × 10−2 | 3.5942 | 860 | 7 | 12 | CDK6, NUAK1, VASH1, HMGA1, ZKSCAN3, NEK6, HMGA2 |
Abbreviations: AFAP1L2, actin filament associated protein 1 like 2; ARRB1, arrestin beta 1; BCL10, B-cell lymphoma/leukaemia 10; CDK6, cyclin-dependent kinase 6; F2RL1, coagulation factor II (thrombin) receptor-like 1; FDR, false discovery rate; GDF2, growth differentiation factor 2; HMGA1, high mobility group AT-hook 1; HMGA2, high mobility group AT-hook 2; IDI1, isopentenyl-diphosphate delta isomerase 1; IFNG, interferon gamma; IL4, interleukin 4; MYD88, myeloid differentiation primary response 88; NEK6, never in mitosis gene a (NIMA)-related kinase 6; NLRP10, nucleotide-binding oligomerization domain (NOD)-like receptor family pyrin domain containing 10; NUAK1, novel (nua) kinase family 1; PATL2, protein associated with topoisomerase 2; TLR4, toll like receptor 4; TLR5, toll like receptor 5; TNF, tumour necrosis factor; VASH1, vasohibin 1; WNT5A, Wnt family member 5A; ZKSCAN3, zinc-finger with Kruppel-associated box (KRAB) and soluble calcium-activated nucleotidase (SCAN) domains.
To evaluate potential associations between differential methylation and cognitive function (e.g., memory, complex attention, psychomotor speed, reaction time, cognitive flexibility) over one year following a diagnosis/treatment for BC, a stepwise selection analysis was used, with each model of the five cognitive domains being adjusted for covariates. No significant associations with acquired/persistent methylation differences were observed for psychomotor speed (covariates in the model included current smoking status and hormone receptor status), reaction time domain (covariates were race, tumour stage, perceived stress scale [PSS], and white cell proportions [change of B cells]), complex attention domain (covariates were age, race, BMI, tumour stage, neoadjuvant status, hospital anxiety and depression scale [HADS], PSS, and change of B cells and monocyte) or cognitive flexibility domain (covariates were age, BMI, tumour stage, current smoking status, neoadjuvant, HADS, PSS, and change of CD4 + T cells and natural killer cells) . However, for memory domain (covariates were age, race, and hormone receptor status), 56 DMPs were identified (P = 4.91 × 10−3) as shown in Figure 1b. For some outcomes, cell proportion differences were identified as significantly contributing to the outcome, including reaction time domain, complex attention domain, and cognitive flexibility domain, while cell proportion differences were not retained by the stepwise procedure for memory domain or psychomotor speed domain models. These 56 CpG positions (at a P-value < .001) (Supplementary Table 2), showed a reduction of DNA methylation at one year post-chemotherapy, with the methylation ratio change being decreased by 4.14% on average (Figure 2a). Seven of these CpG positions (e.g., cg01333080, cg26452989, cg15530112, cg01450807, cg13046524, cg14356530 and cg08034986) were heavily methylated (i.e. B values of greater than 50%) at T1 and T4, based on a classification criteria of greater than 0.5 as having ‘heavily methylated’ status [30]. The top 10 positions showing the largest methylation ratio differences (T1 vs T4) are listed in Table 4. All CpG positions showed significantly negative linear estimate effects, indicating negative estimates of memory domain score changes for every unit increase in methylation ratios. The CpG position (cg26452989) that showed the largest estimated coefficient of memory domain score change for every unit increase in methylation ratio (−327.5, standard error (SE) = 80.9; γ = −0.463, P = 6.11 × 10-8) was localized to the USP6NL gene (Table 4 and Figure 2b). These DMPs included 5 sites localized to gene bodies, 2 sites localized to transcription start site regions, 1 site localized to promoter regions; and 2 sites localized to open sea regions (Table 4). Notably, eight out of the ten positions showing the largest methylation ratio changes were localized to genes involved in either neural functions (ECE2, PPFIBP2) or signalling processes (USP6NL, RIPOR2, KLF5, UBE2V1, DGKA, RPS6KA1). Additional information on all 56 significant DNA methylation positions (P < .005) is shown in supplementary Table 2 and supplementary Figure 1. We also examined putative transcription factors located near 10 top-ranked DMPs. Specificity protein 1 (Sp1), CCAAT/enhancer-binding protein alpha (C/EBPα), and nuclear factor kappa light chain enhancer of activated B cells (NFκB) were examples of transcription factors that were commonly shown. DMPs. Sp1, C/EBPα, and NFκB are involved in encoding proteins related to cellular processes, blood cell differentiation, and inflammation/immunity, respectively. Detailed information is provided in Supplementary Table 3.
