ABSTRACT
The aim of this study was to generate new knowledge on fish epigenetics, assessing the effects of linolenic acid (ALA) conditioning of broodstock in the offspring of the marine fish Sparus aurata. Attention was focused on gene organization, methylation signatures and gene expression patterns of fatty acid desaturase 2 (fads2) and stearoyl-CoA desaturase 1a (scd1a). Blat searches in the genomic IATS-CSIC database (www.nutrigroup-iats.org/seabreamdb) highlighted a conserved exon-intron organization, a conserved PUFA response region, and CG islands at the promoter regions of each gene. The analysed CpG positions in the fads2 promoter were mostly hypomethylated and refractory to broodstock nutrition. The same response was achieved after conditioning of juvenile fish to low water oxygen concentrations, thus methylation susceptibility at individual CpG sites seems to be stringently regulated in fish of different origin and growth trajectories. Conversely, the scd1a promoter was responsive to broodstock nutrition and the offspring of parents fed the ALA-rich diet shared an increased DNA-methylation, mainly in CpG sites neighbouring SP1 and HNF4α binding sites. Cytosine methylation at these sites correlated inversely with the hepatic scd1a expression of the offspring. Co-expression analyses supported that the HNF4α-dependent regulation of scd1a is affected by DNA methylation. The phenotypic output is a regulated liver fat deposition through changes in scd1 expression, which would also allow the preservation of fatty acid unsaturation levels in fish fed reduced levels of n-3 LC-PUFA. Collectively, these findings reveal a reliable mechanism by which parent’s nutrition can shape scd1a gene expression in the fish offspring.
KEYWORDS: Broodstock feeding, DNA methylation, fads2, linolenic acid, nutritional programing, scd1, Sparus aurata
Introduction
Plant-based diets are currently supporting the European aquaculture industry in a scenario of stagnant availability of fish meal (FM) and fish oil (FO) [1]. Thus, farmed gilthead sea bream [2] and European sea bass [3] can grow efficiently from early life stages with less than 10% of marine feed ingredients. These findings are also supported by wide-serum metabolomic approaches, based on ultra-sensitive high liquid chromatography and mass spectrometry [4]. Experimental evidence also indicates that plant-based diets do not represent a safety issue for human consumers due to the low concentration, or reduced carry-over from feeds to fillets, of a wide range of potential harmful compounds including pesticides and mycotoxins [5,6]. However, high inclusion levels of plant-ingredients are often related with a pro-inflammatory condition with a negative impact on fish health and stress resilience [7–10]. Plant-based diets can also alter the normal sex proportion ratio, enhancing the male-female reversal in the protandric gilthead sea bream as the result of less powerful functional females during the reproduction period [11]. Several attempts are underway to mitigate these non-desirable effects, and butyrate supplementation of plant-based diets helps to preserve the sex proportion ratio, contributes to prevent the inflammation of the intestinal epithelium, preserving as well the integrity of the intestinal barrier [11,12]. Supplementation with dietary butyrate also has positive effects on the mucus intestinal proteome and the intestinal microbiome, resulting in improved disease outcomes in fish challenged with bacteria or intestinal parasites [13].
The high replacement of marine feed ingredients by alternative raw materials is, thereby, a complex issue, and the total or high replacement of FO by vegetal oils (VO) can compromise fish health [14] and the nutritive value of fish meat [15,16]. This risk assessment is especially relevant in marine farmed fish, since they have a limited ability to convert α-linolenic acid (ALA, 18:3n-3) into eicosapentaenoic acid (EPA, 20:5n-3) and docosahexaenoic acid (DHA, 22:6n-3) [17]. Fatty acid desaturase 2 (Fads2) catalyzes the first and rate limiting step in this pathway and its regulation and activity has been studied in a wide-range of marine (gilthead sea bream [18], black sea bream [19], European sea bass [20], Japanese sea bass [21], large yellow croaker [22], orange-spotted grouper [23]) and freshwater (rainbow trout [22], striped snakehead [24]) fish species. A main outcome is that diets with low levels of n-3 long chain polyunsaturated fatty acids (n-3 LC-PUFA) up-regulate the expression of fads2, but with differences among fish species that highly reflect the basal activity of their promoters and their differences in LC-PUFA biosynthetic abilities [22,23,25]. Conversely, low n-3 C-PUFA diets lead to increased liver fat deposition in freshwater, salmonids and typically marine farmed fish [26–29], being liver steatosis a common sign of deficiencies in essential fatty acids [30]. It can be argued that this metabolic derangement is primarily the result of an enhanced hepatic lipogenesis, with the involvement of stearoyl-CoA desaturase (Scd), the rate limiting-step in the formation of monounsaturated fatty acids (MUFAs), especially oleate and palmitoleate from stearoyl-CoA and palmitoyl-CoA, respectively. Indeed, in mammals this enzyme increases the fatty acid unsaturation level of lipid depots [31] and is a major regulator of lipid storage in liver and adipose tissue [32–34]. Likewise, fish scd1 is differentially regulated by unsaturated and saturated fats [28,35–37], correlating positively with hepatic fat accumulation [28].
Attempts have been made to drive the lipid metabolism of marine fish by examining the concept of nutritional programming. However, most of these studies have been focused on fads2 with less attention paid to the regulation of scd. A persistent up-regulated expression of fads2 (without data on scd1 regulation) has been reported in European sea bass juveniles fed at the larval stage with n-3 LC-PUFA-deficient diets [20,38,39]. Similar results have been reported in gilthead sea bream larvae, but this procedure is difficult to translate into practice due to the high-associated mortality [40]. Alternatively, the management of early fish nutrition was attempted through parental nutrition, and it is now recognized that the replacement of FO by linseed oil (LO) in broodstock diets improves the growth of offspring fed with low FM/FO diets [41,42]. However, the ultimate mechanisms remain unknown as the enhanced larval expression of fads2 [40] does not appear to be retained in older juveniles [42].
Cytosine methylation at CG dinucleotides (CpG sites) constitutes a stable epigenetic mark that can be transmitted through DNA replication and cell division. In particular, methylation signatures of FADS2 [43,44] and SCD [45] are associated to different gene expression patterns. However, such a clear association remains elusive in non-model fish species [46]. Moreover, most studies in marine fish, including those conducted in gilthead sea bream, have managed the ratio n-3-LC-PUFA/C18 PUFA, but have not yet consider the potential value of C18 PUFA alone as a nutritional programming tool [40–42]. This contrasts with the approaches made in mammals, where there is evidence that ALA supplementation during pregnancy-lactation prevents fatty liver in the adult offspring when challenged with lipogenic diets [47]. Indeed, ALA seems to be more effective than EPA and DHA in triggering this metabolic phenotype later in life [48]. In mice, this occurred along with a decreased expression of both Scd1 and Fads2 [48,49], being the changes in enzyme activity more evident for SCD1 (30%) than for FADS2 (10%) [49]. Hence, in the present study, we focused on the epigenetic effects of ALA in broodstock diet in the offspring of gilthead sea bream, a highly cultured fish in the Mediterranean region. Attention was focused on the gene structure, methylation signatures and expression patterns of fads2 and scd1a, providing new insights in the regulation of lipid metabolism in a typically marine fish.
Results
Ala-rich parental diet affected the nutritional environment of the developing larvae
We fed two broodstock groups with a control and an ALA-rich diet. The fatty acid composition of broodstock diets, eggs and offspring liver is shown in Table 2. Partial replacement of FO by LO in the ALA-rich broodstock diet raised the content of OA, LA and, in particular, ALA. The ALA content increased from 0.9% of total lipids in the control diet to 16.3% of total lipids in the ALA-rich diet. This fatty acid profile was mirrored in eggs, the ALA content being largely increased (4.6-fold), whereas OA and LA remained almost constant. This also applied to ARA, EPA and DHA when comparisons were made between the two types of eggs. In particular, the content of ALA increased from 1.74% to 7.98% of total lipids, in eggs from the control- and the ALA-fed parentals, respectively. On the other hand, the liver fatty acid composition of the offspring mostly reflected the composition of the offspring challenging diet.
Table 2.
Fatty acid (FA) composition of broodstock diets, eggs, and offspring liver.
