Supplemental Digital Content is available in the text.
Keywords: big data, blood pressure, critical care, deep learning, photoplethysmography
Objectives:
Continuous tracking of blood pressure in critically ill patients allows rapid identification of clinically important changes and helps guide treatment. Classically, such tracking requires invasive monitoring with its associated risks, discomfort, and low availability outside critical care units. We hypothesized that information contained in a prevalent noninvasively acquired signal (photoplethysmograph: a byproduct of pulse oximetry) combined with advanced machine learning will allow continuous estimation of the patient’s blood pressure.
Design:
Retrospective cohort study with split sampling for model training and testing.
Setting:
A single urban academic hospital.
Patients:
Three-hundred twenty-nine adult patients admitted to a critical care unit.
Interventions:
None.
Measurements and Main Results:
One hundred thirty-six thousand four-hundred fifty-nine photoplethysmography waveforms of length 30 seconds were used for training (60%), validation (20%), and testing (20%) of the blood pressure estimation network. Each sample had an associated systolic, mean, and diastolic blood pressures extracted from concurrently recorded invasive arterial line waveforms. Blood pressure estimation using photoplethysmography waveforms is achieved using advanced machine learning methods (convolutional neural networks and a Siamese architectural configuration) calibrated for each patient on a single, first available photoplethysmography sample and associated blood pressure reading. The average estimation bias error was 0.52, 0.1, and –0.76 mm Hg for diastolic, mean, and systolic blood pressure, respectively, with associated mean absolute errors of 4.11, 5.51, and 7.98 mm Hg. If used to identify clinically important changes in blood pressure from the initial baseline, with a threshold of a 10 mm Hg increase or decrease in blood pressure, our algorithm shows an accuracy of 85%, 78%, and 74% for diastolic, mean, and systolic blood pressure, respectively. We also report the network’s performance in detecting systolic and diastolic hypo- or hypertension with accuracies ranging from 86% to 97%.
Conclusions:
Using advanced machine learning tools, we show that blood pressure estimation can be achieved using a common noninvasively recorded signal, the photoplethysmography. Such tools can allow for better monitoring of patients that do not have invasively recorded blood pressure, both in the critical care setting and on inpatient wards.
Critically ill patients present a unique treatment challenge as they exhibit rapid fluctuations in their physiologic state. To allow accurate appreciation of the patient’s state and prompt detection of deteriorations patients are continuously monitored with various vital signs collected and displayed on the bedside monitor (1). Of these, the most routinely measured are the electrocardiogram (ECG), respiratory impedance, photoplethysmography as part of pulse oximetry and finally invasive blood pressure (BP) in a subset of the patients. BP tracking specifically is essential in critically ill patients to identify deterioration and help guide treatment in a goal-directed fashion. Invasive continuous BP monitoring is often implemented in these patients, usually through a catheter inserted in a peripheral or central artery. However, arterial cannulation is an invasive maneuver with a potential for adverse effects which are all associated with increased morbidity and costs. BP can also be monitored noninvasively using a cuff-based method, yet, these methods have been associated with measurement errors and are not continuous in nature.
Photoplethysmography is an optically obtained plethysmogram that can be used to detect blood volume changes in the microvascular tissue beds and is often obtained by using a pulse oximeter which illuminates the skin to measure changes in light absorption and correlate them to the arterial hemoglobin’s saturation (2). The interaction between the light and the tissue is complex and contains information on the instantaneous blood volume in the capillary bed, the heart rate, respiratory rate, and even the BP.
The obvious need for noninvasive methods to continuously monitor BP accurately, together with observations that both the photoplethysmography and ECG signals contain information on the former led to many attempts to use these signals to estimate BP, both in critically ill patients and in the ambulatory settings (3–6). Many of these previously published studies used machine learning either based on multiple specific extracted features from the ECG, photoplethysmography or both such as the pulse transit time (3, 7–11), or even completely data-driven advanced methods utilizing deep networks (3, 12–16). Importantly, to the best of our knowledge, many of these studies focused on noncritically ill patients or used datasets that are small containing tens of patients. Furthermore, some studies used photoplethysmography signals in which samples from the same patients were used both for training and test datasets with significant overlap between the sets in terms of patient source—a form of label leakage inserting a bias and preventing generalization to a hitherto unseen cohort (we used a single sample for calibration as detailed below). A table summary of 21 studies using either photoplethysmography, ECG or both to estimate BP is presented in the supplementary material (Supplemental Digital Content 1, http://links.lww.com/CCX/A151).
