Supplemental Digital Content is available in the text.
Keywords: coronavirus disease 2019, registry
Objectives:
The coronavirus disease 2019 pandemic has disproportionally strained intensive care services worldwide. Large areas of uncertainly regarding epidemiology, physiology, practice patterns, and resource demands for patients with coronavirus disease 2019 require rapid collection and dissemination of data. We describe the conception and implementation of an intensive care database rapidly developed and designed to meet data analytic needs in response to the coronavirus disease 2019 pandemic—the multicenter, international Society of Critical Care Medicine Discovery Network Viral Infection and Respiratory Illness Universal Study.
Design:
Prospective cohort study and disease registry.
Setting:
Multinational cohort of ICUs.
Patients:
Critically ill patients with a diagnosis of coronavirus disease 2019.
Interventions:
None.
Measurements and Main Results:
Within 2 weeks of conception of the Society of Critical Care Medicine Discovery Network Viral Infection and Respiratory Illness Universal Study, study leadership was convened, registry case report forms were designed, electronic data entry set up, and more than 250 centers had submitted the protocol for institutional review board approval, with more than 100 cases entered.
Conclusions:
The Society of Critical Care Medicine Discovery Network Viral Infection and Respiratory Illness Universal Study provides an example of a rapidly deployed, international, pandemic registry that seeks to provide near real-time analytics and information regarding intensive care treatments and outcomes for patients with coronavirus disease 2019.
The coronavirus disease 2019 (COVID-19) pandemic has introduced unprecedented challenges to healthcare systems worldwide. Due to the effects of COVID-19 on the respiratory system, geographic areas affected by the pandemic have experienced large surges in critically ill patients who require intensive care and multiple organ system support (1, 2). In addition, case reports of medications hypothesized to reduce viral replication or systemic inflammation have spurred widespread off-label use without the usual level of evidence that has long been accepted in modern medicine, resulting in critical shortages of medications and frequently missing the opportunity to evaluate potential benefits as well as risks of such drugs (3). Large-scale data that enables rapid communication of patient characteristics, treatment strategies, and outcomes during a pandemic response would support nimble organizational planning and evaluation of effective critical care practices. We describe a novel ICU database rapidly designed in response to the COVID-19 pandemic to allow for near real-time data collection, analysis, and display—the multicenter, international Society of Critical Care Medicine (SCCM) Discovery Network Viral Infection and Respiratory Illness Universal Study (VIRUS).
DATABASE CONSORTIUM FORMATION
The SCCM formed the Discovery, the Critical Care Research Network, in 2016 to provide a central resource to link critical care clinical investigators to scale up research in critical illness and injury, providing both centralized networking and tangible resources including data storage and management, statistical support, grant writing assistance, and other project management needs. Remarkably, the social media platform Twitter (4) played a role in bringing together the VIRUS Discovery Consortium—quickly linking a group of critical care clinician-investigators (A.J.W., O.G., R.K.), statisticians (M.O.H.), pharmacists (S.B.), researchers (V.B.), and SCCM staff (V.K.K.) with a strong interest in quickly establishing the multicenter, international SCCM COVID-19 VIRUS registry. Dissemination of information regarding the registry occurred via the SCCM website (5); daily Twitter posts advertising the importance, availability, and purpose of the registry; and word of mouth. Within 14 days of the initial Twitter post bringing together the VIRUS Leadership Team, the Consortium ~250 sites across North America, South America, East, South, and Western Asia, Africa, and Europe had submitted institutional review board (IRB) applications for participation in the registry (Fig. 1); Table 1 shows the number of sites reaching different landmarks to study participation during the first 2 weeks after a call for sites was announced. By week 2 after announcing the formation of the registry, data from more than 100 cases had been uploaded. The rapid enrollment of sites in the absence of external funding or support for the project is indicative of nearly universal enthusiasm across the international critical care community to collaborate across borders and silos in order to quickly learn from accumulating experience with COVID-19.
TABLE 1.
Timeline of Enrollment After the Initial Call for Enrollment in Society of Critical Care Medicine Viral Infection and Respiratory Illness Universal Study Coronavirus Disease 2019 Registry Project
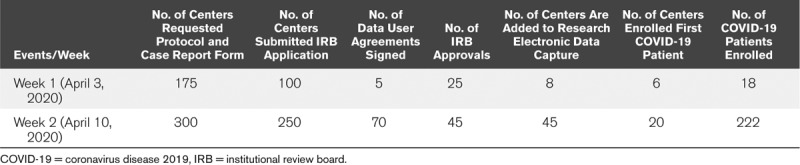
Figure 1.