| Broodstock diet |
Eggs |
Offspring liver |
||||
|---|---|---|---|---|---|---|
| FA (%) | Control | ALA-rich | Control | ALA-rich | Control | ALA-rich |
| 14:0 | 6.65 | 4.43 | 3.76 | 3.00 | 1.62 | 1.64 |
| 14:1n-7 | 0.10 | 0.06 | 0.04 | 0.03 | 0.01 | 0.02 |
| 14:1n-5 | 0.23 | 0.16 | 0.12 | 0.10 | 0.03 | 0.03 |
| 15:0 | 0.33 | 0.26 | 0.30 | 0.28 | 0.15 | 0.14 |
| 15:1n-5 | 0.04 | 0.03 | 0.03 | 0.03 | 0.02 | 0.02 |
| 16:0ISO | 0.07 | 0.05 | 0.05 | 0.05 | 0.02 | 0.02 |
| 16:0 | 13.50 | 12.35 | 20.06 | 19.28 | 15.00 | 14.61 |
| 16:1n-7 | 7.13 | 4.32 | 6.00 | 4.76 | 2.37 | 2.46 |
| 16:1n-5 | 0.12 | 0.10 | 0.09 | 0.09 | 0.12 | 0.13 |
| 16:2n-6 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 16:2n-4 | 0.38 | 0.26 | 0.21 | 0.17 | 0.07 | 0.08 |
| 17:0 | 0.22 | 0.15 | 0.16 | 0.13 | 0.09 | 0.09 |
| 16:3n-4 | 0.15 | 0.13 | 0.22 | 0.19 | 0.20 | 0.20 |
| 16:3n-3 | 0.18 | 0.14 | 0.11 | 0.09 | 0.04 | 0.04 |
| 16:3n-1 | 0.04 | 0.03 | 0.08 | 0.08 | 0.01 | 0.01 |
| 16:4n-3 | 0.37 | 0.28 | 0.07 | 0.06 | 0.04 | 0.05 |
| 16:4n-1 | 0.01 | 0.02 | 0.01 | 0.01 | - | - |
| 18:0 | 1.76 | 2.92 | 3.86 | 4.03 | 5.77 | 5.54 |
| 18:1n-9 | 11.08 | 14.85 | 21.60 | 20.88 | 41.02 | 40.51 |
| 18:1n-7 | 2.93 | 2.33 | 3.06 | 2.73 | 2.56 | 2.53 |
| 18:1n-5 | 0.38 | 0.23 | 0.22 | 0.15 | 0.07 | 0.06 |
| 18:2n-9 | 0.04 | 0.03 | 0.24 | 0.20 | 2.17 | 2.47 |
| 18:2n-6 | 5.57 | 9.92 | 10.25 | 11.50 | 11.47 | 11.38 |
| 18:2n-4 | 0.07 | 0.05 | 0.11 | 0.11 | 0.07 | 0.07 |
| 18:3n-6 | 0.15 | 0.10 | 0.33 | 0.29 | 1.93 | 2.20 |
| 18:3n-4 | 0.06 | 0.03 | 0.11 | 0.10 | 0.06 | 0.07 |
| 18:3n-3 (ALA) | 0.87 | 16.31 | 1.74 | 7.98 | 5.48 | 5.58 |
| 18:3n-1 | 0.01 | 0.01 | 0.00 | 0.01 | 0.00 | 0.00 |
| 18:4n-3 | 1.64 | 1.11 | 0.66 | 0.54 | 1.41 | 1.61 |
| 18:4n-1 | 0.06 | 0.03 | 0.08 | 0.07 | 0.03 | 0.03 |
| 20:0 | 0.17 | 0.19 | 0.09 | 0.09 | 0.16 | 0.15 |
| 20:1n-9 | 0.40 | 0.33 | 0.29 | 0.19 | 0.13 | 0.13 |
| 20:1n-7 | 12.38 | 6.76 | 2.18 | 1.24 | 0.72 | 0.69 |
| 20:1n-5 | 0.63 | 0.35 | 0.20 | 0.14 | 0.08 | 0.07 |
| 20:2n-9 | 0.00 | 0.00 | 0.08 | 0.07 | 0.63 | 0.64 |
| 20:2n-6 | 0.26 | 0.13 | 0.35 | 0.31 | 0.26 | 0.24 |
| 20:3n-9 | 0.02 | 0.03 | 0.02 | 0.02 | 0.01 | 0.01 |
| 20:3n-6 | 0.05 | 0.04 | 0.15 | 0.13 | 0.24 | 0.25 |
| 20:4n-6 (ARA) | 0.39 | 0.34 | 0.62 | 0.62 | 0.37 | 0.36 |
| 20:3n-3 | 0.09 | 0.08 | 0.17 | 0.28 | 0.22 | 0.21 |
| 20:4n-3 | 0.32 | 0.23 | 0.53 | 0.43 | 0.31 | 0.35 |
| 20:5n-3 (EPA) | 6.28 | 4.83 | 4.57 | 4.05 | 1.19 | 1.31 |
| 22:1n-11 | 15.65 | 8.59 | 0.77 | 0.43 | 0.21 | 0.22 |
| 22:1n-9 | 1.63 | 0.95 | 0.25 | 0.15 | 0.27 | 0.25 |
| 22:4n-6 | 0.05 | 0.04 | 0.05 | 0.05 | 0.05 | 0.05 |
| 22:5n-6 | 0.04 | 0.03 | 0.04 | 0.03 | 0.11 | 0.11 |
| 22:5n-3 | 0.46 | 0.34 | 1.98 | 1.87 | 0.50 | 0.55 |
| 22:6n-3 (DHA) | 7.05 | 6.01 | 14.09 | 14.53 | - | - |
General gene organization is conserved in fads2 and scd1a
Searches in the IATS-CSIC genomic database of gilthead sea bream (http://nutrigroup-iats.org/seabreamdb) revealed that the gilthead sea bream fads2 gene comprises 13 exons (Figure 1(a)), and searches in the GenBank database shown the same exon-intron organization in zebrafish (Gene ID: 140,615), Japanese flounder (Gene ID: 109,623,584), yellow catfish (Gene ID: 113,660,090), barramundi perch (Gene ID: 108,886,851) and California yellowtail (Gene ID: 111,666,947), though one exon appears to be lost in higher vertebrates (e.g., humans, mouse).
Figure 1.
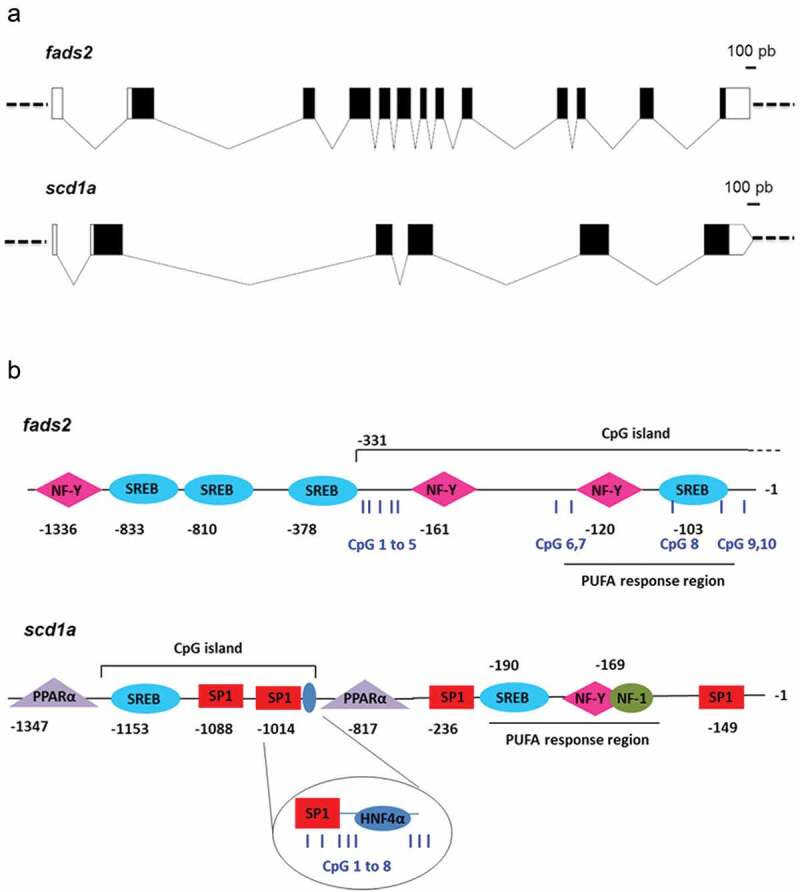
Gene organization (a) and TFBS and CpG islands (b) in promoters of fads2 and scd1a genes of the gilthead sea bream. Boxes in (A) represent exons and connecting lines represent introns. Open boxes mean non-coding regions, black boxes mean coding regions. In (B), numbers indicate position relative to TSS (+1), assumed to be the first base of first exon. Examined CpG positions for cytosine methylation by pyrosequencing are indicated as blue vertical bars and numbered from 1 to 10 for fads2, and from 1 to 8 for scd1a.
The scd1a gene organization is also highly conserved among vertebrates, being composed by six exons in gilthead sea bream (Figure 1(a)), higher vertebrates (e.g., chicken, human, gorilla, wild boar) and other fish species, such as zebrafish (Gene ID: 386,661), cod (ENSGMOG00000000395), channel catfish (Gene ID: 108,263,594) and turbot (ENSSMAG00000002283).
Key TFBS for conserved functionality in fads2 and scd1a proximal promoters
Different regulatory features of these genes, such as TFBS and CpG islands, were described by in silico analyses. A PUFA response region (−120 to −93) composed by one SREBP site and one NF-Y site was identified in the proximal fads2 promoter (Figure 1(b)). This regulatory region, in addition to a more distal NF-Y site (−161), is highly conserved in the fads2 promoter of fish species (Supplementary Figure 1a), and lie within a CpG island that extends into the first fads2 exon of gilthead sea bream (Figure 1(b)). One additional NF-Y site in combination with three SREBPs (−1336 to −378) were found more distal, flanking the proximal CpG island of the fads2 promoter (Figure 1(b)).
In the case of gilthead sea bream scd1a promoter, a PUFA response region (−190 to −160), comprising SREBP, NF-Y and NF-1 (nuclear factor 1) binding sites, was flanked by two SP1 sites (Figure 1(b)). This PUFA response region lies within a non-CpG island region that is highly conserved in the lineage of teleosts (Supplementary Figure 1b). At more distal positions, a less conserved region containing one SREBP site, two SP1 sites, and one HNF4α site were predicted to lie within a CpG island, flanked by two PPARα binding sites (−1347 to −817) (Figure 1(b)).
Methylation of CpG sites in the proximal fads2 promoter was unaffected by nutritional programming and low oxygen concentration exposure
To evaluate the effect of an ALA-rich environment during early development on DNA methylation of selected regulatory regions of fads2 and scd1a, we further analysed by bisulphite pyrosequencing liver samples from the two offsprings at the age of 6 month, and after feeding a common ALA-enriched offspring diet. Among CpG sites analysed in the fads2 promoter, CpG1 to 5 were far from any TFBS, CpG6 and 7 were adjacent to an NF-Y site, CpG8 and 9 were within a SREBP site, and CpG10 was adjacent to the same SREBP site (Figure 1(b), Supplementary Figure 2a). The cytosine methylation at these positions was not altered by the broodstock nutrition (Figure 2(a)), remaining this region in a general hypomethylated state. However, the pattern was to decrease the DNA methylation level downstream CpG3. Sites 8, 9, and 10, associated with a SREBP site, were particularly hypomethylated irrespective of the parent’s diet (Figure 2(a)). Cytosine methylation at these positions did not correlate with gene expression (R CpG1 to 10 = −0.02 to −0.45, P = 0.30 to 0.95).
Figure 2.
