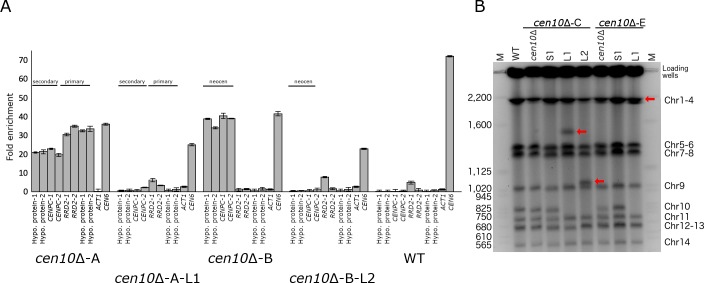
Figure 6. Chromosome fusion results in neocentromere inactivation and karyotype reduction.
(A) Neocentromeres are inactive after chromosomal fusion. For each neocentromere two qPCR primer pairs located in genes spanned by the neocentromere in cen10∆-A and cen10∆-B mutant were used in a ChIP-qPCR experiment. Analyzed is the CENP-A enrichment of 1) a cen10∆ mutant, 2) a large colony derived from the cen10∆ mutant, and 3) the wild-type strain. Centromere 6 (CEN6) was included as a positive control, and actin was included as a negative control. Data are shown for cen10∆-A, cen10∆-A-L1, cen10∆-B, cen10∆-B-L2, and wild type. For cen10∆-A and cen10∆-A-L1 mutants, the chromosomal regions investigated are indicated according to the primary and secondary CENP-A peaks of the cen10∆-A mutant. The cen10∆-B mutant has only one CENP-A-enriched region which co-localized with the secondary CENP-A peak of cen10∆-A and this region is labeled with neocen in cen10∆-B and cen10∆-B-L1. Error bars show standard deviation. (B) PFGE analysis shows that the band corresponding to chromosome 10 was lost in the large colonies and instead larger bands appear due to the fusion of chromosome 10 with other chromosomes. cen10∆ deletion mutants and small colonies derived from 37°C show a wild-type karyotype. Chromosome 10 of the large colonies was fused to chromosome 13, 10, or 1, respectively. Due to limitations of PFGE conditions, the chromosome 10–chromosome 1 fusion did not separate from chromosomes 2, 3, and 4. The positions of the fused chromosomes are indicated with arrows.