In this study, we examined whether photoplethysmography signals acquired as part of routine care in a critical care setting can be used to track the BP of patients and reliably detect clinically relevant fluctuations in BP over time. To that end, we used an openly available dataset of physiologic waveforms and developed an advanced machine learning algorithm based on a Siamese architecture of a convolutional neural network. These networks were trained on photoplethysmography samples 30 seconds in length from a subset of patients and performance was tested on samples from a separate group. We also hypothesized that utilizing for each patient, only the photoplethysmography signal and a calibration single BP reading that correlates to the first available photoplethysmography sample will allow better accuracy without sacrificing generalizability. We demonstrate the feasibility of such an approach that has the potential to improve care and monitoring for critically ill patients and even to be extended to additional scenarios.
MATERIALS AND METHODS
Dataset and Preprocessing
A reliable, large dataset of photoplethysmography sample signals paired with their BP values was compiled using the Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC-II) waveform database (17) containing deidentified health-related data associated with patients who stayed in critical care units of the Beth Israel Deaconess Medical Center. Patient demographics in this dataset can be found elsewhere (18). MIMIC database creation, access and study was approved by the Institutional Review Boards of Beth Israel Deaconess Medical Center (Boston, MA) and the Massachusetts Institute of Technology (Cambridge, MA). Requirement for individual patient consent was waived as all data were deidentified, and there is no impact on clinical care. We focused on two variables solely for our dataset: BP waveform signals that were recorded concurrently with photoplethysmography raw signals at 125 Hz (Fig. 1, A and B for sample traces). Photoplethysmography and BP signals were divided to 30 seconds segments, to allow a balance between a minimal required sample length to derive informative features, BP stability and the potential for future real-time tracking (19–21). As this is a proof-of-concept study, we opted for preprocessing and samples were excluded if either of the following conditions applied: 1) Noisy photoplethysmography signals as defined by a threshold cutoff on the derived total power of the autocorrelation function—thus removing signals that were not strictly periodic as the photoplethysmography is expected to be, or extreme BP fluctuations within a 30 seconds window (reducing the sample from an initial 2,701,154 samples to 194,648 samples); 2) Physiologically improbable or extremely rare BP values (diastolic < 30 mm Hg or > 120 mm Hg, systolic < 55 mm Hg or > 185 mm Hg, reducing the samples to 175,368); and 3) All data from patients with less than 100 adequate samples or those who more than 95% of their data was filtered due to conditions 1–2 (reducing the samples to 155,018). All noisy signal filtering was performed automatically without human reviewing. Furthermore, as our proposed method relies on estimating BP changes relative to a reference initial sample used for calibration, we removed for each patient subsequent samples that varied more than ± 50 mm Hg from the first available sample as such samples were very rare and did not allow for reliable training of the models using this database (reducing the samples slightly to 136,459).
Figure 1.
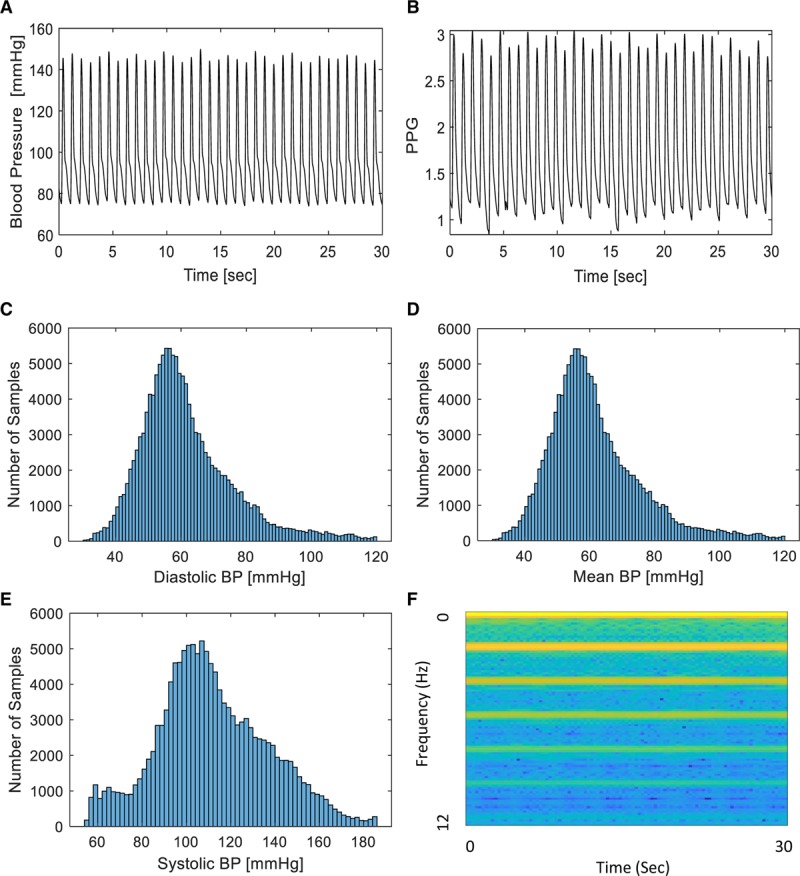
Signal samples and architecture overview. A, Arterial blood pressure (BP) trace from a single patient recorded at 125 Hz. B, Photoplethysmography (PPG) trace from the same patient, recorded at 125 Hz. C, Histogram of samples included in this study with a calculated diastolic BP as denoted on the x-axis. D, Histogram of samples included in this study with a calculated mean BP as denoted on the x-axis. E, Histogram of samples included in this study with a systolic BP as denoted on the x-axis. F, Sample spectrogram of the PPG signal.