Figure 2.
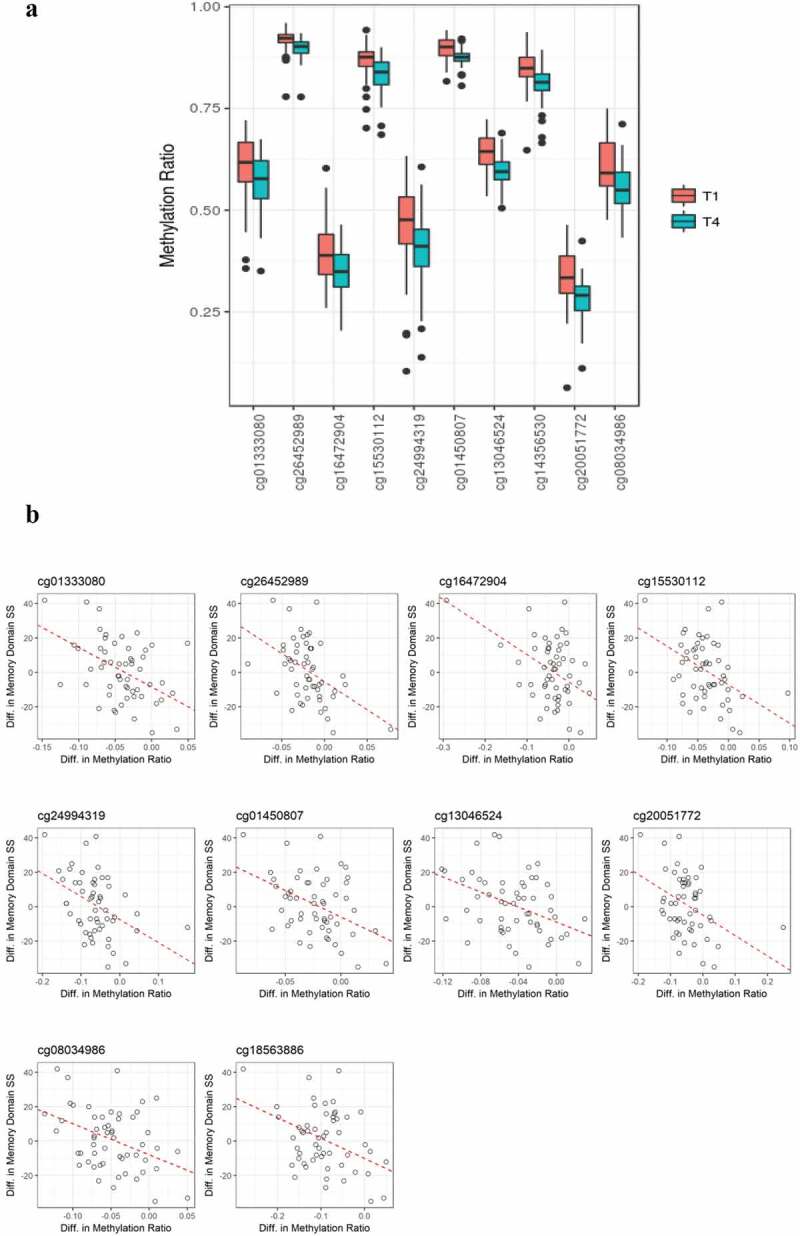
(a). Boxplot showing methylation ratios at T1 and T4 on ten significantly differentially methylated CpG sites. All of these sites showed reduced methylation at T4. Black dots present outliers of the methylation ratios. (b). Directional associations between changes in methylation ratios and changes of memory domain scores at 10 CpG positions. Significant inverse correlations were observed between differences in DNA methylation values and differences in memory domain standardized scores.
Table 4.