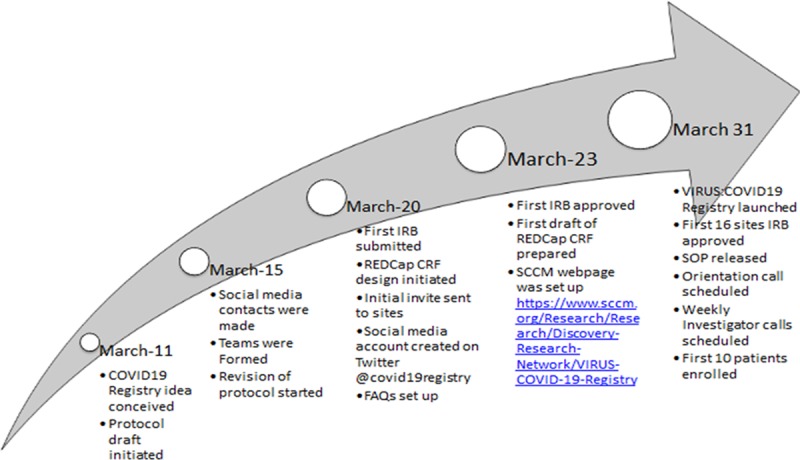
Timeline of Society of Critical Care Medicine (SCCM) Discovery Viral Infection and Respiratory Illness Universal Study (VIRUS) Coronavirus Disease 2019 (COVID-19) database design and development. CRF = case report form, FAQ = frequently asked question, IRB = institutional review board, REDCaP = Research Electronic Data Capture, SOP = standard operating procedure.
DATA ELEMENTS
The overarching purpose of the SCCM VIRUS Discovery Database is to accelerate learning with regards to the epidemiology, physiology, and best practices in response to the COVID-19 pandemic. The de-identified, HIPAA compliant database was developed to capture both core data collection fields containing clinical information collected for all patients, and an enhanced data set of daily physiologic, laboratory, and treatment information collected by sites with available research support and/or infrastructure to allow for more intensive data collection (see data overview in Fig. 2, and detailed Case Report Forms in Appendix 1, Supplemental Digital Content 1, http://links.lww.com/CCX/A163). The case report forms were adapted from the World Health Organization templates data collection forms (6), with edits to focus on an ICU-specific context. Case report forms went through rapid iterative editing to balance feasibility, efficiency, and comprehensiveness, with input from multiple clinical specialties.
Figure 2.
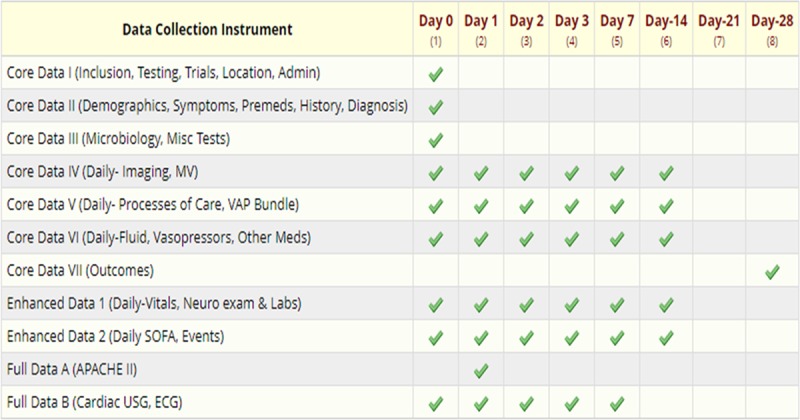
Overview of data collection for Society of Critical Care Medicine Discovery Viral Infection and Respiratory Illness Universal Study Coronavirus Disease 2019 database. APACHE = Acute Physiology and Chronic Health Evaluation, ECG = electrocardiogram, MV = mechanical ventilation, SOFA = Sequential Organ Failure Assessment, USG = Ultrasonography, VAP = ventilator-associated pneumonia.