After all the steps above for filtering, we remained with 136,459 samples which belong to 329 different patients that we used for our tool development and testing. We divided the filtered samples into three different sets: 60% training set, 20% validation set, and 20% test set. Each of these contained samples from different patients with no overlap between patients in the sets. For each 30-second-long photoplethysmography signal sample, we extracted from the concurrently recorded BP waveform the mean and median systolic BP (SBP) and diastolic BP values and these were used as training and test labels. Figure 1C–E shows the distribution of diastolic BP, mean BP, and SBP values for the samples used in this study. Supplemental Figure 1 (Supplemental Digital Content 2, http://links.lww.com/CCX/A152) shows the distributions of BPs in the prefiltered dataset.
Development of Deep Neural Network Models
The core task of our deep network models was to use photoplethysmography waveforms samples of length 30 seconds to estimate the patient’s instantaneous SBP and diastolic BP. As noted above, BP labels were derived for this study from concurrently recorded invasive arterial BP defined as the true value or training/test label. We opted to use deep networks for this task: a completely data-driven approach that does not rely on human-oriented feature extraction. The chosen input for our training methods were photoplethysmography-derived spectrograms (Fig. 1F shows a sample spectrogram), as a more robust way of representing the raw signals, since the signals are smoothed, and many features were problematic and unreliable to extract. Furthermore, spectrograms capture features from both the time and frequency domains and are less sensitive to signal phase shifts. Spectrograms were computed using the short-time Fourier transform of subwindows of length 6 seconds with a Hamming window and with 95% overlap between adjacent subwindows.
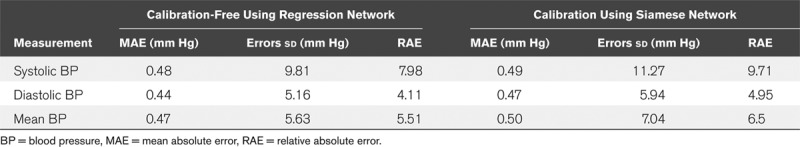
The deep network architectures that classically lends itself to processing fixed-size input such as spectrograms are convolutional neural networks. We did need to adapt these to performing regression rather than classification by changing the last layers of the networks to linear layers with one output channel and using the BP values as regression targets. Another important parameter, we adjusted to our task was the loss function, from cross-entropy, as is customary in classification networks, to L1–loss, which is more suitable for solving regression problems and aims for minimizing the minimum absolute error used as a measure of BP estimation accuracy. Due to its simplicity and the good results it yielded, the specific architecture we chose to use for our network is inspired by the AlexNet architecture (22), which consists of five convolutional layers and three fully connected layers. The first two convolutional layers and the fifth convolutional layer are followed by max pooling layers, while the third and fourth convolutional layers are connected directly. The output of these feature extraction layers goes into a series of two fully connected layers. Finally, the second fully connected layer feeds into a linear regression layer. Rectified linear unit activation function is applied after all convolution and fully connected layers. For regularization, a batch normalization layer follows every convolutional layer and a dropout layer precedes the first two fully connected layers. The optimizer we chose for our network was Adam, and batch size for trainings was 32 (chosen after careful examination of batch size and optimizer type as hyperparameters). Using this approach and testing on samples derived from different patients than those in the training set resulted in mean absolute error (MAE) for estimation of 4.95 mm Hg for diastolic BP, MAE of 6.5 mm Hg for mean BP, and an MAE of 9.71 mm Hg for SBP with mean bias error of 0.22, 0.67, and –1.25 mm Hg, respectively (see Table 1 for additional performance statistics).
TABLE 1.