Ten top-ranked differentially methylated positions and the relevant information (controlled by FDR q < 0.1).
| Methylation ratio difference (T4-T1) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Methylation site | P-value | Q-value | Δβ | t (P-value) |
Estimate (SE) of memory domain score change for every unit increase in methylation ratio | Chromo-some | Chromo-some location | Distance* | Nearest gene | Functional Genomic Distribution of DMPs+ | |
| cg01333080 | 0.000047 | 0.058493 | −0.0358 | −4.80 (1.19 × 10−5) |
−230.5 (51.7) | chr3 | 183,994,568 | 0 | ECE2 | Gene Body | |
| cg26452989 | 0.000181 | 0.078529 | −0.0197 | −6.23 (6.11 × 10−8) |
−327.5 (80.9) | chr10 | 11,581,379 | 0 | USP6NL | Gene Body | |
| cg16472904 | 0.000249 | 0.081212 | −0.0433 | −6.16 (7.73 × 10−8) |
−159.8 (40.5) | chr11 | 7,597,402 | 0 | PPFIBP2 | Gene Body | |
| cg15530112 | 0.000330 | 0.083026 | −0.0357 | −7.74 (1.87 × 10−10) |
−224.2 (58.1) | chr14 | 53,929,908 | 309,861 | DDHD1 | Open Sea | |
| cg24994319 | 0.000371 | 0.083667 | −0.0673 | −8.29 (2.26 × 10−11) |
−133.9 (35.1) | chr6 | 25,027,586 | 0 | RIPOR2 | Promoter | |
| cg01450807 | 0.000460 | 0.084692 | −0.0222 | −6.88 (4.98 × 10−9) |
−307.9 (82.1) | chr13 | 73,715,706 | 64,029 | KLF5 | Open Sea | |
| cg13046524 | 0.000476 | 0.084836 | −0.0461 | −10.18 (1.96 × 10−14) |
−217.8 (58.2) | chr20 | 48,728,642 | 0 | UBE2V1 | Gene Body | |
| cg14356530 | 0.000673 | 0.087461 | −0.0379 | −7.50 (4.62 × 10−10) |
−207.9 (57.3) | chr9 | 99,064,450 | 15 | HSD17B3 | TSS | |
| cg20051772 | 0.000841 | 0.088787 | −0.0544 | −6.55 (1.78 × 10−8) |
−118.3 (33.3) | chr12 | 56,325,015 | 0 | DGKA | TSS | |
| cg08034986 | 0.000860 | 0.088904 | −0.0517 | −9.34 (4.27 × 10−13) |
−180.5 (50.9) | chr1 | 26,860,677 | 0 | RPS6KA1 | Gene Body | |
Abbreviations: DGKA, diacylglycerol kinase alpha; ECE2, endothelin converting enzyme 2; FDR, false discovery rate; HSD17B3, hydroxysteroid 17-beta dehydrogenase 3; KLF5, Kruppel like factor 5; RIPOR2, rho family-interacting cell polarization regulator 2; RPS6KA1, ribosomal protein S6 kinase alpha 1; UBE2V1, ubiquitin-conjugating enzyme E2 variant 1; USP6NL, ubiquitin specific peptidases N-terminal-like protein.
*Distance from the gene is denoted in basepairs; entries with values of ‘0’ indicate that the site is localized to the gene.
+Functional location of the differentially methylated position was determined by the distance. Open sea regions are isolated CpG sites in the genome that do not have a specific functional designation. TSS = transcription start site.
Discussion
Given that the results of emerging data suggest that dynamic changes in methylation (specifically, 5-methylcytosine) have implications in the regulation of gene expression involved in learning and consolidation of memory [31,32], it seems feasible that acquired epigenetic changes could contribute to the cognitive dysfunction frequently reported as an adverse ‘side effect’ with BC and its treatment [6,33]. In our analysis, 2,199 DMPs were identified between the baseline and one-year follow-up post-chemotherapy specimens, with 56 of these positions showing significant associations with the memory domain values. This latter observation is of particular interest since memory was the only cognitive domain that did not show improvement between the T1 and T4 specimens (lacked ‘recovery’ increases). Moreover, when the data was examined on an individual basis, which was feasible due to our longitudinal study design, significant inverse correlations were observed between differences in memory scores and differences in methylation patterns, with decreases in memory scores being associated with gains in methylation (as can be seen with gene ‘silencing’).