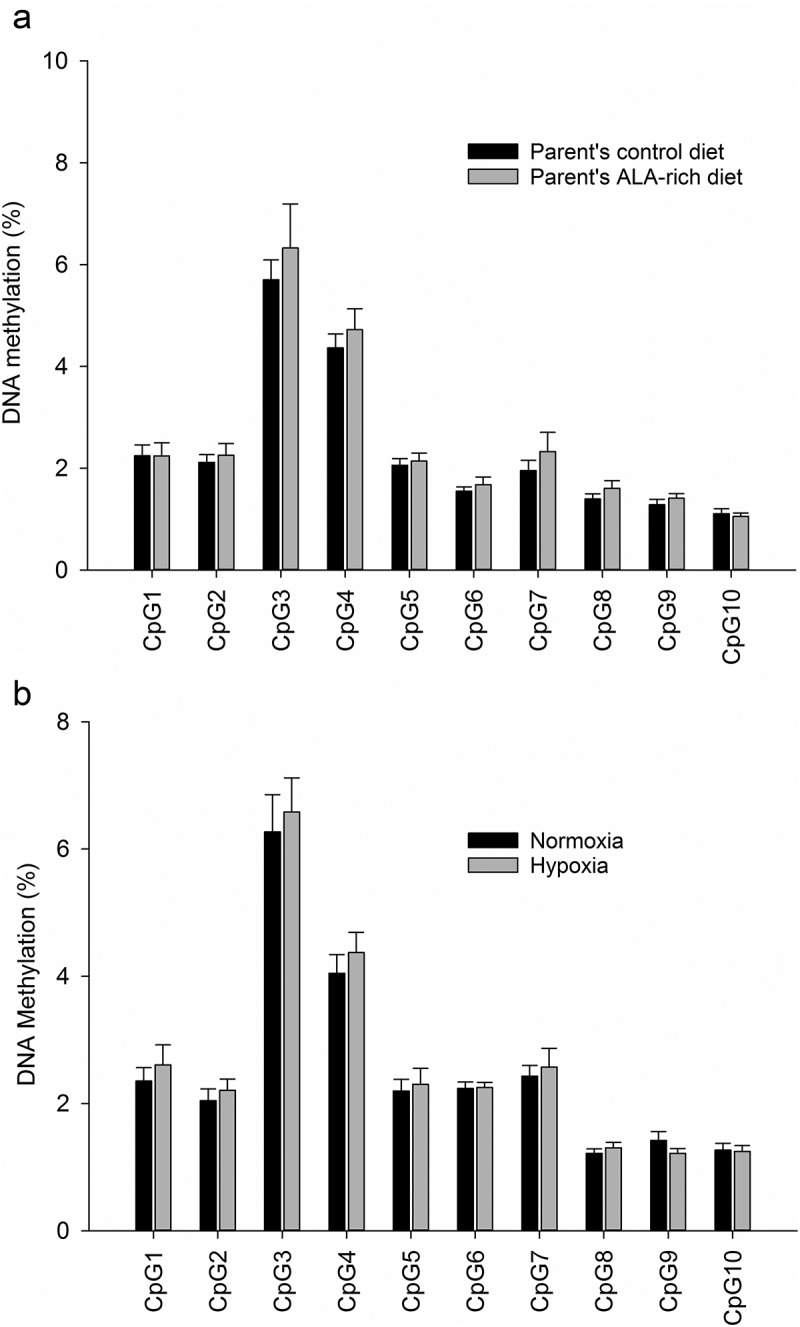
Cytosine methylation in examined promoter regions of fads2 in juveniles of the gilthead sea bream fed a low FM/FO diet, whose parents were fed with a control or an ALA-rich diet (a), or exposed to different oxygen concentration availability (b). Data represent the mean ± sem of 9 and 12 individuals per condition in A and B, respectively. No differences were found for CpG sites analysed by between groups by Student t-test (P > 0.05).
To examine how prone this region of the fads2 proximal promoter is able to change its site-specific methylation by other environmental conditions, we analysed the same CpG sites in fishes subjected to normoxic or moderate hypoxic conditions (~40% oxygen saturation). In higher vertebrates, hypoxia is well known to increase methylation at some loci by decreasing DNA demethylase activity, while decreases methylation at other loci by inhibiting DNA methyltransferases or the availability of methyl donors. As shown in Figure 2(b), the general pattern of cytosine methylation was not altered by the availability of oxygen during the juvenile stage, being the methylation pattern in this separate experiment nearly identical to that observed in the offspring’s of control and ALA-rich fed parents (i.e., highest methylation at CpG3 and lowest at CpGs 9 and 10) (Figure 2(a,b)).
Methylation of CpG sites in the scd1a promoter is affected by ALA-rich parent’s diet
A total of eight CpG sites within or flanking two conserved regulatory motifs were also analysed in the scd1a promoter of the offsprings with different broodstock nutrition (Figure 1(b)). CpG1 to 3 were within an SP1 site, CpG4 to 8 were downstream this site, with CpG5 and 6 flanking an HNF4α binding site (Figure 1(b), Supp. Figure 2b), being their methylation pattern nutritionally regulated (Figure 3(a)). The ALA enrichment of parent’s diets produced a general increase in the methylation level of CpG sites 3 to 8 that was statistically significant for positions 3, 5, 6 and 7 (Figure 3(a)).
Figure 3.
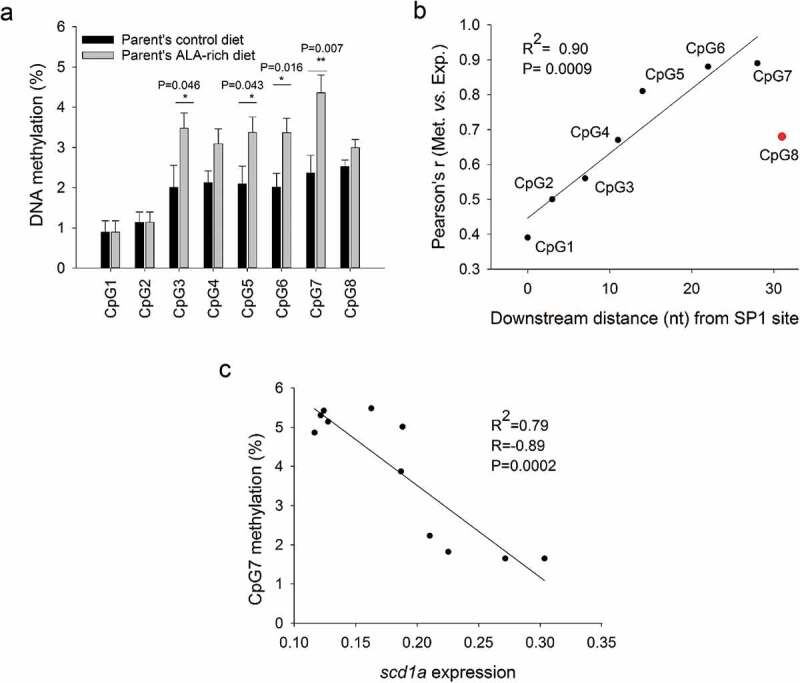
Cytosine methylation in examined promoter regions of scd1a in juveniles of the gilthead sea bream fed a low FM/FO diet, whose parents were fed with a control or an ALA-rich diet. Data represent the mean ± sem of nine individuals per condition. Differences found between groups by Student t-test (P > 0.05) are indicated (a). Correlation between gene expression and the methylation level of CpGs at different positions relative to an SP1 site within the scd1a promoter; correlation is maximal for CpG6 and 7 (b). Correlation between methylation at CpG7 in the scd1a gene promoter and hepatic scd1a gene expression. Significance for correlation is shown (c).
Methylation of CpGs downstream an SP1 site inversely correlated with scd1a expression
To disclose the relationship between the methylation patterns observed and the hepatic expression of scd1a, the association between cytosine methylation at individual CpG sites and scd1a expression in the two offspring groups was investigated by means of Pearson correlation analysis (Figure 3(b)). The analysis was performed with paired data (i.e., methylation-expression) from the same individuals. Methylation at CpG1 and 2 (centred in the SP1 site) were poorly correlated with scd1a expression. The correlation slightly increased for CpG3 (the SP1 outer cytosine), and then continuously rose with maximal correlation for CpG6 and 7. These two CpG sites were adjacent to an HNF4α binding site (Figure 1(b), Supp. Figure 2b). The correlation between cytosine methylation and scd1a expression sharply dropped for CpG8, only three nt downstream CpG7. Thus, CpG7 was considered the most informative site when individual data on methylation and scd1a expression were plotted each other, yielding a statistically significant negative correlation (R = −0.89, P < 0.0002) (Figure 3(c)).
ALA-rich parent’s diet led to offspring with lower scd1a expression and a trend for less hepatic fat storage
The offspring expression level of fads2 was not statistically modified (Mann-Whitney U = 116.5, P = 0.49) by ALA broodstock conditioning. In contrast, consistent changes in scd1a gene expression and methylation were found when individual data were grouped per dietary condition. Hence, the ALA broodstock diet increased significantly the methylation level of the scd1a promoter, and CpG7 site in particular, in the offspring livers (Figure 4(a)), leading to a significant reduction in hepatic scd1a gene expression in six-month-old fish fed low FM/FO diets (Figure 4(b)). Additionally, the trend (P = 0.22) for these animals was to reduce hepatic fat depots, which was accompanied by a reduced individual variability in liver lipid content respect to the offspring from the control broodstock group (Figure 4(c)).
Figure 4.
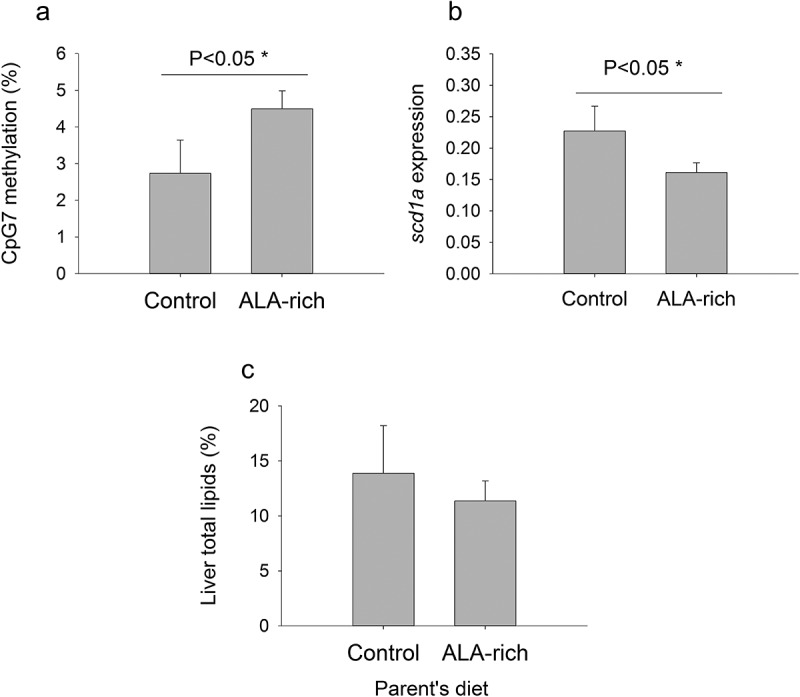
Effect of parent’s diet (Control and ALA-rich) on CpG7 methylation (a), scd1a gene expression (b), and liver total lipids (c) in juvenile offspring’s after fed a low FM/FO diet. Data represent the mean ± sem of 5–6 individuals per condition. Significance for one-tailed t-test is shown in A and B. No statistical significant differences were found in liver total lipids (Mann-Whitney Rank Sum Test, P > 0.05), but the trend is a small median value with a low range of variation in the offspring of ALA-fed parents.