Performance Summary of the Regression Networks
Improving Estimation Using Calibration and Siamese Networks
We sought to improve our estimation accuracy by calibrating the deep networks using a single BP recording associated with the first available photoplethysmography sample for each patient. The underlying assumption was that using a single calibration sample will allow us on the one hand to exploit patient-specific features captured by the photoplethysmography while retaining a generalizable and potentially deployable method that does not require multiple samples from each patient or exhaustive retraining the net for each patient. Thus, the goal was to design a network that uses a photoplethysmography sample to estimate the change in SBP and diastolic BP relative to a reference BP value (that could theoretically also be derived from a noninvasive cuff-based single measure) and associated reference photoplethysmography spectrogram used for calibration. The main idea behind this technique suggests that the physiologic characteristics of each patient are embedded within its photoplethysmography spectrogram. Basically, we adapted a Siamese network architecture and devised a dual network that was fed two inputs: one which includes a reference photoplethysmography signal (the first available sample for that patient) and associated systolic and diastolic readings and the second input was a later acquired photoplethysmography signal. The output of this network is an estimate of the change in SBP, mean BP, and diastolic BP relative to the reference value based on the current photoplethysmography signal’s spectrogram. Siamese networks are neural networks containing two identical subnetwork components (23–25), meaning the Siamese network uses the same architecture and parameters while working in tandem on two different input vectors to compute a comparable output vector. This architecture was used in the past for applications such as face recognition, signature verification, and matching queries with indexed documents. Our architecture differed from classic Siamese network by being a regression neural network and by its final layer. We used the same architecture we used in the regression network, but while the regression network uses the features extracted by its final fully connected layer to estimate BP, our final fully connected layer extracts a vector which is the result of the subtraction of the patient’s representative photoplethysmography spectrogram features, as recognized by the neural network up to this layer, from its current photoplethysmography spectrogram features. This subtraction yields the “Differences” vector between these photoplethysmography spectrograms and used to estimate the change in BP relative to the calibration initial sample. Figure 2 shows an overview of the developed architecture and analysis pipeline. The Siamese networks architecture indeed provided an estimation improvement while still retaining a very applicable and clinically useful approach and the results reported in the next section are based on it.
Figure 2.
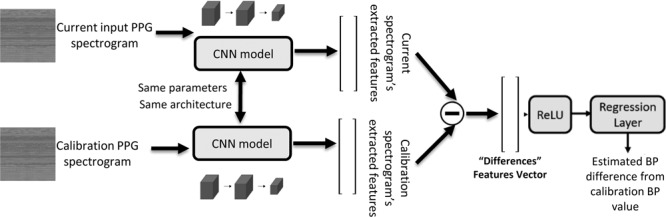
Outline of the Siamese architecture and analysis workflow. Two copies of the same neural network share parameters and are used to compare the current photoplethysmography (PPG)-derived spectrogram to the calibration sample (the first available sample) and is trained to extract features that enable estimating the change in blood pressure (BP) (systolic, mean, and diastolic) from the current value relative to the calibration sample. CNN = convolutional neural network, ReLU = rectified linear unit.
RESULTS
Data (photoplethysmography and BP waveforms) from a total of 329 patients were included for analyses, all derived from the MIMIC-II waveform database (17, 26). Each photoplethysmography sample was 30-second-long with a total of 136,459 samples divided into training, validation and testing (60%, 20%, 20% or 81,629, 27,320, 27,510, respectively). Data from a single patient were not shared between training, validation or testing sets, thus ensuring and accurately capturing the generalization potential of our method. For each tested patient and recorded photoplethysmography sample, we estimated BP changes relative to a single calibration sample using a deep network with Siamese architecture as detailed in the methods and shown in Figure 2. The calibration sample was always the first available photoplethysmography sample for that patient and its associated recorded SBP, mean BP, and diastolic BP. Acquiring a calibration BP and estimating the change from that reference using the deep network and a photoplethysmography sample allowed us to track the BP over time as can be seen, for example, in Figure 3A for a single patient. Figure 3B shows the 2D histogram, normalized per true change; that is, row-wise so that the sum of probabilities in each row is of estimated versus actual true mean BP fluctuations for the entire test dataset, with a maximal fluctuation size of 50 mm Hg. As can be seen, our method is accurate in estimating the magnitude of relatively small fluctuations while for larger ones it is accurate in detecting the direction of change but not the exact magnitude, probably due to the relative scarcity of such fluctuations in the training dataset. Figure 3C shows similarly the estimated versus true recorded mean BP (normalized similarly) demonstrating the network’s precision over a wide range of BPs, with a tendency to underestimate the mean BP for higher values (mean BP above 100 mm Hg). The network’s accuracy can be appreciated by examining the estimation errors distribution. Figure 3, D and E, show these distributions for diastolic and systolic pressures, respectively. For SBP, the mean estimation error is –0.76 ± 12.96 mm Hg (sd), for mean BP it is 0.1 ± 7.87 mm Hg, while for diastolic BP, the mean estimation error is 0.52 ± 6.59 mm Hg. Examining the error distributions, we also find that 95% and 88% of the errors are smaller than 10 mm Hg in magnitude for diastolic and systolic, respectively. Table 1 summarizes the regression networks’ accuracy (both with and without patient-specific calibration) using MAE and the relative absolute error (RAE) defined by: 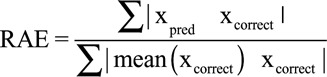 . Of note, the reduction in MAE achieved by using a Siamese network configuration was statistically significant (p < 10–20, Wilcoxon rank-sum test) for diastolic BP, SBP, and mean BP estimation.