One of the 10 top-ranked DMPs (P < .001) showing a significant association with memory was a site localized to the ECE2 gene, which is expressed in neural (as well as other) tissues [34,35]. The results of previous studies suggest that ECE2 is an important amyloid-β-peptide (Aβ) degrading enzyme [36–38]. Interestingly, this gene was the most significantly downregulated gene associated with Aβ accumulation and clearance among a gene set evaluated in specimens collected from patients with AD compared to controls and other conditions [37]. ECE2 is also potentially related to learning and memory. Notably, mice lacking ECE2 exhibited poor learning and memory functions as evidenced by poor performance on the Morris water maze test [34]. Given the association of gene expression in neural tissue for this gene, one might expect that associations related to compromises in cognitive function would result from hypermethylation (gene silencing) rather than hypomethylation for this region. However, in a study of breast cancer tumours by Hon et al. (2012), genes within regions showing global DNA hypomethylation were largely silenced because the acquisition of global DNA hypomethylation was tightly coupled to the repression of chromatin domains, leading to decreased gene expression, as well as alterations in histone modifications [9]. While our results are correlative, rather than functionally based, given the findings of the Hon et al.’s study, it is quite plausible that one could see hypomethylation of the ECE2 gene associated with lack of memory improvement (and speculate that this could relate to alternations in bi-allelic gene expression).
Other genes of interest among the top 10 DMPs (Table 4) included the RIPOR2 gene and the PPFIBP2 gene. The RIPOR2 gene encodes a small G protein that has been associated with hearing loss/deafness. While we did not evaluate hearing loss in this study, it is intriguing to note that hearing loss is a side effect that has been recurrently reported following chemotherapy for a variety of cancers [39,40]; however, this gene has not been clearly associated with cognition. The PPFIBP2 gene is thought to function in neuronal synapse and axon guidance, but it has not been clearly associated with cognitive function or adverse side effects reported following chemotherapy. In addition to the 10 top-ranked sites, the total subset of 56 significant DMPs included sites localized to genes involved in neurotransmission/neural chemical synapses (KCNH4; CHRNB4); inflammatory responses (IKBKE); gait or axonal neuropathy (IER5); and DNA replication/damage response/repair (MCM6; NUAK1) (Supplementary Table 2). Specifically, NUAK1 is known to play a role in regulating tau levels by phosphorylation in mouse model, Drosophila, and human cell systems, in which the reduction of Nuak1 levels or activity may reverse tau accumulation in tauopathies, such as AD and progressive supranuclear palsy (PSP) [41]. While MCM6 is not directly related to cognitive dysfunction, the last 2 genes are interesting for this cohort, since the women who received TAC and TC chemotherapy regimens showed significantly increased frequencies of chromosomal instability in their T4 specimens compared to their T1 specimens [42].
An assessment of functional ontology annotations, which was completed for the full set of 2,199 DMPs, identified enrichment for 5 areas of biological function, with two of these 5 being associated with the modulation of chemokines (‘positive regulation of chemokine biosynthetic process’ and ‘regulation of IL-8 production’). Chemokines are known to play a major role in creating an environment which predisposes to cancer and the development of cancer-related inflammation by leukocyte recruitment, neo-angiogenesis, invasion, and tumour cell proliferation and survival [43,44]. Iwamoto et al. (2011) revealed that ‘chemokine signaling’ and ‘IL-8 signaling’ pathways were significantly associated with chemotherapy response in BC through a gene set analysis [45]. Notably, cytokines have been demonstrated to have a relationship with cognitive dysfunction [46–48]. Lyon et al. (2016) also reported that multiple cytokines had significant associations with psychomotor speed, complex attention, executive function, verbal memory, cognitive flexibility, and visual memory during and after chemotherapy treatment [21]. Another important biological function that was identified was regulation of cellular senescence. Cellular senescence can arise in response to therapy-induced genotoxic stress through epigenetic alterations, such as telomere shortening or DNA damage [49] and has been observed in BC specimens from patients who received neoadjuvant chemotherapy [50]. We have also speculated that cellular senescence could contribute to chemotherapy-related side effects such as cognitive dysfunction [7].