Lower scd1a expression in fish from ALA-rich fed parents was not due to lower TF expression but rather to impaired NF4α binding
TFs are key determinant of gene activity. Thus, to better discern the effect of DNA methylation at the scd1a promoter, we analysed the expression of TFs with key roles in lipid metabolism and scd1a regulation in particular. Considered globally, the broodstock nutrition was not effective in our experimental model to change the hepatic expression of any of the analysed TFs (lxrα, pparα, pparβ, pparδ, sp1, nf-yb, serbp1 and hnf4α) (Figure 5(a)). Interestingly, analysed individually, the expression levels of hnf4α and scd1a were positively correlated in the offspring from the control group (Figure 5(b)), being this correlation lost in the offspring from the ALA conditioned broodstock (Figure 5(c)). Conversely, a similar analysis revealed that the expression of sp1, the other TF with a binding site in the neighbouring of the differentially methylated positions, was not correlated with scd1a expression in the offspring from neither the control (R = 0.003, P = 0.99) nor the ALA conditioned broodstock (R = 0.47, P = 0.06).
Figure 5.
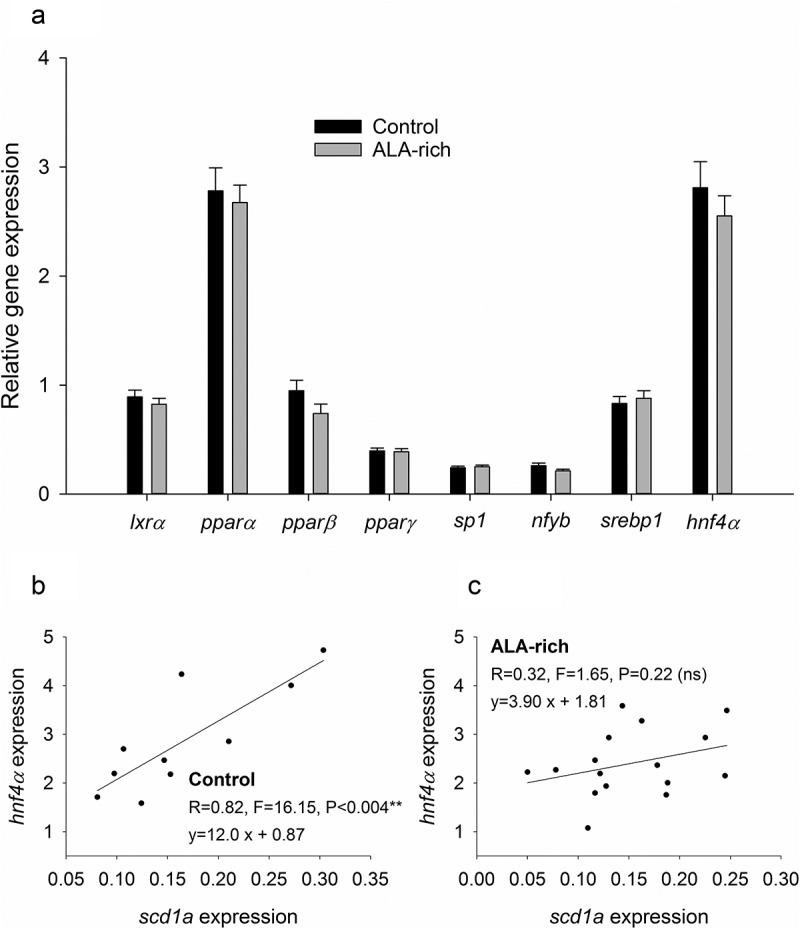
Gene expression of key transcription factors for hepatic lipid metabolism in the livers of offspring fish from control diet and ALA-rich diet fed parents (a). Data represent the mean ± sem of 18 individuals per condition. No significant differences were found in the hepatic expression of these TFs by Student t-test (P > 0.05). Correlation between hnf4a and scd1a gene expression in offspring from control (b) and ALA-rich (c) fed parents.
Discussion
Different studies in fish have revealed how the early environment can have long lasting-effects on the later phenotype and on the ability to adapt to environmental conditions later in life (reviewed by [50]). Such early stimuli refer among others, to temperature at larval stage [51], cold-shock and air-exposure to embryos and larvae [52], early acute hypoxia at embryo stage, broodstock or fry methionine deficient diets [53], and first feeding with high carbohydrates [54], a mix of plant proteins [55] or soya-based diets [56,57]. There is also experimental evidence for the nutritional programming of lipid metabolism in both fish [20,38–42], and higher vertebrates [48,49], though the epigenetic mechanisms regulating the expression of key genes such as fads2 and scd1 remains elusive in fish, and in marine fish in particular. To solve this gap of knowledge, we first analysed the gene structure and promoter organization of fads2 and scd1a genes, and then correlated gene expression patterns with changes in methylation signatures at promoter regions. In this regard, it is noteworthy that cytosine methylation at several positions within the fads2 proximal promoter was neither affected by ALA broodstock conditioning nor by oxygen availability during juvenile fish stages. Conversely, epigenetic marks resulting from broodstock nutrition persisted 6 months after hatching in the promoter of scd1a, correlating negatively the methylation level at specific CpG sites with the hepatic scd1a expression.
Gene organization and conserved regulatory elements in proximal fads2 and scd1a promoters
The fads2 is conserved in gilthead sea bream and a wide-range of fish as a gene of more than 6.7 kb in length and 13 exons. One of these exons was apparently lost in the vertebrate lineage of humans and rodents [58], whereas the predicted PUFA response region remained through vertebrate evolution as a highly conserved regulatory region in fish [23] and humans [59], containing SREB and NF-Y sites, within a CpG island close to the TSS at positions −120 to −93 in gilthead sea bream, or at positions −283 to +1 in humans [59].
Like fads2, scd1 is conserved as a gene of more than 5.7 kb in length, with six exons in a wide range of other vertebrates, including gilthead sea bream (this work), chicken, human, gorilla, grey short-tailed opossum and wild boar [60]. Of note, several well-known conserved regulatory elements (e.g., SP1, NF-Y, NF1, SREBP, PPARα) of the SCD1 promoter in mammals [61] were also retrieved in the scd1a promoter of gilthead sea bream. Likewise, a PUFA response region is present in the scd1 proximal promoter of both gilthead sea bream (this work) and mammals [34,61]. Thus, it appears conclusive that gene organization and essential functional elements in the proximal promoters (herein considered 250 pb upstream TSS [62]) of both gilthead sea bream fads2 and scd1a genes are highly conserved through the evolution of the teleost lineage.
Constrains to programme fish fads2
In mice, methylation of the CpG island at the promoter region of Fads2 negatively correlates with the expression of the gene [43], though it appears that the human FADS2 expression can also be regulated by methylation at four distal CpG sites in a non-CpG island region [44] or by methylation at a CpG island that extends to the beginning of the first intron [63]. In the case of gilthead sea bream, the analysed CpG sites (−331 to −87) within a CpG island at the proximal promoter region appear highly refractory to programing with ALA, but we cannot totally exclude that other, more distal CpG sites, would be susceptible to nutritional programming. Also, it is not known if the high LC-PUFA in the parental diets of our experimental design is related to the hypomethylated PUFA response region observed in both offsprings. In any case, it appears that the methylation pattern at individual CpG sites of the fads2 proximal promoter region is stringently regulated in gilthead sea bream. This is supported by the same methylation pattern in fish of different genetic origin and growth trajectories in the indoor experimental facilities of IU-ECOAQUA (Canary Islands, broodstock trial) and IATS-CSIC (West Mediterranean, hypoxia trial). From our results, it is also conclusive that the PUFA responsive region of fads2 in the proximal promoter region is not prone to being methylated in gilthead sea bream, which would favour basal expression and the up-regulated expression of fads2 in fish fed low levels of n-3 LC-PUFA. Conversely, a study in mice showed that perinatal manipulation of ALA intake alters Fads2 promoter methylation in offspring livers [64], whereas we showed that the activity of the fish fads2 promoter under our experimental conditions would be more affected by the activity/availability of TFs. Certainly, the regulatory effects of SREBP-1 and PPAR-α in response to dietary fatty acids are quite different in rainbow trout (freshwater), Japanese sea bass (euryhaline) and yellow croaker (marine), which may contribute to the differential LC-PUFA biosynthesis abilities (via fads2 regulation) among fish that have adapted to different ambient salinity [22], and act in concert to other differences such as the presence of a SP1 site in the promoter of freshwater species that is absent in marine fish [23]. Likewise, the nutritional condition of European sea bass larvae with high- or low-PUFA diets did not change the methylation level of 28 CpG sites examined within the fads2 promoter [25]. Conversely, in Japanese sea bass, the methylation level of the fads2 promoter (as the mean of 55 CpG sites) increased in fish fed n-3 LC-PUFA rich diets, being coupled this response to a lower fads2 gene expression [21]. Intriguingly, the examined CpG sites in that study were not in key TFBS and thus their relation with gene expression is not yet understood.
Fish scd1a can be programmed through broodstock nutrition
Sequence analysis revealed that the PUFA response region of the gilthead sea bream scd1a gene lie in a non-CpG island region, while other key TFBS were found at distal positions, in a CpG island 1014 pb upstream the TSS. Interestingly, we found that several CpG sites (−1014 to −984) within this CpG island are susceptible to metabolic programming by broodstock nutrition, affecting offspring scd1a gene expression. In agreement with these observations, dietary manipulations in mice also modified the methylation signature of Scd1 in two adjacent CpG sites within a CpG island at −916 to −614 positions relative to TSS [45], though the values observed in mice were higher than those in fish. At the same time, more proximal CpG islands in mice showed no variation [45]. This seems not unusual, as studying the possibility to programme the response to hypoxia and dietary methionine deficiencies in rainbow trout, it was observed that the methylation level of targeted genes was consistently low (0–3%) in proximal positions (until −600 bp), being more susceptible to be methylated the CpG sites at more distal positions [53].