. Of note, the reduction in MAE achieved by using a Siamese network configuration was statistically significant (p < 10–20, Wilcoxon rank-sum test) for diastolic BP, SBP, and mean BP estimation.
Figure 3.
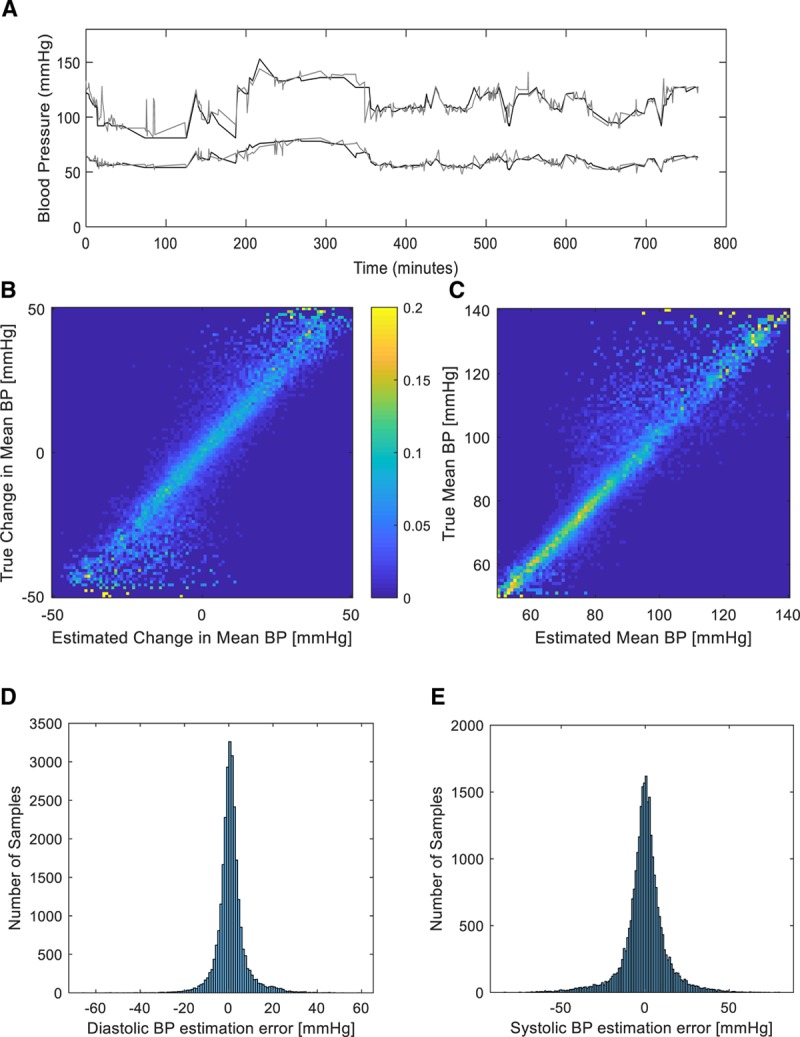
Blood pressure (BP) tracking and estimation performance. A, Sample trace of systolic and diastolic true (as measured by the invasive arterial line, depicted in black) BP readings from one patient over several hours and the associated estimated BP (depicted in gray) derived from the phtoplethysmographic signal and the Siamese estimation network. B, Two-dimensional histogram (normalized per true change, i.e., row-wise) of estimated versus actual true mean BP fluctuations for the entire test dataset, with a maximal fluctuation size of 50 mm Hg. Probability is coded using a color bar as shown. C, Two-dimensional histogram (normalized per true value, i.e., row-wise) of estimated versus actual true mean BP for the entire test dataset. Probability is coded similarly to (C). D, Histogram of estimation errors for diastolic BP as derived for the full test dataset. E, Histogram of estimation errors for systolic BP as derived for the full test dataset.
We tested the detection of clinically significant changes in BP relative to a baseline—for example, using the noninvasively recorded photoplethysmography signal to detect a change in BP larger than a threshold of 10 mm Hg. Figure 4A–C shows the normalized confusion matrices for detection of a change of either an increase or decrease in BP (diastolic, mean, and systolic, respectively) with a threshold of 10 mm Hg in each direction. The total accuracy for detecting a 10 mm Hg fluctuation (and its direction) was 85%, 78%, and 74% for diastolic BP, mean BP, and SBP, respectively. Figure 4D for the same figure depicts the accuracy as a function of the fluctuation threshold.
Figure 4.