This study has several strengths, including, but not limited to, the longitudinal design that controls for inter-individual demographic and genetic factors that could confound the interpretation of the genome-wide DNA methylation and cognition data [51]. However, it also has limitations, including a relatively small sample size. Additionally, we are reporting the results of a single cohort and not yet have replication of these results in an independent cohort which could potentially limit the generalizability of these findings. Another limitation of this study is that the analysis was restricted to DNA methylation changes, but did not include an assessment of: (1) LINE-1 methylation patterns or of histone (or other epigenetic) modifications; or (2) gene expression patterns. Future studies that explore these latter attributes for the targeted loci identified in this analysis could provide additional insight regarding the association of these genes with memory in women following a diagnosis and treatment for BC. Lastly, another limitation of this study is the question of the relevance of peripheral blood DNA methylation changes to alterations that might arise in the brain (e.g., cognitive function). Given that brain tissues cannot be non-invasively obtained for assessments, researchers studying potential epigenetic influences on behavioural traits have evaluated peripheral tissues as a proxy for changes that might be relevant to those that are also present in the brain [52–55]. In their comparison of live human brain to concurrently collected peripheral tissues, Braun et al. (2019) noted relatively high overall correlations in DNA methylation values between brain and blood specimens, but did caution that correlation patterns can vary for individual CpGs [56].
In conclusion, this study provides novel evidence of an association between changes acquired in DNA methylation patterns and recovery of memory function following chemotherapy in women with early-stage BC. Clearly, future studies are needed to confirm these results and to better assess if this observation reflects a direct versus indirect relationship. However, if confirmed, this finding could lead to enhancement in our understanding of the mechanisms contributing to ‘chemo-brain’ in BC survivors.
Funding Statement
This work was supported by the National Institute of Nursing Research (NINR) under R01NR012667 to D.E.L. and C.K.J. and F32NR018367 to G.S.Y.; and the National Cancer Institute (NCI) under P30CA016058 to K.J.A.
Acknowledgement
The authors acknowledge the contributions of Noran Aboalela, PhD, for coordinating the preparation and transit of the DNA specimens to the HudsonAlpha Institute for Biotechnology.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here.
References
- [1].Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics. CA Cancer J Clin. 2016;66(4):271–289. [DOI] [PubMed] [Google Scholar]
- [2].Klemp JR, Myers JS, Fabian CJ, et al. Cognitive functioning and quality of life following chemotherapy in pre- and peri-menopausal women with breast cancer. Support Care Cancer. 2018;26(2):575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Conroy SK, McDonald BC, Smith DJ, et al. Alterations in brain structure and function in breast cancer survivors: effect of post-chemotherapy interval and relation to oxidative DNA damage. Breast Cancer Res Treat. 2013;137(2):493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pomykala KL, Ganz PA, Bower JE, et al. The association between pro-inflammatory cytokines, regional cerebral metabolism, and cognitive complaints following adjuvant chemotherapy for breast cancer. Brain Imaging Behav. 2013;7(4):511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Starkweather A, Kelly DL, Thacker L, et al. Relationships among psychoneurological symptoms and levels of C-reactive protein over 2 years in women with early-stage breast cancer. Support Care Cancer. 2017;25(1):167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang XM, Walitt B, Saligan L, et al. Chemobrain: a critical review and causal hypothesis of link between cytokines and epigenetic reprogramming associated with chemotherapy. Cytokine. 2015;72(1):86–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lyon D, Elmore L, Aboalela N, et al. Potential epigenetic mechanism(s) associated with the persistence of psychoneurological symptoms in women receiving chemotherapy for breast cancer: a hypothesis. Biol Res Nurs. 2014;16(2):160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Smith AK, Conneely KN, Pace TW, et al. Epigenetic changes associated with inflammation in breast cancer patients treated with chemotherapy. Brain Behav Immun. 2014;38:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hon GC, Hawkins RD, Caballero OL, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22(2):246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang S, Wang Y, Gu Y, et al. Specific breast cancer prognosis-subtype distinctions based on DNA methylation patterns. Mol Oncol. 2018;12(7):1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Park SY, Kwon HJ, Lee HE, et al. Promoter CpG island hypermethylation during breast cancer progression. Virchows Arch. 2011;458(1):73–84. [DOI] [PubMed] [Google Scholar]