Studies in dairy cows highlighted that dietary manipulations led to changes in hepatic SCD1 expression by regulating both the abundance of SREBP1c and DNA methylation around the SREBP1c binding site [65]. However, studies in mice suggested that the main effect is exerted by the methylation level [45]. Indeed, we demonstrated that broodstock feeding with ALA did not alter the expression of several TFs, though we cannot exclude changes in TF activity by means of post-translational modifications (e.g., phosphorylation, sumoylation, ubiquitination, acetylation, glycosylation, and methylation) [66]. In our study, responsive CpG sites were within a 20 nt region downstream a binding site for SP1, which is known to be involved in SCD1 transcriptional regulation [67]. However, it remains to be understood the functional significance of this finding in gilthead sea bream, because changes in DNA-methylation may disturb SP1 binding or the binding of HNF4α downstream this site. Regarding the first possibility, methylation in central positions within SP1 site poorly affects SP1 binding, while binding impairment is bigger if methylation occurs at bordering positions [68], and strongest if occurs in adjacent positions (e.g., even 13 nt away [69]). Our sp1 and scd1a co-expression analysis, however, suggested that the binding of this TF was not impaired by methylation at bordering or adjacent positions. Regarding the second possibility, it is known that SP1 cooperates with other TF several nt downstream the site (e.g., even 31 nt away [70]). Moreover, SP1 and HNF4α can even physically interact to produce synergism when they bound simultaneously to DNA [71]. Interestingly, studies in mice indicate that DNA methylation of CpG sites (−838 and −833), in the proximity of hepatocyte nuclear factors (HNFs) binding sites, are of special relevance for Scd1 transcriptional regulation [45]. Notably, a specific haplotype of HNF4α has been associated with SCD1 activity in humans [72]. Likewise, in the present study, co-expression analysis of hnf4α and scd1a suggests a lipogenic role of hnf4α that might allow an enhanced offspring expression of scd1a in gilthead sea bream juveniles from non-ALA conditioned parents, which might be attenuated by an increased methylation near the HNF4α binding site in the offspring of ALA-fed parents.
Phenotypic outputs of fish scd1a programming
Our data on liver fat content suggest that the offspring from ALA-rich fed parents have reduced hepatic lipogenesis and liver fat deposition when feeding VOs. However, the metabolic consequences of this nutritional programing may be more clearly observed, and perhaps more valuable, in fish feeding strong lipogenic diets. In this study, the challenge diet for juvenile offspring was a low FM/FO diet but with an EPA and DHA content similar to that found in both parent’s diets to ensure that any epigenetic modification found in the offspring would be only ascribed to the early environment. This is especially critical for scd1a, since this enzyme is highly responsive to dietary changes in n-3 LC-PUFA. This is part of the adaptive response that prevent the lipotoxic effect of saturated fatty acids, favouring their conversion to more safety stored mono-unsaturated fatty acids [73], also contributing to preserve the unsaturation level of membrane phospholipids [74]. This is supported herein by revisiting the gene expression profiles of fish fed semi-synthetic diets formulated to be deficient in n-3 LC-PUFA [30] or practical diets with a maximized replacement of FM and FO without a negative impact on growth potentiality [2]. Certainly, in this scenario, the gene expression profile of selected markers of lipid metabolism, including fatty acid elongases (elovl1, elovl4, elov5, elovl6) and desaturases (fads2, scd1a, scd1b) highlighted the highest responsiveness of scd1 and secondly elovl6 in fish feeding low n-3 LC-PUFA diets, which is especially evident with the appearance of signs of nutritional deficiencies (Supp. Figure 3). The scd1a is also highly responsive in gilthead sea bream to changes in nutrient availability [75,76]. More recently, experimental data supported a differential regulation of scd1a in liver and skeletal muscle in a fish strain with a high growth potentiality [11]. In other fish species, it is also well known that lipogenic conditions enhance scd1 expression (carp [35], tilapia [28], pond loach [37]), along with hepatic fat accumulation [28]. Collectively, these results point out that scd1a expression contributes to mitigate the signs of n-3 LC-PUFA deficiencies, but at the same time, that it is very important to limit its up-regulation to avoid the dysregulation of hepatic metabolism and the lipotoxic effects of excessive fat accumulation. In agreement with this notion, targeted disruption of the mice Scd1 gene reduced body adiposity and provide resistance to diet-induced weight gain [77], as well as reduced fat accumulation in liver [33], and among livestock, lower SCD1 expression have been associated with fed efficiency and lean growth in cattle [78]. However, the relationship between SCD1 expression and fat accumulation in humans remains controversial, as vary between obese and lean subjects [79], and between carbohydrate-rich [73] and high-fat diets [80].
Concluding remarks
This study revealed constrains to program the expression of fads2 by broodstock nutrition in a typically marine fish, such as gilthead sea bream. In contrast, results of DNA methylation and gene expression profiles of scd1a support for the first time in fish a reliable epigenetic mechanism by which parent’s nutrition can shape scd1a gene expression in the offspring. In particular, we showed that ALA enrichment in parents diet lead to offspring fish with increased methylation at a regulatory region in the scd1a promoter, leading to less gene expression, and likely to less fat accumulation under highly dietary lipogenic conditions, often associated to a reduced supply of n-3 LC-PUFA. We are still far to establish the ultimate mechanisms that regulate the promoter activity of scd1 in farmed fish, but importantly, gene structure analysis highly support a conserved exon-intron organization and functional regulatory elements in proximal promoter regions, which was reinforced by the co-regulated expression of scd1a and hnf4α. However, it remains to be explored how the offspring response is further modulated by a combined effect of changes in dietary ALA and EPA/DHA levels, affecting liver but also other target tissues. For instance, in rats, maternal high-fat feeding during suckling alters the DNA methylation of Scd1 promoter in adipose tissue and programmes visceral adiposity [81]. Additionally, it is now known that SCD1 expression is regulated in pigs by miRNAs [82], and this opens new alternative epigenetic regulatory mechanisms of lipid metabolism that remains mostly unexplored in fish.
Materials and methods
All procedures were carried out according to the European Directive (2010/63/EU) on the protection of animals for scientific purposes, at Fundación Canaria Parque Científico Tecnológico (FCPCT), University of Las Palmas de Gran Canaria (Canary Islands, Spain), and Spanish laws (Royal Decree RD53/2013) on the handling of experimental animals.
Experimental diets and composition analysis
Two broodstock FM-based diets were formulated to be isoproteic and isolipidic with a different fatty acid profile (Table 1). In the broodstock conditioning diet, FO was partially replaced by LO, increasing several folds (x 18.7) the content of ALA in comparison to the control FM/FO diet. The content of oleic acid (OA, 18:1n-9) and linoleic acid (LA, 18:2n-6) increased two times, whereas the arachidonic acid (ARA, 20:4n-6), EPA and DHA were kept in the same order of magnitude. The diet used for challenging the offspring was a low FM and lipid-enriched diet, also with a high ALA content, but with reduced EPA and DHA levels that remained above the n-3 LC-PUFA requirements of gilthead sea bream juveniles (Table 1). Analyses of proximate and fatty acid composition of diets, eggs and offspring livers were performed as elsewhere [42].
Table 1.
Formulation and proximate composition of broodstock diets (supplied 1 month before egg collection) and 6-month offspring diet (supplied for 60 days). One broodstock diet was an ALA rich diet; the challenge diet for all juveniles was also ALA-rich ensuring any observed effect of ALA to be ascribed to early development. The three diets have similar ARA, EPA, and DHA composition to avoid confounding effects, thus ensuring any effect to be attributed to the ALA content of parents diet.
| Parents diets |
Offspring diet |
||
|---|---|---|---|
| Control diet | ALA-rich diet | Challenge diet | |
| Raw material (%) | |||
| Meals from marine sourcesa | 50.0 | 50.0 | 5.0 |
| Fish meal alternative protein sourcesb | 54.5 | ||
| Rapeseed meal cakec | 11.3 | ||
| Sunflower cake | 13.2 | 13.2 | |
| Soya caked | 10.0 | 10.0 | |
| Fish oile | 8.0 | 2.4 | 3 |
| Linseed oilf | - | 5.6 | |
| Vegetable oil mixg | 13 | ||
| Wheath | 9.9 | 9.9 | 6.9 |
| Corn gluten 60i | 7.0 | 7.0 | |
| Vitamin & mineral premixj | 1.1 | 1.1 | 6.3 |
| Biochemical composition | |||
| (% of dry matter) | |||
| Moisture | 9.1 | 8.8 | 6.5 |
| Protein (crude) | 56.3 | 56.1 | 57.2 |
| Lipids (crude) | 17.2 | 17.1 | 21.8 |
| Ash | 8.6 | 8.5 | 6.7 |
| Energy – gross (MJ kg−a) | 21.2 | 21.2 | 22.5 |
| Fatty acids (% of total fatty acids) | |||
| 18:3n-3 (ALA) | 0.9 | 16.3 | 14.9 |
| 20:4n-6 (ARA) | 0.39 | 0.34 | 0.31 |
| 20:5n-3 (EPA) | 6.28 | 4.83 | 3.81 |
| 22:6n-3 (DHA) | 7.05 | 6.01 | 4.49 |
aBroodstock diets contain Fish meal NA LT 70 (Feed Service, Bremen, Germany), Fish meal SA 68 (Feed Service, Bremen, Germany), Krill meal (Qrill, Aker BioMarine, Fjordalleen, Norway) Squid meal (Rieber and Son, Bergen, Norway); offspring diet only contains Fish meal SA 68 super prime (Feed Service, Bremen, Germany)
bBlood meal spray (Daka, Denmark), soya protein concentrates 60% (Svane Shipping, Denmark), corn gluten meal 60 (Cargill, Netherlands), wheat gluten (Cargill, Netherlands).
cEmmelev, Denmark
dForty-eight high protein solvent extract (Svane Shipping, Denmark)
eSouth American, Superprime (Feed Service, Bremen, Germany)
fLinseed (Ch. Daudruy, France)
gLinseed (2.6%) (Ch. Daudruy, France), rapeseed (5.2%) (Emmelev, Denmark) and palm oils (5.2%) (Cargill, Netherlands).
hCargill, Netherlands
iCargill, Netherlands
jSupplied the following vitamins (mg/kg): A 3.8, D 0.05, E 102.4, K3 9.8, B1 2.7, B2 8.3, B6 4.8, B12 0.25, B3 24.8, B5 17.2, folic acid 2.8, H 0.14, C 80; minerals (mg/kg): cobalt 0.94, iodine 0.7, selenium 0.2, iron 32.6. manganese 12, copper 3.2, zinc 67; other (g/kg): taurine 2.45, methionine 0.5, histidine 1.36, cholesterol 1.13. DSM (Netherlands), Evonik Industries (Germany), Deutsche Lanolin Gesellschaft (Germany).