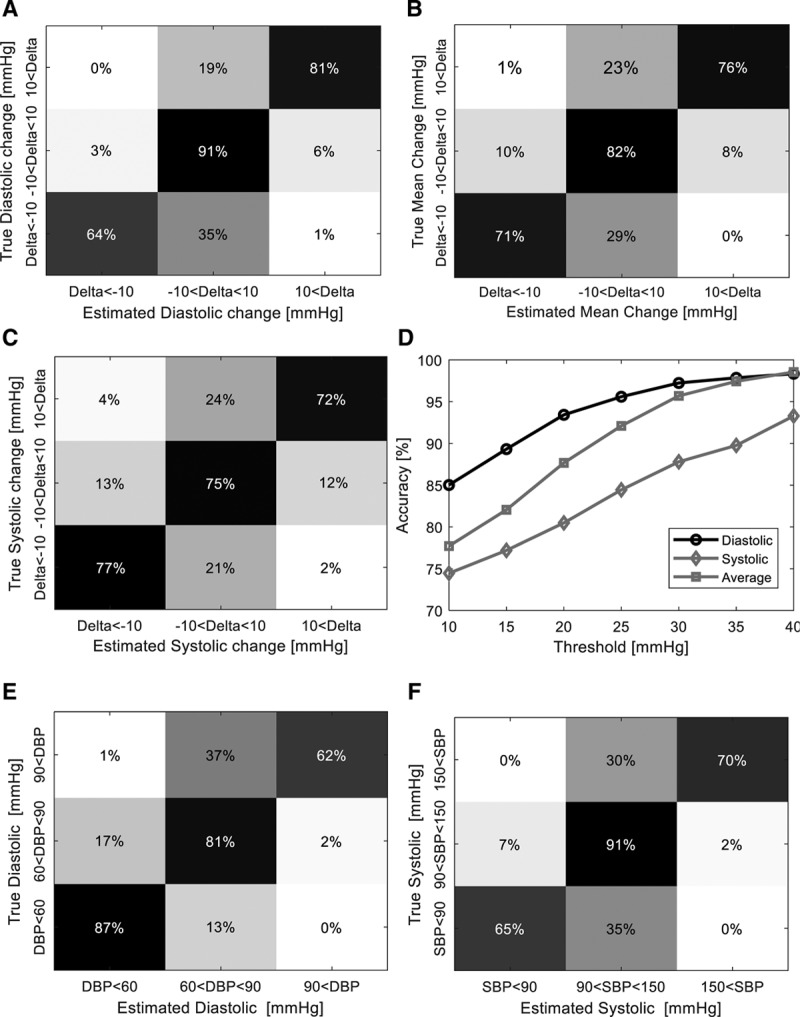
Detection of clinically significant fluctuations. A, Normalized (by true class) confusion matrix showing the conditional probabilities (in percentages) of detecting a diastolic change either less than –10 mm Hg or greater than 10 mm Hg (each row is normalized to the true change summing up to 100%). Probability is denoted using a color scale with black correlating to a probability of 0 and white to a probability of 1. B, Similar normalized confusion matrix as in (A), for mean blood pressure (BP) fluctuations with a similar magnitude. C, Similar normalized confusion matrix as in (A) for systolic BP (SBP) fluctuations with a similar magnitude. D, Accuracy of detecting a clinically relevant fluctuation as a function of the fluctuation magnitude. E, Normalized confusion matrix (normalization similar to (A) showing the sensitivity of detecting diastolic hypotension, normotension, or hypertension as defined by thresholds of less than 60 mm Hg, 60–90 mm Hg, and greater than 90 mm Hg, respectively. F, Normalized confusion matrix (normalization similar to [A]) showing the sensitivity of detecting systolic hypotension, normotension, or hypertension as defined by thresholds of less than 90 mm Hg, 90–150 mm Hg, and greater than 150 mm Hg, respectively. DBP = diastolic blood pressure.
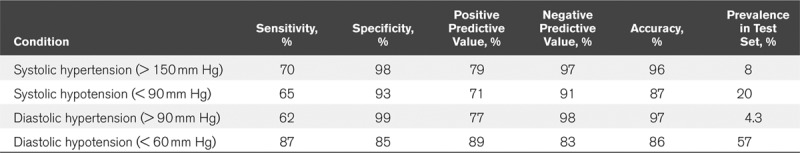
We also examined the utility of our regression network to detect hypotensive and hypertensive events, as can be seen in the normalized (by class) confusion matrices in Figure 4, E and F. Diastolic hypotension, defined as a value below 60 mm Hg can be detected with a sensitivity of 87%, while systolic hypotension (SBP < 90 mm Hg) was detected at a sensitivity 65% and systolic hypertension (SBP > 150 mm Hg) with a sensitivity of 70%. Table 2 details summary statistics of detection sensitivities, specificities, and accuracies for these hypo-and hypertensive events with accuracies for each task ranging from 86% to 97%, as examined on the test dataset of 27,510 samples. As can be seen in Supplemental Figure 1 (Supplemental Digital Content 2, http://links.lww.com/CCX/A152) showing the distribution of BP values in the prefiltered dataset, values above 180 mm Hg were too rare to allow training of the regression network. We also tested the stability of the estimation process and noted no decrease in precision for signal lengths of up to 600 minutes (data not shown).