- [12].Holm K, Staaf J, Lauss M, et al. An integrated genomics analysis of epigenetic subtypes in human breast tumors links DNA methylation patterns to chromatin states in normal mammary cells. Breast Cancer Res. 2016;18(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Flanagan JM, Wilson A, Koo C, et al. Platinum-based chemotherapy induces methylation changes in blood dna associated with overall survival in patients with ovarian cancer. Clin Cancer Res. 2017;23(9):2213–2222. [DOI] [PubMed] [Google Scholar]
- [14].Flowers E, Flentje A, Levine J, et al. A pilot study using a multistaged integrated analysis of gene expression and methylation to evaluate mechanisms for evening fatigue in women who received chemotherapy for breast cancer. Biol Res Nurs. 2019;21(2):142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kovalchuk A, Ilnytskyy Y, Woycicki R, et al. Adverse effects of paternal chemotherapy exposure on the progeny brain: intergenerational chemobrain. Oncotarget. 2018;9(11):10069–10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mathews HL, Konley T, Kosik KL, et al. Epigenetic patterns associated with the immune dysregulation that accompanies psychosocial distress. Brain Behav Immun. 2011;25(5):830–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shnorhavorian M, Schwartz SM, Stansfeld B, et al. Differential DNA methylation regions in adult human sperm following adolescent chemotherapy: potential for epigenetic inheritance. PLoS One. 2017;12(2):e0170085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stephens KE, Levine JD, Aouizerat BE, et al. Associations between genetic and epigenetic variations in cytokine genes and mild persistent breast pain in women following breast cancer surgery. Cytokine. 2017;99:203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Feinberg AP, Longo DL.. The key role of epigenetics in human disease prevention and mitigation. N Engl J Med. 2018;378(14):1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Haberman RP, Quigley CK, Gallagher M. Characterization of CpG island DNA methylation of impairment-related genes in a rat model of cognitive aging. Epigenetics. 2012;7(9):1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lyon DE, Cohen R, Chen H, et al. Relationship of systemic cytokine concentrations to cognitive function over two years in women with early stage breast cancer. J Neuroimmunol. 2016;301:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gualtieri CT, Johnson LG. Reliability and validity of a computerized neurocognitive test battery, CNS Vital Signs. Arch Clin Neuropsychol. 2006;21(7):623–643. [DOI] [PubMed] [Google Scholar]
- [24].Scherling C, Collins B, Mackenzie J, et al. Pre-chemotherapy differences in visuospatial working memory in breast cancer patients compared to controls: an FMRI study. Front Hum Neurosci. 2011;5:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Breckenridge LM, Bruns GL, Todd BL, et al. Cognitive limitations associated with tamoxifen and aromatase inhibitors in employed breast cancer survivors. Psychooncology. 2012;21(1):43–53. [DOI] [PubMed] [Google Scholar]
- [26].van Iterson M, Tobi EW, Slieker RC, et al. MethylAid: visual and interactive quality control of large Illumina 450k datasets. Bioinformatics. 2014;30(23):3435–3437. [DOI] [PubMed] [Google Scholar]
- [27].Burnham KP, Anderson DR. Model selection and multimodel inference: A practical-theoretic approach. 2nd ed. New York: Springer-Verlag; 2002. [Google Scholar]
- [28].Gantala SR, Kondapalli MS, Kummari R, et al. Collagenase-1 (−1607 1G/2G), gelatinase-A (−1306 C/T), stromelysin-1 (−1171 5A/6A) functional promoter polymorphisms in risk prediction of type 2 diabetic nephropathy. Gene. 2018;673:22–31. [DOI] [PubMed] [Google Scholar]
- [29].McLean CY, Bristor D, Hiller M, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Keller TE, Han P, Yi SV. Evolutionary transition of promoter and gene body DNA methylation across invertebrate-vertebrate boundary. Mol Biol Evol. 2016;33(4):1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Halder R, Hennion M, Vidal RO, et al. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat Neurosci. 2016;19(1):102–110. [DOI] [PubMed] [Google Scholar]