Broodstock and offspring feeding
Twenty-four brood fish were distributed by couples into 12 tanks (1000 L) in a flow-through system with filtered seawater (37.0 ± 0.5‰, 19.6–21.3°C), strong aeration and natural photoperiod in the experimental facilities of IU-ECOAQUA at the Canary Islands. Each broodstock diet was randomly allocated to six tanks resulting in two broodstock groups (Control group, 1.9–2.4 kg body weight; ALA-rich group, 1.8–2.6 kg body weight), fed twice a day (08:00 and 15:00 h) at 1% for 37 days. Eggs were collected at spawning from the two broodstocks, and the offsprings were reared under standard conditions for 6 months [83]. Then, six-months-old fish from the two broodstock groups were distributed in twelve 500 L tanks and fed (6 days per week) with the challenging offspring diet twice a day (08:00 and 14:00 h) for 60 days. At the end of trial, 24 h-fasted fish were anaesthetized with 10 ppm clove oil/methanol (1:1) in seawater.
In silico analyses
Gene sequences of fads2 and scd1a were searched in the IATS-CSIC genomic database of gilthead sea bream (http://nutrigroup-iats.org/seabreamdb). The retrieved sequences were manually curated and gene structure predictions were made using the Fgenesh Softberry Service (http://www.softberry.com). Overall gene size was corroborated by PCR amplification in order to ensure that size of amplicons matches with those predicted by the in silico analysis. The resulting gene sequences included >3 kb upstream the start codon, and they were uploaded to GenBank under accession numbers MN061683 (fads2) and MN061682 (scd1a). Graphical gene representations were carried out with Exon-Intron Graphic Maker (http://wormweb.org/exonintron). Core promoter regions were predicted using two complementary approaches: Easy Promoter Prediction Program (EP3) (http://bioinformatics.psb.ugent.be/webtools/ep3/), which uses structural features of DNA to identify promoter regions; and MatInspector (www.genomatrix.de), which identifies promoter modules based on predicted transcription factor binding sites (TFBS). Promoter sequence alignment was performed by Pro-Coffee from ExPASy (https://tcoffee.vital-it.ch/apps/tcoffee/do:procoffee). Prediction of TFBS was done by ConTra v3 (http://bioit2.irc.ugent.be/contra/v3/#/step/1), using the TRANSFAC database with sensitivity and accuracy set at core match = 0.95 and matrix match = 0.85. Predictions of CpG islands were done by MethPrimer (http://www.urogene.org/methprimer). Search parameters for CpG islands were: length ≥200, C + G content ≥50%, ratio of observed/expected CpGs ≥0.60 and window size = 100.
DNA isolation and bisulphite conversion
Liver DNA was extracted using the Quick-DNA™ Miniprep Plus Kit (Zymo Research), following the manufacturer’s instructions. Quantity and quality of DNA were assessed by NanoDrop 2000c Spectrophotometer (Thermo Scientific) and DNA integrity was analysed in a 1% agarose gel. Extracted DNA was bisulphite converted using the EZ DNA Methylation Gold bisulphite conversion kit (Zymo Research) following the manufacturer’s instructions.
PCR of bisulphite-converted DNA
Primers were designed using the PyroMark Assay Design 2.0 (Qiagen) to hybridize CpG-free sites at the highest melting temperature (Supplementary Table 1). Reverse primers were labelled with biotin at the 5ʹ-end and bisulphite-converted DNA was amplified by PCR, using the Invitrogen™ Platinum™ Taq Hot-Start DNA Polymerase (Thermo Fisher Scientific) with forward- and reverse-specific primers at 1 µM each in a total volume of 25 µL. The reaction was performed in a Touchgene Gradient Thermal Cycler (Techne) as follows: 95ºC for 5 min, followed by 35 cycles of 95ºC for 45 s, 60ºC for 45 s, and 72ºC for 1.5 min with a final extension at 72ºC for 5 min. PCR products were checked by 1% agarose gels to ensure specificity before pyrosequencing.
DNA pyrosequencing
Quantification of DNA methylation of 10 and 8 CpG sites in the fads2 and scd1a promoters, respectively, was performed by pyrosequencing as described elsewhere [84]. Briefly, pyrosequencing reactions were carried out by means of a Pyro Gold SQA™ Reagent Kit (Qiagen) in a PyroMark Q96 System version 2.0.6 (Qiagen) (see Supplementary Table 1 for pyrosequencing primers). Methylation values were provided per CpG site in offspring juvenile’s liver with different parental nutrition. Additionally, we also assessed the effect of prolonged exposure to low water oxygen concentrations on the DNA methylation pattern of fads2 promoter using tissue samples from a recently published study [85]. Briefly, juvenile fishes (~34 g of body weight) were distributed into six 90 L tanks (25 fish/tank) coupled to a recirculating aquatic system equipped with physical and biological filtration and programmable temperature (25–27ºC) and O2 concentration. Fishes were fed once daily with a commercial diet (EFICO Forte 824, BioMar). After 12 days of acclimation, the water oxygen of three tanks were kept unchanged (normoxia, >5.5 ppm O2; >85% O2 saturation) whereas the water oxygen in the other three tanks was reduced (moderate hypoxia, 3.0 ppm O2; 42-43% O2 saturation). Unionized ammonia was below toxic levels (<0.50 mg/L) in both groups. Fishes were kept for 22 days under these conditions and then, four fish per tank (n = 12/treatment) were anaesthetized with 3-aminobenzoic acid ethyl ester (100 mg/L) and killed by cervical section. Liver samples were taken and immediately snap-frozen in liquid nitrogen and stored at 80ºC until posterior processing. Quantification of DNA methylation was made as described above.
Gene expression profiling
Liver RNA was extracted with the MagMAX-96 total RNA isolation kit (Life Technologies, Carlsbad, CA, USA). The analysed samples were the same than those used from DNA methylation, ensuring valid associations between DNA methylation and gene expression at individual level. Briefly, total RNA yield was 50–100 μg (Abs260/280 nm of 1.9–2.1), and 500 ng were reverse transcribed with random decamers using the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA). Negative control reactions were run without reverse transcriptase. Real-time quantitative PCR (RT-qPCR) was performed to analyse the expression of fads2 and scd1a in combination with a set of selected transcription factors with key roles in lipid metabolism (lxrα, liver x receptor alpha; pparβ and δ, peroxisome proliferator activated-receptor beta and gamma) and in particular, on the transcriptional regulation of fads2 and scd1a (hnf4α; hepatic nuclear factor 4 alpha; nf-y, nuclear factor-Y; pparα; peroxisome proliferator activated-receptor alpha; sp1, specificity protein 1; srebp1, sterol regulatory element-binding protein 1). Each 25 μL-PCR reaction contained 660 pg of total cDNA, SYBR Green Master Mix (Bio-Rad, Hercules, CA, USA) and 0.9 μM primers (Supplementary Table 1). All pipetting operations were performed by an EpMotion 5070 Liquid Handling Robot (Eppendorf, Hamburg, Germany). RT-qPCR was carried out in an Eppendorf Mastercycler Ep Realplex (Eppendorf, Germany) as follows: initial denaturation at 95°C for 3 min, 40 cycles of denaturation for 15 s at 95°C and annealing/extension for 60 s at 60°C. The efficiency of PCR reactions was higher than 90% for all genes. Negative controls without template were included for each primer pair, and the specificity of reactions was confirmed by melting curve analysis (ramping rates of 0.5°C/10 s over a temperature range of 55–95°C). Gene expression was calculated by the delta-delta Ct method, using ß-actin as housekeeping gene.
Statistical analysis
Statistical analyses were performed using SigmaPlot v13 (Systat Software Inc.), with all P-values set to 0.05. Normality and equal variance of data were tested by Shapiro-Wilk and Levene tests, respectively. Methylation at individual CpG sites and expression data between the two dietary or oxygen conditions were analysed by Student’s t-test. When requirements for this analysis were not met, the median values of the two groups were compared by the Mann-Whitney Rank Sum Test (P < 0.05). Pearson correlation analysis was used to relate the gene expression of fads2 and scd1a with the methylation level at individual CpG sites in their promoters, and for co-expression analysis of sp1 and hnf4α with scd1a.
Funding Statement
Research funded by H2020 Food (727610), PerformFISH (Integrating Innovative Approaches for Competitive and Sustainable Performance across the Mediterranean Aquaculture Value Chain).
Acknowledgments
The authors thank M. A. González for technical assistance with gene expression analyses.