TABLE 2.
Detection of Systolic and Diastolic Hypo- and Hypertension
DISCUSSION
We show that continuous noninvasive tracking of SBP and diastolic BP can be achieved using a noninvasively commonly recorded signal, the photoplethysmography waveform. All that is required for accurate tracking are a single calibration photoplethysmography sample with an associated concurrent recorded BP. Estimation is achieved by utilizing the power of advanced machine learning techniques, and more precisely convolutional deep networks, to automatically extract relevant information from 30-second-long samples of photoplethysmography waveforms. Using a custom designed architecture, we demonstrate an estimation bias of less than 1 mm Hg and a relatively narrow error distribution with a MAEs of ~8 mm Hg for SBP and 4.1 mm Hg for diastolic BP: even partially satisfying the published standards for validating blood measurements devices (27, 28). Additionally, the utility of this method in alerting a clinician to a significant fluctuation from a baseline and the accuracy of detecting hypo- and hypertension is demonstrated.
A major strength of our method is that it requires only a single calibration sample, and thus, it is generalizable to new patients without requiring retraining for each new patient as the test and training samples did not overlap in terms of the patients from which these samples were recorded. This study shows that relevant information about the patient’s BP can be extracted from the photoplethysmography spectrogram, emphasizing both the advantage of recording all physiologically derived waveforms from the bedside monitors and demonstrating the power of advanced machine learning to aid in bedside patient care.
Obvious limitations of this study are that all waveforms were derived from critically ill patients admitted to one hospital, the need for heavy preprocessing to exclude noisy or a-periodic photoplethysmography samples (limiting the ability for uninterrupted continuous estimation if this pipeline would have been deployed as is) and the fact that prediction accuracy was not tested using multiple sensor types and vendors. Furthermore, we limited fluctuations estimation to changes under ± 50 mm Hg as larger fluctuations were very rare in out training dataset, and similarly, we could not identify in this dataset enough events of extreme hyper- or hypotension to allow accurate detection of these events. Also, at present, additional patient-level information such as demographics, ongoing treatments with vasoactive drugs, or underlying diagnoses were not available to us—the effects of these we plan to examine in future work. Working toward deployment, it is possible to test and refine our method to deal with these scenarios as well as extend our estimation algorithm to use additional signal types in more diverse patient populations.
In conclusion, this is a proof-of-concept study demonstrating how using advanced machine learning tools, we can estimate BP from a common noninvasively recorded signal, the photoplethysmography. We show the potential advantage of using deep learning to extract information from physiologic waveforms captured by the bedside monitors and furthermore, the importance of calibrating such models to the specific characteristics of each patient. In the future, these and similar tools can allow for better monitoring of patients that do not have invasively recorded BP, both in the critical care setting and on inpatient wards.
Footnotes
This work was performed at Technion - Israel Institute of Technology.
Mr. Schlesinger and Mr. Vigderhouse contributed equally to this work.
Supplemental digital content is available for this article. Direct URL citations appear in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
The authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.De Georgia MA, Kaffashi F, Jacono FJ, et al. Information technology in critical care: Review of monitoring and data acquisition systems for patient care and research. ScientificWorldJournal. 2015; 2015:727694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alian AA, Shelley KH. Ehrenfeld JM, Cannesson M. Photoplethysmography Analysis of the pulse oximeter waveform. Monitoring Technologies in Acute Care Environments. 2014, New York, NY: Springer, 165–178 [Google Scholar]
- 3.Ding X, Yan BP, Zhang YT, et al. Pulse transit time based continuous cuffless blood pressure estimation: A new extension and a comprehensive evaluation. Sci Rep. 2017; 7:11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peter L, Noury N, Cerny M. A review of methods for non-invasive and continuous blood pressure monitoring: Pulse transit time method is promising? Irbm. 2014; 35:271–282 [Google Scholar]
- 5.Nye R, Zhang Z, Fang Q. Continuous non-invasive blood pressure monitoring using photoplethysmography: A review. 2015 International Symposium on Bioelectronics and Bioinformatics (ISBB) IEEE, 2015, Tsinghua University, Beijing, China, 176–179 [Google Scholar]