DATA GOVERNANCE, RESEARCH, AND REPORTING
Many challenges exist when initiating an international collaboration in multicenter clinical data sharing. The SCCM VIRUS Discovery Network will have a four pillar open science approach to data reporting and sharing. First, all centers have open access to their data for internal quality assurance and pilot studies. Second, summary count data will be displayed on the SCCM VIRUS website (https://www.sccm.org/Research/Research/Discovery-Research-Network/VIRUS-COVID-19-Registry) as an interactive dashboard that will provide public reporting of real-time updates with regards to case counts, ICU resource use, and outcomes. Third, with appropriate data use agreements, investigators will be able apply to use the pooled multicenter data for independent research questions. Fourth, the SCCM COVID-19 research team will identify urgent questions of clinical effectiveness, submit study protocols for independent methodological peer review, and design rigorous observational causal inference approaches (e.g., appropriate missing data methods, target trial emulation, use of directed acyclic graphs for covariate selection, quantitative sensitivity analyses [7]) paired with data visualizations that produce real-time results displayed on the Dashboard for immediate dissemination. We seek to facilitate a timely, democratized, and crowd-sourced discovery process, similar to ICU databases such as Medical Information Mart for Intensive Care (MIMIC-III) (8). The SCCM VIRUS Discovery Network will encourage that all research projects using Consortium data post pre-prints in noncommercial archives, further facilitating rapid-reporting of research findings necessary for nimble response to a pandemic.
LESSONS LEARNED
We learned many lessons in a short period while setting up an international ICU registry during a pandemic. Strategies that worked to facilitate rapid progress included a strong social media presence, open communications and data harmonization with other research networks (e.g., National Heart, Lung, and Blood Institute Prevention and Early Treatment of Acute Lung Injury Network), responsive IRBs that identified the critical need for rapid approval of de-identified data collection in the setting of a pandemic, use of the established database infrastructure Research Electronic Data Capture (9) for construction of harmonized case report forms, as well as an academic-professional society partnership that facilitated rapid processing of data use agreements, and early set up of a central website for communication of study materials, frequently asked questions, and standard operating procedures. Early review of literature and comparative research (10) of previous outbreaks was helpful in preparing the research resources; early reports from sentinel countries guided targeting of relevant data fields. In addition, assembly of a multidisciplinary leadership team enabled multiple stakeholder engagement and shared responsibility and mentorship across training levels. Finally, a daily reminder to focus on the goals of the database—ICU practices, physiology, and outcomes for patients with COVID-19—helped to mitigate scope creep and allow for timely completion of the data infrastructure. Strategies that may have improved the process included the use of a central IRB, funding to support local data entry, and a preexisting team able to “flip the switch” on an existing infrastructure to immediately respond to a crisis.
FUTURE DIRECTIONS
Much of the world was relatively unprepared for the rapidly spreading COVID-19 pandemic. Four days after the pandemic was recognized and declared by the World Health Organization, we assembled an ad hoc team to initiate the registry of critically ill patients with COVID-19 described herein. It is our profound hope that a similar registry will not be required in the future. However, it is likely that we will be applying lessons learned from COVID-19 to future pandemics. Our experience of quickly initiating the SCCM Discovery VIRUS Registry and moving from conception to data accrual within less than a month has taught us several valuable lessons—most important being that clinicians across the world want to donate their time for the greater good. As we continue to accrue data into the SCCM Discovery VIRUS COVID-19 Registry, we anticipate that newly established infrastructure and networks will enable more nimble responses to data collection and discovery that allow us to learn from the past, and be better prepared for future pandemics.
Supplementary Material
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
Dr. Harhay is partially supported by National Institutes of Health/National Heart, Lung, and Blood Institute grant R00 HL141678. The remaining authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.Guan W-J, Ni Z-Y, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020. Feb 28. [online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020; 395:1054–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowland C.FDA Authorizes Widespread Use of Unproven Drugs to Treat Coronavirus, Saying Possible Benefit Outweighs Risk. Washington Post. 2020. Available at: https://www.washingtonpost.com/business/2020/03/30/coronavirus-drugs-hydroxychloroquin-chloroquine/. Accessed March 30, 2020.
- 4.Walkey AJ. Twitter. 2020. Available at: https://twitter.com/WalkeyAllan/status/1239187821400920064. Accessed March 15, 2020
- 5.SCCM VIRUS COVID-19 Registry. 2020. Available at: https://www.sccm.org/Research/Research/Discovery-Research-Network/VIRUS-COVID-19-Registry. Accessed March 31, 2020
- 6.ISARIC COVID-19 Clinical Research Resources. 2020. Available at: https://isaric.tghn.org/covid-19-clinical-research-resources/. Accessed March 31, 2020
- 7.Lederer DJ, Bell SC, Branson RD, et al. Control of confounding and reporting of results in causal inference studies. Guidance for authors from editors of respiratory, sleep, and critical care journals. Ann Am Thorac Soc. 2019; 16:22–28 [DOI] [PubMed] [Google Scholar]
- 8.Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016; 3:160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris PA, Taylor R, Thielke R, et al. Research Electronic Data Capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009; 42:377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah A, Kashyap R, Tosh P, et al. Guide to understanding the 2019 Novel Coronavirus. Mayo Clin Proc. 2020; 95:646–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