Author Contributions Statement
JPS and MI coordinated and designed the study; ST and MJZ conducted the broodstock crosses; MI formulated the experimental diets; ST and HX raised larvae, grown the juvenile fish and conducted the feeding trial; ST performed biochemical analyses; EP and PSM conducted the methylation and gene expression analyses, FNC performed bioinformatic analyses; EP, PSM, MI and JPS analyzed the data; EP and JPS wrote the manuscript; all authors read, edited and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].FAO. The state of world fisheries and aquaculture 2016 . Contributing to food security and nutrition for all. FAO, Publications Division, Rome; 2016. [Google Scholar]
- [2].Benedito-Palos L, Ballester-Lozano GF, Simó P, et al. Lasting effects of butyrate and low FM/FO diets on growth performance, blood haematology/biochemistry and molecular growth-related markers in gilthead sea bream (Sparus aurata). Aquaculture. 2016;454:8–18. [Google Scholar]
- [3].Kousoulaki K, Sæther B-S, Albrektsen S, et al. Review on European sea bass (Dicentrarchus labrax, Linnaeus, 1758) nutrition and feed management: a practical guide for optimizing feed formulation and farming protocols. Aquac Nutr. 2015;21:129–151. [Google Scholar]
- [4].Gil-Solsona R, Calduch-Giner JA, Nácher-Mestre J, et al. Contributions of MS metabolomics to gilthead sea bream (Sparus aurata) nutrition. Serum fingerprinting of fish fed low fish meal and fish oil diets. Aquaculture. 2019;498:503–512. [Google Scholar]
- [5].Nácher-Mestre J, Ballester-Lozano GF, Garlito B, et al. Comprehensive overview of feed-to-fillet transfer of new and traditional contaminants in Atlantic salmon and gilthead sea bream fed plant-based diets. Aquac Nutr. 2018;24:1782–1795. [Google Scholar]
- [6].Glencross BD, Baily J, Berntssen MHG, et al. Risk assessment of the use of alternative animal and plant raw material resources in aquaculture feeds. Rev Aquacult. 2019. DOI: 10.1111/raq.12347 [DOI] [Google Scholar]
- [7].Pérez-Sánchez J, Estensoro I, Redondo MJ, et al. Mucins as diagnostic and prognostic biomarkers in a fish-parasite model: transcriptional and functional analysis. PLoS One. 2013;8:e65457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Piazzon MC, Galindo-Villegas J, Pereiro P, et al. Differential modulation of IgT and IgM upon parasitic, bacterial, viral and dietary challenges in a perciform fish. Front Immunol. 2016;7:637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Martin SAM, Król E.. Nutrigenomics and immune function in fish: new insights from omics technologies. Dev Comp Immunol. 2017;75:86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Michl SC, Ratten JM, Beyer M, et al. The malleable gut microbiome of juvenile rainbow trout (Oncorhynchus mykiss): diet-dependent shifts of bacterial community structures. PLoS ONE. 2017;12:e0177735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Simó-Mirabet P, Perera E, Calduch-Giner JA, et al. Co-expression analysis of sirtuins and related metabolic biomarkers in juveniles of gilthead sea bream (Sparus aurata) with differences in growth performance. Front Physiol. 2018;9:608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Estensoro I, Ballester-Lozano GF, Benedito-Palos L, et al. Dietary butyrate helps to restore the intestinal status of a marine teleost (Sparus aurata) fed extreme diets low in fish meal and fish oil. PLoS One. 2016;11:e0166564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Piazzon MC, Calduch-Giner JA, Fouz B, et al. Under control: how a dietary additive can restore the gut microbiome and proteomic profile, and improve disease resilience in a marine teleostean fish fed vegetable diets. Microbiome. 2017;5:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Montero D, Izquierdo M. Welfare and health of fish fed vegetable oils as alternative lipid sources to fish oil. In: Turchini G, Wing-Keong N, Tocher D, editors. Fish oil replacement and alternative lipid sources in aquaculture feeds; 2010. p. 439–485. DOI: 10.1201/9781439808634-c14. [DOI] [Google Scholar]
- [15].Izquierdo MS, Montero D, Robaina L, et al. Alterations in fillet fatty acid profile and flesh quality in gilthead seabream (Sparus aurata) fed vegetable oils for a long term period. Recovery of fatty acid profiles by fish oil feeding. Aquaculture. 2005;250:431–444. [Google Scholar]
- [16].Benedito-Palos L, Navarro JC, Sitjà-Bobadilla A, et al. High levels of vegetable oils in plant protein-rich diets fed to gilthead sea bream (Sparus aurata L.): growth performance, muscle fatty acid profiles and histological alterations of target tissues. Br J Nutr. 2008;100:992–1003. [DOI] [PubMed] [Google Scholar]
- [17].Tocher DR, Ghioni C. Fatty acid metabolism in marine fish: low activity of fatty acyl Δ 5 desaturation in gilthead sea bream (Sparus aurata) cells. Lipids. 1999;34:433–440. [DOI] [PubMed] [Google Scholar]
- [18].Seiliez I, Panserat S, Corraze G, et al. Cloning and nutritional regulation of a D6-desaturase-like enzyme in the marine teleost gilthead seabream (Sparus aurata). Comp Biochem Physiol B Biochem Mol Biol. 2003;135:449–460. [DOI] [PubMed] [Google Scholar]
- [19].Jin M, Yuan Y, Lu Y, et al. Regulation of growth, tissue fatty acid composition, biochemical parameters and lipid related genes expression by different dietary lipid sources in juvenile black seabream, Acanthopagrus schlegelii. Aquaculture. 2017;479:25–37. [Google Scholar]
- [20].Vagner M, Robin JH, Zambonino-Infante JL, et al. Ontogenic effects of early feeding of sea bass (Dicentrarchus labrax) larvae with a range of dietary n-3 highly unsaturated fatty acid levels on the functioning of polyunsaturated fatty acid desaturation pathways. Br J Nutr. 2009;101:1452–1462. [DOI] [PubMed] [Google Scholar]
- [21].Xu H, Dong X, Ai Q, et al. Regulation of tissue LC-PUFA contents, Δ6 fatty acyl desaturase (FADS2) gene expression and the methylation of the putative FADS2 gene promoter by different dietary fatty acid profiles in Japanese seabass (Lateolabrax japonicus). PLoS One. 2014;9:e87726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dong X, Tan P, Cai Z, et al. Regulation of FADS2 transcription by SREBP-1 and PPAR-α influences LC-PUFA biosynthesis in fish. Sci Rep. 2017;7:40024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xie D, Fu Z, Wang S, et al. Characteristics of the fads2 gene promoter in marine teleost Epinephelus coioides and role of Sp1-binding site in determining promoter activity. Sci Rep. 2018;8:5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kuah MK, Jaya-Ram A, Shu-Chien AC. The capacity for long-chain polyunsaturated fatty acid synthesis in a carnivorous vertebrate: functional characterisation and nutritional regulation of a Fads2 fatty acyl desaturase with Δ4 activity and an Elovl5 elongase in striped snakehead (Channa striata). Biochim Biophys Acta. 2015;1851:248–260. [DOI] [PubMed] [Google Scholar]
- [25].Geay F, Zambonino-Infante J, Reinhardt R, et al. Characteristics of fads2 gene expression and putative promoter in European sea bass (Dicentrarchus labrax): comparison with salmonid species and analysis of CpG methylation. Mar Genomics. 2012;5:7–13. [DOI] [PubMed] [Google Scholar]
- [26].Caballero MJ, Izquierdo MS, Kjørsvik E, et al. Histological alterations in the liver of sea bream, Sparus aurata L., caused by short- or long-term feeding with vegetable oils. Recovery of normal morphology after feeding fish oil as the sole lipid source. J Fish Dis. 2004;27:531–541. [DOI] [PubMed] [Google Scholar]
- [27].Morais S, Pratoomyot J, Taggart JB, et al. Genotype specific responses in Atlantic salmon (Salmo salar) subject to dietary fish oil replacement by vegetable oil: a liver transcriptomic analysis. BMC Genomics. 2011;12:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ayisi CL, Zhao J-L. Fatty acid composition, lipogenic enzyme activities and mRNA expression of genes involved in the lipid metabolism of Nile tilapia fed with palm oil. Turk J Fish Aquat Sci. 2017;17:405–415. [Google Scholar]
- [29].Houston SJS, Karalazos V, Tinsley J, et al. The compositional and metabolic responses of gilthead seabream (Sparus aurata) to a gradient of dietary fish oil and associated n-3 long-chain PUFA content. Br J Nutr. 2017;118:1010–1022. [DOI] [PubMed] [Google Scholar]
- [30].Ballester-Lozano GF, Benedito-Palos L, Estensoro I, et al. Comprehensive biometric, biochemical and histopathological assessment of nutrient deficiencies in gilthead sea bream fed semi-purified diets. Br J Nutr. 2015;114:713–726. [DOI] [PubMed] [Google Scholar]
- [31].Yew Tan C, Virtue S, Murfitt S, et al. Adipose tissue fatty acid chain length and mono-unsaturation increases with obesity and insulin resistance. Sci Rep. 2015;5:18366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Miyazaki M, Dobrzyn A, Man WC, et al. Stearoyl-CoA desaturase 1 gene expression is necessary for fructose-mediated induction of lipogenic gene expression by sterol regulatory element-binding protein-1c-dependent and -independent mechanisms. J Biol Chem. 2004;279:25164–25171. [DOI] [PubMed] [Google Scholar]
- [33].Chu K, Miyazaki M, Man WC, et al. Stearoyl-coenzyme A desaturase 1 deficiency protects against hypertriglyceridemia and increases plasma high-density lipoprotein cholesterol induced by liver X receptor activation. Mol Cell Biol. 2006;26:6786–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mounier MD. Hormonal and nutritional regulation of SCD1 gene expression. Biochimie. 2011;93:78–86. [DOI] [PubMed] [Google Scholar]
- [35].Polley SD, Tiku PE, Trueman RT, et al. Differential expression of cold- and diet-specific genes encoding two carp liver Δ9-acyl-CoA desaturase isoforms. Am J Physiol. 2003;284:R41–R50. [DOI] [PubMed] [Google Scholar]
- [36].Hsieh SL, Hu CY, Hsu YT, et al. Influence of dietary lipids on the fatty acid composition and stearoyl-CoA desaturase expression in hybrid tilapia (Oreochromis niloticus × O. aureus) under cold shock. Com Biochem Physiol B. 2007;147:438–444. [DOI] [PubMed] [Google Scholar]
- [37].Li Y, Jia Z, Liang X, et al. Growth performance, fatty-acid composition, lipid deposition and hepatic-lipid metabolism-related gene expression in juvenile pond loach Misgurnus anguillicaudatus fed diets with different dietary soybean oil levels. J Fish Biol. 2018;92:17–33. [DOI] [PubMed] [Google Scholar]
- [38].Vagner M, Zambonino-Infante JL, Robin JH, et al. Is it possible to influence European sea bass (Dicentrarchus labrax) juvenile metabolism by a nutritional conditioning during larval stage? Aquaculture. 2007;267:165–174. [Google Scholar]
- [39].Geay F, Santigosa E, Corporeau C, et al. Regulation of FADS2 expression and activity in European sea bass (Dicentrarchus labrax, L.) fed a vegetable diet. Comp Biochem Physiol B. 2010;156:237–243. [DOI] [PubMed] [Google Scholar]
- [40].Turkmen S, Castro PL, Caballero MJ, et al. Nutritional stimuli of gilthead seabream (Sparus aurata) larvae by dietary fatty acids: effects on larval performance, gene expression and neurogenesis. Aquac Res. 2017a;48:202–213. [Google Scholar]