- [32].Sultan FA, Day JJ . Epigenetic mechanisms in memory and synaptic function. Epigenomics. 2011;3(2):157–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Myers JS. Chemotherapy-related cognitive impairment: the breast cancer experience. Oncol Nurs Forum. 2012;39(1):E31–E40. [DOI] [PubMed] [Google Scholar]
- [34].Rodriguiz RM, Gadnidze K, Ragnauth A, et al. Animals lacking endothelin-converting enzyme-2 are deficient in learning and memory. Genes Brain Behav. 2008;7(4):418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pacheco-Quinto J, Eckman CB, Eckman EA. Major amyloid-β–degrading enzymes, endothelin-converting enzyme-2 and neprilysin, are expressed by distinct populations of GABAergic interneurons in hippocampus and neocortex. Neurobiol Aging. 2016;48:83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pacheco-Quinto J, Herdt A, Eckman CB, et al. Endothelin-converting enzymes and related metalloproteases in Alzheimer’s disease. J Alzheimers Dis. 2013;33(Suppl 1):S101–S110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Weeraratna AT, Kalehua A, Deleon I, et al. Alterations in immunological and neurological gene expression patterns in Alzheimer’s disease tissues. Exp Cell Res. 2007;313(3):450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8(6):595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Brock PR, Maibach R, Childs M, et al. Sodium thiosulfate for protection from cisplatin-induced hearing loss. N Engl J Med. 2018;378(25):2376–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Frisina RD, Wheeler HE, Fossa SD, et al. Comprehensive audiometric analysis of hearing impairment and tinnitus after cisplatin-based chemotherapy in survivors of adult-onset cancer. J Clin Oncol. 2016;34(23):2712–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lasagna-Reeves CA, de Haro M, Hao S, et al. Reduction of Nuak1 decreases tau and reverses phenotypes in a Tauopathy Mouse Model. Neuron. 2016;92(2):407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Aboalela N, Lyon D, Elswick RK Jr., et al. Perceived stress levels, chemotherapy, radiation treatment and tumor characteristics are associated with a persistent increased frequency of somatic chromosomal instability in women diagnosed with breast cancer: a one year longitudinal study. PLoS One. 2015;10(7):e0133380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4(7):540–550. [DOI] [PubMed] [Google Scholar]
- [44].Allavena P, Germano G, Marchesi F, et al. Chemokines in cancer related inflammation. Exp Cell Res. 2011;317(5):664–673. [DOI] [PubMed] [Google Scholar]
- [45].Iwamoto T, Bianchini G, Booser D, et al. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J Natl Cancer Inst. 2011;103(3):264–272. [DOI] [PubMed] [Google Scholar]
- [46].Janelsins MC, Mustian KM, Palesh OG, et al. Differential expression of cytokines in breast cancer patients receiving different chemotherapies: implications for cognitive impairment research. Support Care Cancer. 2012;20(4):831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Terrando N, Rei Fidalgo A, Vizcaychipi M, Cibelli M, Ma D, Monaco C, et al . The impact of IL-1 modulation on the development of lipopolysaccharide-induced cognitive dysfunction. Crit Care. 2010;14(3):R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hennessy E, Gormley S, Lopez-Rodriguez AB, et al. Systemic TNF-α produces acute cognitive dysfunction and exaggerated sickness behavior when superimposed upon progressive neurodegeneration. Brain Behav Immun. 2017;59:233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130(2):223–233. [DOI] [PubMed] [Google Scholar]
- [50].Te Poele RH, Okorokov AL, Jardine L, et al. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 2002;62(6):1876–1883. [PubMed] [Google Scholar]
- [51].Schur RR, Boks MP, Rutten BPF, et al. Longitudinal changes in glucocorticoid receptor exon 1F methylation and psychopathology after military deployment. Transl Psychiatry. 2017;7(7):e1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Pang KKL, Sharma M, Sajikumar S. Epigenetics and memory: emerging role of histone lysine methyltransferase G9a/GLP complex as bidirectional regulator of synaptic plasticity. Neurobiol Learn Mem. 2019;159:1–5. [DOI] [PubMed] [Google Scholar]
- [53].Ptak C, Petronis A. Epigenetic approaches to psychiatric disorders. Dialogues Clin Neurosci. 2010;12(1):25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sweatt JD. Neural plasticity and behavior - sixty years of conceptual advances. J Neurochem. 2016;139(Suppl 2):179–199. [DOI] [PubMed] [Google Scholar]
- [55].Hoffmann A, Sportelli V, Ziller M, et al. Epigenomics of major depressive disorders and schizophrenia: early life decides. Int J Mol Sci. 2017;18:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Braun PR, Han S, Hing B, et al. Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Transl Psychiatry. 2019;9(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