- 6.Thangada ND, Garg N, Pandey A, et al. The emerging role of mobile-health applications in the management of hypertension. Curr Cardiol Rep. 2018; 20:78. [DOI] [PubMed] [Google Scholar]
- 7.Wang R, Jia W, Mao ZH, et al. Cuff-free blood pressure estimation using pulse transit time and heart rate. Int Conf Signal Process Proc. 2014; 2014:115–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escobar B, Torres R. Feasibility of non-invasive blood pressure estimation based on pulse arrival time: A MIMIC database study. Computing in Cardiology 2014 IEEE, 2014, Cambridge, MA, 1113–1116 [Google Scholar]
- 9.Simjanoska M, Gjoreski M, Gams M, et al. Non-invasive blood pressure estimation from ECG using machine learning techniques. Sensors. 2018; 18:1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kachuee M, Kiani MM, Mohammadzade H, et al. Cuff-less high-accuracy calibration-free blood pressure estimation using pulse transit time. 2015 IEEE International Symposium on Circuits and Systems (ISCAS) IEEE, 2015, Lisbon, Portugal, 1006–1009 [Google Scholar]
- 11.Hayase T. Blood pressure estimation based on pulse rate variation in a certain period. Sci Rep. 2020; 10:1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sideris C, Kalantarian H, Nemati E, et al. Building continuous arterial blood pressure prediction models using recurrent networks. 2016 IEEE International Conference on Smart Computing (SMARTCOMP) IEEE, 2016, Shenzhen, China, 1–5 [Google Scholar]
- 13.Su P, Ding XR, Zhang YT, et al. Long-term blood pressure prediction with deep recurrent neural networks. 2018 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI) IEEE, 2018, Las Vegas, NV, 323–328 [Google Scholar]
- 14.Lo FP, Li CX, Wang JK, et al. Continuous systolic and diastolic blood pressure estimation utilizing long short-term memory network. Conf Proc IEEE Eng Med Biol Soc. 2017; 2017:1853–1856 [DOI] [PubMed] [Google Scholar]
- 15.Yan C, Li Z, Zhao W, et al. Novel deep convolutional neural network for cuff-less blood pressure measurement using ECG and PPG signals. Conf Proc IEEE Eng Med Biol Soc. 2019; 2019:1917–1920 [DOI] [PubMed] [Google Scholar]
- 16.Slapnicar G, Mlakar N, Lustrek M. Blood pressure estimation from photoplethysmogram using a spectro-temporal deep neural network. Sensors (Basel). 2019; 19:E3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saeed M, Villarroel M, Reisner AT, et al. Multiparameter intelligent monitoring in intensive care II: A public-access intensive care unit database. Crit Care Med. 2011; 39:952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J, Scott DJ, Villarroel M, et al. Open-access MIMIC-II database for intensive care research. Conf Proc IEEE Eng Med Biol Soc. 2011; 2011:8315–8318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aubert XL, Brauers A. Estimation of vital signs in bed from a single unobtrusive mechanical sensor: Algorithms and real-life evaluation. Conf Proc IEEE Eng Med Biol Soc. 2008; 2008:4744–4747 [DOI] [PubMed] [Google Scholar]
- 20.Takahashi N, Kuriyama A, Kanazawa H, et al. Validity of spectral analysis based on heart rate variability from 1-minute or less ECG recordings. Pacing Clin Electrophysiol. 2017; 40:1004–1009 [DOI] [PubMed] [Google Scholar]
- 21.Shdefat AY, Joo MI, Choi SH, et al. Utilizing ECG waveform features as new biometric authentication method. Int J Electr Comput Eng. 2018; 8:658 [Google Scholar]
- 22.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems, 2012, Lake Tahoe, NV, 1097–1105 [Google Scholar]
- 23.Bromley J, Guyon I, LeCun Y, et al. Signature verification using a” siamese” time delay neural network. Advances in Neural Information Processing Systems, 1994, Denver, CO, 737–744 [Google Scholar]
- 24.Chopra S, Hadsell R, LeCun Y. Learning a similarity metric discriminatively, with application to face verification. 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05) IEEE, 2005, San Diego, CA, 539–546 [Google Scholar]
- 25.Neculoiu P, Versteegh M, Rotaru M. Learning text similarity with siamese recurrent networks. Proceedings of the 1st Workshop on Representation Learning for NLP, 2016, Berlin, Germany, 148–157 [Google Scholar]
- 26.Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016; 3:160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stergiou GS, Palatini P, Asmar R, et al. European Society of Hypertension Working Group on Blood Pressure Monitoring. Recommendations and practical guidance for performing and reporting validation studies according to the universal standard for the validation of blood pressure measuring devices by the Association for the Advancement of Medical Instrumentation/European Society of Hypertension/International Organization for Standardization (AAMI/ESH/ISO). J Hypertens. 2019; 37:459–466 [DOI] [PubMed] [Google Scholar]
- 28.Stergiou GS, Alpert B, Mieke S, et al. A universal standard for the validation of blood pressure measuring devices: Association for the Advancement of Medical Instrumentation/European Society of Hypertension/International Organization for Standardization (AAMI/ESH/ISO) collaboration statement. Hypertension. 2018; 71:368–374 [DOI] [PubMed] [Google Scholar]