- [41].Izquierdo MS, Turkmen S, Montero D, et al. Nutritional programming through broodstock diets to improve utilization of very low fishmeal and fish oil diets in gilthead sea bream. Aquaculture. 2015;449:18–26. [Google Scholar]
- [42].Turkmen S, Zamorano MJ, Fernández-Palacios H, et al. Parental nutritional programming and a reminder during juvenile stage affect growth, lipid metabolism and utilisation in later developmental stages of a marine teleost, the gilthead sea bream (Sparus aurata). Br J Nutr. 2017b;118:500–512. [DOI] [PubMed] [Google Scholar]
- [43].Devlin AM, Singh R, Wade RE, et al. Hypermethylation of Fads2 and altered hepatic fatty acid and phospholipid metabolism in mice with hyperhomocysteinemia. J Biol Chem. 2007;282:37082–37090. [DOI] [PubMed] [Google Scholar]
- [44].Hoile SP, Clarke-Harris R, Huang RC, et al. Supplementation with N-3 long-chain polyunsaturated fatty acids or olive oil in men and women with renal disease induces differential changes in the DNA methylation of FADS2 and ELOVL5 in peripheral blood mononuclear cells. PLoS ONE. 2014;9:e109896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Schwenk RW, Jonas W, Ernst SB, et al. Diet-dependent alterations of hepatic Scd1 expression are accompanied by differences in promoter methylation. Horm Metab Res. 2013;45:786–794. [DOI] [PubMed] [Google Scholar]
- [46].Best C, Ikert H, Kostyniuk DJ, et al. Epigenetics in teleost fish: from molecular mechanisms to physiological phenotypes. Comp Biochem Physiol B Biochem Mol Biol. 2018;224:210–244. [DOI] [PubMed] [Google Scholar]
- [47].Hollander KS, Tempel Brami C, Konikoff FM, et al. Dietary enrichment with alpha-linolenic acid during pregnancy attenuates insulin resistance in adult offspring in mice. Arch Physiol Biochem. 2014;120:99–111. [DOI] [PubMed] [Google Scholar]
- [48].Shomonov-Wagner L, Raz A, Leikin-Frenkel A. Alpha linolenic acid in maternal diet halts the lipid disarray due to saturated fatty acids in the liver of mice offspring at weaning. Lipids Health Dis. 2015;14:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Leikin-Frenkel A, Shaish A, Harats D. Maternal dietary ALA prevent atherogenic lipid disarray in the offspring via fetal gene programming. Abstracts/Atherosclerosis. 2017;263:e29ee110. [Google Scholar]
- [50].Panserat S, Marandel L, Seiliez I, et al. New insights on intermediary metabolism for a better understanding of nutrition in teleosts. Annu Rev Anim Biosci. 2019;7:195–220. [DOI] [PubMed] [Google Scholar]
- [51].Anastasiadi D, Vandeputte M, Sánchez-Baizán N1, et al. Dynamic epimarks in sex-related genes predict gonad phenotype in the European sea bass, a fish with mixed genetic and environmental sex determination. Epigenetics. 2018;13:988–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Moghadam HK, Johnsen H, Robinson N, et al. Impacts of early life stress on the methylome and transcriptome of Atlantic salmon. Sci Rep. 2017;7:5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Veron V, Marandel L, Liu J, et al. DNA methylation of the promoter region of bnip3 and bnip3l genes induced by metabolic programming. BMC Genomics. 2018;19:677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Geurden I, Mennigen J, Plagnes-Juan E, et al. High or low dietary carbohydrate: proteinratios during first feeding affect glucose metabolism and intestinal microbiota in juvenile rainbow trout. J Exp Biol. 2014;217:3396–3406. [DOI] [PubMed] [Google Scholar]
- [55].Geurden I, Borchert P, Balasubramanian MN, et al. The positive impact of the early-feeding of a plant-based diet on its future acceptance and utilization in rainbow trout. PLoS One. 2013;8:e83162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Perera E, Yúfera M. Soybean meal and soy protein concentrate in early diet elicit different nutritional programming effects on juvenile zebrafish. Zebrafish. 2016;13:61–69. [DOI] [PubMed] [Google Scholar]
- [57].Kemski M, Wick M, Dabrowski K. Nutritional programming effects on growth and reproduction of broodstock and embryonic development of progeny in yellow perch (Perca flavescens) fed soybean meal-based diets. Aquaculture. 2018;497:452–461. [Google Scholar]
- [58].Park WJ. The biochemistry and regulation of fatty acid desaturases in animals (Chapter 5). In: Polyunsaturated fatty acid metabolism, Burdge GC editor. Academic Press and AOCS Press; 2018. p. 87–100. ISBN 978-0-12-811230-4. DOI: 10.1016/B978-0-12-811230-4.00005-3. [DOI] [Google Scholar]
- [59].Nara TY, He WS, Tang C, et al. The E-box like sterol regulatory element mediates the suppression of human delta-6 desaturase gene by highly unsaturated fatty acids. Biochem Biophys Res Commun. 2002;296:111–117. [DOI] [PubMed] [Google Scholar]
- [60].Wu X, Zou X, Chang Q, et al. The evolutionary pattern and the regulation of stearoyl-CoA desaturase genes. Biomed Res Int. 2013;2013:856521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Tabor DE, Kim JB, Spiegelman BM, et al. Identification of conserved cis-elements and transcription factors required for sterol-regulated transcription of stearoyl-CoA desaturase 1 and 2. J Biol Chem. 1999;274:20603–20610. [DOI] [PubMed] [Google Scholar]
- [62].Lodish H, Berk A, Zipursky SL, et al. Molecular cell biology. 4th ed. New York: W. H. Freeman;2000. Section 10.4, Regulatory Sequences in Eukaryotic Protein-Coding Genes. Available from: https://www.ncbi.nlm.nih.gov/books/NBK21745/ [Google Scholar]
- [63].Lupu DS, Cheatham CL, Corbin KD, et al. Genetic and epigenetic transgenerational implications related to omega-3 fatty acids. Part I: maternal FADS2 genotype and DNA methylation correlate with polyunsaturated fatty acid status in toddlers: an exploratory analysis. Nutr Res. 2015;35:939–947. [DOI] [PubMed] [Google Scholar]
- [64].Niculescu MD, Lupu DS, Craciunescu CN. Perinatal manipulation of alpha-linolenic acid intake induces epigenetic changes in maternal and offspring livers. Faseb J. 2013;27:350–358. [DOI] [PubMed] [Google Scholar]
- [65].Xu TL, Seyfert HM, Shen XZ. Epigenetic mechanisms contribute to decrease stearoyl-CoA desaturase 1 expression in the liver of dairy cows after prolonged feeding of high-concentrate diet. J Dairy Sci. 2018;101:2506–2518. [DOI] [PubMed] [Google Scholar]
- [66].Filtz TM, Vogel WK, Leid M. Regulation of transcription factor activity by interconnected post-translational modifications. Trends Pharmacol Sci. 2014;35:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Mauvoisin D, Prevost M, Ducheix S, et al. Key role of the ERK1/2 MAPK pathway in the transcriptional regulation of the Stearoyl-CoA Desaturase (SCD1) gene expression in response to leptin. Mol Cell Endocrinol. 2010;319:116–128. [DOI] [PubMed] [Google Scholar]
- [68].Clark SJ, Harrison J, Molloy PL. Sp1 binding is inhibited by (m)Cp(m)CpG methylation. Gene. 1997;195:67–71. [DOI] [PubMed] [Google Scholar]
- [69].WG Z, Srinivasan K, Dai Z, et al. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter. Mol Cell Biol. 2003;23:4056–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Safe S, Kim K. Nuclear receptor-mediated transactivation through interaction with Sp proteins. Prog Nucleic Acid Res Mol Biol. 2004;77:1–36. [DOI] [PubMed] [Google Scholar]
- [71].Kardassis D, Falvey E, Tsantili P, et al. Direct physical interactions between HNF-4 and Sp1 mediate synergistic transactivation of the apolipoprotein CIII promoter. Biochemistry. 2002;41:1217–1228. [DOI] [PubMed] [Google Scholar]
- [72].Mar-Heyming R, Miyazaki M, Weissglas-Volkov D, et al. Association of stearoyl-CoA desaturase 1 activity with familial combined hyperlipidemia. Arterioscler Thromb Vasc Biol. 2008;28:1193–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Silbernagel G, Kovarova M, Cegan A, et al. High hepatic SCD1 activity is associated with low liver fat content in healthy subjects under a lipogenic diet. J Clin Endocrinol Metab. 2012;97:E2288–E2292. [DOI] [PubMed] [Google Scholar]
- [74].Rodriguez-Cuenca S, Whyte L, Hagen R, et al. Stearoyl-CoA desaturase 1 is a key determinant of membrane lipid composition in 3T3-L1 adipocytes. PLoS One. 2016;11:e0162047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Benedito-Palos L, Calduch-Giner JA, Ballester-Lozano GF, et al. Effect of ration size on fillet fatty acid composition, phospholipid allostasis and mRNA expression patterns of lipid regulatory genes in gilthead sea bream (Sparus aurata). Br J Nutr. 2013;109:1175–1187. [DOI] [PubMed] [Google Scholar]
- [76].Benedito-Palos L, Ballester-Lozano G, Pérez-Sánchez J. Wide-gene expression analysis of lipid-relevant genes in nutritionally challenged gilthead sea bream (Sparus aurata). Gene. 2014;547:34–42. [DOI] [PubMed] [Google Scholar]
- [77].Ntambi JM, Miyazaki M, Stoehr JP, et al. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc Natl Acad Sci USA. 2002;99:11482–11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Tizioto PC, Coutinho LL, Oliveira PS, et al. Gene expression differences in Longissimus muscle of Nelore steers genetically divergent for residual feed intake. Sci Rep. 2016;6:39493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Stefan N, Peter A, Cegan A, et al. Low hepatic stearoyl-CoA desaturase 1 activity is associated with fatty liver and insulin resistance in obese humans. Diabetologia. 2008;51:648–656. [DOI] [PubMed] [Google Scholar]
- [80].Miyazaki M, Flowers MT, Sampath H, et al. Hepatic stearoyl-CoA desaturase-1 deficiency protects mice from carbohydrate-induced adiposity and hepatic steatosis. Cell Metab. 2007;6:484–496. [DOI] [PubMed] [Google Scholar]
- [81].Butruille L, Marousez L, Pourpe C, et al. Maternal high-fat diet during suckling programs visceral adiposity and epigenetic regulation of adipose tissue stearoyl-CoA desaturase-1 in offspring. Int J Obes (Lond). 2019. DOI: 10.1038/s41366-018-0310-z [DOI] [PubMed] [Google Scholar]
- [82].Zhang S, Shen L, Xia Y, et al. DNA methylation landscape of fat deposits and fatty acid composition in obese and lean pigs. Sci Rep. 2016;6:35063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Turkmen S, Hernández-Cruz CM, Zamorano MJ, et al. LC-PUFA profiles in parental diets induce long-term effects on growth, fatty acid profiles, expression of fatty acid desaturase 2 and selected immune system-related genes in the offspring of gilthead seabream. Br J Nutr. 2019;1–14. DOI: 10.1017/S0007114519000977 [DOI] [PubMed] [Google Scholar]
- [84].Pineda B, Diaz-Lagares A, Pérez-Fidalgo JA, et al. A two-gene epigenetic signature for the prediction of response to neoadjuvant chemotherapy in triple-negative breast cancer patients. Clin Epigenetics. 2019;11:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Martos-Sitcha JA, Simó-Mirabet P, de Las Heras V, et al. Tissue-specific orchestration of gilthead sea bream resilience to hypoxia and high stocking density. Front Physiol. 2019. DOI: 10.3389/fphys.2019.008402019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


